 ,1, 肖钢2, 官春云
,1, 肖钢2, 官春云 ,1
,1Regulation of GT and GATA Transcription Factors on Promoter Function of BnA5.FAD2 and BnC5.FAD2 Genes in Brassica napus
LIU Fang ,1, XIAO Gang2, GUAN ChunYun
,1, XIAO Gang2, GUAN ChunYun ,1
,1通讯作者:
收稿日期:2018-07-13接受日期:2018-09-25网络出版日期:2018-12-26
| 基金资助: |
Received:2018-07-13Accepted:2018-09-25Online:2018-12-26

摘要
关键词:
Abstract
Keywords:
PDF (3237KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
刘芳, 肖钢, 官春云. GT和GATA转录因子对甘蓝型油菜BnA5.FAD2和 BnC5.FAD2启动子功能的调控[J]. 中国农业科学, 2018, 51(24): 4603-4614 doi:10.3864/j.issn.0578-1752.2018.24.002
LIU Fang, XIAO Gang, GUAN ChunYun.
0 引言
【研究意义】油酸是多种脂肪酸的前体,通常存在于内质网中,以油酸-CoA、油酸-PLA、油酸-DAG、油酸-PC和油酸-TAG等形式参与油酸代谢。脂肪酸去饱和酶基因(FAD2)是负责将油酸转化成亚油酸的关键基因,甘蓝型油菜中位于A5和C5染色体上的FAD2对于油菜种子中油酸含量具有重要影响[1]。目前,高油酸育种及其形成机理已然成为各类油料作物的研究热点[2]。通过研究其转录调控机制,为高油酸油菜育种提供分子理论基础。【前人研究进展】目前,植物脂肪酸去饱和酶主要在转录水平被调控[3],而且FAD2的转录水平受外界刺激影响。例如,在低温诱导下,棉花FAD2转录物水平增加[4];酿酒酵母中油酸调控基因OLE1的转录水平会被外源性不饱和脂肪酸抑制,但在缺氧条件下转录水平会增加[5]。真核生物的转录过程十分复杂,需要多种转录因子参与,转录因子多与启动子上的特异序列结合,起始转录或调节转录水平。GATA家族转录因子通过识别并结合GATA元件调节基因的转录水平。GATA家族转录因子广泛存在于动、植物和真菌中,对(T/A)GATA(A/G)序列具有很高的亲和力。1991年首次发现GATA-1,与GATA-2、GATA-3、GATA-4、GATA-5、GATA-6组成GATA家族[6]。GATA元件通常位于启动子、转录起始位点的上游和增强子中[7]。在植物中,发现含有GATA序列的I-box元件存在于rbcs、cab和nia启动子的上游,对光和昼夜节律有响应[8]。GT转录因子通常与GT元件结合,调节基因表达以响应内、外部刺激。目前,仅在植物中发现GT转录因子[9],与之结合的GT元件通常是富含T和A的核心序列[10]。GT转录因子最初在豌豆核中被发现,与GT-1元件结合[11]。研究表明GT元件可以参与光调节[12,13],参与野生大麦水分胁迫的调节[14]。GT元件的拷贝数以及拷贝之间的间隔对启动子的转录功能都有影响[15]。不同的GT转录因子与GT元件结合后,会发挥不同的调节功能[16]。LE GOURRIEREC等[17]发现GT-1转录因子可以结合并稳定酵母细胞中的TF IIA-TBP-TATA复合物以调节基因表达,认为GT-1转录因子可以与转录起始复合物结合直接参与转录调节。【本研究切入点】目前,高油酸育种已成为研究热点,对于FAD2的研究也较普遍,但对于转录水平调控的研究鲜有报道。【拟解决的关键问题】本研究通过敲除部分启动子片段来研究2个启动子BnA5.FAD2和BnC5.FAD2的同源区域,并找到控制基因表达的关键启动子区域。通过PlantCARE网站分析,获得潜在的调控元件,借助PlantTFDB网站筛选可能与之结合的转录因子,继而分别在酵母和拟南芥中验证启动子序列和转录因子的互作关系,为高油酸油菜育种奠定基础。1 材料与方法
1.1 试验材料
甘蓝型油菜湘油15种在湖南农业大学耘园基地大田,于2017年3月至5月取不同生长时期的种子。拟南芥(Arabidopsis thaliana,Col-0)由湖南农业大学油料作物研究所提供,种在培养箱内,24℃/16 h光照,22℃/8 h黑暗培养。pEASY-T1载体、大肠杆菌T1感受态购于北京全式金公司。酵母菌种Y187(-Trp/-Leu/-His)由山东农业大学作物生物学国家重点实验室馈赠,酵母单杂交报告载体pHIS2、pGADT7载体、PRI101载体为湖南农业大学油料研究所保存。本研究中所用引物均由南京金斯瑞公司合成。1.2 BnA5.FAD2和BnC5.FAD2启动子同源区的分析
BnA5.FAD2启动子区分为4个不同的区域,分别命名为A(-1—-213 bp)、B(-214—-225 bp)、C(-226—-314 bp)和D(-315—-685 bp),分别构建载体HM-DCBA:GUS/GFP(-685—-1 bp+内含子)、HM-CBA:GUS/GFP(-314—-1 bp+内含子)、HM-BA:GUS/GFP(-225—-1 bp+内含子)和HM-A(-213—-1 bp +内含子)(图1-a)。BnC5.FAD2启动子区分为4个不同的区域,分别命名为A′(-1—-215 bp)、B′(-216—-227 bp)、C′(-228—-312 bp)和D′(-313—-671 bp),分别构建载体HM-D′C′B′A′:GUS/GFP(-671—-1 bp+内含子)、HM-C′B′A′:GUS/GFP(-312—-1 bp+内含子)、HM-B′A′:GUS/GFP(-227—-1 bp+内含子)和HM-A′(-215—-1 bp+内含子)(图1-b)。分别敲除C、C′区域,构建植物表达载体(图1-c和图1-d)。分别敲除GT元件和GATA元件核心序列,构建植物表达载体(图2-a和图2-b)。载体构建中所用引物见电子附表1。所有构建的载体通过冻融法[18]转入农杆菌菌株GV3101中。然后通过花序浸染法转化野生拟南芥[19]。在含有50 mg·L-1潮霉素B的MS平板上筛选转基因植株,然后移植到土壤中培养。从T2代转基因拟南芥种子中提取总蛋白质,通过Western blot检测GFP蛋白含量,具体操作参考刘睿洋等[20]方法。1.3 GATA家族和GT家族基因的筛选及克隆
利用PlantTFDB网站分析,从甘蓝型油菜中获得31个GATA转录因子基因,其中19个完整序列,与油菜数据库(1.4 酵母单杂交鉴定启动子与转录因子的互作
根据Infusion试剂盒(TaRaKa,Japan)说明书,将BnA5.FAD2和BnC5.FAD2启动子克隆到pHIS2.0载体中的GAL4-DNA结合结构域中,将GATA家族和GT家族转录因子编码区序列与pGADT7载体中的GAL4激活结构域融合。将pHIS2-A5-promoter和pHIS2-C5-promoter载体分别转化到营养缺陷型酵母菌株Y187,筛选出阳性菌株后将其涂布到SD-Trp固体平板和含不同浓度3-AT(0、10、20、30、40、50、60、70、80、90和100 mmol·L-1)的固体SD-Trp-His培养基上,30℃培养3—5 d,观察其生长情况,筛选最适3-氨基-1,2,4-唑(3-AT)生长浓度。按照电子附表2的组合进行酵母转化,验证GATA和GT家族基因的功能。将含有转化子的酵母菌涂在营养缺陷培养基上(SD-Trp、SD-Trp-Leu-His+适当浓度的3-AT),30℃培养3—5 d。观察酵母生长情况。
1.5 在拟南芥中的分析转录因子与启动子的互作情况
为了鉴定转录因子基因Bna010243-1、Bna026124-1、Bna026124-2、Bna010915和Bna013749的功能,将其分别克隆并与pRI101载体连接,筛选标记为卡那霉素,构建载体时将血细胞凝集素(HA)标签与转录因子基因融合,同时转入载体pRI101,便于进一步检测是否转化成功。构建载体命名为pRI101-Bna010243-1∷HA、pRI101-Bna026124-1∷HA、pRI101-Bna026124- 2∷HA、pRI101-Bna010915∷HA和pRI101-Bna013749∷HA。将以上载体分别转化已含有BnA5.FAD2和BnC5.FAD2启动子片段的拟南芥。经50 mg·L-1潮霉素B+70mg·L-1卡那霉素的MS平板筛选转基因拟南芥,然后移植到土壤中继续培养。从T2代转基因拟南芥种子中提取总蛋白质通过Western blot检测HA蛋白含量,用于判断转录因子是否成功转化拟南芥,通过Western blot检测GFP蛋白含量用于判断转录因子对于启动子功能的影响,从而判断转录因子是否与启动子序列互作,具体操作同1.2。2 结果
2.1 GT和GATA顺式作用元件在调节BnA5.FAD2和BnC5.FAD2转录中的作用
湖南农业大学油料研究所前期克隆了BnA5.FAD2和BnC5.FAD2启动子序列[21,22],二者约680 bp序列(BnA5.FAD2,686 bp;BnC5.FAD2,676 bp)保持91%的同源性,这一同源区域对于基因表达量的贡献巨大。BnA5.FAD2和BnC5.FAD2启动子区分别分为4个区域A(A′)、B(B′)、C(C′)和D(D′),构建植物表达载体,转化拟南芥,采用Western blot技术检测报告基因GFP的蛋白丰度。结果显示,当区域D和D′缺失时,GFP蛋白的丰度增加,表明该区域可能存在一些负向调控元件,抑制基因表达。随着区域C和C′的进一步缺失,GFP蛋白丰度下降,预示该区域中存在一些用于控制基因表达的正调控元件;进一步敲除区域B和B′后,HM-A(A′)的GFP蛋白丰度一定程度的高于HM-BA(B′A′),即GFP蛋白丰度呈现增加的现象,说明B(B′)区存在一些负调控元件调节基因表达(图1-e和图1-f)。可见,BnA5.FAD2启动子C(-226—-314 bp)区域和BnC5.FAD2启动子C′(-228—-312 bp)区域是保持基因高度表达的关键区域。为了进一步验证C(-226—-314 bp)区域和C′(-228—-312 bp)的功能,分别敲除这两个区域并构建载体-C和-C′(图1-c和图1-d)。结果发现,区域C和C′缺失时,GFP蛋白质丰度均降低(图1-g和图1-h)。表明BnA5.FAD2启动子的-226—-314 bp和BnC5.FAD2启动子的-228—-312 bp是控制甘蓝型油菜种子中BnFAD2-A5和BnFAD2-C5表达的关键区域。
图1
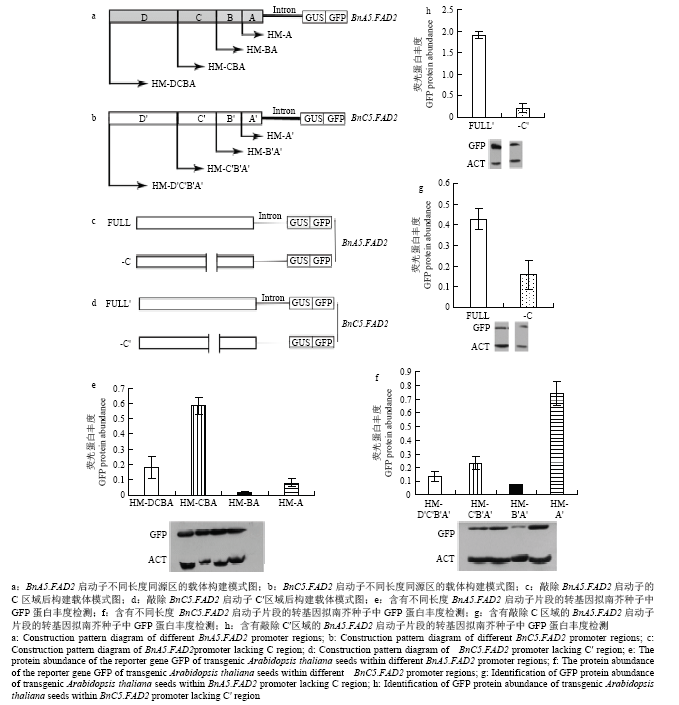 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1 BnA5.FAD2和BnC5.FAD2启动子不同区域的功能分析
Fig. 1Functional analysis of BnFAD2-A5 and BnFAD2-C5 promoters in different regions
BnC5.FAD2启动子的-226—-314 bp和BnC5.FAD2启动子的-228—-312 bp仅存在一个碱基差异,经PlantCARE软件预测发现这两个区域均存在GT和GATA顺式作用元件,分别为GT家族和GATA家族转录因子的结合位点。分别敲除2个元件并构建载体-GT和-GATA与-GT′和GATA′(图2-a和图2-b),转化拟南芥进行功能分析。结果显示,分别单独敲除BnC5.FAD2启动子的GT和GATA顺式作用元件后,均会造成GFP蛋白丰度显着下降(图2-d),表明BnC5.FAD2启动子的GT和GATA顺式作用元件在调节基因转录中起重要作用。分别单独敲除BnA5.FAD2启动子的GT和GATA顺式作用元件后,GFP蛋白丰度显著下降(图2-c),表明BnA5.FAD2启动子中GT和GATA顺式作用元件在调节基因转录中起重要作用。
图2
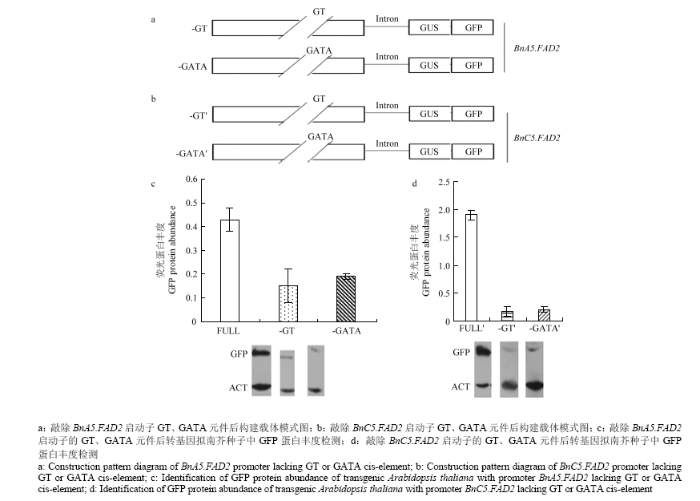 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2敲除GT和GATA元件后BnA5.FAD2和BnC5.FAD2启动子的功能分析
Fig. 2Identification analysis by knocking out the GT or GATA cis-element in promoter BnA5.FAD2 and BnC5.FAD2
2.2 GATA家族和GT家族基因的表达分析和蛋白序列分析
通过qRT-PCR分析转录因子表达规律(图3),发现GATA转录因子家族中Bna010243的表达规律与BnA5.FAD2一致,而Bna006754、Bna015605和Bna026124的表达规律与BnC5.FAD2一致;GT转录因子家族中Bna013749的表达规律与BnA5.FAD2一致,Bna010915的表达规律与BnC5.FAD2一致。图3
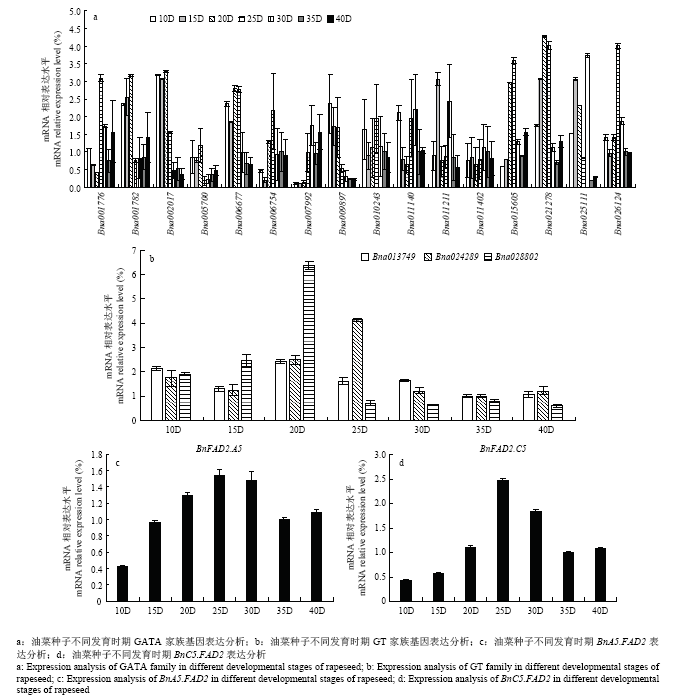 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3GATA和GT家族基因的表达分析
Fig. 3Expression analysis of GATA family and GT family
以甘蓝型油菜湘油15基因组和cDNA基因组序列为模板,克隆GATA家族和GT家族基因,分别为Bna006754(903 bp,301)、Bna010243-1(594 bp,198)、Bna010243-2(567 bp,189)、Bna015605(963 bp,321)、Bna026124-1(546 bp,182)、Bna026124-2(582 bp,194)、Bna010915(681 bp,227)和Bna013749(1 239 bp,413)。GATA家族蛋白均具有保守的单个18个残基的锌指结构(CX2CX18CX2C),与其他作物中GATA蛋白结构相同(图4-a),GT家族中Bna013749蛋白和Bna010915蛋白的3个α-螺旋构成Trihelix结构,而且每个串联重复区具有1个色氨酸残基,符合W-Xn-W-Xn-W的结构特点,Bna010915蛋白第3个α-螺旋中的保守W被F替代(图4-b),与GT-2家族的结构特点相同[23],可能属于GT-2亚家族成员。
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4GATA和GT家族蛋白序列分析
Fig. 4Protein sequence analysis of GATA and GT family
2.3 GATA和GT家族在酵母中的功能分析
为了排除报告基因基底水平表达的影响,需确定适当浓度的3-AT,排除干扰。结果表明,pHIS2-A5- promoter的最佳浓度为50 mmol·L-1,pHIS2-C5-promoter动子的最佳浓度为35 mmol·L-1(图5-a)。将共转化的酵母菌液涂布在SD-Trp-Leu(SD-TL)和SD-Trp- Leu-His(SD-TLH)+3-AT(35 mmol·L-1/50 mmol·L-1)的固体培养基上,并在30℃条件下孵育。观察生长情况3—5 d(图5-b),所有共转化的酵母组合物可以在SD-T-L固体培养基平板上正常生长。在SD-TL-H+ 50 mmol·L-1 3-AT三缺板上,与pHIS2-A5-promoter共转化酵母的Bna010243-1、Bna026124-1、Bna026124-2、Bna010915、Bna013749可以生长,即表明Bna010243-1、Bna026124-1、Bna026124-2、Bna010915和Bna013749可以与BnA5.FAD2启动子相互作用。在SD-T-L-H+35 mmol·L-1 3-AT三缺板上,与pHIS2-C5-promoter共转化酵母的Bna010243-1、Bna026124-1、Bna026124-2、Bna010915和Bna013749可以生长,即表明BnC5.FAD2启动子也可与Bna010243-1、Bna026124-1、Bna026124-2、Bna010915和Bna013749相互作用。图5
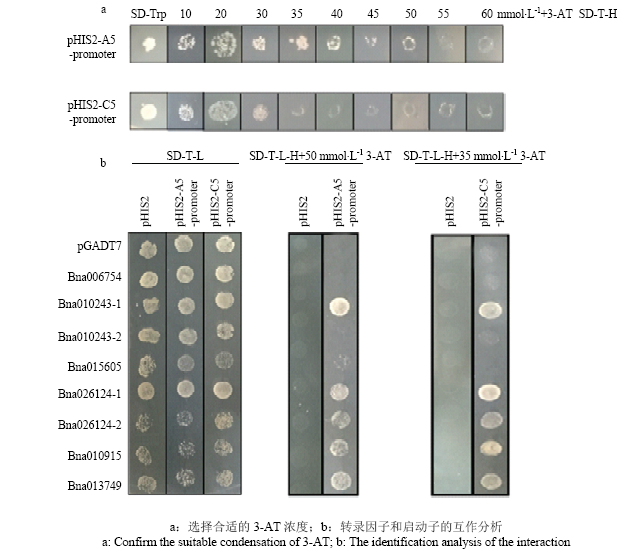 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5酵母单杂交分析转录因子和启动子BnFAD2-A5和BnFAD2-C5的互作
Fig. 5Analysis of the interaction between transcription factors and promoter BnFAD2-A5 or BnFAD2-C5 using yeast one-hybrid method
2.4 GATA和GT家族在拟南芥中与BnA5.FAD和BnC5. FAD2启动子互作分析
将GATA转录因子家族的Bna010243-1、Bna026124-1和Bna026124-2分别构建到pRI101载体中,然后分别转化具有HM-CBA、HM-C′B′A′、-GATA或-GATA′的转基因拟南芥,(图6-i和图6-j);将GT转录因子家族的Bna010915和Bna013749分别构建到pRI101载体中,转化具有HM-CBA、HM-C′B′A′、-GT和-GT′的转基因拟南芥(图6-k和图6-l)。经western blot检测证明转录因子均转染拟南芥,并且可以正常表达(图6-m)。检测报告基因GFP蛋白丰度的变化,鉴定转录因子与BnA5.FAD2或BnC5.FAD2启动子互作与否(图6),当拟南芥含有GATA家族转录因子基因之一和HM-CBA或HM-C′B′A′时,GFP蛋白质丰度有不同程度的增加(图6-a和图6-e)。但当GATA顺式作用元件被敲除时,GFP蛋白丰度没有显著变化(图6-b和图6-f)。说明转录因子基因Bna010243-1、Bna026124-1和Bna026124-2可以与BnA5.FAD2或BnC5.FAD2启动子发生相互作用,而且作用位点在GATA元件这一位置,而且互作的结果均是提高基因表达量。当Bna010915和HM-CBA或HM-C′B′A′共转化拟南芥时,相对于仅含有HM-CBA或HM-C′B′A′的拟南芥,GFP蛋白丰度略有增加(图6-c和图6-g);当基因Bna013749和HM-CBA或HM-C′B′A′共转化到拟南芥中时,GFP蛋白丰度降低(图6-c和图6-g);当GT顺式元件被敲除时,共转化之后的拟南芥GFP蛋白丰度没有显著变化(图6-d和图6-h)。推测Bna010915可能作为一个正调控因子,Bna013749作为一个负调控因子,二者均可与BnA5.FAD2或BnC5.FAD2启动子区的GT元件相互作用,调节基因的表达水平。图6
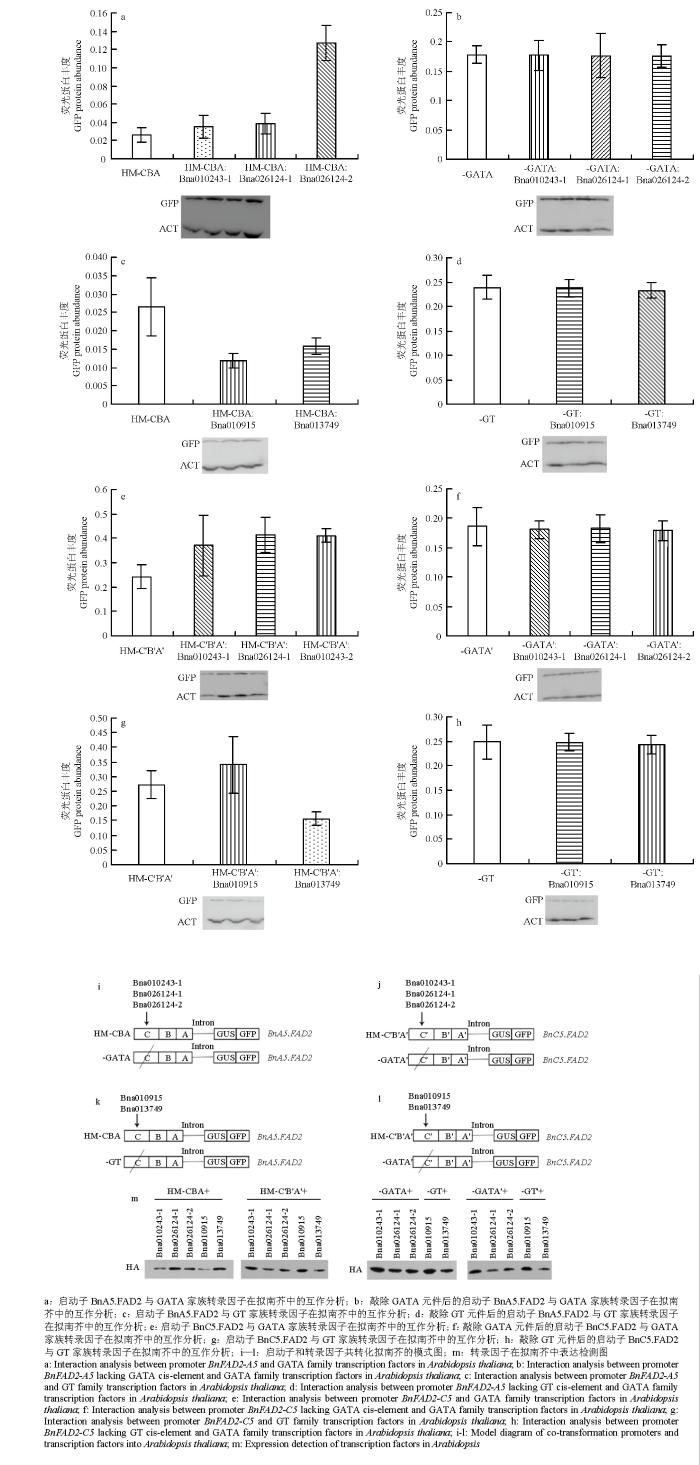 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6转录因子和启动子在拟南芥中的互作分析
Fig. 6The interaction analysis of transcription factor and promoters in Arabidopsis thaliana
3 讨论
3.1 GT转录因子与启动子互作功能分析
转录水平的调控在植物基因调控中起重要作用,这一过程涉及多种顺式作用元件和反式作用因子。启动子上的顺式作用元件与特定的转录因子结合能够调节基因转录的起始,转录效率以及时空特异性[24]。本研究预测到2个潜在顺式元件GATA和GT,与之结合的转录因子为GATA家族和GT家族转录因子。GT元件首先在豌豆叶中的rbcS-3A启动子中被鉴定为光反应元件[11],但在多数启动子中功能不明确。本研究发现当GT元件或GATA元件分别被单独敲除时,报告基因GFP的表达量出现不同程度的降低,初步说明GT和GATA可能与其相应的转录调控因子结合后调控基因的表达上调。在水稻phyA低表达的成熟组织中,属于GT家族的OsGT-2转录因子可以上调phyA的表达量,将水稻启动子phyA转入烟草原生质体中分析,发现OsGT-2也是表现为上调因子的功能[25]。GT元件结合GT转录因子可以调节基因转录水平以抵抗不利环境[26]。GT家族基因属于Trihelix家族,Trihelix家族是植物所特有的[27],而且trihelix结构域具有高度保守性[28]。本研究发现,GT家族基因Bna013749和Bna010915均具有trihelix结构,且符合W-Xn-W-Xn-W结构特点,其中Bna010915蛋白第3个α-螺旋中的保守W被F替代,与GT-2家族的结构特点相同,可能属于GT-2亚家族成员。研究发现ASIL1属于trihelix家族的新的亚科,作为拟南芥种子基因中的负调节因子[29]。本研究发现,Bna013749对于BnA5.FAD2和BnC5.FAD2启动子也起到负调控的作用,与ASIL1功能相似,推测其可能与ALIL1属于同一亚科。
3.2 GATA转录因子与启动子的互作功能分析
GATA家族转录因子识别GATA元件并与WGATAR(W是T或A;R是G或A)序列结合[30]。 该家族的DNA结合结构域通常具有广泛存在于真核生物中的Ⅳ锌指(CX2CX17-20CX2C)的结构[31]。本文中克隆的GATA转录因子基因都具有Ⅳ锌指(CX2CX18CX2C)(图4),BnA5.FAD2和BnC5.FAD2启动子上含有WGATAR序列,推测本研究中克隆的GATA转录因子可能与BnA5.FAD2和BnC5.FAD2启动子上的GATA序列相互作用,参与基因表达的调控,但需要进一步的试验验证。酵母单杂交试验以及转录因子Bna010243-1、Bna026124-1、Bna026124-2与部分启动子序列(HM-CBA或HM-C′B′A′)共转染拟南芥的试验结果表明,Bna010243-1、Bna026124-1、Bna026124-2可以与BnA5.FAD2和BnC5.FAD2启动子中GATA元件相互作用(图6),而且3个转录因子均表现为正向调控的功能。4 结论
GATA家族的转录因子Bna010243-1、Bna026124- 1、Bna026124-2可以与BnA5.FAD2和BnC5.FAD2启动子中GATA元件相互作用,增强基因的表达水平。GT家族的转录因子Bna010915可以与BnA5.FAD2和BnC5.FAD2基动子中的GT元件发生互作,正向调节基因表达;Bna013749通过与启动子BnA5.FAD2和BnC5.FAD2启动子的GT元件发生互作,负向调节基因表达。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.3724/SP.J.1006.2011.01342URL [本文引用: 1]

Rapeseed oil is a major edible oil source for human consumption. Its quality depends on its fatty acid composition. Among fatty acids, 18-C unsaturated fatty acids are most crucial. QTL mapping will pave a way for molecular breeding and cloning genes for fatty acid biosynthesis in rapeseed. In this study, the zero-erucic rapeseed parents HOP (high oleic) and Xiangyou 15 (low oleic) were used to construct a F2 population consisting of 189 individual plants. The construction of a genetic map containing 342 SSR markers and the mapping of QTLs for the 18-C unsaturated fatty acid contents were carried out. Two sampling methods, i.e., half-seed sampling from F2 single seeds and bulk-seed sampling from F2 individual plants (F3 seeds), were used to determine the fatty acid contents by gas chromatography. The correlations of the fatty acid contents between the two sampling methods as well as most of the correlations between these fatty acids were significant. Totally two major QTLs for oleic acid content were detected in the linkage groups (LG) of A5 and C5, which could explain 60-70% of the variance for oleic acid content. The superior one of the two QTLs was mapped in LG A5 and tightly linked to the FAD2 gene. The other one which located in LG C5 was a major QTL newly found for oleic acid content. The marker closest to this QTL was found to be homologous to the marker on the LG A5, implying the QTL on C5 was also associated with the FAD2 gene. Consistent effect of the two QTLs was confirmed using oleic acid contents determined by the two sampling methods.The QTLs for oleic acid content was found to be the ones for the linoleic acid content, which was consistent with the high significant correlation between the two fatty acids.Nevertheless,one more minor QTL for linoleic acid content with a low LOD value was shown on LG A4 using the bulk-seed method. For linolenic acid content,three major QTLs on LG A4, A5, and C4, respectively, were detected consistently using the determinations of the two sampling methods,totally explaining 72-80% of the variance for linolenic acid content.Furthermore,anotherQTL with a low LOD value,only accounting for 12.42% of variance was detected on LG A4 using the half-seed method.
[本文引用: 1]
DOI:10.1194/jlr.R021436URLPMID:22045929Magsci [本文引用: 1]

The compartmentation of neutral lipids in plants is mostly associated with seed tissues, where triacylglycerols (TAGs) stored within lipid droplets (LDs) serve as an essential physiological energy and carbon reserve during postgerminative growth. However, some nonseed tissues, such as leaves, flowers and fruits, also synthesize and store TAGs, yet relatively little is known about the formation or function of LDs in these tissues. Characterization of LD-associated proteins, such as oleosins, caleosins, and sterol dehydrogenases (steroleosins), has revealed surprising features of LD function in plants, including stress responses, hormone signaling pathways, and various aspects of plant growth and development. Although oleosin and caleosin proteins are specific to plants, LD-associated sterol dehydrogenases also are present in mammals, and in both plants and mammals these enzymes have been shown to be important in (steroid) hormone metabolism and signaling. In addition, several other proteins known to be important in LD biogenesis in yeasts and mammals are conserved in plants, suggesting that at least some aspects of LD biogenesis and/or function are evolutionarily conserved.
DOI:10.1093/jxb/ern065URLPMID:2413273 [本文引用: 1]

Lipid modifying enzymes play a key role in the development of cold stress tolerance in cold-resistant plants such as cereals. However, little is known about the role of the endogenous enzymes in cold-sensitive species such as cotton.Delta 12fatty acid desaturases (FAD2), known to participate in adaptation to low temperatures through acyl chain modifications were used in gene expression studies in order to identify parameters of plant response to low temperatures. The induction of microsomaldelta 12fatty acid desaturases at an mRNA level under cold stress in plants is shown here for first time. Quantitative PCR showed that though bothdelta 12 omega 6fatty acid desaturase genes FAD2-3 and FAD2-4 identified in cotton are induced under cold stress, FAD2-4 induction is significantly higher than FAD2-3. The induction of both isoforms was light regulated, in contrast a third isoform FAD2-2 was not affected by cold or light. Stress tolerance and light regulatory elements were identified in the predicted promoters of both FAD2-3 and FAD2-4 genes. Di-unsaturated fatty acid species rapidly increased in the microsomal fraction isolated from cotton leaves, following cold stress. Expression analysis patterns were correlated with the observed increase in both total and microsomal fatty acid unsaturation levels suggesting the direct role of the FAD2 genes in membrane adaptation to cold stress.
DOI:10.1016/j.bbalip.2006.06.010URLPMID:16920014 [本文引用: 1]

Saccharomyces cerevisiae forms monounsaturated fatty acids using the ER membrane-bound 9 fatty acid desaturase, Ole1p, an enzyme system that forms a double bond in saturated fatty acyl CoA substrates. Ole1p is a chimeric protein consisting of an amino terminal desaturase domain fused to cytochrome b 5. It catalyzes the formation of the double bond through an oxygen-dependent mechanism that requires reducing equivalents from NADH. These are transferred to the enzyme via NADH cytochrome b 5 reductase to the Ole1p cytochrome b 5 domain and then to the diiron-oxo catalytic center of the enzyme. The control of OLE1 gene expression appears to mediated through the ER membrane proteins Spt23p and Mga2p. N-terminal fragments of these proteins are released by an ubiquitin/proteasome mediated proteolysis system and translocated to the nucleus where they appear to act as transcription coactivators of OLE1. OLE1 is regulated through Spt23p and Mga2p by multiple systems that control its transcription and mRNA stability in response to diverse stimuli that include nutrient fatty acids, carbon source, metal ions and the availability of oxygen.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
DOI:10.1002/j.1460-2075.1987.tb02542.xURLPMID:3678200 [本文引用: 1]

Pea nuclear extracts were used in gel retardation assays and DNase I footprinting experiments to identify a protein factor that specifically interacts with regulatory DNA sequences upstream of the pea rbcS-3A-gene. This factor, designated GT-1, binds to two short sequences (boxes II and III) in the -150 region that are known to function as light-responsive elements (LREs) in transgenic tobacco. Binding of GT-1 to homologous sequences further upstream (boxes II and III in the -220 region) indicates that these boxes comprise the redundant LRE that functions in vivo when boxes II and III are deleted. In both box II and box II, methylation interference experiments demonstrate that two adjacent G residues are critical for GT-binding. Single Gs present in boxes III and III are also important. Since GT-1 is present in nuclear extracts from leaves of light-grown and dark-adapted pea plants, its regulatory role does not depend on de novo synthesis. Thus if GT-1 binds differentially in vivo it must be postranslationally modified or sterically blocked from binding by another factor in response to light.
DOI:10.1016/S0092-8674(00)00113-6URLPMID:11057894 [本文引用: 1]

The subject is reviewed under the headings: signals from the environment, signalling in a biophysical context, signalling in development, sources of innovation and novelty in the evolution of plant signal transduction pathways, and conclusions. Particular attention is paid to the origins of novelty in the evolution of signalling pathways in plants.
DOI:10.1016/0165-3806(94)90139-2URLPMID:8206981 [本文引用: 2]

Abstract Nuclear genes encoding plastid ribosomal proteins are more highly expressed in leaves than in roots. This leaf-specific induction seems to be light-independent. We have previously characterized a spinach nuclear factor S1F binding to a cis-element within the rps1 promoter, which negatively regulates both the rps1 and the cauliflower mosaic virus 35S promoters in transient expression assays. Here, we show that the S1F binding site is related to but different from the light-responsive Box II of the pea rbcS-3A promoter, which is recognized by the nuclear factor GT-1. Transgenic plant analyses showed that the S1F site tissue-specifically represses the rps1 promoter in roots as well as in etiolated seedlings. We suggest that the GT-1-related S1F binding site is responsible, at least in part, for the transcriptional repression of rps1 in nonphotosynthetic tissues such as roots.
DOI:10.1046/j.1365-313X.1995.08010025.xURLPMID:7655505 [本文引用: 1]

GT-2 is a DNA-binding protein with high target-sequence specificity toward functionally defined, positively acting cis elements in the rice phytochrome A gene promoter. Using immunocytochemical procedures, it is shown here that GT-2 is localized to the nucleus, consistent with a function in transcriptional regulation. Immunoblot and immunocytochemical analyses show that rice shoots contain higher levels of GT-2 protein than roots, and that no photo-induced changes in GT-2 abundance or spatial distribution are detectable in these tissues, a result consistent with the proposed constitutive activity of GT-2. In both shoots and roots, GT-2 protein is undetectable in meristematic tissue but becomes expressed at later stages of cellular development, consistent with a role in contributing to the pattern of phytochrome A gene expression. By transfecting protoplasts with a series of constructs containing deletion derivatives of GT-2 fused to 尾-glucuronidase (GUS), followed by in situ localization of GUS activity, two independent, functionally active nuclear localization sequences (NLSs) have been identified in GT-2. One NLS resides within each of a pair of previously identified, spatially separate, trihelix motifs in the protein. Sequence inversion and alanine-scanning mutagenesis has identified residues within these NLSs necessary for nuclear localization. Each NLS contains two basic domains separated by 10 amino acids, conforming to the bipartite class of NLS involved in the targeting of numerous other nuclear localized proteins.
DOI:10.1016/0014-4827(92)90166-6URL [本文引用: 1]

A triplet of adjacent, highly similar GT motifs in the phyA promoter of rice functions to support maximal expression of this gene. We have obtained a recombinant clone that encodes a full-length nuclear protein, designated GT-2, which binds specifically to these target sequences. This novel protein contains acidic, basic and proline- + glutamine-rich regions, as well as two autonomous DNA-binding domains, one NH2-terminal and the other COOH-terminal, that discriminate with high resolution between the three GT motifs. A duplicated sequence of 75 amino acids, present once in each DNA-binding domain, appears likely to mediate DNA target element recognition. Each copy of this duplicated protein sequence is predicted to form three amphipathic alpha-helices separated from each other by two short loops. The absence of sequence similarity to other known proteins suggests that this predicted structural unit, which we term the trihelix motif, might be representative of a new class of DNA-binding proteins.
DOI:10.1007/s11103-006-9131-xURLPMID:17238046 [本文引用: 1]

Drought is one of the most severe stresses limiting plant growth and yield. Genes involved in water stress tolerance of wild barley ( Hordeum spontaneoum ), the progenitor of cultivated barley, were investigated using genotypes contrasting in their response to water stress. Gene expression profiles of water-stress tolerant vs. water-stress sensitive wild barley genotypes, under severe dehydration stress applied at the seedling stage, were compared using cDNA-AFLP analysis. Of the 1100 transcript-derived fragments (TDFs) amplified about 70 displayed differential expression between control and stress conditions. Eleven of them showed clear difference (up- or down-regulation) between tolerant and susceptible genotypes. These TDFs were isolated, sequenced and tested by RT-PCR. The differential expression of seven TDFs was confirmed by RT-PCR, and TDF-4 was selected as a promising candidate gene for water-stress tolerance. The corresponding gene, designated Hsdr4 ( Hordeum spontaneum dehydration-responsive), was sequenced and the transcribed and flanking regions were determined. The deduced amino acid sequence has similarity to the rice Rho-GTPase-activating protein-like with a Sec14p-like lipid-binding domain. Analysis of Hsdr4 promoter region that was isolated by screening a barley BAC library, revealed a new putative miniature inverted-repeat transposable element (MITE), and several potential stress-related binding sites for transcription factors (MYC, MYB, LTRE, and GT-1), suggesting a role of the Hsdr4 gene in plant tolerance to dehydration stress. Furthermore, the Hsdr4 gene was mapped using wild barley mapping population to the long arm of chromosome 3H between markers EBmac541 and EBmag705 , within a region that previously was shown to affect osmotic adaptation in barley.
DOI:10.1007/BF02715844URLPMID:16131718Magsci [本文引用: 1]

East Asian Drosophila melanogaster are known for great variation in morphological and physiological characters among populations, variation that is believed to be maintained by genetic drift. To understand the genetic properties of Asian D. melanogaster populations, we initiated a population genetic study of chromosome inversion polymorphisms in hitherto unanalysed population samples from Southeast (SE) Asia. We generally found a high frequency of each of the four common cosmopolitan inversions in comparison to populations from Africa, Asia, and Australia. In contrast to the great phenotypic variation among Asian populations, however, we could not detect differences in inversion frequencies among populations. Furthermore, we observed neither correlations of inversion frequencies with population latitude and longitude, nor evidence for linkage disequilibrium between different inversion loci. We propose two explanations for the observed genetic homogeneity among these SE Asian D. melanogaster populations: (i) the observed pattern simply reflects the retention of ancestral polymorphisms originating from a panmictic population that was once present on a large single landmass (Sundaland), and/or is a consequence of high recent gene flow between populations; and (ii) it is caused by selective forces (e.g. balancing selection).
DOI:10.3724/SP.J.1005.2009.00123URLPMID:19273418 [本文引用: 1]

Abstract GT elements are DNA sequences occurred in tandem repeats in the promoter region of plant genes, which are rich in nucleotides T and A in promoters. A few GT elements and their functions have been characterized in different plants. Factors GT are transcription factors with trihelix motifs for DNA-binding domains and specific binding to GT elements. Presently, GT factors are only observed in plants. The interaction of GT factors and GT elements can regulate the transcrip-tion of related genes and improve resistance of plants or involve in plant morphogenesis. In this article, we summarized the discover, structure, and interaction of GT factors and GT elements.
DOI:10.1046/j.1365-313x.1999.00482.xURLPMID:2020202020120202020202020 [本文引用: 1]

Summary GT-1 belongs to the class of trihelix DNA-binding proteins and binds to a promoter sequence found in many different genes. Data presented in this report show that GT-1 contains a trans-activation function in yeast and in plant cells. However, in tobacco BY-2 protoplasts, this activity functions only when an internal region containing the DNA-binding domain is deleted. Gel-shift and co-immunoprecipitation assays have revealed that GT-1 can interact with and stabilize the TFIIA–TBP–TATA complex. These results suggest that GT-1 may activate transcription through direct inter- action with the transcriptional pre-initiation complex.
DOI:10.1016/0076-6879(87)53060-9URL [本文引用: 1]

This chapter discusses binary Ti vectors for plant transformation and promoter analysis. In recent years, several studies have demonstrated the use of the naturally occurring Ti plasmids as transformation vectors for higher plants. In the technique, exogeneous DNA is either inserted into the tDNA region of the Ti plasmid by homologous recombination using an intermediate vector system or directly into one of the recently designed binary vectors. The binary vector system is a convenient way to transform higher plants, because foreign DNA can first be inserted into the vector and manipulated using E. coli as a host. The chapter illustrates an example of binary Ti vector and the important features of the molecule, pGA482, which is 13.2 kbp long for simple and efficient plant transformation. The chapter describes the two different methods that are used to move the binary vector to Agrobacterium triparental mating and direct DNA transfer. The chapter mentions that clonal transformation is enhanced by the protoplast cocultivation method. Cultured tobacco calli can be transformed by the Ti vector system.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.gene.2014.05.008URLPMID:24811682 [本文引用: 1]

61A transcriptional regulatory region controlling the expression of a FAD2 in Brassica napus was characterized.61BnFAD2A5-1 harbors an intron within its 5'-UTR. This intron demonstrated promoter activity.61Deletion analysis of the BnFAD2A5-1 promoter and intron revealed that the –220 to –1bp is the minimum promoter region, while –220 to –110bp and +34 to +285bp are two important regions conferring high-levels of transcription.61BnFAD2A5-1 transcripts can be induced by light, low temperature, and abscisic acid (ABA).
[本文引用: 1]
DOI:10.1016/j.tplants.2011.12.002URLPMID:22236699Magsci [本文引用: 1]

GT factors are the founding members of the trihelix transcription factor family. They bind GT elements in light regulated genes, and their nature was uncovered in a burst of activity in the 1990s. Study of the trihelix family then slowed. However, interest is now re-awakening. Genomic studies have revealed 30 members of this family in Arabidopsis and 31 in rice, falling into five clades. Newly discovered functions involve responses to salt and pathogen stresses, the development of perianth organs, trichomes, stomata and the seed abscission layer, and the regulation of late embryogenesis. Thus the time is ripe for a review of the genomic and functional information now emerging for this neglected family.
URL [本文引用: 1]

In order to explore the expression characteristic and function of GRAS transcription factor gene Hc SCL13 from Halostachys caspica,the 2 200 bp sequence of promoter was cloned using the genomic walking method. The analysis results using the Plant CARE softwaresuggested that the promoter sequence contained not only the core elements such as CAAT-box and TATA-box,but also some cis-elementrelated to the stress response. The 35 S promoter sequence of the p BI121 expression vector was replaced by the Hc SCL13 promoter sequence toconstruct the fusion expression vector. Then,the fusion expression vector was transformed into Arabidopsis thaliana by flora method and the T1 seedlings were stained by GUS. The results of GUS staining showed that the whole transformed A. thaliana seedlings were stained,indicatingthat the Hc SCL13 gene promoter had expression activity,and it might be constitutive promoter.
DOI:10.2307/3870214URLPMID:8672890 [本文引用: 1]

GT-2 is a novel DNA binding protein that interacts with a triplet of functionally defined, positively acting GT-box motifs (GT1-bx, GT2-bx, and GT3-bx) in the rice phytochrome A gene (PHYA) promoter. Data from a transient transfection assay used here show that recombinant GT-2 enhanced transcription from both homologous and heterologous GT-box-containing promoters, thereby indicating that this protein can function as a transcriptional activator in vivo. Previously, we have shown that GT-2 contains separate DNA binding determinants in its N- and C-terminal halves, with binding site preferences for the GT3-bx and GT2-bx promoter motifs, respectively. Here, we demonstrate that the minimal DNA binding domains reside within dual 90-amino acid polypeptide segments encompassing duplicated sequences, termed trihelix regions, in each half of the molecule, plus 15 additional immediately adjacent amino acids downstream. These minimal binding domains retained considerable target sequence selectivity for the different GT-box motifs, but this selectivity was enhanced by a separate polypeptide segment farther downstream on the C-terminal side of each trihelix region. Therefore, the data indicate that the twin DNA binding domains of GT-2 each consist of a general GT-box recognition core with intrinsic differential binding activity toward closely related target motifs and a modifier sequence conferring higher resolution reciprocal selectivity between these motifs.
DOI:10.1016/S0092-8674(00)00113-6URLPMID:11057894 [本文引用: 1]

The subject is reviewed under the headings: signals from the environment, signalling in a biophysical context, signalling in development, sources of innovation and novelty in the evolution of plant signal transduction pathways, and conclusions. Particular attention is paid to the origins of novelty in the evolution of signalling pathways in plants.
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.tplants.2011.12.002URLPMID:22236699 [本文引用: 1]

GT factors are the founding members of the trihelix transcription factor family. They bind GT elements in light regulated genes, and their nature was uncovered in a burst of activity in the 1990s. Study of the trihelix family then slowed. However, interest is now re-awakening. Genomic studies have revealed 30 members of this family in Arabidopsis and 31 in rice, falling into five clades. Newly discovered functions involve responses to salt and pathogen stresses, the development of perianth organs, trichomes, stomata and the seed abscission layer, and the regulation of late embryogenesis. Thus the time is ripe for a review of the genomic and functional information now emerging for this neglected family.
DOI:10.4161/psb.6.12.18709URLPMID:3337171 [本文引用: 1]

In flowering plants, seed development and seed filling are intricate genetically programmed processes that correlate with changes in metabolite levels and that are spatially and temporally regulated by a complex signaling network mediated mainly by sugars and hormones. ASIL1, a member of the plant-specific trihelix family of DNA-binding transcription factors, was isolated based on its interaction with the GT-element of the Arabidopsis thaliana 2S albumin At2S3 promoter. Mutation of ASIL1 derepressed expression of a subset of embryonic genes resulting in accumulation of 2S albumin and embryo-specific lipids in leaves.1 It was recently reported that mutation of ASIL1 led to early embryo development in Arabidopsis.2 In this study, we demonstrated that ASIL1 acts as a temporal regulator of seed filling. In developing siliques, mutation of ASIL1 led to earlier expression of master regulatory genes LEC2, FUS3 and ABI3 as well as genes for seed storage reserves. Moreover, the 12S globulin accumulated to a much higher level in the developing seeds of asil1鈥1 compared with wild type. To our knowledge, this is the first evidence that ASIL1 not only functions as a negative regulator of embryonic traits in seedlings but also contributes to the maintenance of precise temporal control of seed filling. Thus, ASIL1 represents a novel component of the regulatory framework controlling embryonic gene expression in Arabidopsis.
DOI:10.1007/s002399910012URLPMID:10684344Magsci [本文引用: 1]

The GATA -binding transcription factors comprise a protein family whose members contain either one or two highly conserved zinc finger DNA-binding domains. Members of this group have been identified in organisms ranging from cellular slime mold to vertebrates, including plants, fungi, nematodes, insects, and echinoderms. While much work has been done describing the expression patterns, functional aspects, and target genes for many of these proteins, an evolutionary analysis of the entire family has been lacking. Herein we show that only the C-terminal zinc finger (Cf) and basic domain, which together constitute the GATA -binding domain, are conserved throughout this protein family. Phylogenetic analyses of amino acid sequences demonstrate distinct evolutionary pathways. Analysis of GATA factors isolated from vertebrates suggests that the six distinct vertebrate GATAs are descended from a common ancestral sequence, while those isolated from nonvertebrates (with the exception of the fungal AREA orthologues and Arabidopsis paralogues) appear to be related only within the DNA-binding domain and otherwise provide little insight into their evolutionary history. These results suggest multiple modes of evolution, including gene duplication and modular evolution of GATA factors based upon inclusion of a class IV zinc finger motif. As such, GATA transcription factors represent a group of proteins related solely by their homologous DNA-binding domains. Further analysis of this domain examines the degree of conservation at each amino acid site using the Boltzmann entropy measure, thereby identifying residues critical to preservation of structure and function. Finally, we construct a predictive motif that can accurately identify potential GATA proteins.
[D].
[本文引用: 1]
[D].
[本文引用: 1]
