 ,, 刘宁, 张玉萍, 徐铭, 刘硕, 张玉君, 马小雪, 刘威生
,, 刘宁, 张玉萍, 徐铭, 刘硕, 张玉君, 马小雪, 刘威生 ,辽宁省果树科学研究所,辽宁营口 115009
,辽宁省果树科学研究所,辽宁营口 115009Genetic Diversity of the Prunus salicina L. from Different Sources and Their Related Species
WEI Xiao, ZHANG QiuPing ,, LIU Ning, ZHANG YuPing, XU Ming, LIU Shuo, ZHANG YuJun, MA XiaoXue, LIU WeiSheng
,, LIU Ning, ZHANG YuPing, XU Ming, LIU Shuo, ZHANG YuJun, MA XiaoXue, LIU WeiSheng ,Liaoning Institute of Pomology, Yingkou 115009, Liaoning
,Liaoning Institute of Pomology, Yingkou 115009, Liaoning通讯作者:
收稿日期:2018-09-28接受日期:2018-12-10网络出版日期:2019-02-13
| 基金资助: |
Received:2018-09-28Accepted:2018-12-10Online:2019-02-13
作者简介 About authors
魏潇,Tel:15902483962;E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1030KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
魏潇, 章秋平, 刘宁, 张玉萍, 徐铭, 刘硕, 张玉君, 马小雪, 刘威生. 不同来源中国李(Prunus salicina L.)的多样性与近缘种关系[J]. 中国农业科学, 2019, 52(3): 568-578 doi:10.3864/j.issn.0578-1752.2019.03.017
WEI Xiao, ZHANG QiuPing, LIU Ning, ZHANG YuPing, XU Ming, LIU Shuo, ZHANG YuJun, MA XiaoXue, LIU WeiSheng.
0 引言
【研究意义】中国李(Prunus salicina L.)起源于我国,是世界上栽培最广泛的李亚属(Prunophora Subgenus)李组(Euprunus Section)植物,均为二倍体(2n=16)[1]。在我国,南自广东、广西和云贵高原,北至黑龙江省牡丹江等地均有中国李栽培,地区分布广泛[2]。与中国李相比,近缘种杏李(Prunus simonii)和乌苏里李(Prunus ussuriensis)却属于局域种(或类型),分布范围非常有限。研究中国李不同居群间的遗传结构以及近缘种在中国李扩散过程中的作用,对地方品种资源的深入挖掘和新品种选育具有重要意义。【前人研究进展】前人已经利用RAPD标记[3,4]和ISSR标记[5]对李亚属不同物种间的亲缘关系进行了研究。杏李(P. simonii)是原产于我国华北地区的栽培种,常被推测为普通杏和中国李种间杂交形成[6],也有人认为是中国李的变种[5]。乌苏里李(P. ussuriensis)是生长在我国黑龙江等地的一种野生种,其具有极强的抗寒能力[2],但FAUST等[7]认为其是中国李的一个变种,而非独立种。然而,杏李、乌苏里李与同域分布的中国李之间的基因交流情况还有待深入研究。有关中国李不同品种群间演化关系的报道较少。郭忠仁[8]通过孢粉学观察认为中国李的传播是从南向北进行的,南方中国李最原始。乔玉山[9]将中国李品种分为南方品种群和北方品种群两大类。基于分子标记的多样性将中国李划分为3种不同类群,即东北品种群、华北品种群和南方品种群[10,11]。陈圣林等[12]对三峡库区的14份中国李品种进行分析后,发现该区域地方品种与国外引进品种存在明显差异。事实上,国外栽培品种是我国某地区栽培李经由日本传到美国后与近缘种美洲李(Prunus amercana)或樱桃李(Prunus cerasifera)杂交改良所形成[13],但是从中国何地引入并没有确切记录。【本研究切入点】以往对中国李的研究仅局限于中国李内部多样性比较和简单的类群划分,有关中国李不同品种群间的多样性差异、群体结构以及近缘种在中国李驯化中的作用还不清楚。【拟解决的关键问题】本研究以近缘种为对照,分析中国李驯化扩散过程中不同品种群间的多样性差异,明确近缘种在栽培驯化中的作用。1 材料与方法
试验于2015年5月至2016年7月在辽宁省果树科学研究所国家果树种质熊岳李杏圃与辽宁省北方果树资源与育种重点实验室进行。
1.1 试材及取样
本试验材料共48份,其中美洲李(Prunus amercana,2n=16)、樱桃李(P. cerasifera,2n=16)和普通杏(P. armeniaca,2n=16)各取1份材料为组外对照。依据中国李初级核心种质筛选结果[14],从不同来源的中国李材料中选取各地生产中主要栽培品种,其中华北品种群5份、东北品种群11份、南方品种群17份、国外育成品种5份,共计38份。另外,近缘种包括5份杏李(P. simonii,2n=16)和2份乌苏里李(P. ussuriensis,2n=16)(表1)。Table 1
表1
表1供试材料信息
Table 1
| 居群 Population | 编号 Code | 种质名称 Accessions | 来源 Origin | 类群 Group | 编号 Code | 种质名称 Name | 来源 Origin | |
|---|---|---|---|---|---|---|---|---|
| 华北品种(HB) Northern varieties | 1 | 香蕉李 Xiangjiaoli | 北京 Beijing | 25 | 连平三华李 Lianpingsanhuali | 广东 Guangdong | ||
| 2 | 帅李 Shuaili | 山东 Shandong | 26 | 从化三华李 Conghuasanhuali | 广东 Guangdong | |||
| 3 | 昌黎晚红 Changliwanhong | 河北 Hebei | 27 | 长圹三华李 Changkuangsanhuali | 广东 Guangdong | |||
| 4 | 玉皇李 Yuhuangli | 甘肃 Gansu | 28 | 上海芙蓉李 Shanghaifurongli | 上海 Shanghai | |||
| 5 | 平顶香 Pingdingxiang | 山东 Shandong | 29 | 永泰芙蓉李 Yongtaifurongli | 福建 Fujian | |||
| 东北品种 (DB) Northeast varieties | 6 | 康什 Kangshi | 吉林 Jilin | 30 | 福建芙蓉李 Fujianfurongli | 福建 Fujian | ||
| 7 | 奎丰 Kuifeng | 东北品种实生 Seedling of northeast varieties | 31 | 福州青奈 Fuzhouqingnai | 福建 Fujian | |||
| 8 | 矮甜李Aitianli | 黑龙江Heilongjiang | 32 | 银醉李Yinzuili | 江苏Jiangsu | |||
| 9 | 黄干核Huangganhe | 吉林Jilin | 33 | 天目蜜李Tianmumili | 浙江Zhejiang | |||
| 10 | 延吉李Yanjili | 吉林Jilin | 国外品种(EX) Foreign varieties | 34 | 安哥诺Angeleno | 美国USA | ||
| 11 | 矮化李Aihuali | 吉林Jilin | 35 | 百班克Baibanke | 美国USA | |||
| 12 | 矬李Cuoli | 黑龙江Heilongjiang | 36 | 布尔班克Burbank | 美国USA | |||
| 13 | 小离核Xiaolihe | 吉林Jilin | 37 | 黑宝石Black Diamond | 美国USA | |||
| 14 | 小黄李Xiaohuangli | 黑龙江Heilongjiang | 38 | 幸运Fortune | 美国USA | |||
| 15 | 大叶伏李Dayefuli | 辽宁Liaoning | 杏李(XL) | 39 | 扁艳Bianyan | 山东Shandong | ||
| 16 | 大叶砧木 Dayezhenmu | 小黄李实生 Seedling of Xiaohuangli | P. simonii | 40 | 熊岳香扁 Xiongyuexiangbian | 辽宁 Liaoning | ||
| 南方品种 (NF) Southern varieties | 17 | 青冬李 Qingdongli | 四川 Sichuan | 41 | 昌黎香蕉李 Changlixiangjiaoli | 河北 Hebei | ||
| 18 | 红冬李Hongdongli | 四川Sichuan | 42 | 黄牛心李Huangniuxinli | 不详 Unknown | |||
| 19 | 巴塘李Batangli | 四川Sichuan | 43 | 昌黎香扁Changlixiangbian | 河北Hebei | |||
| 20 | 庐山李Lushanli | 江西 Jiangxi | 乌苏里李(WU) P. ussuriensis | 44 | 海底亚克夫 Haidiyakefu | 黑龙江Heilongjiang | ||
| 21 | 竹丝李Zhusili | 广东Guangdong | 45 | 乌苏里李 Wusulili | 黑龙江Heilongjiang | |||
| 22 | 西瓜李 Xiguali | 江西 Jiangxi | 樱桃李 P. cerasifera | 46 | 新疆樱桃李 Xinjiangyingtaoli | 新疆 Xinjiang | ||
| 23 | 铜盘奈 Tongpannai | 广东 Guangdong | 美洲李 P. amercana | 47 | 锦西牛心李 Jinxiniuxinli | 美国 USA | ||
| 24 | 利源三华李 Liyuansanhuali | 广东 Guangdong | 普通杏 P. armeniaca | 48 | 金妈妈 Jinmama | 甘肃 Gansu |
新窗口打开|下载CSV
1.2 DNA提取及SSR基因型检测
于2015年春季,采集不同样品幼嫩叶片,并用硅胶干燥保存。采用DOYLE等[15]的改良CTAB法提取总基因组DNA;利用紫外分光光度计和1%浓度的琼脂糖凝胶电泳检测DNA质量和浓度,并稀释至20 ng·μL-1。本研究中,参照SCHUELKE等[16]设计带有M13接头的SSR引物(Tailed primer M13 microsatellite markers,TP-M13-SSR):即正向引物的3′末端增加一段序列为TGTAAAACGACGGCCAGT的接头引物;反向引物序列不变。22对SSR引物均来自Prunus属公共图谱不同染色体上,具体信息见表2。PCR反应采用20 μL反应体系,其中DNA模板5 μL、2×Taq Master Mix(北京康为世纪生物公司)10 μL、带M13接头的正向引物0.4 μL、通用M13荧光引物1.6 μL、反向引物2 μL。PCR扩增反应程序如下:在95℃下解链5 min;整个PCR反应共40个循环,前15个循环为94℃ 45 s,55℃ 45 s,72℃ 45 s,后25个循环为94℃ 45 s,52℃ 45 s,72℃ 45 s;最后,在72℃下延伸10 min,保存于4℃。
Table 2
表2
表2SSR位点的多样性指数
Table 2
| 位点 Locus | 连锁群LG | 位置Sites | Ao | Ne | I | Ho | He | F | Nm |
|---|---|---|---|---|---|---|---|---|---|
| SSRPACITA5[17] | G1 | 13.6 | 7 | 3.207 | 1.429 | 0.667 | 0.688 | 0.031 | 0.388 |
| EPDCU5100[18] | G1 | 14.5 | 6 | 3.734 | 1.406 | 0.729 | 0.732 | 0.004 | 0.363 |
| PGS1.24[19] | G1 | 58.4 | 19 | 9.125 | 2.473 | 0.667 | 0.890 | 0.251 | 0.374 |
| BPPCT028[19] | G1 | 60.7 | 10 | 6.055 | 1.990 | 0.688 | 0.835 | 0.177 | 0.387 |
| pchgms1[20] | G1 | — | 9 | 4.571 | 1.730 | 0.792 | 0.781 | -0.013 | 0.461 |
| PceGA025[21] | G1 | 77.4 | 21 | 12.031 | 2.696 | 0.500 | 0.917 | 0.455 | 0.412 |
| CPSCT039[17] | G2 | 35.1 | 15 | 8.678 | 2.371 | 0.958 | 0.885 | -0.083 | 0.449 |
| CPDCT028[22] | G2 | 48.6 | 21 | 9.521 | 2.583 | 0.813 | 0.895 | 0.092 | 0.376 |
| CPDCT016[23] | G3 | 28.4 | 21 | 11.157 | 2.656 | 0.833 | 0.910 | 0.085 | 0.299 |
| CPDCT045[18] | G3 | 46.4 | 14 | 7.211 | 2.231 | 0.833 | 0.861 | 0.033 | 0.519 |
| CPSCT034[22] | G4 | 1.8 | 13 | 4.347 | 1.854 | 0.708 | 0.770 | 0.080 | 0.455 |
| CPPCT030[23] | G4 | 16.8 | 16 | 7.189 | 2.273 | 0.688 | 0.861 | 0.201 | 0.326 |
| CPSCT011[22] | G5 | 5.2 | 19 | 9.253 | 2.559 | 0.667 | 0.892 | 0.253 | 0.529 |
| PGS1.21[19] | G5 | 26.7 | 8 | 3.050 | 1.330 | 0.500 | 0.672 | 0.256 | 0.212 |
| CPSCT004[23] | G5 | 30.7 | 5 | 3.504 | 1.399 | 0.833 | 0.715 | -0.166 | 1.502 |
| CPDCT027[18] | G5 | — | 3 | 1.889 | 0.703 | 0.333 | 0.471 | 0.292 | 0.233 |
| BPPCT025[21] | G6 | 56.4 | 18 | 5.654 | 2.176 | 0.583 | 0.823 | 0.291 | 0.324 |
| BPPCT029[24] | G6 | 80.2 | 16 | 10.355 | 2.500 | 0.708 | 0.903 | 0.216 | 0.426 |
| CPPCT006[22] | G7 | 9.5 | 17 | 8.727 | 2.413 | 0.875 | 0.885 | 0.012 | 0.523 |
| pchcms4[21] | G7 | 29.6 | 7 | 2.503 | 1.170 | 0.500 | 0.600 | 0.167 | 0.313 |
| CPSCT018[22] | G8 | 0 | 19 | 8.471 | 2.414 | 0.750 | 0.882 | 0.150 | 0.382 |
| CPDCT008[24] | G8 | — | 14 | 3.266 | 1.695 | 0.458 | 0.694 | 0.339 | 0.158 |
| 整体Whole | — | — | 298 | 143.5 | 2.002 | 0.686 | 0.798 | 0.142 | 0.428 |
新窗口打开|下载CSV
上机前对PCR产物进行变性和纯化[25]。将纯化的PCR产物按比例稀释30倍后,取1 μL纯化后的PCR产物、0.12 μL Liz-500 Size-Standard内标和7 μL的甲酰胺进行混合,在95℃下变性5 min,并立即冷却10 min。利用ABI 3730 DNA遗传分析仪进行荧光电泳检测,并利用ABI GeneMapper V3.0软件与Peak Scanner Software V1.0软件对电泳检测结果进行基因型分型整理。
1.3 数据整理及分析
利用GenAlEx 6.41软件整理基因型数据,计算不同位点或不同居群间的多样性指数、AMOVA(analysis of molecular variance)分析以及不同居群间的Nei’s遗传距离和遗传分化系数等[26]。采用NTSYS-pc 2.1软件对48份材料构建聚类分析图;利用Structure 2.2软件计算不同居群间的遗传结构差异[27],选择混合祖先模型(ancestry admixture models)和相关等位变异频率模型(allele frequencies correlated model)运算。设定种群分类亚群数(K值)为1到10变化,每次种群划分重复运行10次,每次运行迭代次数(markov chain monte carlo,MCMC)为100 000,运行步长(length of burnin peroid)为100 000。依据EVANNO等[28]的方法,利用每个K值所对应的LnPD(D)值计算不同分类亚群的ΔK,并绘制ΔK趋势图。按照EARL等[29]的方法,利用CLUMPP软件进行重复抽样,使用DISTRUCT软件绘制柱形图。
2 结果
2.1 SSR位点的多态性比较
在本研究中,利用22对SSR标记检测了48份不同种质的基因型,共获得298个等位变异,每位点平均检测到13.54个。不同位点上检测到的等位变异数量明显不同,其中PceGA025、CPDCT016和CPDCT028位点检测到的等位变异数最多,均为21个;而CPDCT027位点仅检测到3个等位变异(表2)。有效等位变异(Ne)变异范围为1.889(CPDCT027位点)—12.03(PceGA025位点),整个群体有143.50个,平均有效等位变异数为6.52。与检测到的总等位变异数相比,有51.8%的等位变异属于稀有等位变异。Shannon’s多样性指数(I)变异范围从PceGA025位点的2.969到CPDCT027位点的0.703,整个群体不同位点上平均Shannon’s多样性指数为2.002,但大多数位点上的Shannon’s多样性指数均较小(表2)。观察杂合度(Ho)变异范围从CPSCT039位点的0.958到CPDCT027位点的0.333,整个群体不同位点上的平均值为0.686;而期望杂合度(He)变异范围从PceGA025位点的0.917到CPDCT027位点的0.471,整个群体不同位点上的平均值为0.798(表2)。大多数SSR位点上的Ho值小于He值,表明供试材料在大多数位点上表现出杂合度不足。从Wright’s固定系数F值可以看出,多数位点符合Hardy-Weinberg平衡(接近于0)。除了CPSCT004位点的基因流Nm值大于1外,其余SSR位点上的基因流值均小于1,表明不同SSR位点上的等位变异在供试材料间存在着遗传分化。
2.2 不同品种群间的多样性比较
从表3可以看出,不同品种群间的多样性存在明显差异。通过平均有效等位变异(Ne)和平均Shannon’s多样性指数(I)可以看出,南方品种群的多样性最高,其次为东北品种群;而杏李的多样性最低,且明显低于华北品种群。通过观察杂合度和期望杂合度可以看出,南方品种群和东北品种群具有较高的遗传多样性,而华北品种群与国外品种群相对较低。杏李多样性最低,且观察杂合度远远高于期望杂合度,这种杂合度过量可能与居群间的基因交流相关。该居群固定系数F=-1,表明该居群内繁衰退明显,且与外居群间无任何基因交流。Table 3
表3
表3不同品种群体间的多样性比较
Table 3
| 居群 Population | Ao | Ne | I | Ho | He | F |
|---|---|---|---|---|---|---|
| HB | 77 | 2.743 | 0.977 | 0.645 | 0.527 | -0.203 |
| DB | 141 | 4.035 | 1.509 | 0.686 | 0.712 | 0.024 |
| NF | 191 | 5.141 | 1.709 | 0.682 | 0.736 | 0.074 |
| EX | 83 | 2.874 | 1.129 | 0.718 | 0.624 | -0.165 |
| XL | 39 | 1.773 | 0.536 | 0.773 | 0.386 | -1.000 |
新窗口打开|下载CSV
2.3 不同品种群间的遗传分化
为了分析不同品种群间的多样性差异来源以及近缘种对栽培品种群的影响,通过AMOVA分析,不同品种群间的遗传差异达到极显著水平(F6,40=3.79,P<0.01),但是大部分遗传多样性仍主要来源于居群内变异(69%),居群间的遗传变异仅有31%。遗传分化系数Fst是衡量不同居群间的遗传分化程度,一般认为,当Fst大于0.05时,居群间存在着中等遗传分化;当Fst大于0.15时居群间存在较大的遗传分化[30]。通过表4可以看出,不同栽培中国李居群间均存在着适当的遗传分化,其中国外品种群与南方品种群的遗传分化程度最小;而乌苏里李与所有中国李品种群存在较大的遗传分化;杏李则与北方品种群间有较小的遗传分化,与其余品种群间存在较大的遗传分化程度。同时,Nei’s遗传距离显示乌苏里李相对东北品种群、杏李相对北方品种群均存在较小的距离。
Table 4
表4
表4不同居群间的Nei’s遗传距离(右上角)与遗传分化系数Fst值(左下角)比较
Table 4
| HB | DB | NF | EX | XL | WU | |
|---|---|---|---|---|---|---|
| HB | - | 0.590 | 0.706 | 0.564 | 0.190 | 1.492 |
| DB | 0.126 | - | 0.627 | 0.627 | 0.767 | 0.412 |
| NF | 0.146 | 0.089 | - | 0.366 | 0.916 | 1.313 |
| EX | 0.140 | 0.105 | 0.068 | - | 0.779 | 1.529 |
| XL | 0.086 | 0.189 | 0.207 | 0.214 | - | 1.622 |
| WU | 0.288 | 0.116 | 0.216 | 0.256 | 0.365 | - |
新窗口打开|下载CSV
2.4 基于核SSR的聚类分析
基于SSR基因分型结果对所有中国李及其近缘种进行Jaccard遗传相似系数检测。以普通杏、美洲李和樱桃李为外组对照,对45份中国李及近缘种进行聚类分析,构建树状亲缘关系图。在UPGMA聚类图(图1)中,当阈值为0.83时,将供试材料划分为5个类群,第I类群包括杏李和所有华北品种群材料;第II类主要为以布尔班克为基础的国外育成品种,同时也包括浙江一带的天目蜜李、银醉李等大果型品种;第III类全部为我国南方地方品种,包括西南的青冬李等脆肉型品种、广东三华李、福建?李和芙蓉李等类型;第IV类群则包含东北品种和2份乌苏里李;第V类群仅有1份庐山李,其与其他品种聚类距离最远;自聚类树根部依次为组外对照材料普通杏、美洲李和樱桃李。树状聚类图表明,国外育成的中国李品种与我国南方地方品种具有较近的亲缘关系;而北方品种群与杏李关系密切;东北品种群与乌苏里李关系紧密。图1
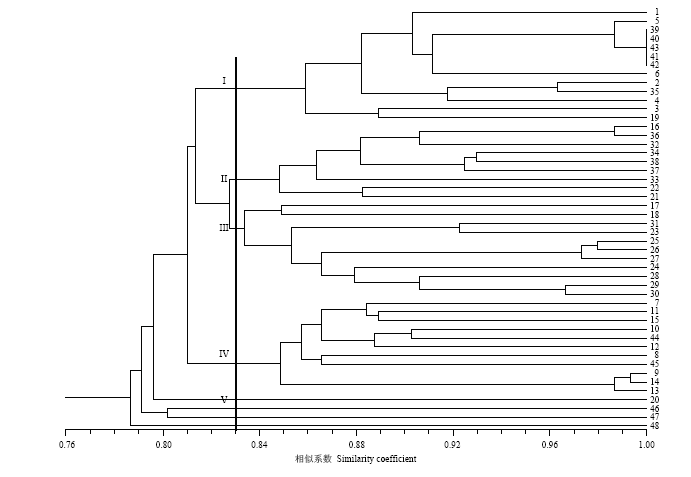 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1基于SSR标记和UPGMA聚类分析48份材料的树形图
Fig. 1A dendrogram of 48 accessions obtained by UPGMA cluster analysis based on SSR markers
2.5 不同来源种质间的群体结构
利用混合祖先模型和等位变异发生频率模型推测中国李不同种质间的群体结构。根据EVANNO等[28]的方法,绘制?K变化趋势图(图2)推测最佳分类群数。当群体数K=4时,?K值最大,认为本研究的中国李种质资源划分为4类较为合理,即华北品种群、东北品种群、南方小果脆肉品种群、南方大果品种群。为了更清楚地阐明不同种质材料间的遗传关系,比较了在不同假设分类群条件下,供试材料的分类划分情况,如图3所示。当K=2时,杏李和来自华北的中国李品种(绿色)与其他供试材料分离为两个类群(红色);当K=3时,乌苏里李和来自东北地区的中国李品种(蓝色)从其余材料中分离出来,独立为新类群,这表明华北品种群和东北品种群分别与杏李、乌苏里李具有相似的血统或相近的血缘关系;当K=4时,来自南方的中国李品种被划分为2个类群,第一组为小果脆肉型品种群(橙色),另一组为大果型品种群(红色)。大多数国外育成品种均属于南方大果型品种群;而南方种质中大部分红肉型种质属于以上2种血缘的混合类型。当K=5时,4种不同来源的中国李中均分离出新的血缘类型(黄色),而杏李和乌苏里李并无此类血统。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2利用作图法推断参试种质的合理组群划分
Fig. 2Reasonable groups number of tested accessions inferred by mapping method
图3
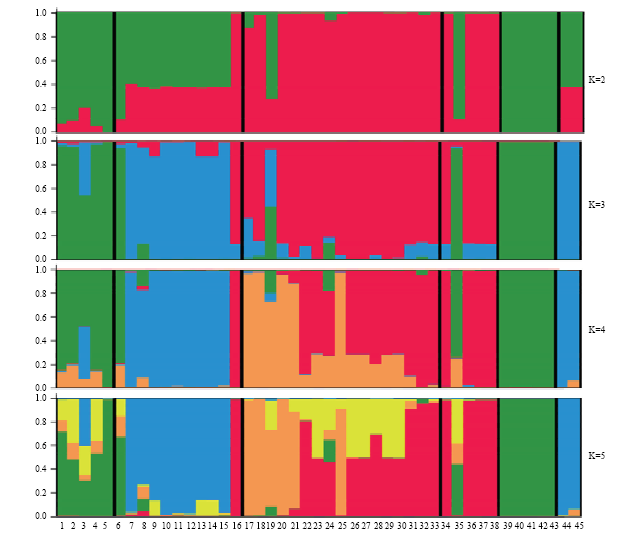 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3中国李种质资源的群体结构
Fig. 3The genetic structure of the plum accessions
3 讨论
3.1 不同来源中国李的多样性比较
在本研究中,22对SSR引物共检测到298个等位变异,平均每个SSR位点检测到13.54个,平均有效等位变异数(Ne)为6.52,平均多数性指数为2.0,平均观察杂合度(Ho)为0.686,平均期望杂合度(He)为0.798。本研究中检测到中国李的多样性略高于孙萍等[31]在47份中国李检测的结果(Na=10.85,Ne=4.81,Ho=0.746,He=0.774);也高于CARRASCO等[32]检测到的(Na=12.1,Ne=5.2)。但是,CARRASCO等[32]的研究中的Ho和He值略高于本研究检测结果,且Ho大于He,这可能与该研究中选用的中国李多为育成品种,含有较多的近缘种有关。陈红等[33]通过ISSR标记认为贵州地区栽培的中国李多样性极为丰富(I=0.508),本研究中SSR数据显示南方品种群的Shannon’s多样性指数为1.709。本研究通过不同地理来源居群间的多样性比较(表3),发现我国南方品种群的多样性(Ne=5.141,He=0.736)远高于其他居群,而东北品种群、国外品种群、华北品种群的多样性依次降低。郁香荷等[34]分析了405份来自全国各地的中国李资源果实大小变异后,发现单果重变异系数为47.09%,其中南方种质的单果重变异最大,而国外品种群和华北品种群的变异系数最小。结合树状聚类图,发现大部分南方种质资源被聚在第III类群中,包括西南的青冬李等脆肉李、广东三华李、福建?李和芙蓉李等类型,但是第II类群中仍有南方品种天目蜜李、银醉李,第I类群有巴塘李,第V类群仅由庐山李组成(图2)。这进一步表明,我国南方种质资源多样性较为丰富,但是这可能受南方品种群取样量大的影响。一般认为,中国李(P. salicina)起源于我国南方长江流域[13]。因此,南方品种群多样性丰富也可能与中国李起源中心有关。在李种质资源考察过程中,笔者在云南、四川一带发现了野生李林的生长,推测我国西南地区可能是中国李的原始驯化中心。
3.2 中国李不同品种群的形成与近缘种关系
在我国,中国李具有悠久的栽培历史,形成了许多优良地方品种群或类型。乔玉山[9]根据RAPD和ISSR标记将中国李品种分为南方品种群和北方品种群两大类。刘威生[10]则依据SSR标记划分为南方品种群、华北品种群和东北品种群3种不同类型,并且将国外育成品种划归到南方品种群。本研究认为中国李可以划分为华北品种群、东北品种群、南方小果脆肉品种群和南方大果品种群(包括国外引进品种)。郭忠仁[8]在孢粉学观察结果上认为中国李的传播是由南向北进行的,且南方李更原始。在本研究中,不同居群间的多样性是从南方品种群、东北品种群、国外品种群、华北品种群的多样性依次降低。东北品种群的多样性(I=1.509)仅次于南方品种群,明显高于华北品种群(I=0.977),这可能与生长在东北地区的乌苏里李(P. ussuriensis)有关。在本研究中,乌苏里李与东北品种群中国李的遗传距离和遗传分化系数明显小于其他品种群,这也进一步表明乌苏里李对东北品种群形成具有重要作用。郁香荷等[34]发现东北品种群果实大小显著低于华北品种群,这可能是该品种群为了适应高纬度的严寒气候降低了果实大小的选择压力。因此,推测东北品种群在提高适应性方面融入了野生近缘种乌苏里李的基因。在本研究中,杏李(P. simonii)的固定系数为-1,表现出极端的完全自交群体,且杏李的基因型高度杂合(Ho=0.773)(表3)。但是,杏李的多样性极低,且与华北品种群遗传分化系数最小。因此,认为杏李是华北品种群中国李高度驯化后的特化类型,且该类型是通过无性繁殖进行传播的[35]。
3.3 南方中国李对现代育成品种形成的作用
SSR标记的亲缘关系分析能够提高育种过程中的亲本选配效率。为了适应果品市场质量需求,从上世纪末至本世纪初以来,我国先后从美国引进大果硬肉型品种,如幸运(Fortune)、安哥诺(Angeleno)和黑宝石(Black Diamond)等具有较强的耐贮运特性,并且这些品种在我国部分地区逐渐成为主栽品种。本研究SSR数据显示国外品种群多样性较低,且与南方品种群的遗传距离最小,仅为0.068。在左力辉等[11]和孙萍等[31]的研究中,也认为国外育成品种与我国南方品种亲缘关系较近。事实上,中国李被世界各国广泛栽培是从我国某地部分李品种通过日本传播到美国开始的[13]。为了提高栽培品种的本地适应性,国外育种者利用本土美洲李(P. amercana)、樱桃李(P. cerasifera)与仅有的少量中国李进行种间杂交,先后培育出百班克(Baibanke)、美丽李(Beauty)和福摩萨(Formosa)等系列种间杂交品种,这些材料成为国内外现代李育种的主要亲本来源[13]。在本研究中,国外育成品种与南方江浙一带的大果型品种聚类在一起,而与美洲李和樱桃李的关系较远,这可能是由于多代多次杂交后国外育成品种保留了较少的美洲李或樱桃李血缘。另外,以上结果也暗示国外育成品种的间接亲本可能来源于我国南方江浙一带,而我国育种亲本则多选择华北地方品种进行相互杂交,这为我国李育种提供了重要借鉴。本研究选用的标记种类和样本量有限,为此在后续研究中将通过大样本取样和全基因组变异检测(如GBS测序)等途径对以上结论进行进一步验证。4 结论
南方品种群位于中国李栽培驯化中心,其多样性最为丰富;江浙一带的南方大果型种质对国外品种群的形成具有重要影响。在华北品种群中,通过高度驯化和人工选择形成一类以无性繁殖为基础的、果实香气浓郁的杏李(P. simonii)类型。东北品种群形成则与乌苏里李(P. ussuriensis)基因渗透有关。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1007/s11295-009-0226-9URL [本文引用: 1]

The genus Prunus contains the subgenus Prunus incorporating the European plums (section Prunus ), the North American plums (section Prunocerasus ) and the apricots (section Armeniaca ). In section Prunus , there are approximately 20 species, which occur in three levels of ploidy, diploid \( \left( {2n = 2x = 16} \right) \) , tetraploid \( \left( {2n = 4x = 32} \right) \) and hexaploid \( \left( {2n = 6x = 48} \right) \) . Despite a clear distinction between section Prunus and the other sections, phylogenetic relationships between species within the section are unclear. We performed a phylogenetic analysis on members of the section Prunus and three outgroup species using sequence data from four single-copy phylogenetically informative chloroplast DNA regions ( atp B- rbc L, mat K, rpl 16, and trn L- trn F). After alignment, the analysed regions totalled 4,696 bp of sequence, containing 68 parsimony-informative sites and 14 parsimony-informative indels. Data were analysed using both maximum parsimony and Bayesian likelihood and phylogenetic trees were reconstructed. The analyses recovered trees with congruent topologies and similar levels of statistical support for relationships between taxa. They confirmed that species belonging to section Prunus form a monophyletic clade within Prunus . The section is resolved into four well-supported clades, which correspond to the geographical distribution of the species. The hexaploid species could not be resolved into distinct species clades but formed a well-supported group separate from the tetraploid species, highlighting the distinct evolutionary origins of the different polyploid groups. The close relationship between the hexaploids and Prunus divaricata , Prunus cerasifera and Prunus ursina indicates the former may have derived from an ancestor of P. cerasifera and its allies.
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1080/14620316.2006.11512057URL [本文引用: 1]

One hundred-and-eight accessions of plum and apricot, including seven plum species (Prunus salicina, P. simonii, P. ussuriensis, P. domestica, P. cerasifera, P. nigra and P. spinosa), seven accessions of wild European plum from Xinjiang, two related species (P. armeniaca and P. tomentosa) and eight hybrids between plum and apricot were examined using random amplified polymorphic DNA (RAPD) markers. Cluster analysis using the unweighted paired group method of arithmetic means (UPGMA) showed four main clusters: a group comprising apricot (P. armeniaca) and hybrids between apricot and plum; a Chinese plum-type group; a European plum group (P. domestica); and an out-group including P. ussuriensis, P. cerasifera, P. nigra, P. spinosa and P. tomentosa. Data also revealed that P. simonii should be a variant of P. salicina, and P. ussuriensis is an independent species. P. cerasifera and P. spinosa were distinct from the European plum group, disproving the hypothesis that P. domestica originated from natural hybrids of P. cerasifera and P. spinosa.
DOI:10.1007/s10658-007-9180-2URL [本文引用: 2]

ABSTRACT Inter-simple sequence repeat (ISSR) markers were used to evaluate genetic similarity and interrelationship among 104 plum (Prunus L. spp.) and related accessions from the Chinese National Germplasm Repository for Plums and Apricots and the Tianshan Germplasm Repository for Wild Fruit Resources, including six plum species (Prunus salicina Lindl., Prunus simonii Carr., Prunus ussuriensis Kov. et Kost., Prunus domestica L., Prunus cerasifera Ehrh., and Prunus spinosa L.), two related species [apricot (Prunus armeniaca L.) and nanking cherry (Prunus tomentosa Thunb.)], eight putative hybrids between plum and apricot (plumcot), and six accessions of wild European plum (P. domestica). Out of the 42 ISSR primers, 12 were selected, which generated 103 markers in total, 99 of which were polymorphic. Possible accession-specific ISSR bands or patterns were also found. Some possible synonyms or homonyms were clarified or discussed, and closely related accessions such as bud mutants were discriminated. Based on the unweighted pair group method with arithmetic mean (UPGMA) analysis and principal coordinate analysis (PCoA) using the Jaccard coefficient, two different dendrograms were constructed - one including accessions grouped by species and one with all 104 accessions - and a two-dimensional plot was obtained. Three groups were formed in both dendrograms and PCoA plot: Group I including apricot ('Yinxiangbai') and plumcot types; Group II containing Asia-originated diploid species [e.g., P. cerasifera, P. ussuriensis, P. tomentosa, and Chinese plum-types (i.e., P. salicina and its hybrids)]; and Group III involving European-origin polyploid species (e.g., P. spinosa and P. domestica) and recently found wild European plum accessions in China. The dendrogram with accessions grouped by species implied that 1) plumcot types had closer relatedness with apricot than with plum; 2) P. simonii should be a variant of P. salicina while P. ussuriensis an independent species; 3) P. domestica was more closely related to P. spinosa than to P. cerasifera. Two accessions of European plum ('89-7-3' and 'Wanhei') were clustered into outgroups in the dendrogram with all 104 accessions, which could been grouped within Group III in the PCoA plot. The distribution of both European plum and Chinese plum-types across respective groups did not reflect the geographic origins. The present study also further confirmed that the wild plants found in Xinjiang of China were P. domestica.
[本文引用: 1]
DOI:10.1002/9780470650585.ch10URL [本文引用: 1]

The article discusses the origin of various plum species, development of plum cultivars throughout the centuries, and present day distribution of production. The first archeological information on wild plums species dates back to Ukraine to 4000 B.C. Between this time and about 300 B.C. only seeds of Damson plum and sloe were found in graves. Prunus domestica, the garden plum or prune, and the Japanese plum have no wild ancestors. The origin of garden plum is likely to be hybrid and developed or found in eastern Turkey. Because of the hybrid origin there are several subgroups of this species including the Reine-Claude and Yellow egg plums. Another species, P. insititia, includes the Mirabelle and St. Julien plum groups. The origin and development of all these plums are detailed. There are several American species which were sparingly used by the Indians as fruit in their diet. The improvement of American species started early in the 19th century and continued until about 1960. Luther Burbank combined many plum species and cultivars and developed a new group of plums which are the basis of the plum production today. In contrast the prune industry is based on an ancient cultivar, 'Agen', that dates back to time of the Crusades and was imported to California in 1856 under the name of 'French Prune'. Present production of plums worldwide is estimated at 7.2 million tons. United States production is about one million tons, 60% of which is used for dried prune production.
[D].
[本文引用: 2]
[D].
[本文引用: 2]
[D].
[本文引用: 2]
[D].
[本文引用: 2]
[D].
[本文引用: 2]
[D].
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 4]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
DOI:10.1007/s001220051499URL [本文引用: 4]

Microsatellites have emerged as an important system of molecular markers. We evaluated the potential of microsatellites for use in genetic studies of peach [ Prunus persica (L.) Batsch]. Microsatellite loci in peach were identified by screening a pUC8 genomic library, a 位ZAPII leaf cDNA library, as well as through database searches. Primer sequences for the microsatellite loci were tested from the related Rosaceae species apple ( Malus domestica ) and sour cherry ( Prunus cerasus L.). The genomic library was screened for CT, CA and AGG repeats, while the cDNA library was screened for (CT) n - and (CA) n -containing clones. Estimates of microsatellite frequencies were determined from the genomic library screening, and indicate that CT repeats occur every 100 kb, CA repeats every 420 kb, and AGG repeats every 700 kb in the peach genome. Microsatellite- containing clones were sequenced, and specific PCR primers were designed to amplify the microsatellite- containing regions from genomic DNA. The level of microsatellite polymorphism was evaluated among 28 scion peach cultivars which displayed one to four alleles per primer pair. Five microsatellites were found to segregate in intraspecific peach-mapping crosses. In addition, these microsatellite markers were tested for their utility in cross-species amplification for use in comparative mapping both within the Rosaceae, and with the un- related species Arabidopsis thaliana L.
[本文引用: 3]
DOI:10.1007/s11032-011-9685-4URL [本文引用: 3]

AbstractSharka disease, caused by the plum pox virus (PPV), is one of the major limiting factors for stone fruit production in Europe and America. Attempts to stop the disease through the eradication of infected trees have been unsuccessful. Introgression of PPV resistance for crop improvement is therefore the most important goal in Prunus breeding programs. Due to time- and labour-consuming protocols, phenotyping for sharka is still the major bottleneck in the breeding pipeline. In this context, screening of seedlings at early stages of development and marker-assisted selection (MAS) provide the best solution for enhancing breeding efficiency. In this study, we generated 42 simple sequence repeat (SSR) markers from the peach genome assembly v1.0 and an apricot bacterial artificial chromosome clone identified in the physical map of the PPV resistance locus previously defined in apricot. Using a linkage mapping approach, we found SSR markers tightly linked to PPV resistance trait in all our progenies. Three SSR markers, PGS1.21 PGS1.23 and PGS1.24, showed allelic variants associated with PPV resistance with no recombinants in the crosses analysed. These markers unambiguously discriminated resistant from susceptible accessions in different genetic backgrounds. The results presented here are the first successful application of their use in MAS for breeding resistance in Prunus species.
DOI:10.1046/j.1471-8286.2002.00132.xURL [本文引用: 1]

The sequences of 21 primer pairs of microsatellite loci screened from a genomic library of apricot ( Prunus armeniaca L.) are reported in this study. All the identified microsatellite loci were characterized in a set of 25 apricot cultivars and revealed to be polymorphic with 3–12 alleles per locus. These markers showed to be more informative than isozymes and restriction fragment length polymorphisms (RFLPs) reported in the literature.
[本文引用: 3]
DOI:10.1111/j.1471-8286.2004.00603.xURL [本文引用: 5]

Thirty-five polymorphic microsatellites were developed using a CT/AG enriched genomic library of Japanese plum cv. Santa Rosa. Twenty-seven of them detected a single locus and eight two or more loci. A high level of variability was observed in a set of eight cultivars for the 27 single-locus microsatellites: 5.7 average number of alleles per locus; 73% mean observed heterozygosity and 74% discrimination power. Most SSRs were transferable to peach (85%) and almond (78%).
DOI:10.1111/j.1471-8286.2005.00977.xURL [本文引用: 3]

We report 47 new simple sequence repeats (SSRs) obtained from a CT/AG enriched genomic library of almond cv. Texas (syn. Mission). Forty-two of them were polymorphic in a sample of eight almond cultivars and 31 of these were single-locus. The average values of the number of alleles per locus (6.6), and mean observed (65%) and expected (76%) heterozygosities for these 31 SSRs indicated a high level of variability. All cultivars studied could be individually identified using any one of the five SSRs. Transportability to other Prunus species (peach, sweet cherry, Japanese plum and apricot) was also high (83 100%).
DOI:10.1046/j.1439-0523.2002.00656.xURL [本文引用: 2]

A genomic DNA library enriched with AG/CT repeats has been developed from the peach cultivar 'Merrill O'Henry'. The enrichment method was efficient, with 61% of the clones obtained carrying a microsatellite sequence and a yield of one polymorphic microsatellite every 2.17 sequenced clones. From 35 microsatellites detected, 24 were polymorphic in a set of 25 cultivars including 14 peaches and 11 nectarines. A total of 82 alleles were found with the polymorphic microsatellites, with an average of a 37% of observed heterozygosity. Microsatellites with a high number of repeats were generally those having the largest number of alleles. All cultivars except two ('Spring Lady' and 'Queencrest') could be individually distinguished with the markers used. Just three selected microsatellites were enough for the discrimination of 24 out of the 25 possible genotypes. Cluster analysis grouped all nectarines in a single cluster. Peaches, with 75 of the 82 alleles found, were more variable than nectarines, with only 64. Microsatellites appear to be powerful and suitable markers for application in peach genetics and breeding.
[本文引用: 1]
[本文引用: 1]
DOI:10.1111/j.1471-8286.2005.01155.xURL [本文引用: 1]

genalex is a user-friendly cross-platform package that runs within Microsoft Excel, enabling population genetic analyses of codominant, haploid and binary data. Allele frequency-based analyses include heterozygosity, F statistics, Nei's genetic distance, population assignment, probabilities of identity and pairwise relatedness. Distance-based calculations include amova , principal coordinates analysis (PCA), Mantel tests, multivariate and 2D spatial autocorrelation and twogener . More than 20 different graphs summarize data and aid exploration. Sequence and genotype data can be imported from automated sequencers, and exported to other software. Initially designed as tool for teaching, genalex 6 now offers features for researchers as well. Documentation and the program are available at http://www.anu.edu.au/BoZo/GenAlEx/
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
