 ,1, 张靖男1, 崔顺立1, Charles Y.Chen2, 穆国俊1, 侯名语1, 杨鑫雷
,1, 张靖男1, 崔顺立1, Charles Y.Chen2, 穆国俊1, 侯名语1, 杨鑫雷 ,1,*, 刘立峰
,1,*, 刘立峰 ,1,*
,1,*QTL mapping and QTL × Environment interaction analysis of pod and seed related traits in cultivated peanut (Arachis hypogaea L.)
MENG Xin-Hao ,1, ZHANG Jing-Nan1, CUI Shun-Li1, Charles Y. Chen2, MU Guo-Jun1, HOU Ming-Yu1, YANG Xin-Lei
,1, ZHANG Jing-Nan1, CUI Shun-Li1, Charles Y. Chen2, MU Guo-Jun1, HOU Ming-Yu1, YANG Xin-Lei ,1,*, LIU Li-Feng
,1,*, LIU Li-Feng ,1,*
,1,*通讯作者: *杨鑫雷, E-mail:peanut@hebau.edu.cn;刘立峰, E-mail:liulifeng@hebau.edu.cn
收稿日期:2020-09-22接受日期:2021-03-19网络出版日期:2021-04-08
| 基金资助: |
Corresponding authors: *E-mail:peanut@hebau.edu.cn;E-mail:liulifeng@hebau.edu.cn
Received:2020-09-22Accepted:2021-03-19Published online:2021-04-08
| Fund supported: |
作者简介 About authors
E-mail:mxinhao1994@126.com

摘要
花生荚果、种子性状与产量紧密相关, 是重要的农艺性状。为挖掘与荚果、种子性状紧密连锁的分子标记, 本研究以大果品种冀花5号和小果美国资源M130组配衍生的315个家系RIL8群体为材料, 利用SSR、AhTE、SRAP和TRAP等标记构建了一张包含363个多态性位点的遗传连锁图谱。该图谱共包含21个连锁群, 总长为1360.38 cM, 标记间平均距离为3.75 cM。利用完备区间作图法对2017—2018年5个环境的荚果、种子相关性状进行数量性状基因座(quantitative trait locus, QTL)分析, 共鉴定到97个与荚果、种子性状相关的QTL, 可解释的表型变异为2.36%~12.15%, 分布在A02、A05、A08、A09、B02、B03、B04、B08和B09等9条染色体上。其中, 9个与荚果长相关, 13个与荚果宽相关, 14个与荚果厚相关, 11个与种子长相关, 14个与种子宽相关, 13个与种子厚相关, 13个与百果重相关, 10个与百仁重相关; 4个主效QTL分别为qPWA08.1、qPTA08.3、qPTA08.4和qSWB08.5, 可解释的表型变异分别为10.02%、11.06%、12.15%和11.97%; 45个稳定表达的QTL在3个以上环境可被重复检测; 连锁群A02、A08、B02、B04和B08上存在QTL聚集区。另外, 检测到15对上位性QTL, 可解释的表型变异为10.23%~51.84%。研究结果将为花生荚果、种子性状的分子标记辅助育种提供重要的理论依据。
关键词:
Abstract
Pod and seed traits are important agronomy traits, which are closely related to yield in cultivated peanut (Arachis hypogaea L.). In the present study, to identify molecular markers closely linked to pod and seed traits, a RIL8 population with 315 families was developed that derived from Jihua 5 with large pod and M130 with small pod of US germplasm. A genetic linkage map containing 363 polymorphic loci was constructed using SSR, AhTE, SRAP, and TRAP markers. All polymorphic loci were mapped on 21 linkage groups, which spanned 1360.38 cM with an average distance of 3.75 cM. Subsequently, a total of 97 QTLs for pod and seed traits were identified by ICIM method at five environments from 2017 to 2018, explaining the phenotypic variations of 2.36%-12.15%, and located on A02, A05, A08, A09, B02, B03, B04, B08, and B09 chromosomes. Among them, nine QTLs were detected for pod length, 13 QTLs for pod width, 14 QTLs for pod thickness, 11 QTLs for seed length, 13 QTLs for seed width, 13 QTLs for hundred-pod weight, 10 QTLs for hundred-seed weight. Four QTLs with major effect were detected, including qPWA08.1, qPTA08.3, qPTA08.4, and qSWB08.5, which explained the phenotypic variations of 10.02%-12.15%. Furthermore, 45 stable QTLs were repeatedly detected in more than three environments. QTL clusters were detected on A02, A08, B02, B04, and B08 chromosomes, respectively. In addition, 15 epistatic QTLs were identified that explaining phenotypic variation of 10.23%-51.84%. These results will provide an important theoretical basis for molecular marker-assisted breeding of pod and seed traits in peanut.
Keywords:
PDF (4158KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
孟鑫浩, 张靖男, 崔顺立, Charles Y.Chen, 穆国俊, 侯名语, 杨鑫雷, 刘立峰. 花生荚果与种子相关性状QTL定位及与环境互作分析[J]. 作物学报, 2021, 47(10): 1874-1890 DOI:10.3724/SP.J.1006.2021.04216
MENG Xin-Hao, ZHANG Jing-Nan, CUI Shun-Li, Charles Y. Chen, MU Guo-Jun, HOU Ming-Yu, YANG Xin-Lei, LIU Li-Feng.
花生(Arachis hypogaea L.)又名落花生, 起源于南美洲, 是热带和亚热带地区种植广泛的油料作物和经济作物, 也是人类食用油的主要来源之一[1], 产量相关性状遗传研究广受关注。花生荚果、种子性状属于重要的产量性状, 是典型的数量性状, 受多基因与环境共同调控, 其遗传规律十分复杂[2]。研究数量性状的遗传机制为花生分子育种提供了新的可能, 而单靠常规育种已无法满足生产上的需要, 需借助分子标记辅助选择技术[3]。分子标记可以对与产量性状密切相关的数量性状进行选择, 提高选择的准确性, 从而加快花生育种进程, 对培育高产花生新品种具有重要意义。数量性状定位是研究数量性状的主要方法之一, 亦是揭示花生荚果、种子相关性状遗传规律的重要途径[4]。RIL群体属于永久性定位群体, 具有遗传稳定、QTL定位及遗传效应分析准确等优点[5]。
利用RIL群体进行多环境花生荚果、种子性状QTL定位的研究已有进展。Luo等[6]利用“徐花13×中花6号”重组自交系群体, 在4个环境下检测到33个与荚果性状相关的QTL, 主要分布在A05、A07、A08等染色体上。Wang等[7]利用“ZH16×sd-H1”构建的RIL群体作为研究材料, 在3个环境下获得30个荚果相关的QTL, 主要分布在B06和B07染色体上。Luo等[8]利用构建的栽培种花生遗传图谱, 在4个环境下重复检测到42个与荚果性状相关的QTL, 主要分布在A05、A06、A09和B05染色体上。李英杰[9]利用已构建的遗传图谱, 对10个产量相关性状进行QTL定位分析, 共检测到8个主效QTL, 单个QTL的PVE为5.66%~28.05%。李振动等[10]以“远杂9102×徐州68-4”衍生的RIL群体为材料, 2年间共检测到41个数量性状位点, 其可解释的表型变异(phenotypic variation explained, PVE)为3.14%~ 18.27%, 有6个位点在2个环境下被稳定检测到。吕维娜等[11]利用“白沙1016×A. monticola”构建的RIL群体为试验材料, 在3个不同的环境下定位到72个与百果重、百仁重等农艺性状相关的数量性状位点。成良强[12]以“富川大花生×ICG6375”衍生的F2群体为研究材料进行QTL定位, 获得68个与荚果长、荚果宽、种子长、种子宽、百果重和百仁重等产量性状相关的QTL。Shirasawa等[13]以2个F2群体为材料, 检测到23个QTL与荚果、种子性状相关, 其PVE为4.8%~28.2%。
目前, 花生荚果、种子相关性状QTL在不同研究间仍存在较大差异, 多环境下稳定的QTL仍较少。因此, 本研究以大果型花生品种冀花5号和小果型美国种质资源M130组配衍生的包含315个家系的RIL群体为材料, 利用SSR、SRAP、TRAP等分子标记构建花生遗传连锁图, 并利用完备区间作图法对5个环境下的8个荚果、种子相关性状进行QTL定位及与环境的互作效应分析。鉴定出的QTL将为荚果和种子性状的遗传解析、图位克隆和遗传改良提供理论参考。
1 材料与方法
1.1 试验材料
以大果型花生品种冀花5号为母本, 小果型美国花生种质资源M130为父本(图1), 采用单粒传法, 构建包含315个家系的F8重组自交系群体为研究材料。2017年分别种植在河北省邯郸市大名县(DM, 35º57′N & 115º09′E)和河北省保定市清苑区(QY, 38º40′N & 115º30′E)。2018年在河北省唐山市迁安市(QA, 39º99′N & 118º70′E)、河北省邯郸市大名县和河北省保定市清苑区等地分别种植亲本和群体。每个家系种植1行, 行长1.5 m, 行距为0.5 m, 株距为0.17 m。每行种植10株。随机区组设计, 2次重复, 常规田间管理。试验材料收获晒干后, 参考姜慧芳等[14]的《花生种质资源描述规范和数据标准》对荚果长(pod length, PL)、荚果宽(pod width, PW)、荚果厚(pod thickness, PT)、种子长(seed length, SL)、种子宽(seed width, SW)、种子厚(seed thickness, ST)、百果重(hundred-pods weight, HPW)、百仁重(hundred-seeds weight, HSW)等8个产量相关性状进行表型鉴定。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1荚果及种子表型图
白色指示条为标尺, 大小为1 cm。
Fig. 1Phenotypes of pods and seeds
The white bar indicates 1 cm.
1.2 DNA提取及引物筛选
在田间取亲本和RIL群体各株系的幼嫩倒三叶, 采用改良SDS-CTAB法[15]提取花生基因组总DNA。利用3964对SSR标记和转座子引物(1.3 基因型统计及连锁图谱构建
群体基因型采用母本带型记为“a”, 父本带型记为“b”, 杂合带型记为“h”的标记方法进行统计。带型模糊不清或数据缺失使用“-”替代。群体基因型通过JoinMap 4.0 [19]进行连锁分析, 设置步长为0.5, LOD > 3, 在LOD值3~10范围内将所得到的标记进行分组, 利用Kosambi函数将重组率转化为连锁距离[20]。采用Mapchart 2.3 [21]绘制遗传连锁图谱。构建好的遗传连锁图谱与Peanut genome resource(1.4 表型统计及QTL定位
采用GraphPad Prism 8(2 结果与分析
2.1 表型统计分析
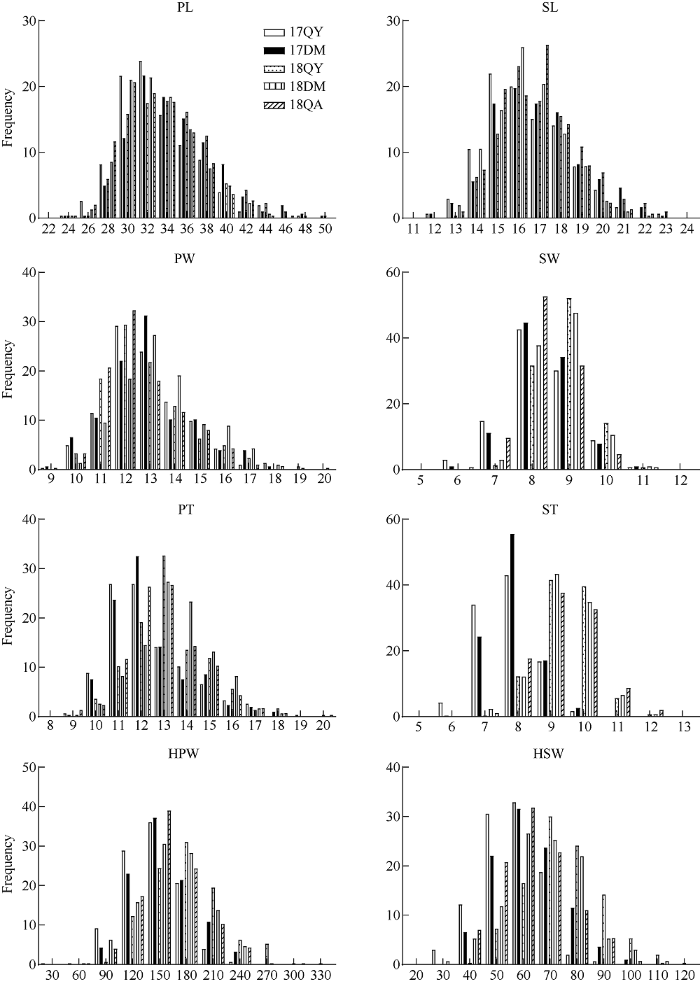
2.1.1 荚果和种子的表型及相关性分析 对2017—2018年花生亲本及其RIL群体各家系荚果长、荚果宽、荚果厚、种子长、种子宽、种子厚、百果重、百仁重等8个性状进行描述性统计分析(表1)和相关性分析(表2)。结果表明, 在5个环境中, 父母本在每个性状均表现出显著差异, RIL群体具有较大的变异范围, 且群体各性状的最小值和最大值均超过亲本, 说明各性状都具有正向超亲和负向超亲优势。群体表型值的偏度和峰度均趋于0, 且w-test均表现出不同程度的显著性, 说明各性状符合正态性(表1和图2), 适合QTL分析。相关性分析结果表明, 各个性状在同一环境下均表现为极显著的正相关关系(P<0.001) (表2), 推测多个性状可能被定位到同一个QTL区间, 即“一因多效”。Table 1
表1
表1亲本及RIL群体的描述性统计结果
Table 1
| 性状 Trait | 环境 Environment | 亲本 Parents | RIL群体 RIL population | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 冀花5号 Jihua 5 | M130 | 最小值 Minimum | 最大值 Maximum | 平均值±标准误 Mean±SD | 变异系数 CV | Shapiro-Wilk (w-test) | 偏度 Skewness | 峰度 Kurtosis | ||
| 荚果长 | 17QY | 43.28±1.46** | 30.25±1.24 | 24.09 | 47.94 | 33.08±3.80 | 0.11 | 0.9592*** | 0.75 | 0.72 |
| PL (mm) | 17DM | 43.57±0.96** | 29.84±0.65 | 24.81 | 49.04 | 34.72±4.13 | 0.12 | 0.9606*** | 0.69 | 0.52 |
| 18QY | 45.33±0.99** | 31.66±1.15 | 23.96 | 49.04 | 34.53±4.18 | 0.12 | 0.9685*** | 0.58 | 0.20 | |
| 18DM | 41.25±2.08** | 28.84±1.06 | 24.80 | 44.60 | 33.23±3.61 | 0.11 | 0.9685*** | 0.46 | -0.18 | |
| 18QA | 43.59±1.03** | 30.18±1.17 | 23.35 | 46.95 | 33.01±3.79 | 0.11 | 0.9723** | 0.53 | 0.13 | |
| 荚果宽 | 17QY | 15.17±0.39** | 12.42±0.50 | 9.12 | 17.88 | 12.86±1.63 | 0.13 | 0.9649*** | 0.54 | 0.24 |
| PW (mm) | 17DM | 15.16±0.97** | 12.81±0.51 | 9.32 | 18.04 | 12.94±1.72 | 0.13 | 0.9597*** | 0.47 | 0.13 |
| 18QY | 14.95±1.03** | 11.90±0.49 | 10.16 | 19.09 | 12.77±1.65 | 0.13 | 0.9327*** | 0.91 | 0.74 | |
| 18DM | 15.40±0.81** | 12.95±1.12 | 9.21 | 19.79 | 13.50±1.73 | 0.13 | 0.9668*** | 0.61 | 0.25 | |
| 18QA | 15.02±0.86** | 12.51±0.79 | 9.79 | 18.20 | 12.68±1.57 | 0.12 | 0.9347*** | 0.87 | 0.53 | |
| 荚果厚 | 17QY | 15.33±0.54** | 12.98±0.43 | 8.51 | 17.17 | 12.30±1.66 | 0.13 | 0.9262*** | 0.85 | 0.22 |
| PT (mm) | 17DM | 15.53±0.90** | 13.2±0.38 | 8.78 | 18.79 | 12.41±1.72 | 0.14 | 0.9254*** | 0.98 | 0.77 |
| 18QY | 15.28±0.65** | 13.19±0.59 | 9.65 | 19.50 | 13.21±1.64 | 0.12 | 0.9647*** | 0.66 | 0.72 | |
| 18DM | 15.39±0.85** | 12.77±0.71 | 9.16 | 17.73 | 13.46±1.58 | 0.12 | 0.982* | 0.04 | -0.14 | |
| 18QA | 15.23±0.95** | 12.86±0.35 | 9.32 | 19.65 | 12.98±1.62 | 0.12 | 0.9698** | 0.6 | 0.79 | |
| 种子长 | 17QY | 19.56±0.64** | 16.38±0.96 | 12.26 | 23.09 | 16.43±1.88 | 0.11 | 0.9691** | 0.54 | 0.24 |
| SL (mm) | 17DM | 19.58±0.38** | 16.12±0.56 | 11.89 | 22.94 | 16.94±2.04 | 0.12 | 0.9728** | 0.38 | -0.15 |
| 18QY | 20.32±0.79** | 17.30±0.79 | 13.34 | 23.33 | 17.17±1.95 | 0.11 | 0.9594 *** | 0.57 | 0.01 | |
| 18DM | 18.79±1.75** | 15.47±0.91 | 12.98 | 22.44 | 16.46±1.66 | 0.10 | 0.9723** | 0.47 | 0.10 | |
| 18QA | 18.18±0.69** | 16.03±0.81 | 13.06 | 22.03 | 16.63±1.65 | 0.10 | 0.9703** | 0.47 | 0.17 | |
| 种子宽 | 17QY | 9.89±0.24** | 8.67±0.63 | 5.89 | 11.02 | 8.29±0.89 | 0.11 | 0.9852* | 0.01 | 0.09 |
| SW (mm) | 17DM | 9.78±0.53** | 8.39±0.22 | 6.33 | 11.19 | 8.37±0.80 | 0.10 | 0.9849* | 0.20 | 0.36 |
| 18QY | 10.42±0.74** | 8.91±0.62 | 7.14 | 10.91 | 8.82±0.65 | 0.07 | 0.9725** | 0.44 | 0.17 | |
| 18DM | 9.37±0.78** | 8.43±0.54 | 6.89 | 11.32 | 8.70±0.67 | 0.08 | 0.9855* | 0.35 | 0.64 | |
| 18QA | 9.49±0.40* | 8.57±0.89 | 6.39 | 10.93 | 8.32±0.68 | 0.08 | 0.9848* | 0.34 | 0.52 | |
| 种子厚 | 17QY | 10.29±0.32** | 9.46±0.45 | 6.00 | 10.04 | 7.78±0.76 | 0.10 | 0.9783* | 0.20 | -0.22 |
| ST (mm) | 17DM | 10.28±0.38** | 9.20±0.37 | 6.18 | 10.39 | 7.96±0.67 | 0.08 | 0.9763* | 0.51 | 0.82 |
| 18QY | 10.45±0.93* | 9.74±0.61 | 7.29 | 12.42 | 9.41±0.77 | 0.08 | 0.9899* | 0.09 | 0.40 | |
| 18DM | 10.13±0.45** | 9.18±0.94 | 6.91 | 11.96 | 9.34±0.82 | 0.09 | 0.9853* | -0.13 | 0.19 | |
| 18QA | 9.83±0.36* | 9.07±0.65 | 6.95 | 11.99 | 9.33±0.90 | 0.10 | 0.9811* | 0.17 | 0.06 | |
| 百果重 | 17QY | 254.03±6.84** | 170.13±0.91 | 71.04 | 239.22 | 144.50±30.93 | 0.21 | 0.9812* | 0.19 | -0.12 |
| HPW (g) | 17DM | 247.18±2.30** | 164.64±3.78 | 79.86 | 245.97 | 157.07±33.15 | 0.21 | 0.9681*** | 0.37 | -0.23 |
| 18QY | 271.28±8.80** | 184.48±2.45 | 94.85 | 325.53 | 180.11±40.66 | 0.23 | 0.9678*** | 0.54 | 0.19 | |
| 18DM | 236.77±6.46** | 155.79±3.57 | 61.32 | 256.43 | 163.31±36.51 | 0.22 | 0.9825* | 0.05 | -0.17 | |
| 18QA | 233.38±1.89** | 159.70±5.95 | 78.28 | 296.87 | 159.77±34.14 | 0.21 | 0.9788* | 0.51 | 0.51 | |
| 百仁重 | 17QY | 105.22±2.17** | 70.04±0.69 | 27.73 | 91.24 | 56.08±11.05 | 0.20 | 0.9859* | 0.05 | -0.11 |
| HSW (g) | 17DM | 100.82±1.49** | 68.83±1.14 | 36.93 | 101.23 | 62.80±12.11 | 0.19 | 0.9719** | 0.42 | 0.07 |
| 18QY | 114.32±2.73** | 76.20±3.11 | 43.73 | 119.87 | 74.77±13.96 | 0.19 | 0.9775* | 0.34 | -0.03 | |
| 18DM | 96.12±2.48** | 63.88±1.06 | 30.07 | 106.51 | 67.38±13.71 | 0.20 | 0.9826* | 0.14 | -0.21 | |
| 18QA | 100.24±1.44** | 63.90±1.65 | 37.21 | 108.20 | 63.25±13.05 | 0.21 | 0.9666 *** | 0.53 | 0.14 | |
新窗口打开|下载CSV
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2不同环境下RIL群体各性状的频率分布图
缩写同
Fig. 2Frequency profile for each trait of RIL population in different environments
Abbreviations are the same as those given in
Table 2
表2
表2不同环境下各性状的相关性分析结果
Table 2
| 环境 Environment | 性状 Trait | 荚果长 PL | 荚果宽 PW | 荚果厚 PT | 种子长 SL | 种子宽 SW | 种子厚 ST | 百果重 HPW |
|---|---|---|---|---|---|---|---|---|
| 17QY | 荚果宽PW | 0.6536*** | ||||||
| 荚果厚PT | 0.7286*** | 0.9226*** | ||||||
| 种子长SL | 0.8522*** | 0.6346*** | 0.7011*** | |||||
| 种子宽SW | 0.2752*** | 0.5291*** | 0.4692*** | 0.5357*** | ||||
| 种子厚ST | 0.3724*** | 0.4974*** | 0.5333*** | 0.5713*** | 0.7453*** | |||
| 百果重HPW | 0.6587*** | 0.6515*** | 0.6454*** | 0.6549*** | 0.5196*** | 0.5614*** | ||
| 百仁重HSW | 0.6242*** | 0.6188*** | 0.6282*** | 0.6737*** | 0.6041*** | 0.5970*** | 0.8280*** | |
| 17DM | 荚果宽PW | 0.6667*** | ||||||
| 荚果厚PT | 0.7353*** | 0.9087*** | ||||||
| 种子长SL | 0.8430*** | 0.6208*** | 0.6751*** | |||||
| 种子宽SW | 0.2599*** | 0.4804*** | 0.3924*** | 0.4370*** | ||||
| 种子厚ST | 0.3991*** | 0.5326*** | 0.6052*** | 0.5025*** | 0.6033*** | |||
| 百果重HPW | 0.6214*** | 0.6927*** | 0.6884*** | 0.6196*** | 0.4634*** | 0.5350*** | ||
| 百仁重HSW | 0.6037*** | 0.6143*** | 0.6171*** | 0.6326*** | 0.5256*** | 0.5961*** | 0.8651*** | |
| 18QY | 荚果宽PW | 0.7442*** | ||||||
| 荚果厚PT | 0.6400*** | 0.9129*** | ||||||
| 种子长SL | 0.8711*** | 0.6054*** | 0.4848*** | |||||
| 种子宽SW | 0.4281*** | 0.6622*** | 0.5653*** | 0.4141*** | ||||
| 种子厚ST | 0.3921*** | 0.5563*** | 0.5647*** | 0.4347*** | 0.6530*** | |||
| 百果重HPW | 0.6801*** | 0.6522*** | 0.5825*** | 0.7041*** | 0.5554*** | 0.6193*** | ||
| 百仁重HSW | 0.7373*** | 0.7132*** | 0.6362*** | 0.7571*** | 0.6475*** | 0.6927*** | 0.8726*** | |
| 18DM | 荚果宽PW | 0.6995*** | ||||||
| 荚果厚PT | 0.6364*** | 0.9102*** | ||||||
| 种子长SL | 0.8786*** | 0.6328*** | 0.5997*** | |||||
| 种子宽SW | 0.4231*** | 0.5523*** | 0.5578*** | 0.4899*** | ||||
| 种子厚ST | 0.3408*** | 0.4766*** | 0.5764*** | 0.4895*** | 0.6946*** | |||
| 百果重HPW | 0.6586*** | 0.6285*** | 0.6474*** | 0.6724*** | 0.6040*** | 0.6302*** | ||
| 百仁重HSW | 0.6676*** | 0.6262*** | 0.6347*** | 0.7130*** | 0.6717*** | 0.7052*** | 0.8606*** | |
| 18QA | 荚果宽PW | 0.7231*** | ||||||
| 荚果厚PT | 0.6760*** | 0.9333*** | ||||||
| 种子长SL | 0.8586*** | 0.6254*** | 0.5920*** | |||||
| 种子宽SW | 0.2765*** | 0.4873*** | 0.4558*** | 0.3500*** | ||||
| 种子厚ST | 0.3117*** | 0.4761*** | 0.5240*** | 0.4215*** | 0.6740*** | |||
| 百果重HPW | 0.5869*** | 0.6571*** | 0.6460*** | 0.6029*** | 0.4700*** | 0.5189*** | ||
| 百仁重HSW | 0.4829*** | 0.5246*** | 0.5032*** | 0.5874*** | 0.5972*** | 0.6089*** | 0.8070*** |
新窗口打开|下载CSV
2.1.2 方差分析 通过2017—2018年的8个荚果、种子相关性状方差分析(表3)可知, 同一环境下各性状在重复间不存在显著性差异(P>0.05); 群体各家系的同一性状存在极显著差异(P<0.01); 同一性状在不同环境下表现出极显著差异(P<0.01); 群体基因型与环境互作(G×E)在不同性状间均呈现极显著差异(P<0.01)。各性状遗传变异系数最小的为荚果长(GCV=12.87%), 最大的为百果重(GCV=17.54%); 荚果、种子相关性状均呈现中等遗传力(54.09%~67.50%), 说明各性状受环境影响较大。综合方差分析结果推测, 荚果、种子相关性状受微效多基因控制的可能性较大, 各性状可能会定位到多个QTL位点。
Table 3
表3
表3RIL群体各性状方差分析及广义遗传力
Table 3
| 变异来源 Source of variation | 自由度 DF | 荚果长 PL (mm) | 荚果宽 PW (mm) | 荚果厚 PT (mm) | 种子长 SL (mm) | 种子宽 SW (mm) | 种子厚 ST (mm) | 百果重 HPW (g) | 百仁重 HSW (g) |
|---|---|---|---|---|---|---|---|---|---|
| 区组间Block/environment | 5 | 2.05 ns | 3.39 ns | 5.87 ns | 2.84 ns | 3.23 ns | 4.53 ns | 29.36 ns | 14.19 ns |
| 基因型Genotype (G) | 314 | 143.74** | 101.27** | 105.53** | 46.78** | 10.30** | 17.39** | 31.78** | 77.12** |
| 环境Environment (E) | 4 | 502.85** | 310.54** | 915.58** | 112.56** | 154.97** | 2337.02** | 349.28** | 1898.64** |
| 基因型×环境G × E | 1196 | 8.79** | 7.36** | 7.53** | 3.82** | 3.08** | 4.54** | 4.30** | 10.18** |
| 遗传变异系数GVC (%) | — | 12.87 | 15.34 | 14.21 | 13.96 | 16.39 | 15.68 | 17.54 | 17.28 |
| 广义遗传力h2B (%) | — | 64.61 | 60.76 | 61.21 | 65.88 | 61.71 | 67.50 | 54.09 | 57.83 |
新窗口打开|下载CSV
2.2 遗传连锁图构建与共线性分析
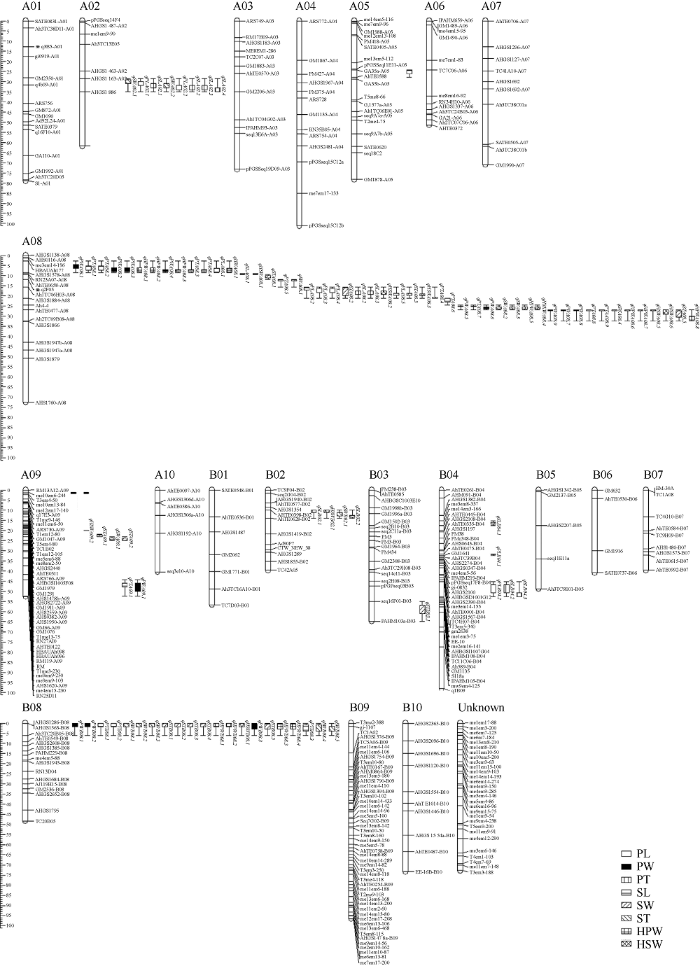
利用SSR、AhTE、SRAP和TRAP等分子标记对亲本进行多态性筛选, 获得640对条带清晰、多态性好的分子标记用于RIL群体基因分型。将分型后的基因型数据通过JoinMap 4.0软件(设置LOD=3.0~10.0)进行连锁分析, 获得1张包含21个连锁群的遗传连锁图谱(图3)。该图谱包含363个标记位点, 单条连锁群长度为39.59~101.05 cM, 包含4~50个分子标记, 标记间平均距离为3.75 cM。其中, 标记位点最少的染色体为B06 (4个), 标记位点最多的染色体为B09 (50个), 29个标记位点未匹配到染色体上, 命名为Unknown连锁群。与高密度图谱进行共线性分析(图4)可知, 84个标记与整合图谱符合, 分布在A01 (11个)、A02 (3个)、A04 (5个)、A05 (2个)、A07 (5个)、A08 (5个)、A09 (5个)、A10 (4个)、B01 (3个)、B02 (5个)、B03 (4个)、B04 (7个)、B05 (1个)、B06 (1个)、B07 (4个)、B08 (6个)、B09 (8个)、B10 (5个)染色体上。除A05染色体外, 其他染色体上的标记顺序均存在位置颠倒变化。此外, A03和A06染色体上的标记与整合图谱无共线性标记。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3花生遗传连锁图谱
缩写同
Fig. 3Genetic linkage map of peanut
Abbreviations are the same as those given in
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4本研究遗传图谱与整合图谱的共线性
Fig. 4Collinearity between genetic map and integrated map of this study
2.3 QTL定位
2.3.1 加性QTL分析 荚果、种子性状QTL定位分析表明, 在2017—2018年5个环境下共检测到97个QTL (表4和图3), 分布在A02、A05、A08、A09、B02、B03、B04、B08等8条染色体上, 其贡献率在2.36%~12.15%, LOD值在2.52~11.01, 加性效应为-10.993~12.309。其中, 主效QTL有4个, 2个与荚果厚相关(qPTA08.3和qPTA08.4), 位于A08染色体HBAUAh177~AhTE0658标记区间, PVE分别为11.06% (LOD=11.01)和12.15% (LOD=9.44); 1个与荚果宽相关(qPWA08.1), 位于A08染色体HBAUAh177~AhTE0658标记区间, PVE为10.02% (LOD=8.35); 1个与种子宽相关(qKWB08.5), 位于B08染色体AHGS1286~TC20B05标记区间, PVE为11.97% (LOD=9.98)。58个QTL的加性效应值为正, 推测控制性状的基因可能来源于母本; 39个QTL的加性效应值为负, 推测控制性状的基因可能来源于父本。本研究共检测到9个QTL聚集区(图3), 分别为A02染色体的AHGS1163~AHGS1886标记区间、A08染色体上的Ah4-4~TC9B08、me3em14- 196~Ah4-4、AhTE0658~TC6H03、TC6H03~AhTE0477和HBAUAh177~AhTE0658标记区间, B02染色体的AHTE0398~CTW_NEW_38标记区间、B04染色体的T3em5-340~me1em3-75标记区间、B08染色体的AHGS1286~TC20B05标记区间, 涉及荚果、种子8个相关性状, 说明这些染色体区段上的基因可能存在“一因多效”的现象。另外, 本研究在3个以上环境重复检测到的稳定QTL有45个(表4和图3), 分布在A02、A08、B04和B08染色体上, 说明这些QTL受环境影响较小。Table 4
表4
表4荚果、种子相关性状QTL定位结果
Table 4
| 性状 Trait | 位点 QTL | 环境Environment | 染色体 Chr. | 位置 Position | 范围 Range | 标记区间 Marker interval | LOD | 可解释的表型变异PVE (%) | 加性效应 Additive | 来源 Variety |
|---|---|---|---|---|---|---|---|---|---|---|
| 荚果长 | qPLA08.5 | 17QY | A08 | 19.317 | 1.395-24.581 | me3em14-196-Ah4-4 | 3.0543 | 4.5412 | 1.0026 | 冀花5号Jihua 5 |
| PL (mm) | qPLB04.2 | 18QY | B04 | 48.814 | 45.2-53.084 | T3em5-340-me1em3-75 | 3.1524 | 4.4841 | -1.052 | M130 |
| qPLA02.2 | 18QY | A02 | 33 | 28.586-35.04 | AHGS1163-AHGS1886 | 3.4276 | 4.1455 | -1.009 | M130 | |
| qPLA08.2 | 18QY | A08 | 16.487 | 1.395-24.581 | me3em14-196-Ah4-4 | 3.165 | 5.3035 | 1.142 | 冀花5号Jihua 5 | |
| qPLB04.1 | 18DM | B04 | 31.814 | 29.825-32.253 | PMc348-GM1641 | 2.7216 | 2.4116 | -0.7788 | M130 | |
| qPLA08.3 | 18DM | A08 | 17.43 | 1.395-24.581 | me3em14-196-Ah4-4 | 3.6112 | 5.5059 | 1.1219 | 冀花5号Jihua 5 | |
| qPLA02.1 | 18QA | A02 | 31 | 28.586-35.04 | AHGS1163-AHGS1886 | 3.1466 | 3.8886 | -0.8444 | M130 | |
| qPLA08.4 | 18QA | A08 | 17.43 | 1.395-24.581 | me3em14-196-Ah4-4 | 3.1664 | 6.2318 | 1.0698 | 冀花5号Jihua 5 | |
| qPLA08.1 | 18QA | A08 | 8.403 | 2.674-9.42 | HBAUAh177-AhTE0658 | 3.1246 | 3.7403 | 0.83 | 冀花5号Jihua 5 | |
| 荚果宽 | qPWA08.9 | 17QY | A08 | 27 | 24.581-32.375 | Ah4-4-TC9B08 | 3.0347 | 3.4027 | 0.3487 | 冀花5号Jihua 5 |
| PW (mm) | qPWA08.1 | 17QY | A08 | 5.539 | 2.674-9.42 | HBAUAh177-AhTE0658 | 8.3529 | 10.0181 | 0.6021 | 冀花5号Jihua 5 |
| qPWA09.1 | 17DM | A09 | 47.976 | 44.512-52.51 | AHS1620-me4em15-230 | 3.4111 | 5.6928 | -0.4961 | M130 | |
| qPWA08.5 | 17DM | A08 | 25 | 24.581-32.375 | Ah4-4-TC9B08 | 2.6823 | 2.4856 | 0.3265 | 冀花5号Jihua 5 | |
| qPWB08.1 | 17DM | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 4.5802 | 4.1738 | -0.426 | M130 | |
| qPWA08.2 | 17DM | A08 | 6.971 | 2.674-9.42 | HBAUAh177-AhTE0658 | 7.2488 | 8.0948 | 0.5908 | 冀花5号Jihua 5 | |
| qPWA08.6 | 18QY | A08 | 26 | 24.581-32.375 | Ah4-4-TC9B08 | 3.405 | 4.9727 | 0.3919 | 冀花5号Jihua 5 | |
| qPWA08.7 | 18DM | A08 | 27 | 24.581-32.375 | Ah4-4-TC9B08 | 5.1642 | 6.0632 | 0.4812 | 冀花5号Jihua 5 | |
| qPWB08.3 | 18DM | B08 | 1 | 0-6.125 | AHGS1286-TC20B05 | 4.8423 | 5.3595 | -0.4547 | M130 | |
| qPWA08.3 | 18DM | A08 | 6.971 | 2.674-9.42 | HBAUAh177-AhTE0658 | 6.2926 | 7.5419 | 0.5384 | 冀花5号Jihua 5 | |
| qPWA08.8 | 18QA | A08 | 27 | 24.581-32.375 | Ah4-4-TC9B08 | 4.0778 | 5.4765 | 0.3976 | 冀花5号Jihua 5 | |
| qPWB08.2 | 18QA | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 3.2573 | 3.7144 | -0.3288 | M130 | |
| qPWA08.4 | 18QA | A08 | 7.687 | 2.674-9.42 | HBAUAh177-AhTE0658 | 5.0836 | 6.6321 | 0.439 | 冀花5号Jihua 5 | |
| 荚果厚 | qPTA08.6 | 17QY | A08 | 22.146 | 1.395-24.581 | me3em14-196-Ah4-4 | 4.2717 | 6.0053 | 0.4499 | 冀花5号Jihua 5 |
| PT (mm) | qPTA08.1 | 17QY | A08 | 6.255 | 2.674-9.42 | HBAUAh177-AhTE0658 | 3.8997 | 5.4416 | 0.4305 | 冀花5号Jihua 5 |
| qPTA08.7 | 17DM | A08 | 25 | 24.581-32.375 | Ah4-4-TC9B08 | 4.683 | 5.2195 | 0.434 | 冀花5号Jihua 5 | |
| qPTB08.1 | 17DM | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 3.5352 | 3.7738 | -0.3716 | M130 | |
| qPTA08.2 | 17DM | A08 | 6.255 | 2.674-9.42 | HBAUAh177-AhTE0658 | 4.1823 | 5.3897 | 0.4421 | 冀花5号Jihua 5 | |
| qPTA08.5 | 18QY | A08 | 12 | 9.42-15.535 | AhTE0658-TC6H03 | 3.3146 | 4.8431 | 0.3898 | 冀花5号Jihua 5 | |
| qPTA09.1 | 18DM | A09 | 46.976 | 44.512-52.51 | AHS1620-me4em15-230 | 3.9877 | 5.5482 | -0.4349 | M130 | |
| qPTA08.8 | 18DM | A08 | 27 | 24.581-32.375 | Ah4-4-TC9B08 | 3.8396 | 3.7738 | 0.3577 | 冀花5号Jihua 5 | |
| qPTA05.1 | 18DM | A05 | 25.186 | 20.904-39.785 | me13em5-112-GM1577 | 2.5575 | 2.6714 | -0.3018 | M130 | |
| qPTB08.3 | 18DM | B08 | 2 | 0-6.125 | AHGS1286-TC20B05 | 6.4241 | 6.1373 | -0.4581 | M130 | |
| qPTA08.3 | 18DM | A08 | 7.688 | 2.674-9.42 | HBAUAh177-AhTE0658 | 11.0128 | 11.0554 | 0.6137 | 冀花5号Jihua 5 | |
| qPTA08.9 | 18QA | A08 | 27 | 24.581-32.375 | Ah4-4-TC9B08 | 3.3743 | 4.1636 | 0.3621 | 冀花5号Jihua 5 | |
| qPTB08.2 | 18QA | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 3.3221 | 3.5571 | -0.3361 | M130 | |
| qPTA08.4 | 18QA | A08 | 7.688 | 2.674-9.42 | HBAUAh177-AhTE0658 | 9.437 | 12.1489 | 0.6205 | 冀花5号Jihua 5 | |
| 种子长 | qSLB04.3 | 17QY | B04 | 51.814 | 45.2-53.084 | T3em5-340-me1em3-75 | 3.5098 | 4.5251 | -0.4663 | M130 |
| SL (mm) | qSLA02.4 | 17QY | A02 | 34 | 28.586-35.04 | AHGS1163-AHGS1886 | 4.5943 | 5.4414 | -0.5079 | M130 |
| qSLA02.2 | 17DM | A02 | 33 | 28.586-35.04 | AHGS1163-AHGS1886 | 5.3238 | 7.4048 | -0.6248 | M130 | |
| qSLB04.2 | 18QY | B04 | 49.814 | 45.2-53.084 | T3em5-340-me1em3-75 | 4.3546 | 6.0388 | -0.5601 | M130 | |
| qSLA02.3 | 18QY | A02 | 33 | 28.586-35.04 | AHGS1163-AHGS1886 | 5.7194 | 6.4678 | -0.5774 | M130 | |
| qSLA08.2 | 18QY | A08 | 18.373 | 1.395-24.581 | me3em14-196-Ah4-4 | 4.392 | 6.4226 | 0.576 | 冀花5号Jihua 5 | |
| qSLB02.1 | 18QY | B02 | 10 | 10-29.536 | AHTE0398-CTW_NEW_38 | 2.5587 | 2.3638 | -0.3495 | M130 | |
| qSLB04.1 | 18DM | B04 | 16.405 | 43.26-4.103 | AHTE0001-AHM091 | 3.9425 | 5.5578 | -0.43 | M130 | |
| qSLA08.1 | 18DM | A08 | 9.119 | 2.674-9.42 | HBAUAh177-AhTE0658 | 2.898 | 3.8087 | 0.3432 | 冀花5号Jihua 5 | |
| qSLA02.1 | 18QA | A02 | 32 | 28.586-35.04 | AHGS1163-AHGS1886 | 3.3469 | 3.8192 | -0.3879 | M130 | |
| qSLA08.3 | 18QA | A08 | 18.373 | 1.395-24.581 | me3em14-196-Ah4-4 | 4.1987 | 6.4454 | 0.5045 | 冀花5号Jihua 5 | |
| 种子宽 | qSWB08.1 | 17QY | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 3.3062 | 5.0411 | -0.197 | M130 |
| SW (mm) | qSWB08.2 | 17DM | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 3.2831 | 3.6766 | -0.1703 | M130 |
| qSWA08.2 | 17DM | A08 | 26 | 15.535-26.911 | TC6H03-AhTE0477 | 3.9168 | 4.8359 | 0.1949 | 冀花5号Jihua 5 | |
| qSWA08.4 | 18QY | A08 | 27 | 24.581-32.375 | Ah4-4-TC9B08 | 2.7094 | 3.7658 | 0.1335 | JH5 | |
| qSWB08.4 | 18QY | B08 | 2 | 0-6.125 | AHGS1286-TC20B05 | 5.52 | 7.3282 | -0.188 | M130 | |
| qSWB02.1 | 18QY | B02 | 12 | 10-29.536 | AHTE0398-CTW_NEW_38 | 2.5171 | 3.4186 | -0.1275 | M130 | |
| qSWA09.1 | 18DM | A09 | 23.976 | 23.052-24.982 | RN27A10-AHTE0122 | 2.6959 | 3.1535 | 0.1305 | 冀花5号Jihua 5 | |
| qSWA02.1 | 18DM | A02 | 30 | 28.586-35.04 | AHGS1163-AHGS1886 | 3.4558 | 3.8727 | 0.1449 | 冀花5号Jihua 5 | |
| qSWB08.5 | 18DM | B08 | 3 | 0-6.125 | AHGS1286-TC20B05 | 9.9761 | 11.9698 | -0.2551 | M130 | |
| qSWA08.1 | 18DM | A08 | 12 | 9.42-15.535 | AhTE0658-TC6H03 | 4.1572 | 4.7351 | 0.1598 | 冀花5号Jihua 5 | |
| qSWA02.2 | 18QA | A02 | 32 | 28.586-35.04 | AHGS1163-AHGS1886 | 3.2739 | 4.3875 | 0.1518 | 冀花5号Jihua 5 | |
| qSWA08.3 | 18QA | A08 | 26 | 24.581-32.375 | Ah4-4-TC9B08 | 3.511 | 4.5457 | 0.1544 | 冀花5号Jihua 5 | |
| qSWB08.3 | 18QA | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 4.8277 | 5.6235 | -0.1723 | M130 | |
| qSWB02.2 | 18QA | B02 | 13 | 10-29.536 | AHTE0398-CTW_NEW_38 | 3.055 | 4.4285 | -0.1526 | M130 | |
| 种子厚 | qSTA09.1 | 17QY | A09 | 22.976 | 21.827-23.052 | T1me13-75-RN27A10 | 2.9792 | 4.3897 | 0.1586 | 冀花5号Jihua 5 |
| ST (mm) | qSTB08.1 | 17QY | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 4.1523 | 5.9184 | -0.1856 | M130 |
| qSTB08.5 | 17DM | B08 | 1 | 0-6.125 | AHGS1286-TC20B05 | 3.511 | 5.2695 | -0.1578 | M130 | |
| qSTA08.3 | 18QY | A08 | 19.317 | 1.395-24.581 | me3em14-196-Ah4-4 | 4.3155 | 6.9533 | 0.2296 | 冀花5号Jihua 5 | |
| qSTB08.2 | 18QY | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 4.6686 | 4.8687 | -0.1935 | M130 | |
| qSTA08.4 | 18QY | A08 | 21 | 15.535-26.911 | TC6H03-AhTE0477 | 2.8698 | 3.8907 | 0.1716 | 冀花5号Jihua 5 | |
| qSTA09.2 | 18DM | A09 | 24.976 | 23.052-24.982 | RN27A10-AHTE0122 | 3.8787 | 4.14 | 0.1812 | 冀花5号Jihua 5 | |
| qSTB08.3 | 18DM | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 5.4215 | 5.8448 | -0.2147 | M130 | |
| qSTB03.1 | 18DM | B03 | 59.043 | 27.96-64.668 | GM1954-IPAHM103 | 2.8689 | 3.5976 | 0.1682 | 冀花5号Jihua 5 | |
| qSTA08.1 | 18DM | A08 | 10 | 9.42-15.535 | AhTE0658-TC6H03 | 6.5305 | 7.3459 | 0.24 | 冀花5号Jihua 5 | |
| qSTA08.5 | 18QA | A08 | 29 | 24.581-32.375 | Ah4-4-TC9B08 | 5.1955 | 7.3853 | 0.2579 | 冀花5号Jihua 5 | |
| qSTB08.4 | 18QA | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 2.608 | 3.1638 | -0.1697 | M130 | |
| qSTA08.2 | 18QA | A08 | 18 | 15.535-26.911 | TC6H03-AhTE0477 | 3.5829 | 5.2767 | 0.2178 | 冀花5号Jihua 5 | |
| 百果重 | qHPWA08.8 | 17QY | A08 | 31 | 24.581-32.375 | Ah4-4-TC9B08 | 2.612 | 2.884 | 6.1294 | J冀花5号ihua 5 |
| HPW (g) | qHPWB08.3 | 17QY | B08 | 1 | 0-6.125 | AHGS1286-TC20B05 | 4.2913 | 4.8043 | -7.9731 | M130 |
| qHPWA08.1 | 17QY | A08 | 6.971 | 2.674-9.42 | HBAUAh177-AhTE0658 | 3.942 | 5.0068 | 8.1239 | 冀花5号Jihua 5 | |
| qHPWB08.1 | 17DM | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 5.4732 | 6.1589 | -9.2979 | M130 | |
| qHPWA08.2 | 17DM | A08 | 6.971 | 2.674-9.42 | HBAUAh177-AhTE0658 | 3.361 | 4.5239 | 7.9317 | 冀花5号Jihua 5 | |
| qHPWA08.5 | 18QY | A08 | 26 | 24.581-32.375 | Ah4-4-TC9B08 | 3.7742 | 5.4674 | 10.1994 | 冀花5号Jihua 5 | |
| qHPWA09.1 | 18DM | A09 | 1.574 | 1.243-1.881 | me10em6-244-T3me4-50 | 2.9275 | 4.4348 | 9.1722 | 冀花5号Jihua 5 | |
| qHPWA08.6 | 18DM | A08 | 27 | 24.581-32.375 | Ah4-4-TC9B08 | 4.3701 | 4.589 | 9.3388 | 冀花5号Jihua 5 | |
| qHPWB08.4 | 18DM | B08 | 1 | 0-6.125 | AHGS1286-TC20B05 | 6.3447 | 6.2963 | -10.993 | M130 | |
| qHPWA08.3 | 18DM | A08 | 7.687 | 2.674-9.42 | HBAUAh177-AhTE0658 | 7.3085 | 7.9283 | 12.3093 | 冀花5号Jihua 5 | |
| qHPWA08.7 | 18QA | A08 | 27 | 24.581-32.375 | Ah4-4-TC9B08 | 3.3108 | 4.3355 | 7.727 | 冀花5号Jihua 5 | |
| qHPWB08.2 | 18QA | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 3.1924 | 3.6078 | -7.0792 | M130 | |
| qHPWA08.4 | 18QA | A08 | 8.403 | 2.674-9.42 | HBAUAh177-AhTE0658 | 6.215 | 7.9641 | 10.5143 | 冀花5号Jihua 5 | |
| 百仁重 | qHSWA09.2 | 17QY | A09 | 19.976 | 19.851-20.044 | AHGS0362-AHS1950 | 2.7386 | 3.9653 | 2.281 | 冀花5号Jihua 5 |
| HSW (g) | qHSWA08.6 | 17QY | A08 | 28 | 24.581-32.375 | Ah4-4-TC9B08 | 3.2048 | 4.984 | 2.5511 | 冀花5号Jihua 5 |
| qHSWA08.4 | 18QY | A08 | 26 | 24.581-32.375 | Ah4-4-TC9B08 | 6.1813 | 8.9514 | 4.4051 | 冀花5号Jihua 5 | |
| qHSWB02.1 | 18QY | B02 | 10 | 10-29.536 | AHTE0398-CTW_NEW_38 | 3.4265 | 4.1876 | -3.0213 | M130 | |
| qHSWA08.5 | 18DM | A08 | 27 | 24.581-32.375 | Ah4-4-TC9B08 | 3.116 | 4.3944 | 2.9937 | 冀花5号Jihua 5 | |
| qHSWB08.1 | 18DM | B08 | 0 | 0-6.125 | AHGS1286-TC20B05 | 4.3546 | 5.07 | -3.2325 | M130 | |
| qHSWA08.1 | 18DM | A08 | 9.119 | 2.674-9.42 | HBAUAh177-AhTE0658 | 4.7443 | 5.7178 | 3.4257 | 冀花5号Jihua 5 | |
| qHSWA09.1 | 18QA | A09 | 1.574 | 1.243-1.881 | me10em6-244-T3me4-50 | 2.6987 | 4.1254 | 3.2042 | 冀花5号Jihua 5 | |
| qHSWA08.2 | 18QA | A08 | 18.373 | 1.395-24.581 | me3em14-196-Ah4-4 | 2.6696 | 4.3569 | 3.306 | 冀花5号Jihua 5 | |
| qHSWA08.3 | 18QA | A08 | 20 | 15.535-26.911 | TC6H03-AhTE0477 | 4.2105 | 5.6344 | 3.7483 | 冀花5号Jihua 5 |
新窗口打开|下载CSV
2.3.2 上位性QTL分析 对8个荚果、种子性状在5个环境下进行上位性QTL分析(表5), 共获得15对上位性QTL, 涉及荚果长、荚果宽、荚果厚、种子长、百果重和百仁重等6个性状, 其LOD值为5.09~8.05, PVE为10.23%~51.84%。其中, 控制荚果长、种子长和百仁重的上位性QTL各1对, 其LOD值分别为5.09、7.60和7.30, PVE分别为10.23%、11.61%、13.99%; 控制百果重的上位性QTL有2对, 其LOD值为8.05和5.04, PVE为15.36%、17.75%; 控制荚果宽的上位性QTL有2对, 其LOD值为5.33和5.54, PVE为11.95%~42.46%; 控制荚果厚的上位性QTL有8对, 其LOD值为5.01~7.66, PVE为26.30%~51.84%。
Table 5
表5
表5荚果和种子相关性状的上位性QTL定位结果
Table 5
| 性状 Trait | 上位性QTL名称 Epi-QTL name | 环境 Environment | 染色体 Chr. | 位置1 Position 1 | 标记区间1 Marker interval 1 | 上位性QTL名称 Epi-QTL name | 染色体 Chr. | 位置2 Position 2 | 标记区间2 Marker interval 2 | LOD | 可解释的遗传变异 PVE (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PL (mm) | Epi-qPLA09.1 | 18BD | A09 | 50.976 | AHS1620-me4em15-230 | Epi-qPLA05.1 | A05 | 50 | T2me4-75-seq18C2 | 5.09 | 10.23 |
| PW (mm) | Epi-qPWB08.1 | 17BD | B08 | 5 | AHGS1286-TC20B05 | Epi-qPWA08.2 | A08 | 25 | TC6H03-AhTE0477 | 5.33 | 11.95 |
| Epi-qPWA08.1 | 18BD | A08 | 15.544 | me3em14-196-Ah4-4 | Epi-qPWA08.1 | A08 | 20.26 | me3em14-196-Ah4-4 | 5.54 | 42.46 | |
| PT (mm) | Epi-qPTB09.1 | 17BD | B09 | 5 | T3me2-388-AHGS1576 | Epi-qPTB09.1 | B09 | 10 | T3me2-388-AHGS1576 | 6.05 | 39.57 |
| Epi-qPTA02.3 | 17BD | A02 | 50 | AHGS1886-AHGS1159 | Epi-qPTA02.3 | A02 | 55 | AHGS1886-AHGS1159 | 5.05 | 40.63 | |
| Epi-qPTA08.1 | 17BD | A08 | 10.827 | me3em14-196-Ah4-4 | Epi-qPTA08.1 | A08 | 15.544 | me3em14-196-Ah4-4 | 7.66 | 51.84 | |
| Epi-qPTA03.1 | 17BD | A03 | 40 | AhTE0570-TC4G02 | Epi-qPTA03.1 | A03 | 45 | AhTE0570-TC4G02 | 6.42 | 37.16 | |
| Epi-qPTA10.1 | 17BD | A10 | 20 | AhTE0586-AHGS1192 | Epi-qPTA10.1 | A10 | 25 | AHGS1192-seq3e10 | 6.56 | 38.94 | |
| Epi-qPTA02.2 | 17DM | A02 | 45 | AHGS1886-AHGS1159 | Epi-qPTA02.2 | A02 | 50 | AHGS1886-AHGS1159 | 6.05 | 40.39 | |
| Epi-qPTA08.2 | 17DM | A08 | 15.544 | me3em14-196-Ah4-4 | Epi-qPTA08.2 | A08 | 20.26 | me3em14-196-Ah4-4 | 5.01 | 39.62 | |
| Epi-qPTA02.1 | 18QA | A02 | 10 | pPGSseq14F4-Ah3TC13E05 | Epi-qPTA02.1 | A02 | 15 | Ah3TC13E05-AHGS1463 | 5.37 | 26.30 | |
| SL (mm) | Epi-qSLB08.1 | 17BD | B08 | 0 | AHGS1286-TC20B05 | Epi-qSLA08.1 | A08 | 25 | TC6H03-AhTE0477 | 7.60 | 11.61 |
| HPW (g) | Epi-qHPWB08.1 | 17BD | B08 | 5 | AHGS1286-TC20B05 | Epi-qHPWA08.1 | A08 | 25 | TC6H03-AhTE0477 | 8.05 | 17.75 |
| Epi-qHPWB08.2 | 18DM | B08 | 5 | AHGS1286-TC20B05 | Epi-qHPWA08.2 | A08 | 25 | TC6H03-AhTE0477 | 5.04 | 15.36 | |
| HSW (g) | Epi-qHSWB08.1 | 17BD | B08 | 5 | AHGS1286-TC20B05 | Epi-qHSWA08.1 | A08 | 25 | TC6H03-AhTE0477 | 7.30 | 13.99 |
新窗口打开|下载CSV
2.4 QTL与环境互作效应结果分析
2.4.1 加性QTL与环境互作分析 利用IciMapping 4.2软件加性QTL与环境的互作效应进行分析(表6), 共有6对与环境有互作效应的加性QTL, 与荚果长、种子厚和百果重相关的QTL各1个, 其加性效应值分别为-0.55、-0.37和0.36, 加性遗传贡献率分别为1.280%、0.624%和0.618%, 与环境互作的遗传贡献率分别为0.082%、0.059%和0.014%; 与种子长相关的QTL有3个, 其加性效应值为-0.642~ -0.491, 加性遗传贡献率为1.10%~1.90%, 与环境互作的遗传贡献率为0.046%~0.159%。Table 6
表6
表6荚果和种子相关性状的加性QTL与环境互作结果
Table 6
| 性状 Trait | 位置 Position | 标记区间 Marker interval | LOD | LOD(A) | LOD (AbyE) | 可解释的遗传变异PVE | 可解释的遗传变异PVE(A) | 可解释的遗传变异PVE (AbyE) | 加性效应 Add | AbyE_01 | AbyE_02 | AbyE_03 | AbyE_04 | AbyE_05 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPW | 1.976 | T3me4-50-me10em13-84 | 3.2854 | 3.162 | 0.1234 | 0.6316 | 0.6176 | 0.014 | 0.3661 | -0.006 | -0.0408 | 0.0953 | 0.016 | -0.0645 |
| SL | 16.405 | AHM091-AHTE0001 | 6.4625 | 6.2415 | 0.2211 | 1.1913 | 1.145 | 0.0462 | -0.5154 | 0.1206 | -0.1441 | -0.0132 | 0.1118 | -0.0751 |
| PL | 32.814 | GM1641-Ah3TC39B04 | 7.7019 | 7.1093 | 0.5927 | 1.3629 | 1.2801 | 0.0828 | -0.5506 | 0.1878 | 0.0587 | -0.178 | -0.1467 | 0.0782 |
| SL | 49.814 | T3em5-340-me1em3-75 | 6.2931 | 5.6064 | 0.6866 | 1.2594 | 1.1008 | 0.1586 | -0.4907 | 0.0103 | -0.1087 | -0.282 | 0.2596 | 0.1208 |
| SL | 33 | AHGS1163-AHGS1886 | 10.3475 | 9.9861 | 0.3615 | 1.9775 | 1.9001 | 0.0773 | -0.6416 | 0.027 | 0.0248 | -0.1949 | 0.2023 | -0.0592 |
| ST | 37.706 | GM1954-seq2H08 | 3.4137 | 3.1998 | 0.2139 | 0.683 | 0.6243 | 0.0588 | -0.3667 | -0.011 | -0.2127 | 0.0458 | 0.0907 | 0.0872 |
新窗口打开|下载CSV
2.4.2 上位性QTL与环境互作分析 由表7可知, 共有13对上位性QTL与环境有互作效应, 其中3对与荚果长相关, PVE为1.95%~2.65%, PVE (AAE)为0.007%~0.089%; 与荚果厚相关的QTL有6对, PVE为1.47%~2.81%, PVE (AAE)为0.007%~0.073%; 与种子长和百仁重分别相关的QTL各2对, 其PVE分别为1.286%、4.009%和2.417%、1.489%, PVE (AAE)分别为0.137%、0.024%和0.039%、0.019%。所有和环境互作的QTL, 在不同的环境下均表现出不同的互作效应。
Table 7
表7
表7荚果和种子相关性状的上位性QTL与环境互作效应
Table 7
| Chr.1 | 位置1 Position 1 | 性状 Trait | 标记区间1 Marker interval 1 | Chr.2 | 位置2 Position 2 | 标记区间2 Marker interval 2 | LOD | LOD (AAbyE) | PVE | PVE (AAbyE) | Add1 | Add2 | AddbyAdd | AAbyE_01 | AAbyE_02 | AAbyE_03 | AAbyE_04 | AAbyE_05 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A09 | 45.976 | PL | AHS1620-me4em15-230 | B07 | 0 | TC1A08-TC9H09 | 7.0121 | 0.0728 | 1.9524 | 0.0197 | -0.1218 | 0.0794 | -0.5448 | -0.0493 | -0.0654 | 0.0668 | 0.0148 | 0.0331 |
| A09 | 50.976 | PL | AHS1620-me4em15-230 | A06 | 35 | me7em1-83-me8em16-92 | 7.8477 | 0.0188 | 2.2588 | 0.0066 | 0.0036 | 0.2423 | -0.5959 | 0.0149 | -0.0405 | -0.0235 | 0.0533 | -0.0043 |
| A09 | 45.976 | PL | AHS1620-me4em15-230 | B03 | 31.236 | GM1954-IPAHM103 | 10.0123 | 0.4698 | 2.6465 | 0.0887 | -0.183 | -0.3668 | 0.6229 | -0.1048 | 0.1381 | 0.139 | -0.1102 | -0.0622 |
| A02 | 0 | PT | pPGSseq14F4-Ah3TC13E05 | A05 | 5 | me7em9-96-me13em5-112 | 8.6941 | 0.3066 | 2.3716 | 0.0727 | -0.0142 | -0.1429 | -0.5926 | -0.0159 | -0.1916 | 0.0964 | 0.0257 | 0.0854 |
| A02 | 60 | PT | AHGS1886-AHGS1159 | A03 | 10 | RM17H09-me8em1-286 | 7.1732 | 0.1598 | 1.9488 | 0.0372 | 0.1981 | 0.0409 | -0.5484 | 0.0342 | 0.011 | -0.1155 | -0.0432 | 0.1136 |
| A02 | 45 | PT | AHGS1886-AHGS1159 | A06 | 36.412 | TC7C06-AHTE0372 | 8.959 | 0.1938 | 2.44 | 0.0354 | -0.2324 | 0.4976 | -0.608 | -0.0709 | -0.0324 | -0.069 | 0.11 | 0.0623 |
| A08 | 6.111 | PT | me3em14-196-Ah4-4 | B03 | 2.834 | AHBGSC1003E10-GM1996 | 10.1986 | 0.0214 | 2.8149 | 0.0074 | 0.5756 | 0.21 | 0.6596 | 0.0296 | 0.0139 | 0.0111 | -0.0598 | 0.0053 |
| A08 | 1.395 | PT | me3em14-196-Ah4-4 | A08 | 25 | TC6H03-AhTE0477 | 5.4087 | 0.2251 | 1.4752 | 0.0536 | 0.4861 | 0.1886 | -0.4783 | -0.1029 | 0.175 | -0.0222 | -0.0274 | -0.0225 |
| A08 | 15.544 | PT | me3em14-196-Ah4-4 | B02 | 10 | AHTE0398-CTW_NEW_38 | 6.5397 | 0.3284 | 1.6356 | 0.0709 | 0.7735 | -0.5558 | -0.4935 | 0.0526 | -0.1013 | -0.0825 | -0.0451 | 0.1763 |
| B08 | 0 | SL | AHGS1286-TC20B05 | B03 | 59.043 | GM1954-IPAHM103 | 5.0287 | 0.5997 | 1.2864 | 0.1371 | -0.0958 | -0.1712 | 0.422 | -0.1143 | 0.2612 | -0.0654 | -0.1314 | 0.05 |
| B08 | 0 | SL | AHGS1286-TC20B05 | A08 | 25 | TC6H03-AhTE0477 | 15.7588 | 0.1295 | 4.009 | 0.0244 | -0.1851 | 0.4017 | 0.7983 | 0.106 | 0.0345 | -0.072 | -0.022 | -0.0465 |
| A03 | 35 | HSW | AhTE0570-TC4G02 | B03 | 6.453 | AHGS1940a-AHGS1940b | 9.1572 | 0.2059 | 2.4169 | 0.0351 | -0.2737 | -0.1534 | -0.6022 | 0.0048 | -0.1046 | 0.0963 | -0.0615 | 0.065 |
| A10 | 20 | HSW | AhTE0586-AHGS1192 | B02 | 25 | AHTE0398-CTW_NEW_38 | 5.4877 | 0.1016 | 1.4894 | 0.0194 | 0.0107 | -0.2795 | -0.4755 | 0.0571 | -0.0731 | -0.0229 | 0.0769 | -0.038 |
新窗口打开|下载CSV
3 讨论
花生荚果、种子性状与产量紧密相关, 是重要的农艺性状。本研究选用冀花5号和M130 2个花生品种作为亲本材料, 其生长发育和对环境的适应性均表现良好, 而且2个亲本间与荚果、种子相关的8个性状差异显著(P<0.01)。在构建的RIL群体中, 荚果长、荚果宽、荚果厚、种子长、种子宽、种子厚、百果重和百仁重等8个性状的变异符合正态分布(图2), 且表现出较大的变异范围, 其最大、最小值均超过双亲, 这为构建花生遗传连锁图以及QTL分析奠定了坚实的基础。一般认为, 主效QTL的贡献率大于10%, LOD值越大, QTL越稳定。本研究通过构建遗传连锁图谱, 结合多年多环境的表型数据共检测到97个QTL, 其中, 在A08染色体AHTE0658标记附近检测到与百仁重相关的QTL, 与Lu等[27]的产量QTLs meta分析结果一致。然而, Lu等[27]在B08染色体AHGS2268~ AHGS494区间检测到2个与百果重相关的Meta QTL, 而本研究与百果重相关的QTL则是在B08染色体的AHGS1286~TC20B05区间, QTL标记区间的差异可能是由于图谱之间的标记种类不一致所造成。在与前人[13,28-32]的研究对比中发现, 前人重复检测到的QTL主要集中在A05染色体上, 而本研究定位到的QTL集中分布在A02、A08和B02染色体上, 并且具有较高的贡献率。此外, 4个主效QTL分别分布在A08染色体(qPWA08.1、qPTA08.3、qPTA08.4) HBAUAh177~AhTE0658区间和B08染色体上(qKWB08.5) AHGS1286~TC20B05区间上, 与Lu等[27]的研究结果不同, 说明这些QTL是全新的。本研究发现了9个QTL聚集区, 其中5个QTL聚集区分布在A08染色体上, 涉及所有的荚果、种子相关性状, 且在多个环境下能够被重复检测到, 表明所检测到的QTL是稳定的, 且在A08染色体上与荚果、种子性状相关的QTL鲜有报道。因此, A08染色体是研究花生荚果、种子相关性状十分重要的染色体之一, 可进一步进行分子标记的加密研究。
基因型×环境互作是作物数量性状的普遍属性和遗传育种改良的关注重点, 但在前人[27,28,29,30,31,32]的研究中未进行基因型×环境互作分析, 本研究共获得6个与环境互作的加性QTL和15对上位性QTL, 涉及荚果长、荚果厚、种子长、种子厚、百果重和百仁重等性状, 说明在花生荚果、种子相关性状中存在基因型×环境互作效应。因此, 在利用QTL改良作物品种时, 既需注重QTL的遗传主效应, 还需重视QTL与环境的互作效应[33], 上位性效应对数量性状亦有重要的作用[34]。
随着分子标记的开发应用和QTL定位技术的不断发展, 在分子标记辅助育种中既应该考虑起主效作用的QTL, 又要考虑与其存在上位效应的QTL。然而, 同一性状的QTL 在不同定位群体和不同环境下可能表现不一致, 通过对QTL在不同环境背景下的研究, 有利于定位到受环境影响较小的QTL。不仅考虑到显性和上位性, 同时考虑到与环境的互作效应, 有助于提高分子标记选择的效率。
4 结论
本研究构建了1张包含363个标记位点、21条连锁群、覆盖长度为1360.38 cM的花生遗传连锁图谱。利用该图谱对2017—2018年5个环境下8个荚果、种子相关性状进行QTL定位, 获得97个QTL, 其中, 4个主效QTL分别为qPWA08.1、qPTA08.3、qPTA08.4和qSWB08.5。3个以上环境重复检测到的稳定QTL有45个, A02、A08、B02、B04和B08染色体上存在9个QTL聚集区。对8个荚果、种子相关性状在5个环境下进行上位性QTL定位分析, 检测到15对上位性QTL, 涉及荚果长、荚果宽、荚果厚、种子长、百果重和百仁重等6个性状。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIPMID [本文引用: 1]

Many of the world's most important food legumes are grown in arid and semi-arid regions of Africa and Asia, where crop productivity is hampered by biotic and abiotic stresses. Until recently, these crops have also suffered from a dearth of genomic and molecular-genetic resources and thus were 'orphans' of the genome revolution. However, the community of legume researchers has begun a concerted effort to change this situation. The driving force is a series of international collaborations that benefit from recent advances in genome sequencing and genotyping technologies. The focus of these activities is the development of genome-scale data sets that can be used in high-throughput approaches to facilitate genomics-assisted breeding in these legumes.
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOIPMID [本文引用: 2]

Background: Peanut (Arachis hypogaea) is an autogamous allotetraploid legume (2n = 4x = 40) that is widely cultivated as a food and oil crop. More than 6,000 DNA markers have been developed in Arachis spp., but high-density linkage maps useful for genetics, genomics, and breeding have not been constructed due to extremely low genetic diversity. Polymorphic marker loci are useful for the construction of such high-density linkage maps. The present study used in silico analysis to develop simple sequence repeat-based and transposon-based markers.;Results: The use of in silico analysis increased the efficiency of polymorphic marker development by more than 3-fold. In total, 926 (34.2%) of 2,702 markers showed polymorphisms between parental lines of the mapping population. Linkage analysis of the 926 markers along with 253 polymorphic markers selected from 4,449 published markers generated 21 linkage groups covering 2,166.4 cM with 1,114 loci. Based on the map thus produced, 23 quantitative trait loci (QTLs) for 15 agronomical traits were detected. Another linkage map with 326 loci was also constructed and revealed a relationship between the genotypes of the FAD2 genes and the ratio of oleic/linoleic acid in peanut seed.;Conclusions: In silico analysis of polymorphisms increased the efficiency of polymorphic marker development, and contributed to the construction of high-density linkage maps in cultivated peanut. The resultant maps were applicable to QTL analysis. Marker subsets and linkage maps developed in this study should be useful for genetics, genomics, and breeding in Arachis. The data are available at the Kazusa DNA Marker Database (http://marker.kazusa.or.jp).
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
PMID [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIPMID

Molecular markers and genetic linkage maps are pre-requisites for molecular breeding in any crop species. In case of peanut or groundnut (Arachis hypogaea L.), an amphidiploid (4X) species, not a single genetic map is, however, available based on a mapping population derived from cultivated genotypes. In order to develop a genetic linkage map for tetraploid cultivated groundnut, a total of 1,145 microsatellite or simple sequence repeat (SSR) markers available in public domain as well as unpublished markers from several sources were screened on two genotypes, TAG 24 and ICGV 86031 that are parents of a recombinant inbred line mapping population. As a result, 144 (12.6%) polymorphic markers were identified and these amplified a total of 150 loci. A total of 135 SSR loci could be mapped into 22 linkage groups (LGs). While six LGs had only two SSR loci, the other LGs contained 3 (LG_AhXV) to 15 (LG_AhVIII) loci. As the mapping population used for developing the genetic map segregates for drought tolerance traits, phenotyping data obtained for transpiration, transpiration efficiency, specific leaf area and SPAD chlorophyll meter reading (SCMR) for 2 years were analyzed together with genotyping data. Although, 2-5 QTLs for each trait mentioned above were identified, the phenotypic variation explained by these QTLs was in the range of 3.5-14.1%. In addition, alignment of two linkage groups (LGs) (LG_AhIII and LG_AhVI) of the developed genetic map was shown with available genetic maps of AA diploid genome of groundnut and Lotus and Medicago. The present study reports the construction of the first genetic map for cultivated groundnut and demonstrates its utility for molecular mapping of QTLs controlling drought tolerance related traits as well as establishing relationships with diploid AA genome of groundnut and model legume genome species. Therefore, the map should be useful for the community for a variety of applications.
DOIURL [本文引用: 4]
DOIURL [本文引用: 2]
DOIURL [本文引用: 1]
DOIPMID [本文引用: 1]

SSR-based QTL mapping provides useful information for map-based cloning of major QTLs and can be used to improve the agronomic and quality traits in cultivated peanut by marker-assisted selection. Cultivated peanut (Arachis hypogaea L.) is an allotetraploid species (AABB, 2n = 4× = 40), valued for its edible oil and digestible protein. Linkage mapping has been successfully conducted for most crops, and it has been applied to detect the quantitative trait loci (QTLs) of biotic and abiotic traits in peanut. However, the genetic basis of agronomic and quality-related traits remains unclear. In this study, high levels of phenotypic variation, broad-sense heritability and significant correlations were observed for agronomic and quality-related traits in an F 2:3 population. A genetic linkage map was constructed for cultivated peanut containing 470 simple sequence repeat (SSR) loci, with a total length of 1877.3 cM and average distance of 4.0 cM between flanking markers. For 10 agronomic traits, 24 QTLs were identified and each QTL explained 1.69-18.70 % of the phenotypic variance. For 8 quality-related traits, 12 QTLs were identified that explained 1.72-20.20 % of the phenotypic variance. Several QTLs for multiple traits were overlapped, reflecting the phenotypic correlation between these traits. The majority of QTLs exhibited obvious dominance or over-dominance effects on agronomic and quality traits, highlighting the importance of heterosis for breeding. A comparative analysis revealed genomic duplication and arrangement of peanut genome, which aids the assembly of scaffolds in genomic sequencing of Arachis hypogaea. Our QTL analysis results enabled us to clearly understand the genetic base of agronomic and quality traits in cultivated peanut, further accelerating the progress of map-based cloning of major QTLs and marker-assisted selection in future breeding.
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
DOIURL [本文引用: 1]
