 ,1,2, 张旭1,3, 吴磊1, 何漪1, 张平平1, 马鸿翔
,1,2, 张旭1,3, 吴磊1, 何漪1, 张平平1, 马鸿翔 ,1,*, 孔令让
,1,*, 孔令让 ,2,*
,2,*Genetic analysis for yield related traits of wheat (Triticum aestivum L.) based on a recombinant inbred line population from Ningmai 9 and Yangmai 158
JIANG Peng ,1,2, ZHANG Xu1,3, WU Lei1, HE Yi1, ZHANG Ping-Ping1, MA Hong-Xiang
,1,2, ZHANG Xu1,3, WU Lei1, HE Yi1, ZHANG Ping-Ping1, MA Hong-Xiang ,1,*, KONG Ling-Rang
,1,*, KONG Ling-Rang ,2,*
,2,*通讯作者:
收稿日期:2020-06-18接受日期:2020-10-15网络出版日期:2021-05-12
| 基金资助: |
Received:2020-06-18Accepted:2020-10-15Online:2021-05-12
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1312KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
姜朋, 张旭, 吴磊, 何漪, 张平平, 马鸿翔, 孔令让. 宁麦9号/扬麦158重组自交系群体产量性状的遗传解析[J]. 作物学报, 2021, 47(5): 869-881. doi:10.3724/SP.J.1006.2021.01051
JIANG Peng, ZHANG Xu, WU Lei, HE Yi, ZHANG Ping-Ping, MA Hong-Xiang, KONG Ling-Rang.
高产是小麦育种的重要目标, 小麦产量由单位面积穗数、每穗粒数及粒重构成, 三者的平衡协调发展决定着产量的最终实现[1]。小麦单位面积穗数取决于基本苗、分蘖数和分蘖成穗率, 是获得高产的基础, 分蘖数与最终成穗数是小麦群体对外界环境适应的结果。穗粒数与穗数往往呈显著负相关关系, 即增加穗粒数会降低单位面积穗数。千粒重则相对独立, 多数研究表明其对产量的直接贡献最大, 在群体条件下增加单穗粒重是增加群体产量潜力的关键[2]。千粒重主要受加性效应的控制, 在产量因素中遗传力最高, 可达59%~80% [3]。小麦产量及构成要素为数量性状, 呈现连续变异, 受微效多基因调控[4]。
分子生物学的发展为复杂性状分子定位和标记辅助育种提供了有力工具, 小麦产量及构成要素性状的遗传定位研究已有诸多报道, Mason等[5]对不同播期下的小麦产量性状进行调查, 在3B、5B、7D染色体上检测到3个穗数QTL, 在2D和5A染色体上检测到2个千粒重QTL。Hu等[6]以包含4个RIL群体的巢式作图群体为材料, 在8个环境中进行了产量性状调查, 鉴定到8个多环境稳定的位点, 并利用自然群体进行了验证。Tura等[7]在32个环境中对一个DH群体开展了产量性状评价, 分别鉴定到24个和27个产量、千粒重QTL, 并对其中2个主效位点QYld.aww-1B.2与QTgw.aww-1B进行了精细定位。由于产量及其构成因素的复杂性, 产量性状的定位结果存在较大差异。
随着基因组学的发展, 分子标记从RFLP、RAPD和SSR等发展到近年来的SNP标记。SNP具有遗传稳定、数量多、分布广等特点, 并且适于高通量检测, 基于其开发的9k、90k、660k等基因型芯片集合了成千上万个SNP标记, 大大提高了遗传图谱的质量和密度[8,9,10]。基于SNP位点差异, 还可开发特异匹配引物末端碱基的KASP (Kompetitive Allele Specific PCR)标记, 可对SNP位点进行精准的双等位基因判断, 并具有高通量的特点, 适于大量样本的分子标记检测, 与育种选择相契合, 在育种中具有广泛应用前景[11]。
宁麦9号与扬麦158分别是江苏省农业科学院与江苏省里下河地区农业科学研究所育成的高产优质抗病小麦品种, 不仅曾在生产上具有较大的推广面积, 而且是重要的育种亲本, 近10年长江中下游冬麦区国家区试与江苏省淮南区试各有36个和34个品种通过审定, 含有宁麦9号或扬麦158的遗传背景的材料均为27个, 占比近80%。在产量性状方面, 宁麦9号多穗、多粒, 但千粒重偏低, 扬麦158穗粒数不及宁麦9号, 但千粒重较高, 二者在产量构成因素方面存在差异[12,13]。本研究对来源于宁麦9号与扬麦158的重组自交系群体进行多环境的产量性状评价, 并结合高密度遗传图谱, 试图解析其产量性状的遗传的分子机制, 为长江中下游麦区产量性状的分子标记辅助选择育种提供理论依据和技术支撑。
1 材料与方法
1.1 试验材料与数据采集
以宁麦9号×扬麦158构建重组自交系(RIL)群体(F2:8), 包括282个家系。2015—2016、2016—2017和2017—2018连续3个生长季将RIL群体及其亲本种植于江苏省农业科学院六合基地, 为方便描述, 以收获年份2016、2017与2018分别表示3个环境。采用增广设计[14], 2个亲本及随机选取的30个材料种植2次重复, 其余材料1次重复, 3行小区种植, 每行60粒, 行长1.6 m, 行距0.25 m, 常规栽培管理。灌浆后期调查每个小区中间行穗数, 并换算为公顷穗数; 收获时先取10个长势均匀的麦穗进行混合脱粒, 并统计总粒数, 进一步换算为单穗粒数; 将整小区进行收获测产, 并换算为公顷产量; 最后利用万深籽粒考种机(SC-A1, 浙江杭州)进行千粒重测定。另同时测量139份小麦高代品系材料的千粒重, 用于后续验证。
1.2 数据处理与QTL分析
表型初步统计采用Microsoft Excel 2016, 相关分析、方差分析及t检测采用SPSS 19.0。遗传力计算依据He等[15]的方法, 计算公式为h2 = δg2/(δg2 + δg*y2/y +δe2/ry)。其中δg2表示基因型方差, δg*y2表示基因型与环境互作方差, δe2为误差, y代表环境数目, r代表重复数。采用Illumina 90K芯片获取基因型。遗传图谱覆盖21条染色体, 包含41个连锁群, 2285个bin标记, 总长为3022 cM [16]。采用IciMapping 4.1软件的完备区间作图法(inclusive composite interval mapping, ICIM)进行QTL定位[17], walking step设为0.1 cM, LOD阈值设为2.5。
1.3 KASP标记的开发
根据SNP位点和侧翼序列设计PCR扩增引物, 开发KASP分子标记。每个标记设计2条SNP特异性引物(F1/F2)和一条通用引物(R), F1尾部添加能够与FAM荧光结合的特异性序列5′-GAAGGTGACCA AGTTCATGCT-3′, F2尾部添加能够与HEX荧光结合的特异性序列5′-GAAGGTCGGAGTCAACGGAT T-3′。利用Polymarker (从RILs群体中随机挑选46份材料进行幼嫩叶片取样, 采用CTAB法[18]提取基因组DNA。KASP反应总体系为5 μL, 包含2×KASP Master Mix 2.5 μL、KASP Assay Mix (引物混合工作液) 0.07 μL、浓度为20 ng μL?1的模板DNA 2.43 μL。KASP反应程序为, 第一步: 94℃, 15 min; 第二步: 94℃, 20 s, 61~55℃, 1 min, 每个循环降低0.6℃, 共进行10个循环; 第三步: 94℃, 20 s, 55℃, 1 min, 共进行26个循环, PCR在购买自LGC公司型号为Hydrocycler 16的水浴PCR仪中进行。通过KASP荧光分析仪(LGC公司型号为PHERAstar plus)扫描分析PCR结果。
2 结果与分析
2.1 产量及其三要素的表型分析
在连续3年的试验中(表1), 宁麦9号穗粒数均高于扬麦158, 而千粒重低于扬麦158, 而穗数与产量在不同环境中表现不一致, 同时穗数与产量的变异系数明显高于穗粒数与千粒重。从遗传力来看, 产量仅为0.18, 穗数为0.13, 说明此两性状受环境影响较大, 方差分析结果同样证实了这一点(表2), 穗数与产量在不同环境中均呈极显著正相关(表3)。穗粒数与千粒重呈负相关, 基因型与环境对穗粒数和千粒重有极显著影响, 穗粒数的遗传力为0.37, 不同环境间相关性不稳定, 与产量的相关性也不稳定; 千粒重的遗传力为0.60, 不同环境间均表现一致, 与产量呈极显著正相关。此外, 方差分析表明, 基因型与年份的互作对这4个性状均无显著影响。Table 1
表1
表1产量及其三要素的表型统计
Table 1
| 性状 Trait | 年份 Year | 宁麦9号 Ningmai 9 | 扬麦158 Yangmai 158 | 重组自交系群体 RIL population | 遗传力 Heritability | ||||
|---|---|---|---|---|---|---|---|---|---|
| 最大值 Max. | 最小值 Min. | 平均值 Mean | 标准差 SD | 变异系数 CV (%) | |||||
| 穗数 Spike number (×104 hm-2) | 2016 | 197.30 | 292.21 | 729.27 | 137.36 | 343.77 | 84.10 | 24.46 | 0.13 |
| 2017 | 549.45 | 412.09 | 779.22 | 252.25 | 497.33 | 96.53 | 19.41 | ||
| 2018 | 479.52 | 534.47 | 816.68 | 249.75 | 495.31 | 94.39 | 19.06 | ||
| 穗粒数 Kernel number per spike | 2016 | 69.00 | 53.33 | 75.14 | 32.00 | 52.53 | 7.18 | 13.68 | 0.37 |
| 2017 | 65.00 | 58.40 | 82.80 | 37.60 | 63.31 | 7.98 | 12.60 | ||
| 2018 | 52.20 | 46.60 | 74.00 | 31.00 | 44.17 | 6.36 | 14.40 | ||
| 千粒重 1000-kernel weight (g) | 2016 | 32.55 | 35.48 | 47.63 | 21.75 | 34.98 | 3.90 | 11.16 | 0.60 |
| 2017 | 30.98 | 32.94 | 51.96 | 25.37 | 35.55 | 4.50 | 12.67 | ||
| 2018 | 40.40 | 47.01 | 50.62 | 31.51 | 41.79 | 3.27 | 7.82 | ||
| 产量 Yield (kg hm-2) | 2016 | 3238.43 | 3887.78 | 7251.08 | 1506.83 | 3903.92 | 940.63 | 24.09 | 0.18 |
| 2017 | 6018.98 | 4920.08 | 11,163.83 | 2006.33 | 6492.86 | 1752.69 | 26.99 | ||
| 2018 | 6993.00 | 8658.00 | 12,321.00 | 3330.00 | 7128.55 | 1420.14 | 19.92 | ||
新窗口打开|下载CSV
Table 2
表2
表2产量及其三要素的方差分析(F值)
Table 2
| 项目 Item | 穗数 Spike number (×104 hm-2) | 穗粒数 Kernel number per spike | 千粒重 1000-kernel weight (g) | 产量 Yield (kg hm-2) |
|---|---|---|---|---|
| 基因型 Genotype | 0.78 | 1.92** | 2.64** | 0.86 |
| 年份 Year | 214.54** | 754.62** | 415.05** | 320.18** |
| 基因型×年份 Genotype×Year | 0.64 | 1.21 | 1.06 | 0.64 |
新窗口打开|下载CSV
Table 3
表3
表3产量及其三要素的相关分析
Table 3
| 性状 Trait | 环境 Environment | 穗数 Spike number (×104 hm-2) | 穗粒数 Kernel number per spike | 千粒重 1000-kernel weight (g) | 产量 Yield (kg hm-2) |
|---|---|---|---|---|---|
| 穗数 Spike number (×104 hm-2) | 2016 | (0.136*) | |||
| 2017 | (0.128*) | ||||
| 2018 | (-0.042) | ||||
| 穗粒数 Kernel number per spike | 2016 | -0.154** | (0.081) | ||
| 2017 | -0.157** | (0.344**) | |||
| 2018 | -0.054 | (-0.014) | |||
| 千粒重 1000-kernel weight (g) | 2016 | -0.004 | -0.131* | (0.316**) | |
| 2017 | -0.025 | -0.284** | (0.364**) | ||
| 2018 | -0.171** | -0.146** | (0.288**) | ||
| 产量 Yield (kg hm-2) | 2016 | 0.169** | -0.077 | 0.289** | (0.242**) |
| 2017 | 0.418** | -0.201** | 0.439** | (0.052) | |
| 2018 | 0.312** | 0.306** | 0.085 | (0.120*) |
新窗口打开|下载CSV
2.2 产量及其三要素的QTL分析
综合3年表型数据, 检测到9个控制穗数的QTL, 分布在1A、1B、2B、4B、5A、7A共6条染色体上, 表型变异解释率为3.45%~10.39% (表4和图1); 检测到8个控制穗粒数的QTL, 分布在2B、3B、4A、5B、6B、7A共6条染色体上, 表型变异解释率为3.76%~8.66%; 检测到10个控制千粒重的QTL, 分布在1A、1B、2B、3A、4A、4B、5A、5B、7A、7B共10条染色体上, 表型变异解释率为3.10%~9.43%; 检测到12个控制产量的QTL, 分布在1B、2A、4B、5A、5B、6A、6B、7A、7B共10条染色体上, 表型变异解释率为3.23%~11.79%。Table 4
表4
表4产量及其三要素的QTL定位
Table 4
| 性状 Traits | 序号No. | QTL | 环境 Environment | 遗传位置 Genetic position (cM) | 物理区间 Physical interval (Mb) | 区间 Interval | LOD | 表型贡献率 PVEa (%) | 加性效应 AEb | 前人研究中的产量相关性状 Yield-related traits in previous studies |
|---|---|---|---|---|---|---|---|---|---|---|
| 穗数 Spike number | 1 | Qsn-1A | 2018 | 101.45-101.95 | 543-574 | Tdurum_contig96086_74-RFL_Contig3683_1784 | 4.85 | 7.54 | -27.00 | 产量、穗粒数、单穗产量、收获指数[19] Yield, kernel number per spike, kernel weight per spike, harvest index[19] |
| 2 | Qsn-1B.1 | 2016 | 6.35-11.15 | 614-616 | wsnp_BE590634B_Ta_2_5-Tdurum_contig10362_555 | 4.03 | 5.11 | 19.22 | 产量[20] Yield[20] | |
| 3 | Qsn-1B.2 | 2016 | 15.95-20.55 | 630-641 | BS00069044_51-wsnp_Ex_rep_c78209_74462664 | 7.40 | 10.39 | -29.21 | 穗长、千粒重、穗粒数[21,22] Spike length, 1000-kernel weight, kernel number per spike[21,22] | |
| 4 | Qsn-1B.3 | 2017 | 97.75-98.25 | 678-680 | BobWhite_c4482_73-Tdurum_contig52086_342 | 2.97 | 3.45 | -19.25 | 千粒重[23] 1000-kernel weight[23] | |
| 5 | Qsn-2B | 2017 | 78.05-79.35 | 781-783 | Ku_c2936_1987-RAC875_c18928_455 | 3.21 | 3.68 | 19.78 | — | |
| 6 | Qsn-4B | 2016 | 14.95-15.95 | 603-620 | BS00039402_51-Excalibur_rep_c109675_738 | 3.44 | 3.62 | 14.41 | — | |
| 7 | Qsn-5A | 2017 | 49.45-50.35 | 505-520 | tplb0056b09_1000-BS00085826_51 | 6.27 | 7.56 | 28.32 | 千粒重、穗粒数[23] 1000-kernel weight, kernel number per spike[23] | |
| 8 | Qsn-7A.1 | 2017 | 38.25-39.45 | 688-697 | Tdurum_contig31699_276-wsnp_Ku_c4035_7363089 | 3.18 | 4.48 | 24.15 | 千粒重[23] 1000-kernel weight[23] | |
| 9 | Qsn-7A.2 | 2017 | 108.35-111.1 | 730-735 | BS00069019_51-BS00022284_51 | 3.74 | 4.66 | -22.28 | — | |
| 穗粒数 Kernel number per spike | 10 | Qkn-2B | 2016 | 68.25-69.35 | 768-776 | Kukri_c24939_378-RAC875_rep_c83950_222 | 3.69 | 5.84 | -1.75 | — |
| 12 | Qkn-3B.1 | 2018 | 105.45-105.85 | 146-151 | RAC875_c45525_131-IAAV1079 | 6.10 | 8.66 | -3.41 | — | |
| 11 | Qkn-3B.2 | 2017 | 62.95-65.35 | 724-742 | BobWhite_c2937_1426-BS00040739_51 | 3.08 | 4.42 | -1.88 | 穗数、产量[5] Spike number, yield[5] | |
| 13 | Qkn-4A | 2017 | 4.05-14.95 | 664-683 | BS00066066_51-IAAV5722 | 2.76 | 5.03 | 1.76 | 穗粒数、穗数、千粒重[23,24] Kernel number per spike, spike number, 1000-kernel weight[23,24] | |
| 14 | Qkn-5B.1 | 2018 | 91.25-93.35 | 615-618 | Tdurum_contig85060_121-Kukri_rep_c101622_604 | 3.78 | 5.30 | 1.60 | 产量[25] Yield[25] | |
| 15 | Qkn-5B.2 | 2017 | 129.35-130.05 | 682-698 | Excalibur_c6346_266-IAAV8023 | 3.06 | 4.40 | -1.77 | 千粒重、穗粒数[23] 1000-kernel weight, kernel number per spike[23] | |
| 16 | Qkn-6B | 2016 | 49.35-50.95 | 96-141 | RAC875_c23812_187-wsnp_Ku_c12559_20252463 | 3.02 | 5.18 | 1.64 | — | |
| 17 | Qkn-7A | 2018 | 0-7.05 | 670-680 | Ku_c62742_888-wsnp_Ku_c42539_50247333 | 2.68 | 3.76 | -1.40 | 千粒重、穗粒数、穗数[23] 1000-kernel weight, kernel number per spike, spike number[23] | |
| 千粒重 1000-kernel weight | 18 | Qtkw-1A | 2016 | 47.05-48.95 | 31-33 | BS00084022_51-wsnp_Ex_c31983_40709866 | 3.08 | 3.60 | 0.76 | 千粒重、单穗产量、穗粒数[23,26] 1000-kernel weight, kernel weight per spike, kernel number per spike[23,26] |
| 19 | Qtkw-1B | 2017/2018 | 35.25-39.55 | 640-653 | IAAV1683-IAAV565 | 3.68 | 5.80 | -1.15 | 穗长、千粒重、穗粒数[21] Spike length, 1000-kernel weight, kernel number per spike[21] | |
| 20 | Qtkw-2B | 2018 | 51.45-52.05 | 748-750 | CAP8_c8516_542-BS00094578_51 | 8.09 | 9.43 | 1.06 | 千粒重[24] 1000-kernel weight[24] | |
| 21 | Qtkw-3A | 2016 | 0-6.15 | 9-10 | Excalibur_c10014_307-RAC875_c36922_829 | 3.29 | 3.71 | 0.88 | — | |
| 22 | Qtkw-4A | 2016/2017 | 3.75-19.75 | 661-683 | BS00066066_51-BS00066891_51 | 5.55 | 6.42 | -1.02 | 穗粒数、穗数、千粒重[23,24] Kernel number per spike, spike number, 1000-kernel weight[23,24] | |
| 23 | Qtkw-4B | 2018 | 4.85-5.05 | 427-428 | GENE-2847_1060-wsnp_Ex_rep_c70265_69211592 | 7.90 | 8.82 | -1.02 | 千粒重[27] 1000-kernel weight[27] | |
| 24 | Qtkw-5A | 2016 | 55.85-56.55 | 537-539 | BS00096758_51-RAC875_c103967_76 | 7.51 | 8.85 | 1.20 | 千粒重、穗粒数[23] 1000-kernel weight, kernel number per spike[23] | |
| 25 | Qtkw-5B | 2018 | 67.15-70.95 | 586-587 | Kukri_c45964_273-RAC875_c43383_483 | 2.88 | 3.10 | -0.65 | 穗粒数[28] Kernel number per spike[28] | |
| 26 | Qtkw-7A | 2017/2018 | 48.55-49.55 | 261-285 | RFL_Contig428_290-RAC875_c16624_970 | 4.21 | 4.57 | -0.79 | — | |
| 27 | Qtkw-7B | 2016 | 0-4.95 | 15-25 | wsnp_CAP7_c44_26549-Ex_c101666_634 | 4.74 | 5.99 | -0.99 | 产量、小穗数[20,28] Yield, the spikelet number per spike[20,28] | |
| 产量 Yield | 28 | Qyd-1B.1 | 2017 | 37.55-38.65 | 236-433 | BobWhite_c4147_1429-BS00033681_51 | 5.40 | 5.33 | 441.75 | 千粒重、单穗产量、产量[29,30] 1000-kernel weight, kernel weight per spike, yield[29,30] |
| 29 | Qyd-1B.2 | 2017 | 65.55-73.25 | 480-498 | Ra_c68984_1882-Tdurum_contig81102_102 | 4.27 | 4.17 | -611.83 | — | |
| 30 | Qyd-2A | 2018 | 47.85-55.05 | 653-706 | BS00090569_51-BS00029332_51 | 3.87 | 6.19 | 332.86 | 单株产量、小穗数、千粒重、穗粒数[31] Kernel weight per plant, the spikelet number per spike, 1000-kernel weight, kernel number per spike[31] | |
| 31 | Qyd-4B | 2017 | 68.85-70.45 | 101-168 | BS00071792_51-Tdurum_contig28490_463 | 3.26 | 3.23 | 337.82 | 穗粒数、单穗产量、千粒重[5,23,32] Kernel number per spike, kernel weight per spike, 1000-kernel weight[5,23,32] | |
| 32 | Qyd-5A.1 | 2016 | 13.35-14.55 | 389-420 | BS00086963_51-BS00001322_51 | 4.11 | 7.91 | 242.37 | 千粒重、穗粒数[23] 1000-kernel weight, kernel number per spike[23] | |
| 33 | Qyd-5A.2 | 2017 | 53.35-54.45 | 511-540 | wsnp_Ex_c49211_53875575-CAP7_c1404_72 | 10.88 | 11.79 | 642.73 | 千粒重、穗粒数[23] 1000-kernel weight, kernel number per spike[23] | |
| 34 | Qyd-5B.1 | 2018 | 45.15-46.15 | 545-547 | Tdurum_contig25513_195-wsnp_Ex_c8659_14515623 | 3.59 | 4.45 | -418.08 | 产量[27] Yield[27] | |
| 35 | Qyd-5B.2 | 2017 | 113.05-113.85 | 663-665 | BobWhite_c36154_81-Kukri_c41_858 | 4.87 | 4.80 | -285.41 | — | |
| 36 | Qyd-6A | 2017 | 35.35-38.2 | 609-617 | Excalibur_c96749_512-Excalibur_c98257_136 | 3.82 | 3.88 | -408.26 | 产量、千粒重[20,23] Yield, 1000-kernel weight[20,23] | |
| 37 | Qyd-6B | 2018 | 53.35-53.75 | 144-148 | BobWhite_c1905_98-wsnp_Ku_c5891_10414090 | 3.08 | 3.88 | -281.40 | — | |
| 38 | Qyd-7A | 2018 | 33.25-34.25 | 83-109 | Excalibur_rep_c101407_222-BS00011072_51 | 2.92 | 3.60 | -256.01 | 穗重、千粒重、穗粒数、穗数[23,33-34] Ear weight per tiller, 1000-kernel weight, kernel number per spike, spike number[23,33-34] | |
| 39 | Qyd-7B | 2018 | 133.15-133.95 | 743-745 | Tdurum_contig51105_1538-tplb0040b02_681 | 4.36 | 5.48 | 313.01 | 产量、千粒重[20,23] Yield, 1000-kernel weight[20,23] |
新窗口打开|下载CSV
图1
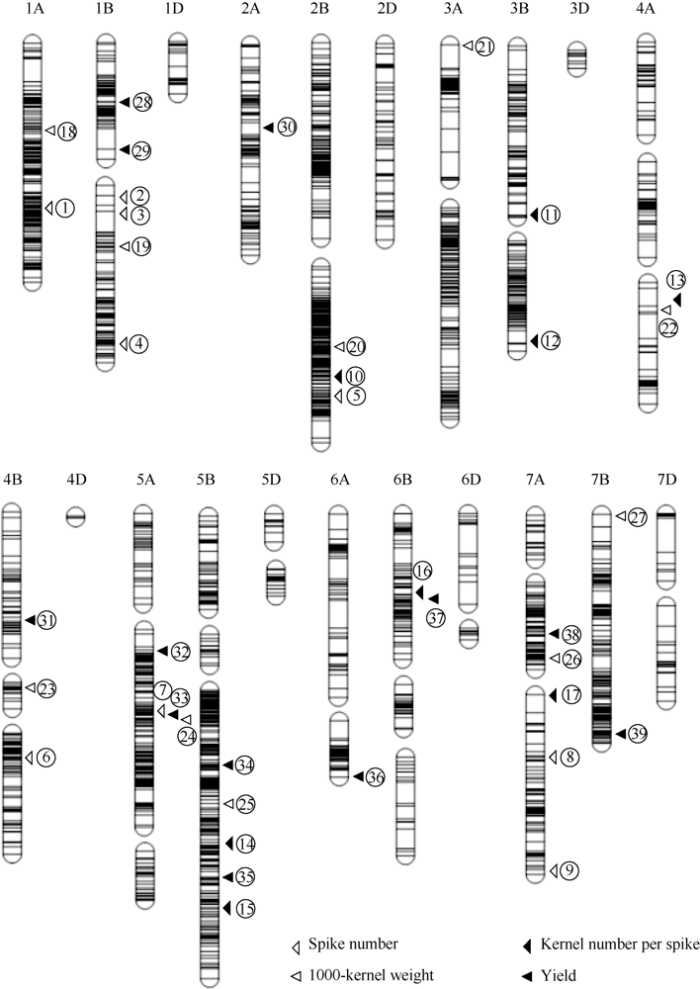 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1产量及其三要素的QTL定位
图中数字与
Fig. 1QTL mapping for yield and its components
The numbers in the figure are consistent with the sequence numbers of QTL in
在上述QTL定位结果中, 控制千粒重的QTL-Qtkw-1B、Qtkw-4A及Qtkw-7A等3个QTL能够被重复检测到。根据连锁图谱位置分析, 4A染色体上的穗粒数QTL Qkn-4A与千粒重QTL Qtkw-4A, 5A染色体上穗数QTL Qsn-5A、千粒重QTL Qtkw-5A与产量QTL Qyd-5A, 6B上穗粒数QTL Qkn-6B与产量QTL Qyd-6B定位区间重合或相近。进一步分析其加性效应, 4A染色体上的高穗粒数QTL和高千粒重QTL分别来自宁麦9号与扬麦158, 5A染色体上的高穗数、穗粒数和产量QTL均来源于宁麦9号, 6B染色体上的高穗粒数和产量QTL分别来自宁麦9号与扬麦158。
2.3 千粒重的QTL聚合效应分析
在产量及其构成要素中, 千粒重的遗传力最高。宁麦9号/扬麦158的282个重组自交系群体中在3年中被重复检测到的千粒重QTL有3个, 分别位于1B、4A和7A染色体上(表4), 从不同QTL对千粒重的影响看, 后代中存在Qtkw-1B、Qtkw-4A及Qtkw-7A单一位点时, Qtkw-4A对千粒重的影响最显著也最稳定(表5), 从增加千粒重的效应看, Qtkw-4A亦优于Qtkw-1B及Qtkw-7A, 不同等位变异最高可产生2.07 g的千粒重差异。当后代中聚合2个位点后, 2个位点的组合效应优于单个位点, Qtkw-1B+ Qtkw-4A的组合最稳定, 且高于其他两种组合, 在不同年份可使千粒重提高2.76~3.44 g, 而Qtkw-1B+ Qtkw-7A、Qtkw-4A+Qtkw-7A两种组合无显著差异。后代中聚合3个位点后, 对千粒重的效应显著提高, 2017年甚至产生超过5 g的千粒重差异。Table 5
表5
表5千粒重QTL不同等位变异的t检测
Table 5
| QTL | 环境 Environment | 优势等位变异 Favorable alleles (g) | 非优势等位变异 Unfavorable alleles (g) | 差值 Difference (g) | t值 t-value | P值 P-value |
|---|---|---|---|---|---|---|
| Qtkw-1B | 2016 | 35.21 (121) | 34.56 (133) | 0.65 | 1.36 | 0.18 |
| 2017 | 36.47 (121) | 34.62 (133) | 1.85 | 3.31 | <0.01 | |
| 2018 | 42.37 (121) | 40.95 (133) | 1.42 | 3.56 | <0.01 | |
| Qtkw-4A | 2016 | 35.68 (110) | 33.71 (98) | 1.97 | 3.86 | <0.01 |
| 2017 | 36.48 (110) | 34.41 (98) | 2.07 | 3.41 | <0.01 | |
| 2018 | 42.38 (110) | 40.68 (98) | 1.7 | 3.94 | <0.01 | |
| Qtkw-7A | 2016 | 34.78 (168) | 35.03 (78) | -0.25 | -0.47 | 0.64 |
| 2017 | 36.31 (168) | 34.46 (78) | 1.85 | 2.97 | <0.01 | |
| 2018 | 42.02 (168) | 41.30 (78) | 0.72 | 1.38 | 0.17 | |
| Qtkw-1B+Qtkw-4A | 2016 | 36.62 (46) | 33.48 (51) | 3.14 | 4.22 | <0.01 |
| 2017 | 37.03 (46) | 33.59 (51) | 3.44 | 4.11 | <0.01 | |
| 2018 | 43.32 (46) | 40.56 (51) | 2.76 | 4.24 | <0.01 | |
| Qtkw-1B+Qtkw-7A | 2016 | 35.42 (74) | 35.48 (34) | -0.06 | -0.09 | 0.93 |
| 2017 | 37.51 (74) | 33.58 (34) | 3.93 | 4.26 | <0.01 | |
| 2018 | 42.82 (74) | 40.79 (34) | 2.03 | 2.98 | <0.01 | |
| Qtkw-4A+Qtkw-7A | 2016 | 35.46 (68) | 33.73 (34) | 1.73 | 2.11 | 0.04* |
| 2017 | 37.42 (68) | 34.56 (34) | 2.86 | 3.04 | <0.01 | |
| 2018 | 42.27 (68) | 40.51 (34) | 1.76 | 2.85 | <0.01 | |
| Qtkw-1B+Qtkw-4A+Qtkw-7A | 2016 | 36.55 (26) | 33.73 (12) | 2.82 | 2.14 | 0.04* |
| 2017 | 38.42 (26) | 33.08 (12) | 5.34 | 5.22 | <0.01 | |
| 2018 | 43.10 (26) | 41.16 (12) | 1.94 | 1.55 | 0.14 |
新窗口打开|下载CSV
2.4 千粒重QTL的KASP标记开发与利用
为了更好地对3个千粒重QTL位点进行育种利用, 基于其区间标记序列开发了适于高通量分型的KASP标记。首先从3个标记区间各选择2~3个同源性较低的SNP标记进行引物设计(附表1), 经过扩增验证, Kukri_c42354_263、IAAV5722与IACX1477扩增效果良好(附图1), 可应用于Qtkw-1B、Qtkw-4A与Qtkw-7A等3个位点的育种选择。进一步利用开发成功的3个KASP标记对139份小麦高代品系材料进行基因分型, 并结合表型数据进行统计分析(表6), 发现3个标记在千粒重选择中均具有显著作用, 2个标记的选择效果优于单标记选择, 而聚合3个标记后, 产生了超过6 g的千粒重组间差异, 与遗传效应分析结果(2.3)基本一致。Qtkw-4A与Qtkw-7A两个位点多呈现来源于扬麦158的优势等位变异, 而Qtkw-1B未表现出明显的选择倾向。
Table S1
附表1
附表1千粒重的KASP标记序列
Table S1
| SNP | QTL | F1 (5°-3°) | F2 (5°-3°) | R (5°-3°) |
|---|---|---|---|---|
| Kukri_c18109_682 | Qtkw-1B | AACGACGCCTGGTTTTCCTCA | CGACGCCTGGTTTTCCTCG | TCCGAGATGGACGAGGAACTG |
| IAAV1683 | Qtkw-1B | TGTTCTTACCTGAACTTGTGTGACA | GTTCTTACCTGAACTTGTGTGACG | CTGAAGCTCTGATATCATCATCATCC |
| Kukri_c42354_263 | Qtkw-1B | TCGACGATACTGGCGGTGTA | TCGACGATACTGGCGGTGTG | TGATCGCAAAGTGCATTATGTT |
| BS00066891_51 | Qtkw-4A | TGGACGGATGGTTGATGGCTCG | ATGGACGGATGGTTGATGGCTCA | TATAAGCTACTACCGCTCCGGC |
| IAAV5722 | Qtkw-4A | AGCTGCAACCCATCCTCT | AGCTGCAACCCATCCTCC | TGTGACCTACTCGATGTTCATAAC |
| Excalibur_rep_c70004_275 | Qtkw-7A | GGTGTTCTCCCTGGTAGCTCA | GGTGTTCTCCCTGGTAGCTCG | CTTCTGGCACCTGCTTTCATCG |
| RFL_Contig428_290 | Qtkw-7A | GGATGTCCCGGTTGGAGAAGAC | GGATGTCCCGGTTGGAGAAGAT | GACAAGTACGGCCCCATCTTCT |
| IACX1477 | Qtkw-7A | CCAAAATGGCTTGTCAAATGAGATA | CCAAAATGGCTTGTCAAATGAGA TG | GCTTGACCCTTAGCTCATGA |
新窗口打开|下载CSV
附图1
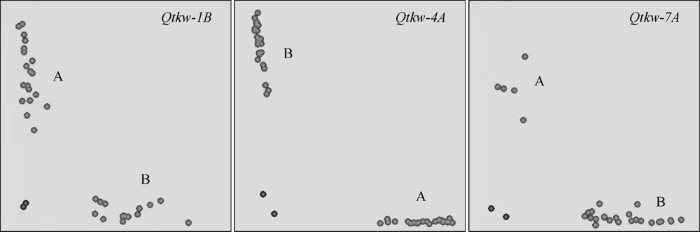 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图1千粒重QTL的KASP标记开发
A表示宁麦9号等位变异类型, B表示扬麦158等位变异类型。
Fig. S1Development of KASP markers for the QTLs for 1000-kernel weight
A indicates the allele of Ningmai 9 and B indicates the allele of Yangmai 158.
Table 6
表6
表6千粒重KASP标记的效用检测
Table 6
| QTL | 优势等位变异 Favorable alleles (g) | 非优势等位变异 Unfavorable alleles (g) | 差值 Difference (g) | t值 t-value | P值 P-value |
|---|---|---|---|---|---|
| Qtkw-1B | 48.32 (62) | 45.92 (77) | 2.40 | -3.79 | <0.01 |
| Qtkw-4A | 47.67 (100) | 45.26 (39) | 2.41 | 3.40 | <0.01 |
| Qtkw-7A | 47.47 (122) | 43.53 (17) | 3.95 | -4.15 | <0.01 |
| Qtkw-1B+Qtkw-4A | 48.73 (49) | 44.49 (26) | 4.23 | 4.61 | <0.01 |
| Qtkw-1B+Qtkw-7A | 48.43 (60) | 43.32 (15) | 5.11 | -4.73 | <0.01 |
| Qtkw-4A+Qtkw-7A | 47.67 (98) | 43.01 (15) | 4.66 | -4.59 | <0.01 |
| Qtkw-1B+Qtkw-4A+Qtkw-7A | 48.73 (49) | 42.69 (13) | 6.03 | -5.27 | <0.01 |
新窗口打开|下载CSV
3 讨论
未来全球对小麦的需求仍将呈增长趋势, 进一步提高单产潜力仍然是小麦育种最基本和最重要的目标[35]。分子标记辅助选择用于小麦育种的研究已有20多年的历史, 但在育种中应用较好的是抗病和品质性状相关标记, 产量相关性状的分子标记研究仍有待加强[36,37]。本研究以长江中下游品种的骨干亲本宁麦9号和扬麦158杂交构建的重组自交系群体为材料, 连续3年对单位面积产量、单位面积穗数、穗粒数和千粒重进行QTL定位分析, 共定位到39个QTL, 研究结果验证了产量及其构成因素遗传的复杂性, 性状受微效多位点控制。与前人研究比较(表4), 控制千粒重的Qtkw-2B与Qtkw-4B, 控制产量的Qyd-5B.1与前人定位的QTL一致[24,27]; Qsn-1A、Qsn-1B.1与Qsn-1B.2等此前报道与产量性状相关, 但并未明确与穗数有关[19,20,21,22]; 同样, 类似的QTL还有控制穗粒数的Qkn-3B.2与Qkn-5B.1[5,25], 控制千粒重的Qtkw-5B与Qtkw- 7B[20,28]和控制产量的Qyd-5A.1与Qyd-5A.2[23]。本研究检测到了一些的新的QTL, 分别是控制穗数的Qsn-2B、Qsn-4B与Qsn-7A.1, 控制穗粒数的Qkn-2B、Qkn-3B.1与Qkn-6B, 控制千粒重的Qtkw-3A与Qtkw-7A, 控制产量的Qyd-1B.2、Qyd-5B.2与Qyd-6B, 这些位点此前均未曾报道与产量性状有关。通过QTL在遗传图谱中的定位分析与比对, 发现3个QTL簇, 分别位于4A、5A及6B染色体上, 4A和5A染色体区段与产量及其构成要素均相关, 6B染色体区段与穗粒数、产量相关, 且此前未被报道。4A区段调控穗粒数与千粒重的加性效应分别来源于宁麦9号与扬麦158, 与两个亲本对应的多粒、大粒特征一致, 在选择时可根据育种目标合理选择亲本; 5A区段的加性效应均来源于宁麦9号, 在产量育种中可选择宁麦9号的优良等位变异; 6B区段控制高产的加性效应来源于扬麦158, 可选择扬麦158的优良等位变异在高产育种中应用。
在产量构成因素中, 不同地区、不同时期及不同高产品种的产量结构类型差异很大, 程顺和等[38]提出应将穗数维持在一定水平上, 主攻粒重。宋健民等[39]分析了近年来山东省通过审定的55个小麦品种产量性状的变化趋势, 结果显示产量呈显著上升趋势, 穗数与穗粒数变化不显著, 千粒重显著增加; 沈磊和赵凯铭[40]根据河南省2001—2014年间审定的202个品种的统计数据, 发现穗数与穗粒数较为稳定, 千粒重呈上升趋势; 蒋进等[41]对四川省2008—2018年育成的100个品种进行分析, 发现产量水平逐年升高, 穗数、千粒重呈下降趋势, 穗粒数呈上升趋势, 而其中高产品种穗数与千粒重均处于较高的水平; 姚国才等[42]以长江中下游麦区育成的和大面积应用的小麦品种为材料进行分析, 在产量水平由低中产到较高产时, 穗数增加较多, 千粒重次之, 穗粒数变化较小, 而在高产组次中, 穗数基本稳定, 每穗粒数增幅较大, 千粒重明显提高。上述研究表明, 千粒重是当前高产育种的重点改良因素。在产量三要素的遗传中, 穗数的遗传力最低, 国外研究报道中有的低至0.070和0.046[38], 多项研究结果表明穗数的遗传力在0.2以下[43,44], 本研究结果与其相近; 前人研究还表明, 穗数与产量的相关性较高[45,46], 但易受环境条件影响, 因而产量的遗传力也不高, 本研究检测到的穗数和产量QTL尽管在同一环境中可同时定位到, 但受环境影响不稳定, 因此在不同环境中未能重复检测到。与前人研究类似, 本研究表明基因型与年份分别影响着产量及其三要素性状, 基因型与年份(环境)的互作对产量性状的影响在不同研究中有不同结果[49,50,51], 本研究中基因型与年份互作对产量及其三要素的影响不显著, 说明产量及其三要素不是随着年份的变化而呈线性变化, 反映了产量及其三要素受基因型及环境影响的复杂性。
在产量构成因素中, 千粒重具有较高的遗传力, 在本研究中为0.60, 与前人报道相近[47,48], 因此, 可通过分子标记辅助选择对千粒重性状进行改良。研究检测到3个稳定的千粒重QTL, 遗传效应分析显示其对千粒重均有显著影响。与前人研究比较发现, Qtkw-1B附近包含穗长、穗粒数的控制位点[21], Qtkw-4A附近区域存在着穗数和穗粒数的控制位点[23,24], Qtkw-7A是一个新鉴定到的千粒重QTL。
近些年来, 分子标记辅助选择在小麦育种中的应用越来越广泛, 特别是适于高通量分型的KASP标记, 其与大规模育种筛选策略完美契合, 在育种中具有良好的应用前景。本研究成功开发了3个千粒重QTL相连锁的KASP标记, 并在育种材料中进行了初步应用, 选择效果较好, 3个QTL用于标记辅助选择对提高千粒重有明显效果, 且多位点聚合的选择效果优于单一QTL的选择; 但多位点选择在提高选择效果的同时, 增加了工作量, 减少了中选材料, 需结合实际育种规模选择相应的育种策略。
4 结论
结合3个环境的产量数据, 鉴定到控制穗数、穗粒数、千粒重及产量的QTL分别为9、8、10与12个, 其中穗数、穗粒数和产量的遗传力较低, 千粒重的遗传力较高, 可进行遗传选择。针对3个稳定的千粒重QTL, 进一步成功开发出相连锁的KASP标记, 并在育种材料中进行了初步应用, 3个QTL对提高千粒重有明显效果, 且多位点聚合的选择效果优于单一QTL的选择。本研究结果可应用于小麦千粒重的遗传改良。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 6]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
[本文引用: 10]
[本文引用: 6]
[本文引用: 3]
[本文引用: 32]
[本文引用: 8]
[本文引用: 3]
[本文引用: 2]
[本文引用: 5]
[本文引用: 5]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
