 ,1, Nadil Shah1, 周元委2, 侯照科1, 龚建芳1, 刘珏3, 尚政伟1, 张磊1, 战宗祥1, 常海滨4, 傅廷栋1, 朴钟云
,1, Nadil Shah1, 周元委2, 侯照科1, 龚建芳1, 刘珏3, 尚政伟1, 张磊1, 战宗祥1, 常海滨4, 傅廷栋1, 朴钟云 ,5,*, 张椿雨
,5,*, 张椿雨 ,1,*
,1,*Breeding of a novel clubroot disease-resistant Brassica napus variety Huayouza 62R
LI Qian ,1, Nadil Shah1, ZHOU Yuan-Wei2, HOU Zhao-Ke1, GONG Jian-Fang1, LIU Jue3, SHANG Zheng-Wei1, ZHANG Lei1, ZHAN Zong-Xiang1, CHANG Hai-Bin4, FU Ting-Dong1, PIAO Zhong-Yun
,1, Nadil Shah1, ZHOU Yuan-Wei2, HOU Zhao-Ke1, GONG Jian-Fang1, LIU Jue3, SHANG Zheng-Wei1, ZHANG Lei1, ZHAN Zong-Xiang1, CHANG Hai-Bin4, FU Ting-Dong1, PIAO Zhong-Yun ,5,*, ZHANG Chun-Yu
,5,*, ZHANG Chun-Yu ,1,*
,1,*通讯作者:
收稿日期:2020-04-2接受日期:2020-07-2网络出版日期:2020-07-11
| 基金资助: |
Received:2020-04-2Accepted:2020-07-2Online:2020-07-11
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (6984KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李倩, Nadil Shah, 周元委, 侯照科, 龚建芳, 刘珏, 尚政伟, 张磊, 战宗祥, 常海滨, 傅廷栋, 朴钟云, 张椿雨. 抗根肿病甘蓝型油菜新品种华油杂62R的选育[J]. 作物学报, 2021, 47(2): 210-223. doi:10.3724/SP.J.1006.2021.04086
LI Qian, Nadil Shah, ZHOU Yuan-Wei, HOU Zhao-Ke, GONG Jian-Fang, LIU Jue, SHANG Zheng-Wei, ZHANG Lei, ZHAN Zong-Xiang, CHANG Hai-Bin, FU Ting-Dong, PIAO Zhong-Yun, ZHANG Chun-Yu.
根肿病是由芸薹根肿菌(Plamodiophora brassicae Woron)侵染引起的一种世界性土传病害。该病原菌专性侵染十字花科植物, 包括油菜、大白菜、小白菜、甘蓝、萝卜、花椰菜、芥菜等在内的多种栽培和野生种[1]。被该病原菌侵染后, 植物根系细胞经过非正常扩增和分裂, 从而产生肿瘤, 导致营养与水分的吸收被限制, 地上部分植株发黄并枯萎, 生长发育不良, 作物产量和品质均受到严重影响。根肿菌休眠孢子在土壤中存活时间长(可长达15年以上), 其传染性强、传播途径广、传播速度快、防治难度大[2,3,4]。十字花科油菜是我国最重要的油料作物, 长江流域常年种植面积约667万公顷, 所产优质菜籽油占国产植物油的50%以上。近年, 我国油菜生产受根肿病影响较大, 据不完全统计发病面积约在66.7万公顷(约占总种植面积的10%), 并且随着机械化程度的不断提高, 油菜根肿病在我国有大面积爆发的趋势, 特别是我国所有甘蓝型油菜品种均不抗根肿病, 因此油菜产业面临严重威胁。
已有研究表明, 选育并种植抗病品种是防治根肿病最经济、最有效的途径。目前应用最广泛的根肿病抗源材料主要为欧洲饲用芜菁(Brassica rapa ssp. rapifera, AA, 2n=20), 包括‘ECD01-04’、‘Gelria R’、‘Siloga’、‘Debra’以及‘Milan White’等[5,6]。日韩等国研究人员以芜菁为抗源材料, 培育出一些抗根肿病的大白菜品种。进一步在白菜中QTL定位了多个抗病位点, 主要包括Crr1、Crr2、Crr3、Crr4、CRa、CRb、CRc、CRk等[7,8,9], 这些位点分布在不同的染色体上。目前已成功分离克隆的位点有CRa与Crr1a, 这2个基因均编码TIR-NB-LRR结构蛋白, 能够识别病原菌并引起植物的免疫反应[10,11]。CRa与CRb均定位于A03染色体, 物理位置紧密连锁, 最新研究认为CRa和CRb是同一个抗病位点[12]。
由于国外油菜根肿病发生较早, 在抗病育种方面也率先取得了一些进展。Diederichsen等[13]利用抗病芜菁和抗病甘蓝材料进行种间杂交, 人工合成了抗病甘蓝型油菜品种Mendel, 经室内和田间抗病性鉴定均表现抗病, 并发现该抗性至少由2个不连锁的显性基因控制。加拿大在油菜抗根肿病育种方面也开展了一些工作, 培育出了油菜抗病新品种[14]。我国油菜抗根肿病的育种工作起步较晚, 主要集中在对现有主栽品种或资源材料的抗病性筛选方面[15,16,17,18], 而利用芜菁等高抗资源材料改良我国油菜品种的工作还鲜有报道。本实验室战宗祥博士等人以含多个抗病位点的芜菁ECD04为父本与优良甘蓝型油菜常规品种华双5号杂交, 结合回交育种策略及分子标记辅助选择手段, 成功将ECD04中的抗病位点PbBa8.1转育到优良油菜常规品种华双5号中, 育成了我国首个抗根肿病甘蓝型油菜常规新品系[19]。
本研究以含有CRb抗病位点, 对多种生理小种(2、4、7、10号)均具较好抗性的大白菜CR Shinki为供体父本, 与国审甘蓝型油菜优良杂交种华油杂62的波里马恢复系Bing409为母本, 通过杂交、回交以及分子标记辅助选择和抗病性鉴定等策略, 将CRb抗病位点精准导入到Bing409中, 创建了抗根肿病的新恢复系Bing409R, 并在此基础上配制了抗根肿病杂交油菜新品种华油杂62R, 为抵抗油菜根肿病, 稳定和保障我国油菜产业持续健康发展提供有力保障。
1 材料与方法
1.1 材料
供体亲本(父本)为含有CRb位点(位于A03染色体)的抗根肿病大白菜材料CR Shinki (AA, 2n=20), 根据威廉姆斯寄主鉴别系统鉴定结果, 该位点对根肿菌2号、4号、7号和10号生理小种具有优异抗性; 感病轮回亲本(母本)为波里马细胞质雄性不育(Pol.CMS)三系杂交种华油杂62 (国审)的恢复系Bing409, 同时在本研究作为感病对照材料。1.2 方法
1.2.1 技术路线 亲本材料于2013年10月种植于沈阳农业大学日光温室, 2014年2月完成授粉获得F1, 并利用4号生理小种进行室内接菌鉴定。2014年5月于沈阳农业大学播种, 8月完成F1代植物的回交, 10月获得含有492株单株的BC1群体。为加快育种进程, 从BC1代开始, 利用与抗病位点CRb紧密连锁的分子标记进行前景选择筛选含有CRb位点的单株, 淘汰无CRb的植株; 再利用均匀覆盖甘蓝型油菜A基因组的123个多态性标记(附表1), 对含CRb抗病前景位点的植株进行遗传背景筛选, 保留背景回复率高的植株。经过3次回交1次自交后, 于2016年春获得CRb位点纯合、背景恢复率在95%以上的抗病植株, 命名为Bing409R。2016年夏繁, 利用华油杂62的不育系与Bing409R配制了抗病新组合(命名为华油杂62R), 于2016年秋将Bing409R和华油杂62R种植在不同病区(枝江、黄山等)进行田间抗病性鉴定。技术路线如图1所示。Table S1
附表1
附表1用于遗传背景筛选的多态性分子标记
Table S1
| 染色体 Chr. | 标记名称 Marker name | 物理位置 Physical location (Mb) | 正向引物 Forward primer (5°-3°) | 反向引物 Reverse primer (5°-3°) | |
|---|---|---|---|---|---|
| A01 | niab_ssr113 | 366,648 | CAAAAAGTTGCGGTCAATCT | CCTCCAAAGCTCAATCACTG | |
| sau_um356 | 2,137,110 | CATCTTCGTCTCTCCATCACCT | GTATGGTAGGAGGAGAGTTCGCT | ||
| cnu_ssr134 | 4,481,886 | TCTCTTTGCCATCGTCGTTTC | CCCCTCAAACTGAGCAGTCAA | ||
| niab_ssr032 | 7,684,077 | TTCTCCCCATCCTCTCATCTTA | ACCCACAACCAACAAAATCTTC | ||
| cnu_mBRPGM0190 | 9,861,961 | GAGATCCAATAGCGAGCACA | TGTGTTATCGGGTGAAGTGG | ||
| 染色体 Chr. | 标记名称 Marker name | 物理位置 Physical location (Mb) | 正向引物 Forward primer (5°-3°) | 反向引物 Reverse primer (5°-3°) | |
| A01 | ACMP00456 | 13,955,674 | CTCTACAAGCCGCAGAGAGA | CATCCACCACAGAGATTGCT | |
| ACMP00617 | 15,044,996 | AACTCAATGCTCTTCGCTCA | CCTTCTGCTGCTTCCAAGAT | ||
| ACMP00297 | 16,999,515 | CAGATATTGCCCAAACACCA | CTTCGCTTCTGAGCTTTCCT | ||
| sau_um586 | 20,719,752 | AGGGTGATGATGATGTACAGGAG | CTTCGTTAATTCTTCCACCCAG | ||
| cnu_ssr125 | 26,899,174 | TCCAAGTGAAGGATAATGCTCGT | TGTACAATGGGGATGTTGTGG | ||
| A02 | sau-um619 | 1,404,160 | GAGAGCTCACTTGTGCTTCTTGT | CCGATCAAGTTATTCGCTCTCT | |
| cnu_ssr116 | 3,025,687 | GCCATGGTGGTGGGATTAGA | TGGTCGAACGTGTGTCGATAAT | ||
| niab_ssr143 | 5,142,784 | GATATGTTTCCAAACAAGTCAA | AAATTCGACCCCTTTTCG | ||
| sau_um434 | 7,390,390 | AGCTAGGGATGAGAAGAAGACCG | TCTTTGTGTCCGTCAAGAGTCC | ||
| cnu_ssr036 | 10,214,155 | AGCACAGTCAAAACTCAGAATGA | AGGCATGCGCTCTCTATCTC | ||
| cnu_mBRPGM1391 | 11,315,073 | GGTTTAGAGTGGTGCGGAAT | CCGGGCCTAAAGTAATCAAA | ||
| cnu_mBRPGM0182a | 14,588,355 | TGGAGTAGAAGCGTCGTCTG | CCTTCCTCCTCTCAGTTCCA | ||
| cnu_mBRPGM1527a | 16,911,280 | GCAAGCACGGAGACTAAACA | GACGGTGAAGCTGTACGAAG | ||
| cnu_mBRPGM0813 | 18,824,804 | GTTCCATAGGGCGTTCACAT | CCGATGATTCTCTCAATCAGG | ||
| cnu_ssr447 | 21,870,396 | TGGTGTCAAACGGACAGAAA | ATACTCGGCTCAAACCGTCA | ||
| CB10416 | 23,408,087 | GCTGTTGCTGTAGGTTTGA | GAGCCAGCGTTGATAAGA | ||
| A03 | niab_ssr115 | 1,049,474 | CGGTGTATACCGAACGAGAA | AAACCCAATCAACCCCTTTA | |
| ACMP00292 | 2,920,902 | GGGTTGCTGGTTTAGCTGTT | TGAATCCGCTGAACTCTCTG | ||
| cnu_mBRPGM0240 | 5,317,208 | GAGGGAAAGAGGACAATACGA | TCATCGAGAAACGAAGGGTA | ||
| cnu_ssr173 | 8,444,756 | TGTATTCCATTATTTCCGACTAACCT | CCGCATTTTAAAAACGTGAGAAA | ||
| cnu_ssr290 | 9,860,984 | CGATTTTGCCATTGTCTAAGC | TGAAGACACGTTGGTTGAACA | ||
| ACMP00755 | 11,006,524 | GTCATCGCAAGAGGACAAGA | AAAGCTCCATCAAAGCACCT | ||
| cnu_ssr098 | 14,357,780 | TGCGACCCAAGTAGGTGAAAC | TGTCTCTCGCTCATTCATCCAA | ||
| cnu_ssr327 | 17,867,509 | TTCTTGACCAAAAGAATCATGG | CTAACACGGGGAAAAGCAGA | ||
| ACMP00186 | 19,379,932 | GCCTCCCTGACTTGTACCTC | TTCAATGCGCCAGTTAAGAC | ||
| ACMP00410 | 20,585,363 | CTCAAAGCATGAACGTGGAC | CCTCCCTTGATTTGTGGAGT | ||
| sau_um146 | 22,875,333 | CTCGCAAAATCCCTTCTTCG | CATATCGCTCGAGTTGCAGA | ||
| cnu_ssr526 | 23,351,679 | TCCGAGAAGCACACAAAGAA | TGACCATTTTCTGCCATTCA | ||
| TCR079 | 23,692,426 | TGACGTTCAATCAAAGCCTGA | TTTAGCAATCAAATGCAAATTCAA | ||
| cnu_ssr492 | 23,747,774 | TCGAGGTGGTTACAATCCAA | CAATGCGGATCTACCTCTCA | ||
| cra_id011 | 24,345,511 | GTCGGATTTCTTTCTACACG | TGAACCTATCTTCCTCAACG | ||
| cra_ssr015 | 24,345,899 | CGGCCTCCGGAAATTTATTA | TGGGAGGACCTCTCTCTTCTT | ||
| Cra_RT1 | 24,351,487 | ACGTAAAGAAGCTGACCGGAGAC | AGGCTTAACAACAGTTCCAGATT | ||
| cra_ssr017 | 24,355,996 | GTGTGACCGCACTGTTGTTT | AGTTTGACCCAAACGCATAA | ||
| sau_um026 | 25,648,956 | AGTGGCTCCCAGGAGGATAATA | CTTGGAGAAGAGAAACTTGGGC | ||
| cnu_ssr241 | 27,414,883 | AATGCTGTGTCCATGACCAA | CGGGCATCCACCTAATTTGT | ||
| At2g35530 | 29,980,641 | CAGAGTAACTGGTTATGCCCGTC | CAATAGGGATAAACCTGGAGACAG | ||
| ACMP00563 | 31,183,993 | CGGAGATGGGATTAAGGAGA | AAAGATCGTGTGGGTGGAAT | ||
| A04 | BrID10929 | 251,988 | CAATTTGGGAAGACAGTTCT | GATTCGTTGATATAAGGCCA | |
| BrID10321 | 2,417,875 | TGTGTTTCCTAGTGTGTTGG | ATCAGTCTGAGGGTTCATCA | ||
| BrID101249 | 3,990,390 | TTGCATAGCACATGTAGGAG | GAACGTCTACTTATGGAGAACA | ||
| BrID10645 | 7,441,204 | GCAGAGGAAAACAATCAGAC | AAGCGTCGACTTGAAATCT | ||
| ACMP00073 | 8,674,696 | TTCAACCACACCGACAAACT | CTGACGGAGTCCCTGTACCT | ||
| ACMP00356 | 11,811,660 | TGAGGTCTACAGCCCAAGTG | AATGGAGATCGTGTGCAAAG | ||
| 染色体 Chr. | 标记名称 Marker name | 物理位置 Physical location (Mb) | 正向引物 Forward primer (5°-3°) | 反向引物 Reverse primer (5°-3°) | |
| A04 | ACMP00744 | 12,860,981 | CCTGTTCCAATCCCATTCTT | AGATCCCTGACGGGTTTATG | |
| ACMP00281 | 15,028,874 | TCGACTTTGACGAAGACTGG | AGTCCTCATGGCATCAAACC | ||
| hri_Mbrms195 | 17,105,559 | AATACTTTCTGAAGTTGTCCGCTAA | AACCTACGCAAGATGCTTCTACTT | ||
| cnu_ssr005 | 18,776,905 | AGGAGTCTCGTTCCGTGAGA | TTGAAATTAAGTCGAGCAAACAA | ||
| A05 | cnu_ssr387 | 236,456 | ACTCGAACCATTCTGGCAAA | GGAACGACTTCCTCCCGTAT | |
| niab_ssr017 | 2,501,334 | GGTTAAGCAGACGATGGAAGTAA | TATAGGGGAAATCATCTCAAGCA | ||
| cnu_ssr381 | 4,003,395 | TGCTTTTAACCAAACTCAAACG | TGCAGAGAGGCAAGTTTCAA | ||
| sau_um392 | 5,882,422 | GCCAGTTTCGTCTTCTTCTCTG | GAAGTCACACCCCCATCTCTATC | ||
| BrID90500 | 6,762,400 | ATGGGCTTTGTTTGGGTAAA | TGAAAGAGACAAGCTGGCAA | ||
| BrID10825 | 9,949,084 | CAATGTGTTCGATGGAATG | TAGTGTGCGAACAGATTCAG | ||
| ACMP00841 | 13,132,010 | CAGTGGCAACAACATCAACA | CTTCACATCGTCTCCACCAC | ||
| BrID101239 | 13,226,921 | TCCACACAGAAGATAACTGGT | GAGATGACATTTTCGCTGAC | ||
| cnu_ssr458 | 16,415,511 | GGGGTGAATCTTGGATGAGG | CTGACGGATTCCCAACGAAT | ||
| sau_um366 | 18,549,687 | TTCTCCTCGTATCACCACTCCT | GCCTACGTCTTCTACAGCGAGAT | ||
| niab_ssr082 | 20,079,421 | CATTTCCCCGTGACTATCTG | CGTCTTCATCTCAATCTCGC | ||
| sau_um551 | 21,619,807 | GTCCATCTCCTACCTGCTCCA | GTTTTGAGCCGAATAATGGTTG | ||
| hri_mBRMS007 | 23,268,922 | AAATTGTTTCTCTTCCCCAT | GTGTTAGGGAGCTGGAGAAT | ||
| A06 | BrID10395 | 1,513,120 | GCTGACATGTACCTTTTGAA | CATCTAAGACCGAGTCAAGC | |
| sau_um278 | 3,092,702 | GAGAACAAGAGGAGGACGCTT | CCGGAGGCAAGTATCCATAAG | ||
| niab_ssr134 | 5,621,445 | CGCAGCCTTTTGCTTCT | TTGCTCTCCTGCAGCTTG | ||
| cnu_mBRPGM1016a | 6,689,917 | TGGAGATGGCTGTTGTTGAT | AGCAGATGTCGGGAATAACC | ||
| niab_ssr049 | 7,659,576 | GAGGAATTAACGGCGTCTTG | CAGTCGCCACTACCTGGTTT | ||
| ACMP00739 | 10,965,852 | GGTGACTGTTCCTCATCAGC | ATCCCTATCCAAACCTCTCG | ||
| BrID10847 | 14,198,854 | TCATTGCCTTACTTTGTGAC | CTGACACAGGTGAATCAACA | ||
| BrID10849 | 14,265,494 | AAAGATCTGTGGAATCATGG | GGCAAACATGGGTTGATA | ||
| BrID10627 | 15,630,795 | AACACAGATCCAATCTAGCG | GTCCTTAGGCCAAGCATT | ||
| ACMP00692 | 17,268,545 | CGAGTTGCAGAGCCAAGTAG | AACGTAACGCTTCCTCTTCC | ||
| sau-um121 | 18,555,402 | GAACCTAACGAAAGGCACCAC | AGTGAGGGTAGACAGGGAGAGAG | ||
| cnu_ssr220 | 21,896,695 | ATCAGAACCGAATCCGACCA | CAATGGTTGCAATGTTATTTGGA | ||
| sau-um415 | 22,971,940 | ACGCAAAGAGAGCGAAAGAGTC | GGTCTTAATCGCATGGAATCCG | ||
| sau-um616 | 23,447,329 | ATTACCTATTGACCCCACACCAC | GACGTAGAACAAGTGAGAAGGGA | ||
| A07 | ACMP00261 | 1,018,274 | AAGCCTCGACTTTGGAAGAT | ATCTCCGTCTGGTCTCGTCT | |
| At2g06510 | 2,516,307 | GATCGGGTTAAGTCAGGACA | ATGGTCTCCATGTTCAGCAC | ||
| ACMP00785 | 4,675,451 | TGGCTCTGTTTCCTTCTCCT | AAGGGATTGATCGGAACTTG | ||
| BrID10283 | 8,743,728 | CGGTTAGGTCGTAACTCGTA | CTCTTCATACGCAAGTCTTT | ||
| BrID10487 | 11,667,757 | TCCCTTACAAGTTCATGGAT | ATGGTAGCAACAAAACCAGT | ||
| nia_m063 | 12,010,219 | GAAGAAACTCGGTGGGGAGT | AAAGAGTTCCGAAAATGGGC | ||
| cnu_ssr511 | 14,701,383 | TGTGGACGAGAAACTGAGGA | TGAGATACTGGTGCGTGTGG | ||
| cnu_ssr044 | 16,404,302 | TGTTTTGATCTTTACTGTTTTTGGA | AATGTTTTTATATCACTATTGCCAAAT | ||
| cnu_ssr048 | 18,226,238 | TTCTCCATGCTGTTCAATTCAC | CATGAAAAATCGACCTTATTCCA | ||
| cnu_ssr516 | 20,127,600 | ACTTGCCTTAGCCAGCAGCG | AAGATTTTGTGTTGTGGTCTGGTGA | ||
| cnu_ssr1566 | 22,103,544 | GTCAGACTCGGATGGCTTG | TAGGAGCAGTTGGTTCAGCA | ||
| A08 | ACMP00659 | 1,364,408 | CCGCCTCAATCTCTAGCTTT | CTCGTTCACCACCTCTGCTA | |
| ACMP00551 | 2,716,097 | CAGCAATGGTGGACATCTCT | AACAAGACCGGAACCATCTC | ||
| 染色体 Chr. | 标记名称 Marker name | 物理位置 Physical location (Mb) | 正向引物 Forward primer (5°-3°) | 反向引物 Reverse primer (5°-3°) | |
| A08 | BrID90197 | 4,203,640 | TGTTCAAACTTCCCACCCTC | GTGAGCCCAAAAGTTCCTGA | |
| BrID10839 | 6,657,012 | AGGAACAATACCCATTTGTG | GGTGTTTGTGTGTTCGGTA | ||
| BrID101199 | 7,356,083 | ATGATGGAGATGGACATTTG | TGTATCGGCAGAAGAATCTC | ||
| sau_um192 | 10,064,084 | TCCCTCCTCTCTACGTCTTCTTC | CTTCTCTGTAACGGGCTTTGAC | ||
| sau_um044 | 14,603,121 | CGATTCATCCATCCATCACC | CTATAGGACCGATCCCGTCTTT | ||
| cnu_ssr432 | 16,431,751 | CAAACCTCGTCCTAAGCAGAA | ACCTGAAGATGACCCAGACG | ||
| sau_um077 | 17,469,120 | CTGATCCTCGAAGAAGACAGTGC | CTTCATGCACTTGAGGAGTCG | ||
| cnu_ssr176 | 18,202,437 | TGTAAGTCACGTTCGGTTTGCT | AGGCATGTATGGAGATGTAGAGTGA | ||
| BRMS-198 | 21,521,373 | CGAGAGCAGTTAGGAAGCTTATAGA | AGAGATACTCTGTCCTCCACCTCTT | ||
| A09 | nia_m010 | 1,815,031 | GGTTGACGTCTCATTGTGTTCTT | TAGCTTTGCTCACTTTTCACTCC | |
| sau_um219 | 3,482,038 | CGCAGCTTCCTCTGTATTGCTA | GGCTCTCACCAGAGTCAAGTCTA | ||
| cnu_ssr157 | 5,671,382 | CCGCAGTTGATCCATTAGCC | ACGCTGCATCCACATGAAAC | ||
| sau_um368 | 6,944,590 | AGCCCCGTCTCTTTCACTGTAT | GATCTAGGGTCTCGTCGACTTTG | ||
| nia_m022 | 10,330,441 | CTCTCGTCTCGGAGGATCTAAA | GTGAGAGTGGTTGCTGAGTGAG | ||
| sau_um019 | 13,593,016 | GGTCCTGCCATTCCTATTCTCT | CATGCCACGTCAGCAATATG | ||
| sau_um138 | 16,538,853 | CGCACACCATTTCCACAAAC | GAGATGAATGTGCGTCTCCTG | ||
| ACMP00188 | 18,078,993 | GATTCCTCTCCACGACCATT | TCTCCCAAATCGGTTCTTTC | ||
| sau_um101 | 21,178,401 | GATCTTATCGTGCCCATTGC | CTCCTCATAGGGCTCCTTTTTC | ||
| cnu_ssr598 | 24,440,145 | TTCACCGTCTGCTCTTATCG | CTGCTCCCATACGATCCACT | ||
| cnu_ssr016 | 27,461,366 | GGTGAATGGAATCTTGTCTTGA | CCCAACAATCCCAGAAACAC | ||
| cnu_ssr119 | 30,057,081 | ACACCTACTTGTTTCCATCCAAAT | CGGGTATTTGCGTTGTTTCC | ||
| sau_um187 | 33,166,753 | GTCCTCCTCAACCTCATCATCA | AGTCGAGAGTAACGGGAAGAGAG | ||
| sau_um105 | 36,739,828 | CCTTTCTAATGGGAAGCGGTAG | CTCCCTCTTCGAATTGACTCAC | ||
| A10 | sau_um126 | 2,528,134 | AGAACACGCTCCTAACCATCAG | TAGCTACGAGGCCTTAGAGGGTA | |
| niab_ssr034 | 4,065,070 | GTGCAAGTCAGTGCCAAAGA | CTCGGTGGTTGAGTGAAGGT | ||
| sau_um433 | 7,037,138 | AAGAGTCCACAGCAGGAGATTG | GGGATGAGAAAAAGACAGGTGG | ||
| sau_um216 | 10,127,674 | CTTTCTTCTCTCCGTCGTTCC | AAGGTTAGGGTTAGAGAAGCCG | ||
| niab_ssr123 | 11,313,024 | GGATCTAGAAACCCCTTCACA | ATCTTGTGTCGGGCAGATAA | ||
| Nniab_ssr122 | 12,936,001 | ACTTCTCCGGCTGGATACTC | CCGTTTAAACTTGCGTTTGT | ||
| niab_ssr009 | 15,004,910 | TTCCCAAGCTTGCTGGTACT | GAGATTTCCCTCGCTTGATG | ||
| sau_um310 | 15,341,225 | TCTTTTCCATCTCTCTACCATCATC | CCTATGAGAGGAAGACCGAGACT | ||
新窗口打开|下载CSV
图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1华油杂62恢复系Bing409根肿病抗性遗传改良的技术路线
Fig. 1Genetic improvement route of Huayouza 62 restorer line Bing409 for resistance against clubroot disease
1.2.2 根肿病温室接种体系 温室接种采用菌土法接种。取出-20℃保存的油菜病根, 室温下解冻称量并用匀浆机磨碎, 与风干的草炭土按照1∶20的比例混匀, 25℃密封保存48 h以上[20]。将培养土装到50孔穴盘中并灌足底水, 少量菌土放到穴盘中后, 在菌土上播种1~2粒种子, 25℃培养, 6周后进行表型鉴定并分级。0级为没有根瘤, 1级在侧根处有小根瘤, 2级在主根侧根处有较小根瘤, 3级在主根侧根有较大根瘤, 4级在主根侧根有较大的明显根瘤[21]。
1.2.3 根肿病田间接种及表型鉴定 田间表型鉴定通过选取近几年发病严重且相对比较均一的田块为病圃, 直接分成不同的小区进行直播, 一般在9月下旬播种, 并于6~10周后或在感病对照组有明显受害表型时开始进行表型鉴定, 发病等级划分标准与室内接种相同。
1.2.4 病情指数 病株率(%) = 发病株数/调查总株数×100; 病情指数 = ∑(病株数×相应病害级别)/(调查总株数×最高级别) × 100。校正系数K = 50/对照品种的实际病情指数; 相对病情指数 = K×鉴定材料的病情指数。品种资源的抗病性差异根据相对病情指数进行分类: 免疫(I) = 0; 0<高抗HR≤5; 5<抗R≤10; 10<中抗MR≤20; 20<中感MS≤30; 30<感病S≤50; 50<高感HS≤100[21]。
1.2.5 DNA提取及PCR程序 利用改良CTAB法提取亲本和各世代单株幼嫩叶片DNA, 10 μL的PCR的反应体系, 包含2 ng的DNA模板、正向反向引物各250 nmol L-1、0.25 nmol L-1 dNTP、1 μL Taq酶。PCR反应条件为94℃预变性3 min; 94℃ 1 min, 55℃ 30 s, 72℃ 1 min, 35个循环; 72℃ 10 min; 10℃ 保存。PCR产物在6%的变性聚丙烯酰胺凝胶进行电泳, 结束后进行银染、显影。
1.2.6 前景选择 用于CRb抗病位点前景选择的标记由沈阳农业大学朴钟云教授课题组提供或本实验室开发, Pol.CMS恢复基因前景选择标记由华中农业大学易斌教授提供。经过两亲本间多态性检测, 选择具有多态性的分子标记进行前景选择(表1), 其中CRb_ssr541、CRb_ssr01和CRb_ssr413是位于CRb抗病位点两侧的连锁标记, Rfp_ssr52和Rfp_rt5标记分别是与甘蓝型油菜Pol.CMS恢复基因Rfp的连锁的分子标记和功能标记[22]。需要说明的是, 每一代回交分离群体中含有前景抗病位点CRb的个体, 其恢复基因Rfp都需要进行标记选择, 保留同时具有CRb及Rfp的个体, 继续进行遗传背景筛选, 故本文将Rfp位点的筛选归在前景选择中。
Table 1
表1
表1CRb抗病位点及Rfp恢复基因连锁标记
Table 1
| 标记名称 Marker name | 标记类型 Marker type | 正向引物 Forward primer (5°-3°) | 反向引物 Reverse primer (5°-3°) | 物理位置 Physical location (Mb) |
|---|---|---|---|---|
| CRb_ssr541 | SSR | TGCTTGAGCAGAAACAATATCAA | TTGCGCATCTCTGTTTAGCTT | A03: 23693689 |
| CRb_ssr413 | SSR | ATTGTGCCGTCGGAATTAAA | GATGATTAGAAAAGGTGTCTATTGC | A03: 23762587 |
| CRb_ssr01 | SSR | TCGAGGTGGTTACAATCCAA | CAATGCGGATCTACCTCTCA | A03: 24031219 |
| Rfp_ssr52 | SSR | TCAACAACAACAGCCTTTCG | GGAAGAAGTCGCTTCCTGTG | A09: 34418209 |
| Rfp_rt5 | 功能标记FM | GGGATGCGATCCTGATATTTG | GAGAGAGGCTACAGAACAAACT | A09: 34485601 |
新窗口打开|下载CSV
1.2.7 背景选择 在亲本材料Bing409与CR Shinki中筛选出了225对多态性标记, 通过本地BLAST, 筛选得到物理位置均匀分布于油菜A基因组的124对标记, 且相邻标记间平均物理距离2 Mb左右, 用于回交群体背景回复率鉴定(表2, 附表1)。
遗传背景回复率 = 基因型恢复到轮回亲本背景的标记数/总共标记数。
Table 2
表2
表2均匀覆盖甘蓝型油菜A基因组的标记统计
Table 2
| 染色体 Chr. | 标记数目Marker numbers | 标记间平均间距 Average distance of adjacent markers (Mb) |
|---|---|---|
| A01 | 10 | 2.95 |
| A02 | 11 | 2.30 |
| A03 | 22 | 2.01 |
| A04 | 10 | 2.07 |
| A05 | 13 | 1.92 |
| A06 | 14 | 1.83 |
| A07 | 11 | 2.11 |
| A08 | 11 | 2.02 |
| A09 | 14 | 2.69 |
| A10 | 8 | 1.83 |
| 总数Total | 124 | — |
新窗口打开|下载CSV
1.2.8 农艺性状测定与测验 2017—2018年度, 在黄冈市农业科学院梅家墩试验农场(非根肿病发病区)进行油菜品种产量及主要农艺性状分析试验。供试品种为根肿病抗病品种华油杂62R, 对照品种为长江中游优异品种华油杂12, 每个品种各种植3个重复, 小区面积20 m2。考察的主要产量性状包括株高、有效分枝数、单株有效角果、每角粒数、千粒重及单株产量等, 每一性状每个重复调查植株15株, 最后实测小区产量。采用近红外分析仪检测的方法测定含油量、芥酸及硫苷含量等油菜品质。
2 结果与分析
2.1 F1及BC1的获得、分子标记检测及抗病性鉴定
以国审甘蓝型油菜杂交种华油杂62的Pol. CMS恢复系Bing409为母本与含CRb抗病位点的CR shinki为父本, 杂交获得F1, 以Bing409为轮回亲本回交获得了BC1代材料。先后对F1、BC1及两亲本以根肿菌4号生理小种在温室进行接种鉴定, 接种40 d后调查上述材料的根部受侵染情况, 不同发病等级株数统计见附表2。Table S2
附表2
附表2F1、BC1及两亲本的发病率统计
Table S2
| 发病等级 Disease level | 调查株数Number of plants investigated | |||
|---|---|---|---|---|
| F1 | BC1 | CR Shinki | Bing409 | |
| 4级 Level 4 | 0 | 19 | 0 | 35 |
| 3级 Level 3 | 0 | 0 | 0 | 12 |
| 2级 Level 2 | 0 | 0 | 0 | 0 |
| 1级 Level 1 | 0 | 0 | 0 | 0 |
| 0级 Level 0 | 31 | 24 | 30 | 0 |
新窗口打开|下载CSV
对F1表型鉴定发现, F1对4号生理小种表现出100%抗病, 病情指数为0, 与抗病父本CR shinki表型一致, 而轮回亲本材料Bing409则表现出极度感病, 发病指数高达93.6% (图2-a, b)。表明CR shinki中所含的CRb抗病位点对根肿菌4号生理小种表现为显性抗病。
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2F1及BC1抗病性鉴定
a: F 1接菌表型鉴定; b: F 1病情指数; c: BC1接菌表型鉴定; d: BC1病情指数; e: BC1基因型鉴定。A表示Bing409基因型, B表示CR Shinki基因型, H表示杂合基因型; S表示感病表型, R表示抗病表型。
Fig. 2Clubroot disease resistance identification of F1 and BC1
a: phenotypic identification of F1 generation after P. brassicae pathotype inoculation; b: disease index of F1 generation; c: phenotypic identification of BC1 generation after inoculation; d: disease index of BC1 generation; e: genotypic identification of BC1 generation. A represents genotype of Bing409, B represents genotype of CR Shinki, and H represents heterozygous genotype; S represents susceptibility phenotype, and R represents resistance phenotype.
对BC1群体各单株的表型鉴定时发现, 发病等级呈现出两极分布, 分别为抗病(0级, 24株)以及感病(4级, 19株)(图2-c, d)。卡方测验结果显示, BC1群体抗病、感病分离比符合1∶1分离(N=43, χ2 = 0.58 < χ20.05(1) = 3.84), 符合理论预期。用连锁标记CRb_ssr413(表1)对以上植株进行基因型鉴定(图2-e), 其中A、H、B分别代表Bing409基因型、杂合基因型、抗病CR Shinki基因型。表明BC1群体中所有感病植株的基因型均为A, 抗病植株基因型均为H, 基因型鉴定结果与表型一致。F1及BC1代群体分子鉴定和接种鉴定的结果都表明, CRb抗病位点对我国油菜疫区的4号优势生理小种为显性抗病遗传。
2.2 利用分子标记对BC1F1~BC3F1各世代株系的前景和背景筛选
利用与抗病位点连锁标记, 对BC1F1~BC3F1群体进行前景选择, 筛选出含CRb抗病位点的植株, 然后利用附表1中所列的分子标记对这些单株进行遗传背景分析, 根据每一个标记基因型计算并统计遗传背景回复率, 每一代保留遗传背景回复率较高的株系继续进行回交。图3是BC1F1~BC3F1代含抗病位点的入选株系的遗传背景回复率从低到高排列的柱形图。本试验中BC1群体背景回复率在0.29~0.71之间, 均值为0.49, 低于BC1群体的理论恢复率75%。BC2F1入选单株背景回复率在0.73~0.93之间, BC3F1在0.87~0.97之间。图3
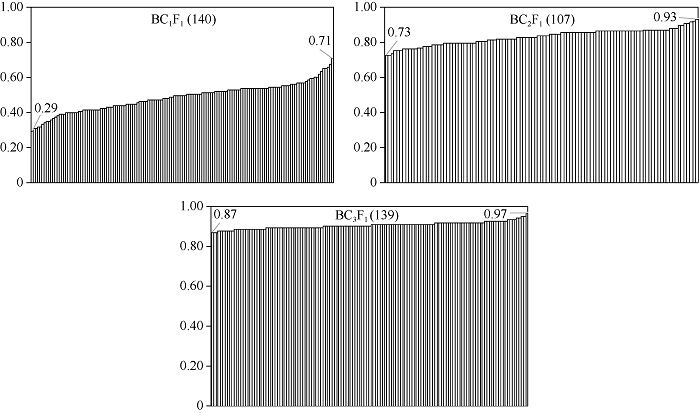 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3BC1F1~BC3F1前景入选单株遗传背景回复率统计
图中括号里面的数字表示当代经过前景选择入选的含有前景抗病位点的单株数。
Fig. 3Genetic background recovery rates of the resistant plants selected from BC1F1 to BC3F1 generations
The numbers in parentheses represent the total plants containing resistant locus after foreground selection.
表3列出了BC1~BC3代各群体单株数、含CRb位点的株数及最后保留的较高遗传背景的单株数。BC1选择轮回亲本基因组回复率>60%的5个单株编号分别为438A、65B、128C、464D、94E, 继续进行下一代回交。由438A回交群体(含46株)、65B回交群体(含68株)、128C回交群体(含118株)组成的BC2群体共232株, 经过前景和背景筛选获得了含有CRb位点、轮回亲本基因组回复率>90%的4个株系, 编号分别为438A1、65B1、65B2和65B3, 各自对应的背景回复率分别为92%、92%、91%和91%。其中, 编号464D和94E的回交后代只进行了抗病位点前景选择, 未进行背景筛选。438A1、65B1、65B2和65B3这4个株系继续回交, 获得了BC3后代总共267株, 经过同样的筛选, 获得含CRb位点、轮回亲本基因组背景回复率高达97%的2个单株, 分别为438A1-1和65B2-2。
Table 3
表3
表3BC1~BC3各世代前景及背景选择概况
Table 3
| 世代 Generation | 抗病位点前景选择Foreground selection | 背景筛选标记数 No. of markers used to background selection | 最终保留株数 No. of selected plants | 背景回复率 Genetic background recovery rates (%) | 用途 Application | ||
|---|---|---|---|---|---|---|---|
| 检测株数 No. of plants tested | 筛选标记数 No. of markers | 入选株数 No. of selected plants | |||||
| BC1F1 | 492 | 5 | 256 | 118 | 5 | 64-71 | 回交Backcross |
| BC2F1 | 232 | 5 | 107 | 36 | 4 | 73-93 | 回交Backcross |
| BC3F1 | 267 | 5 | 139 | 5 | 6 | 87-97 | 回交、自交 Backcross, self-cross |
新窗口打开|下载CSV
2.3 高世代当选株系抗病性鉴定、分子标记分析及主要品质性状检测
2.3.1 高世代分离群体抗病性鉴定及恢复基因的分子检测 本研究在对BC3F1回交后代进行分子标记检测之前, 随机抽取了编号为65B2的BC3F1回交后代用4号生理小种进行了接种鉴定。抗病性鉴定结果显示, BC3F1分离群体中含CRb杂合抗病位点的(H基因型)材料对4号生理小种表现出免疫抗病性(R, 24株), 不含CRb位点的材料(A基因型)的均为感病表型(S, 19株), 抗感基因型与表型相对应(图4-a, b, c), BC3F1抗感表型分离比符合1∶1分离(N = 43, χ2 = 0.58 < χ20.05(1) = 3.84), 这一结果与BC1群体的鉴定结果相吻合。由此可见, 该连锁标记能够准确鉴定抗病位点, 不用每一代都进行抗病性鉴定, 以有效节省时间。图4
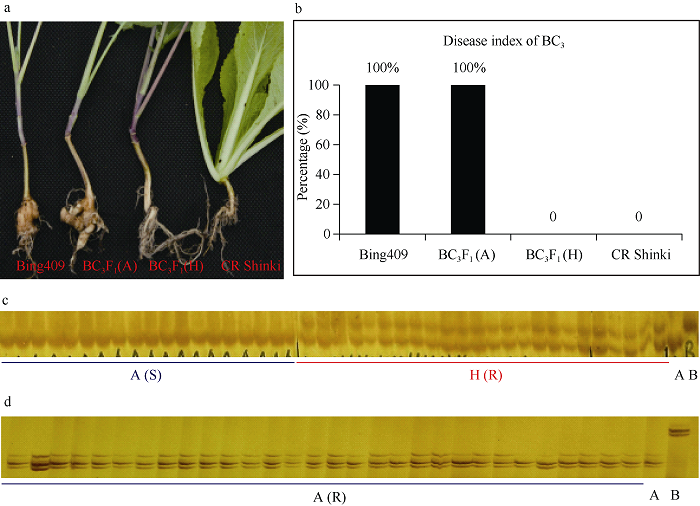 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4BC3抗病性鉴定及恢复基因Rfp位点检测
a: BC3接菌表型鉴定; b: BC3病情指数; c: BC3基因型鉴定; d: BC3抗病植株恢复基因Rfp位点检测。A表示Bing409基因型, B表示CR Shinki基因型, H表示杂合基因型; S表示感病表型, R表示抗病表型。
Fig. 4Clubroot disease resistance evaluation and restore gene Rfp identification of BC3 plants
a: phenotypic identification of BC3 after clubtoot pathogen inoculation; b: disease index of BC3 generation; c: genotypic identification of BC3 generation; d: identification of restore gene Rfp of resistant plants in BC3 generation. A represents genotype of Bing409, B represents genotype of CR Shinki, and H represents heterozygous genotype; S represents susceptibility phenotype, and R represents resistance phenotype.
随后, 利用与波里马恢复基因连锁标记Rfp-ssr52[22], 对65B2的BC3F1回交后代中分离出的部分抗病单株(30株)的基因型进行检测。结果如图4-d所示, 这些单株的基因型均与轮回亲本Bing409的基因型一致(A)。最后将经过抗病性鉴定及恢复基因位点鉴定、遗传背景回复率高的抗病单株65B2-2经自交获得了纯合抗病株系, 并正式命名为Bing409R。
2.3.2 抗病近等基因系材料Bing409R对不同生理小种的抗性评价 本研究在选育Bing409R的过程中, 主要选取了根肿菌4号生理小种进行抗性鉴定, 为了增加结果的可靠性, 还选取了我国油菜根肿病发病较为严重地区的根肿菌, 包括云南、四川、湖北、安徽等省, 对Bing409R进行接菌鉴定。接种我国不同地区根肿菌株数统计见附表3, 表型调查结果见表4。Bing409R对湖北宜昌和枝江、安徽黄山以及四川省不同地区的根肿菌均表现出免疫抗性(I); 对云南不同地区根肿菌的抗性存在较大差异, 分别表现为免疫抗性(I)、抗(R)、中抗(MR)、中感(MS)和高感(HS)等表型; 对湖北恩施巴东的根肿菌表现为高感(HS)。说明我国根肿病生理小种的类型是多样的, 四川地区根肿菌生理小种比较单一, 而云南地区生理小种种类复杂多样。这一结果的获得可为抗病品种的合理应用提供重要依据。
Table S3
附表3
附表3Bing409R抗病材料接种我国不同地区根肿菌株数统计
Table S3
| 根肿菌 P. brassica | 收集地点 Collection place | Bing409R接菌株数Number of inoculated Bing409R plants (R/S) | 对照 Control (R/S) | ||
|---|---|---|---|---|---|
| 重复1 Repeat 1 | 重复2 Repeat 2 | 重复3 Repeat 3 | |||
| Y-TC | 云南腾冲Tengchong, Yunnan | 15 (3/12) | 24 (5/19) | 26 (5/21) | 19 (0/19) |
| Y-XC | 云南楚雄Xiongchu, Yunnan | 19 (16/3) | 25 (19/6) | 25 (21/4) | 25 (0/25) |
| Y-BS | 云南保山Baoshan, Yunnan | 24 (5/19) | 26 (6/20) | 23 (4/19) | 20 (0/20) |
| Y-LC | 云南临沧Lincang, Yunnan | 20 (11/9) | 25 (15/10) | 26 (16/10) | 21 (0/21) |
| Y-DH | 云南德宏Dehong, Yunnan | 20 (4/16) | 26 (6/20) | 24 (5/19) | 25 (0/25) |
| H-ES | 湖北恩施Enshi, Hubei | 17 (4/13) | 26 (6/20) | 20 (3/17) | 22 (0/22) |
| H-YC | 湖北宜昌Yichang, Hubei | 23 (23/0) | 22 (22/0) | 26 (26/0) | 22 (0/22) |
| H-ZJ | 湖北枝江Zhijiang, Hubei | 22 (22/0) | 21 (21/0) | 19 (19/0) | 20 (0/22) |
| S-DY | 四川德阳Deyang, Sichuan | 26 (26/0) | 22 (22/0) | 24 (24/0) | 22 (0/22) |
| S-PZ | 四川彭州Pengzhou, Sichuan | 27 (27/0) | 22 (22/0) | 23 (23/0) | 20 (0/20) |
| S-GH | 四川广汉Guanghan, Sichuan | 25 (25/0) | 23 (23/0) | 23 (23/0) | 25 (0/22) |
| S-CD | 四川成都Chengdu, Sichuan | 24 (24/0) | 20 (20/0) | 19 (19/0) | 25 (0/22) |
| A-HS | 安徽黄山Huangshan, Anhui | 17 (17/0) | 19 (19/0) | 23 (23/0) | 21 (0/22) |
新窗口打开|下载CSV
Table 4
表4
表4Bing409R抗病材料对我国不同地区根肿菌的抗性评价
Table 4
| 根肿菌编号 No. of P. brassica | 收集地点 Collection locations | 409R相对病情指数Relative disease index of 409R | 抗性评价 Resistance evaluation | ||
|---|---|---|---|---|---|
| 重复1 Repeat 1 | 重复2 Repeat 2 | 重复3 Repeat 3 | |||
| Y-TC | 云南腾冲Tengchong, Yunnan | 40 | 40 | 40 | S (40±0) |
| Y-XC | 云南楚雄Xiongchu, Yunnan | 8 | 12 | 8 | R (9±1) |
| Y-BS | 云南保山Baoshan, Yunnan | 40 | 38 | 41 | S (40±1) |
| Y-LC | 云南临沧Lincang, Yunnan | 23 | 20 | 19 | MS (23±1) |
| Y-DH | 云南德宏Dehong, Yunnan | 40 | 38 | 40 | S (39±1) |
| H-ES | 湖北恩施Enshi, Hubei | 38 | 38 | 43 | S (40±1) |
| H-YC | 湖北宜昌Yichang, Hubei | 0 | 0 | 0 | I (0±0) |
| H-ZJ | 湖北枝江Zhijiang, Hubei | 0 | 0 | 0 | I (0±0) |
| S-DY | 四川德阳Deyang, Sichuan | 0 | 0 | 0 | I (0±0) |
| S-PZ | 四川彭州Pengzhou, Sichuan | 0 | 0 | 0 | I (0±0) |
| S-GH | 四川广汉Guanghan, Sichuan | 0 | 0 | 0 | I (0±0) |
| S-CD | 四川成都Chengdu, Sichuan | 0 | 0 | 0 | I (0±0) |
| A-HS | 安徽黄山Huangshan, Anhui | 0 | 0 | 0 | I (0±0) |
新窗口打开|下载CSV
2.3.3 Bing409R不同株系的籽粒品质检测 随机选取65B2-2自交后代衍生出的BC3F2纯合抗病株系自交种子(共9个系, 编号分别为622-22、622-37、622-30、624-04、625-17、630-11、18ZP06、18ZP07和18ZP08), 利用近红外分析仪进行品质检测。由表5可知, 所有被检测株系含油量均与未改良的轮回亲本Bing409 (409S01、409S02和409S03为Bing409不同株系的3个重复)相当, 芥酸及硫苷含量都符合我国“双低油菜”的生产标准。
Table 5
表5
表5BC3F3不同抗病株系品质测定
Table 5
| 抗病株系 Resistant lines | 含油量 Oil content (%) | 芥酸 Erucic acid (%) | 硫甙 Glucosinolate (μmol g-1) |
|---|---|---|---|
| 622-22 | 41.81 | 0.17 | 21.76 |
| 622-37 | 40.05 | 0 | 24.61 |
| 623-30 | 40.99 | 0 | 23.49 |
| 624-04 | 43.63 | 0.34 | 26.54 |
| 625-17 | 41.11 | 0 | 24.32 |
| 630-11 | 40.65 | 0 | 23.33 |
| 18ZP06 | 42.66 | 0 | 23.69 |
| 18ZP07 | 42.44 | 0 | 30.05 |
| 18ZP08 | 41.99 | 0.08 | 28.61 |
| 409S01 (CK) | 41.06 | 0 | 22.76 |
| 409S02 (CK) | 40.38 | 0 | 24.31 |
| 409S03 (CK) | 43.78 | 0 | 23.78 |
新窗口打开|下载CSV
2.4 华油杂62R的田间根肿病抗性鉴定
2016年夏, 将华油杂62的不育系与Bing409R配制了杂交组合, 正式命名为华油杂62R。收获的华油杂62R分别于2016年秋种植于湖北枝江和安徽黄山病区进行抗病性鉴定。苗期(播种后约2个月)田间抗性调查发现, Bing409R及华杂62R在黄山地区表现出全抗(各调查60株), 而对照材料Bing409发病率100% (调查40株); Bing409R及华油杂62R在枝江地区也表现全抗(各调查80株); 进一步基因型分析表明, F1植株(随机检测12株)及Bing409R植株(随机检测7株)均含有CRb抗病位点, 而感病对照Bing409 (随机检测12株)均不含有该位点(图5)。苗后期(播种后约3个半月), 在枝江病区华杂62R与当地不抗病主推对照品种相比, 表现出极强抗性且田间长势强, 明显优于对照(图6)。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5F1枝江病区表型鉴定
华油杂62R (F1)的抗病性明显优于其感病亲本Bing409S。
Fig. 5Phenotypic identification of F1 in Zhijiang diseased fields
The disease resistance of Huayouza 62R (F1) was significantly better than its parent Bing409S.
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6华杂62R新品种在枝江根肿病区的田间表现
Fig. 6Huayouza 62R new lines were integrated and demonstrated in Zhijiang infested fields
2.5 抗根肿病新品种华油杂62R的产量及农艺性状测试
2017—2018年度, 在黄冈市农业科学院梅家墩验农场进行了根肿病抗病新品种华油杂62R的产量及主要农艺性状考察, 以长江中游优良品种华油杂12号为对照, 每个品种各种植3个重复, 小区面积20 m2, 考察主要与产量相关的农艺性状并测定小区产量, 每一性状调查植株15株。株高、有效分枝数、单株有效角果、每角粒数、千粒重、单株产量等主要产量性状的考察结果如表6所示, 其中株高、每角粒数均低于对照华油杂12, 单株有效角果数及千粒重均高于对照华油杂12, 单株产量与对照相比无差异。华油杂62R的小区产量与对照品种华油杂12相比基本相当, 折合单产3183.5 kg hm-2。由此可见, 华油杂62R不仅对根肿菌4号生理小种表现出免疫抗性, 并且还表现出较高的产量生产能力。Table 6
表6
表6抗根肿病杂交种华油杂62R主要产量性状考察及小区产量测定
Table 6
| 品种 Variety | 株高 Plant height (cm) | 有效分枝数 Effective branch number | 单株有效角果数 Silique number per plant | 每角粒数 Seeds per silique | 千粒重 Thousand-seed weight (g) | 单株产量 Yield per plant (g) | 小区产量 Yield per plot (kg) |
|---|---|---|---|---|---|---|---|
| 华油杂12 Huayouza 12 | 170.8 | 5.6 | 162.2 | 20.9 | 3.41 | 11.6 | 6.367 |
| 华油杂62R Huayouza 62R | 161.2 | 5.8 | 176.8 | 18.1 | 3.62 | 11.6 | 6.267 |
新窗口打开|下载CSV
3 讨论
本研究以抗根肿病大白菜CR Shinki为供体, 通过远缘杂交和分子标记筛选, 成功地将甘蓝型油菜国审三系杂交种华油杂62的恢复系Bing409进行了抗性改良(命名为Bing409R)。在改良过程中, 从BC1代开始采用对抗病位点和遗传背景筛选, 并结合适当表型鉴定的策略, 极大缩短了育种时间, 显著提高了育种效率。然后, 以此为基础利用Bing409R与华油杂62的波里马不育系配制杂交组合, 成功选育了甘蓝型油菜抗根肿病杂交新品种华油杂62R, 并完成了品种登记(GDP油菜(2018) 420213), 为我国选育的第一个抗根肿病油菜杂交种。近年来, 我国油菜根肿病的发生面积逐年扩大, 重灾区主要分布在四川、湖北、安徽等地[23], 室内及病区接菌鉴定结果表明, 华油杂62R对我国油菜主产区四川、湖北、安徽等地的根肿菌具免疫或高抗抗性。因此该品种应用范围较大, 产量水平较高, 如在安徽绩溪发病田块产量可达3000 kg hm-2以上, 因此其应用前景广阔。近年来, 虽然华油杂62R年推广面积在30,000~50,000 hm2左右, 但仍远远不能满足油菜生产的需要, 目前全国主要油菜育种单位利用这个抗源正在加紧抗病新品种的选育工作, 这对保障我国油菜产业免受油菜根肿病的威胁具有重要意义。根肿病菌是十字花科专性寄生菌, 生理小种多样, 长期种植含单一抗病基因的品种容易造成抗性丧失[24]。因此, 未来将不同的抗病位点如CRb与PbBa8.1[19]进行聚合, 可培育出对根肿病多个生理小种同时具有抗性的优良油菜品种, 以降低品种抗性丧失的风险。今后, 还需要加大不同抗源材料的创制力度, 以增加抗病基因的遗传多样性, 为抗病位点聚合育种奠定坚实基础。
4 结论
通过对杂交、回交及自交等育种程序, 结合前景和遗传背景的分子标记辅助筛选, 将CRb根肿病抗病位点导入到甘蓝型油菜国审品种华油杂62的父本Pol CMS恢复系Bing409中; 在改良了波里马恢复系Bing409根肿病抗性的基础上, 选育了我国第一个抗根肿病油菜杂交新品种华油杂62R。根肿病抗病性遗传改良并未对抗病新恢复系Bing409R及由其配制的杂交种华油杂62R的产量、品质造成不良影响; Bing409R及华油杂62R对我国四川、湖北、安徽等地区根肿菌生理小种具有免疫抗性。本研究的开展, 为我国油菜抗根肿病育种提供了宝贵的资源, 为我国抵抗油菜根肿病的威胁提供了重要支撑。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1046/j.1365-2559.1998.00319.xURLPMID:9522214 [本文引用: 1]

[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00344-009-9094-7URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00344-009-9100-0URL [本文引用: 1]
[本文引用: 1]
URLPMID:12955203 [本文引用: 1]
DOI:10.1007/s00122-008-0817-0URLPMID:18612625 [本文引用: 1]

A number of clubroot resistant (CR) Chinese cabbage cultivars have been developed in Japan using resistant genes from CR European fodder turnips (B. rapa ssp. rapifera). Clubroot resistance in European fodder turnips are known to be controlled by the combined action of several dominant resistance genes. We have developed three Chinese cabbage clubroot-resistant doubled haploid (DH) lines--T136-8, K10, and C9--which express resistance in different manners against two isolates of Plasmodiophora brassicae, M85 and K04. Depending on the isolates, we identified two CR loci, CRk and CRc. CRk was identified by quantitative trait loci (QTL) analysis of an F(2) population derived from a cross between K10 and Q5. This locus showed resistance to both isolates and is located close to Crr3 in linkage group R3. The other locus, CRc was identified by QTL analysis of an F(2) population derived from a cross between C9 and susceptible DH line, 6R. This locus was mapped to linkage group R2 and is independent from any published CR loci. We developed sequence-tagged site markers linked to this locus.
DOI:10.1007/s00122-003-1577-5URLPMID:14997298 [本文引用: 1]

Clubroot disease, caused by Plasmodiophora brassicae Wor., is highly damaging for Chinese cabbage. The CR (clubroot resistant) Shinki DH (doubled haploid) line of Chinese cabbage carries a single dominant gene, CRb, which confers resistance to the P. brassicae races 2, 4, and 8. An F(2) population derived from a cross between the CR Shinki DH line and a susceptible line, 94SK, was used to map the CRb gene. Inoculation of F(3) families with SSI (single-spore isolate) resulted in a 1:2:1 segregation ratio. Use of the AFLP technique combined with bulked segregant analysis allowed five co-dominant AFLP markers, and four and seven dominant AFLP markers linked in coupling and repulsion, respectively, to be identified. Six of the 16 AFLP markers showing low frequencies of recombination with the CRb locus among 138 F(2) lines were cloned. A reliable conversion procedure allowed five AFLP markers to be successfully converted into CAPS and SCAR markers. An F(2) population (143 plants) was analyzed with these markers and a previously identified SCAR marker, and a genetic map around CRb covering a total distance of 6.75 cM was constructed. One dominant marker, TCR09, was located 0.78 cM from CRb. The remaining markers (TCR05, TCR01, TCR10, TCR08, and TCR03) were located on the other side of CRb, and the nearest of these was TCR05, at a distance of 1.92 cM.
DOI:10.1371/journal.pone.0054745URLPMID:23382954 [本文引用: 1]

Clubroot disease, caused by the obligate biotrophic protist Plasmodiophora brassicae Woronin, is one of the most economically important diseases of Brassica crops in the world. Although many clubroot resistance (CR) loci have been identified through genetic analysis and QTL mapping, the molecular mechanisms of defense responses against P. brassicae remain unknown. Fine mapping of the Crr1 locus, which was originally identified as a single locus, revealed that it comprises two gene loci, Crr1a and Crr1b. Here we report the map-based cloning and characterization of Crr1a, which confers resistance to clubroot in Brassica rapa. Crr1a(G004), cloned from the resistant line G004, encodes a Toll-Interleukin-1 receptor/nucleotide-binding site/leucine-rich repeat (TIR-NB-LRR) protein expressed in the stele and cortex of hypocotyl and roots, where secondary infection of the pathogen occurs, but not in root hairs, where primary infection occurs. Gain-of-function analysis proved that Crr1a(G004) alone conferred resistance to isolate Ano-01 in susceptible Arabidopsis and B. rapa. In comparison, the susceptible allele Crr1a(A9709) encodes a truncated NB-LRR protein, which lacked more than half of the TIR domain on account of the insertion of a solo-long terminal repeat (LTR) in exon 1 and included several substitutions and insertion-deletions in the LRR domain. This study provides a basis for further molecular analysis of defense mechanisms against P. brassicae and will contribute to the breeding of resistant cultivars of Brassica vegetables by marker-assisted selection.Data deposition The sequence reported in this paper has been deposited in the GenBank database (accession no. AB605024).
DOI:10.1007/s11103-012-9971-5URLPMID:23054353 [本文引用: 1]

Clubroot disease is one of the major diseases affecting Brassicaceae crops, and a number of these crops grown commercially, such as Chinese cabbage (Brassica rapa L. ssp. pekinensis), are known to be highly susceptible to clubroot disease. To provide protection from this disease, plant breeders have introduced genes for resistance to clubroot from the European turnip into susceptible lines. The CRa gene confers specific resistance to the clubroot pathogen Plasmodiophora brassicae isolate M85. Fine mapping of the CRa locus using synteny to the Arabidopsis thaliana genome and partial genome sequences of B. rapa revealed a candidate gene encoding a TIR-NBS-LRR protein. Several structural differences in this candidate gene were found between susceptible and resistant lines, and CRa expression was observed only in the resistant line. Four mutant lines lacking clubroot resistance were obtained by the UV irradiation of pollen from a resistant line, and all of these mutant lines carried independent mutations in the candidate TIR-NBS-LRR gene. This genetic and molecular evidence strongly suggests that the identified gene is CRa. This is the first report on the molecular characterization of a clubroot Resistance gene in Brassicaceae and of the disease resistance gene in B. rapa.
DOI:10.1007/s00438-016-1281-1URLPMID:28013378 [本文引用: 1]

To facilitate prevention of clubroot disease, a major threat to the successful cultivation of Chinese cabbage (Brassica rapa L.), we bred clubroot-resistant (CR) cultivars by introducing resistance genes from CR turnips via conventional breeding. Among 11 CR loci found in B. rapa, we identified CRb in Chinese cabbage cultivar 'CR Shinki' as a single dominant gene for resistance against Plasmodiophora brassicae pathotype group 3, against which the stacking of Crr1 and Crr2 loci was not effective. However, the precise location and pathotype specificity of CRb have been controversial, because CRa and Rcr1 also map near this locus. Previously, our fine-mapping study revealed that CRb is located in a 140-kb genomic region on chromosome A03. Here, we determined the nucleotide sequence of an approximately 64-kb candidate region in the resistant line; this region contains six open reading frames (ORFs) similar to NB-LRR encoding genes that are predicted to occur in tandem with the same orientation. Among the six ORFs present, only four on the genome of the resistant line showed a strong DNA sequence identity with each other, and only one of those four could confer resistance to P. brassicae isolate No. 14 of the pathotype group 3. These results suggest that these genes evolved through recent gene duplication and uneven crossover events that could lead to the acquisition of clubroot resistance. The DNA sequence of the functional ORF was identical to that of the previously cloned CRa gene; thus, we showed that the independently identified CRb and CRa are one and the same clubroot-resistance gene.
[本文引用: 1]
DOI:10.1111/j.1364-3703.2011.00729.xURLPMID:21726396 [本文引用: 1]

UNLABELLED: Plasmodiophora brassicae causes clubroot disease in cruciferous plants, and is an emerging threat to Canadian canola (Brassica napus) production. This review focuses on recent studies into the pathogenic diversity of P. brassicae populations, mechanisms of pathogenesis and resistance, and the development of diagnostic tests for pathogen detection and quantification. TAXONOMY: Plasmodiophora brassicae is a soil-borne, obligate parasite within the class Phytomyxea (plasmodiophorids) of the protist supergroup Rhizaria. DISEASE SYMPTOMS: Clubroot development is characterized by the formation of club-shaped galls on the roots of affected plants. Above-ground symptoms include wilting, stunting, yellowing and premature senescence. DISEASE CYCLE: Plasmodiophora brassicae first infects the root hairs, producing motile zoospores that invade the cortical tissue. Secondary plasmodia form within the root cortex and, by triggering the expression of genes involved in the production of auxins, cytokinins and other plant growth regulators, divert a substantial proportion of plant resources into hypertrophic growth of the root tissues, resulting in the formation of galls. The secondary plasmodia are cleaved into millions of resting spores and the root galls quickly disintegrate, releasing long-lived resting spores into the soil. A serine protease, PRO1, has been shown to trigger resting spore germination. PHYSIOLOGICAL SPECIALIZATION: Physiological specialization occurs in populations of P. brassicae, and various host differential sets, consisting of different collections of Brassica genotypes, are used to distinguish among pathotypes of the parasite. DETECTION AND QUANTIFICATION: As P. brassicae cannot be cultured, bioassays with bait plants were traditionally used to detect the pathogen in the soil. More recent innovations for the detection and quantification of P. brassicae include the use of antibodies, quantitative polymerase chain reaction (qPCR) and qPCR in conjunction with signature fatty acid analysis, all of which are more sensitive than bioassays. RESISTANCE IN CANOLA: Clubroot-resistant canola hybrids, recently introduced into the Canadian market, represent an important new tool for clubroot management in this crop. Genetic resistance must be carefully managed, however, as it has been quickly overcome in other regions. At least three resistance genes and one or two quantitative trait loci are involved in conferring resistance to P. brassicae. Root hair infection still occurs in resistant cultivars, but secondary plasmodia often remain immature and unable to produce resting spores. Fewer cell wall breakages occur in resistant hosts, and spread of the plasmodium through cortical tissue is restricted. More information on the genetics of clubroot resistance in canola is needed to ensure more effective resistance stewardship. USEFUL WEBSITES: http://www.canolacouncil.org/clubroot/resources.aspx, http://tu-dresden.de/die_tu_dresden/fakultaeten/fakultaet_mathematik_und_naturwissenschaften/fachrichtung_biologie/botanik/pflanzenphysiologie/clubroot, http://www.ohio.edu/people/braselto/plasmos/
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
DOI:10.1094/PHYTO-11-17-0362-RURLPMID:29996697 [本文引用: 1]

Clubroot disease is an important disease on cruciferous crops caused by Plasmodiophora brassicae infections. The pathotypes have been classified based on the reactions of differential hosts. However, molecular markers of particular pathotypes for P. brassicae are limited. In this study, we found five genetic markers in association with different pathotypes. Different gene expression patterns among different pathotypes (P4, P7, P9, and P11) were assayed according to the transcriptome data. The assay indicated that molecular markers PBRA_007750 and PBRA_009348 could be used to distinguish P11 from P4, P7, and P9; PBRA_009348 and Novel342 could distinguish P9 from P4, P7, and P11; and PBRA_008439 and Novel342 could represent a kind of P4. Polymerase chain reaction cycles ranging from 25 to 30 were able to identify the predominant pathotype in general. Therefore, these molecular markers would be a valuable tool to identify and discriminate pathotypes in P. brassicae population.
