 ,1,*
,1,*Identification and expression analysis of GRF transcription factor family of Chenopodium quinoa
SHI Pi-Biao1, HE Bing2, FEI Yue-Yue1, WANG Jun1, WANG Wei-Yi1, WEI Fu-You1, LYU Yuan-Da2, GU Min-Feng ,1,*
,1,*通讯作者:
收稿日期:2019-03-22接受日期:2019-06-22网络出版日期:2019-07-13
| 基金资助: |
Received:2019-03-22Accepted:2019-06-22Online:2019-07-13
| Fund supported: |
作者简介 About authors
时丕彪,E-mail:1032175660@qq.com。

摘要
关键词:
Abstract
Keywords:
PDF (3002KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
时丕彪, 何冰, 费月跃, 王军, 王伟义, 魏福友, 吕远大, 顾闽峰. 藜麦GRF转录因子家族的鉴定及表达分析[J]. 作物学报, 2019, 45(12): 1841-1850. doi:10.3724/SP.J.1006.2019.94049
SHI Pi-Biao, HE Bing, FEI Yue-Yue, WANG Jun, WANG Wei-Yi, WEI Fu-You, LYU Yuan-Da, GU Min-Feng.
转录因子是植物中最重要的一类调节基因, 参与植物生长、发育、代谢、繁殖、分化等多种生物学过程[1,2,3]。目前在植物中已发现60多个转录因子家族[4], 其中生长调控因子(growth-regulating factor, GRF)是植物特有的一类蛋白, 对植物的生长发育起重要的调控作用[5,6,7]。GRF转录因子在N端区域含有QLQ和WRC两个保守结构域[8,9], QLQ结构域与GRF互作因子GIF (GRF interacting factors)相互作用形成转录激活因子[10], WRC结构域包含一个功能核定位信号NLS (nuclear localization signal)区域和一个与DNA结合的锌指基序[8], 均与GRF转录因子发挥生物学功能密切相关。
在水稻中首次发现了GRF基因OsGRF1, 该基因能够调控赤霉素诱导的水稻茎的伸长[11]。近年来, 随着基因组测序的完成, 越来越多物种中的GRF基因被鉴定和研究[12], 其中, 双子叶模式植物拟南芥中共有9个GRF成员[8], 单子叶植物模式植物水稻中共有12个GRF成员[13]。相关研究表明, 这些GRF基因在生长发育活跃的组织中表达水平较高, 如茎尖、花芽、未成熟的叶片等, 但在成熟的组织或器官中表达量相对较低, 其中多种GRF基因参与了叶片原基细胞增殖的调控和茎尖分生组织器官的分离[14,15,16,17,18,19]。在拟南芥[20,21]和毛果杨[22]中发现GRF基因对花的发育起重要的调控作用; 有些GRF基因还参与拟南芥雌性生殖器官的发育和胚珠的形成[23], 说明它们可能与种子的发育有关。在油菜中, BnGRF2基因能通过调控细胞数目和光合作用来增加油菜籽产油量[24]。
GRF蛋白主要是通过与GIF互作形成蛋白复合体参与这些生物学过程[25,26,27,28], 而GRF的功能有时又会受miR396的调控, 比如拟南芥中有7个AtGRF基因是miR396的靶基因, 其表达受到miR396的调控, 进而对拟南芥根、茎、叶的生长发育产生影响[14]。GRF的表达量越高, 并不是越利于植物的生长发育, 如水稻OsGRF1过量表达引起植株多种生理缺陷, 包括叶片卷曲、开花延迟和心皮发育不完善[11]; 玉米ZmGRF1过表达虽能增加叶片大小, 但降低了玉米可育性[29], ZmGRF10过表达减小了叶片面积[30]; 拟南芥AtGRF9超表达植株的叶片生长受到明显抑制[31]。
藜麦(Chenopodium quinoa Willd.)是一种全营养谷物, 富含人体必需的9种氨基酸[32], 被联合国粮农组织(FAO)认定为一种单体植物即可满足人体基本营养需求的食物[33], 并且藜麦对多种非生物胁迫如盐碱、干旱、霜冻等具有一定的耐受性[34,35,36]。因此, 藜麦已引起了研究人员和消费者的广泛关注。2017年公布了藜麦高质量参考基因组, 促进了藜麦功能基因组学的研究[37]。本研究利用生物信息学手段, 以拟南芥GRF基因家族成员的蛋白质序列为参考, 在藜麦基因组数据库通过BlastP序列比对, 获得藜麦GRF转录因子家族全部成员, 并对其理化性质、基因结构、保守结构域和系统发育关系以及根据RNA-seq数据对藜麦GRF基因在不同组织的表达模式进行分析, 旨在为进一步研究藜麦GRF转录因子的功能及GRF影响植物生长发育的调控机制提供理论指导。
1 材料与方法
1.1 基因组数据和转录组数据的来源
藜麦全基因组数据从Phytozome v12 (1.2 藜麦GRF基因家族成员的鉴定及分析
从拟南芥数据库获取AtGRF基因家族成员的蛋白序列, 并以此为查询对象在藜麦基因组数据库进行Blastp比对, 设E值为10-5, 将得到的藜麦蛋白序列在Pfam (将获取的藜麦GRF基因的氨基酸序列输入ExPasy网站(
1.3 系统进化树的构建
利用ClustalW对藜麦、拟南芥、水稻GRF的氨基酸序列进行多重比对, 使用MEGA 5.05软件及邻接法(Neighbor-Joining, NJ)(bootstrap = 1000)构建无根进化树。1.4 藜麦GRF基因结构分析
根据藜麦GRF基因家族成员的基因组DNA序列及CDS序列, 利用GSDS 2.0 (1.5 藜麦GRF家族成员保守结构域分析
在Pfam分析得到藜麦GRF转录因子保守结构域的氨基酸序列起始位置, 利用IBS 1.0绘制结构域位置图, 并利用ClustalX 2.0软件对结构域氨基酸序列进行比对。1.6 藜麦GRF基因的组织表达分析
从NCBI下载藜麦生长发育过程中各组织器官的RNA-Seq数据, 包括苗龄为6周的藜麦植株的根、茎、叶片、花序和幼苗以及0周龄的成熟干种子, 以此来分析藜麦GRF基因的表达情况。对各成员的表达量取log2TPM, 然后用R绘制藜麦GRF基因家族的组织表达热图。1.7 基因的荧光定量表达分析
采用实时荧光定量PCR进一步确定从表达图谱中筛选出来的组织表达特异性较为明显的6个GRF基因的表达模式, 所取样品与RNA-Seq所选样品一致。使用TIANGEN试剂盒提取RNA, 用Takara的反转录试剂盒合成cDNA第1条链, 以各样品的cDNA为模板, 浓度统一稀释为100 ng μL-1, 进行定量PCR扩增, EF1a为内参基因[38], 各基因特异引物序列见表1。反应体系为20 μL, 包含10 μL SYBR green supermix、5.5 μL ddH2O、正反向引物各1 μL、2.5 μL cDNA。反应条件为95℃预变性10 min; 95℃变性10 s, 60℃退火30 s, 40个循环。采用2-ΔΔCt法计算目的基因的相对表达量。Table 1
表1
表1定量PCR引物序列
Table 1
| 基因 Gene | 正向引物 Forward primer (5°-3°) | 反向引物 Reverse primer (5°-3°) |
|---|---|---|
| EF1a | GTACGCATGGGTGCTTGACAAACTC | ATCAGCCTGGGAGGTACCAGTAAT |
| AUR62001481 | TCCCACCTGAACTCTTCTCTGCT | ACATCTTCCTGGCTCTGGGTCT |
| AUR62002094 | GGAGGCATGGTCGGGTTTGG | GGGATCGTGGACTGGCTGAG |
| AUR62009885 | CCCTTTCACAGCAAGTCAATGGC | AGAGAAGAGTCCAGAAGAAGAGGAAGT |
| AUR62028212 | TGCGCCAAAGAAGCCTACCC | GACGACACAGAAGCCGTCGT |
| AUR62033894 | TAGTGGTGCCTGGTGGTGGT | GCCTTGCATTCCCTGACTGCT |
| AUR62043106 | ACCGGGAAACCCAACATCGG | CCCACCGGCACTCCAATTCA |
新窗口打开|下载CSV
2 结果与分析
2.1 藜麦GRF基因家族成员的鉴定及分析
把拟南芥GRF蛋白序列在藜麦数据库进行Blastp比对, 对所获得的蛋白序列在Pfam数据库进行结构域分析, 筛选出具有QLQ或WRC保守结构域的蛋白序列, 最终鉴定出18个藜麦GRF候选基因(表2)。Table 2
表2
表2藜麦GRF基因家族成员基本信息
Table 2
| 基因名称 Gene name | Scaffold位置及基因方向 Scaffold location and gene direction (bp) | 蛋白长度 Length (aa) | 分子量 MW (kDa) | 等电点 pI |
|---|---|---|---|---|
| AUR62001481 | C_Quinoa_2716: 5162638-5164244 (+) | 248 | 28.17 | 9.15 |
| AUR62002094 | C_Quinoa_4480: 2028959-2032889 (-) | 331 | 36.64 | 8.84 |
| AUR62004030 | C_Quinoa_2370: 7633423-7637087 (-) | 570 | 61.48 | 8.25 |
| AUR62004236 | C_Quinoa_4250: 1136571-1139467 (+) | 362 | 40.73 | 9.32 |
| AUR62006018 | C_Quinoa_1001: 1290301-1292042 (-) | 273 | 29.95 | 5.23 |
| AUR62007068 | C_Quinoa_1971: 832455-834729 (+) | 369 | 39.86 | 8.41 |
| AUR62007538 | C_Quinoa_2646: 201458-205187 (-) | 621 | 67.38 | 8.00 |
| AUR62009885 | C_Quinoa_2493: 4358019-4358318 (-) | 77 | 8.81 | 9.18 |
| AUR62013612 | C_Quinoa_1412: 163344-166379 (-) | 374 | 41.77 | 9.37 |
| 基因名称 Gene name | Scaffold位置及基因方向 Scaffold location and gene direction (bp) | 蛋白长度 Length (aa) | 分子量 MW (kD) | 等电点 pI |
| AUR62019933 | C_Quinoa_1480: 2623570-2625376 (+) | 287 | 30.90 | 6.25 |
| AUR62024537 | C_Quinoa_2876: 5196136-5201404 (+) | 541 | 58.33 | 9.11 |
| AUR62025191 | C_Quinoa_4329: 621270-623166 (-) | 302 | 33.61 | 6.59 |
| AUR62028212 | C_Quinoa_2933: 1647993-1651942 (-) | 325 | 36.02 | 8.69 |
| AUR62028983 | C_Quinoa_2412: 1335673-1337123 (+) | 309 | 34.31 | 6.45 |
| AUR62033547 | C_Quinoa_1776: 1206319-1208711 (-) | 268 | 28.79 | 7.79 |
| AUR62033894 | C_Quinoa_2654: 4420101-4424297 (+) | 532 | 57.33 | 8.97 |
| AUR62035608 | C_Quinoa_2193: 618326-619612 (+) | 283 | 30.81 | 5.43 |
| AUR62043106 | C_Quinoa_1071: 168739-171090 (-) | 270 | 29.22 | 6.75 |
新窗口打开|下载CSV
利用ExPasy对藜麦GRF基因的理化性质分析表明, 不同GRF转录因子的蛋白序列存在较大差异, 氨基酸长度77~621 aa, 蛋白分子量8.81~67.38 kDa, 其中均以AUR62007538最大, AUR62009885最小。等电点以AUR62013612最大, 为9.37, AUR62006018最小, 为5.23; 其中等电点小于7的GRF蛋白有6个, 偏酸性; 其余的12个GRF蛋白等电点均大于7, 偏碱性。
2.2 藜麦GRF基因家族系统发育关系分析
为了更好地了解藜麦GRF基因家族的系统发育关系, 将藜麦(18个GRF基因)、拟南芥(9个GRF基因)和水稻(12个GRF基因)的GRF蛋白序列进行多重比对, 构建无根进化树(图1)。根据进化树分支, 将39个GRF转录因子划分为Class I、II、III、IV四类, 其中Class I和Class IV均有6个藜麦GRF基因, Class II有4个藜麦GRF基因, Class III存在2个藜麦GRF基因。Class III和Class IV只有藜麦和拟南芥GRF分布, 不存在水稻GRF基因, 并且在其他两类中表现出拟南芥与藜麦GRF进化关系相对较近一些, 说明藜麦与拟南芥的GRF转录因子亲缘关系比水稻更近。Class III只有3个基因AtGRF8、AUR62004326和AUR62013612, 说明这2个藜麦GRF基因与拟南芥AtGRF8具有较高的同源性, 在功能上可能也具有相似性。图1
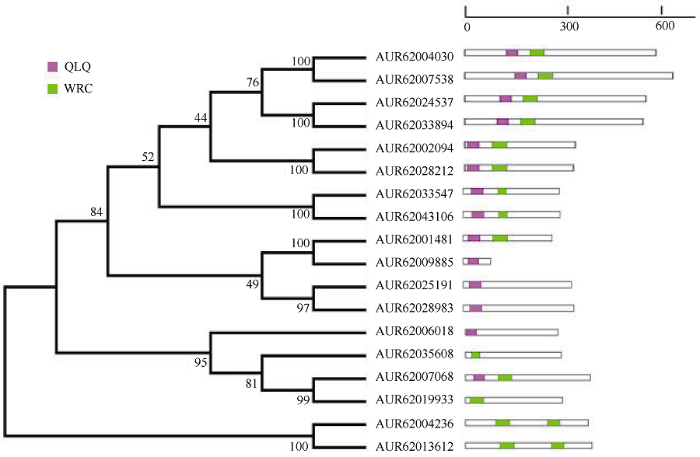 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1藜麦、拟南芥和水稻GRF基因家族系统进化树
Fig. 1Phylogenetic tree of GRF gene family in quinoa, Arabidopsis, and rice
2.3 藜麦GRF基因结构分析
如图2所示, 不同GRF基因的结构存在一定的差异, 除AUR62001481、AUR62009885和AUR62019933具有2个外显子和1个内含子外, 其他成员均含有2~4个内含子及3~5个外显子; 根据目前藜麦基因组注释检测到, 6个GRF基因同时具有5°-UTR(非翻译区)和3°-UTR, 4个GRF基因只含有3°-UTR, 其余8个GRF基因没有非翻译区。图2
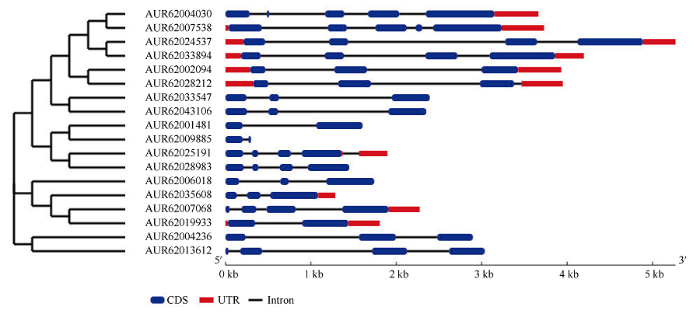 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2藜麦GRF基因家族进化树与基因结构
Fig. 2Phylogenetic tree and gene structure of quinoa GRF gene family
18个GRF基因形成了8个旁系同源基因, 同源基因的结构具有一定的相似性(图2)。如AUR62033547和AUR62043106为一对同源基因, 都含有3个外显子和2个内含子, 基因长度也相似; AUR62024537和AUR62033894为一对同源基因, 均具有4个外显子、3个内含子、5°-UTR及3°-UTR, 但第2个内含子长度差异较大。
2.4 藜麦GRF基因家族保守结构域分析
藜麦GRF转录因子蛋白序列在Pfam上分析结果显示, 18个GRF蛋白均含有QLQ或WRC结构域, 并且两结构域均位于蛋白的N端(图3)。其中, AUR62009885、AUR62025191、AUR62028983和AUR62006018只具有QLQ结构域, AUR62035608、AUR62019933、AUR62004236和AUR62013612只具有WRC结构域, 其他10个GRF转录因子同时具有QLQ和WRC结构域。从图3可以看出, 藜麦GRF同源基因对的蛋白长度及结构域位置有很高的相似性。图3
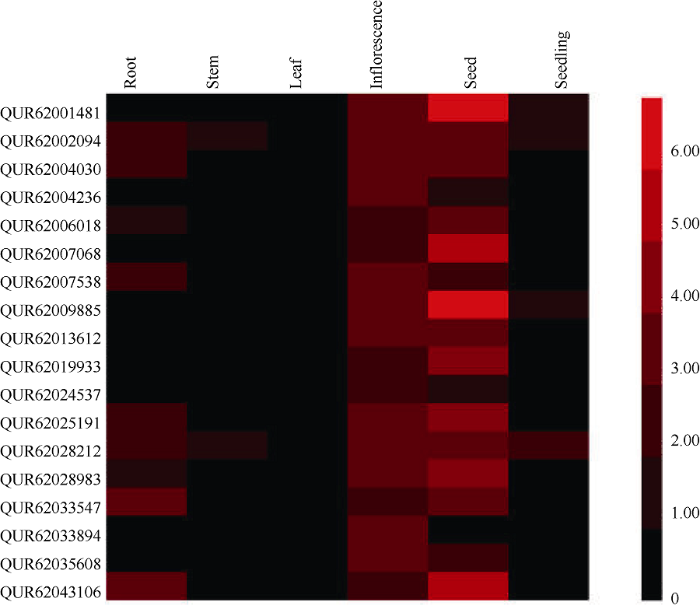 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3藜麦GRF蛋白保守结构域分析
Fig. 3Analysis of conserved domains of GRF proteins in quinoa
为了进一步分析藜麦GRF转录因子QLQ和WRC结构域的保守性, 对2个结构域的氨基酸序列进行多重比对, 发现QLQ结构域由31~35个氨基酸组成, WRC结构域由25~43个氨基酸组成, 并且两结构域中均有多个氨基酸位点是保守不变的(图4)。QLQ结构域的氨基酸序列为QX3LX2Q; WRC结构域含有一个跨越3个半胱氨酸C和1个组氨酸H的C3H模体, 其氨基酸序列为CX9CX10CX2H; 在藜麦中, WRC结构域的保守性比QLQ结构域更高一些。QLQ和WRC结构域中这些氨基酸保守区域可能通过与其他因子相互结合或相互作用, 从而影响藜麦GRF转录因子的激活与抑制。
图4
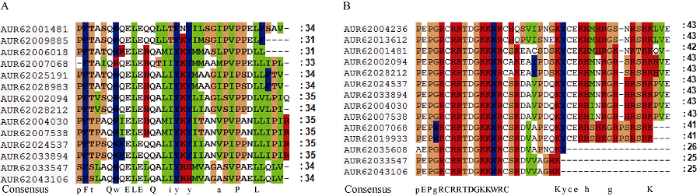 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4藜麦GRF蛋白QLQ (A)和WRC (B)结构域的氨基酸序列比对
Fig. 4Amino acid sequence alignment of QLQ (A) and WRC (B) domain of quinoa GRF proteins
2.5 藜麦GRF基因家族的组织表达分析
为初步分析藜麦GRF基因的功能, 利用RNA-Seq转录组数据对18个GRF基因在6个不同组织器官的表达情况进行分析, 包括6周龄的根、茎、叶片、花序和幼苗以及0周龄的成熟干种子。结果表明, 不同GRF基因在藜麦不同组织器官中的表达量存在明显差异(图5)。除AUR62033894在种子中不表达外, 其他GRF基因均在种子中有较高的表达量, 说明这些基因可能与藜麦种子的发育有重要关系; 各GRF基因在花序中均有表达, 表明它们可能在花序的发育过程中起着重要作用; 有9个GRF基因在根中不表达, 说明它们可能没有参与藜麦根系的发育过程; 所有GRF基因在叶片中均不表达, 可能是因为GRF基因与藜麦叶片的发育关系不大; 除AUR62002094和AUR62028212外, 其他GRF基因均在茎中不表达, 说明这2个基因可能参与了藜麦茎的发育过程; 只有AUR62001481、AUR62002094、AUR62009885和AUR62028212在幼苗中有一定的表达量, 它们可能调控着藜麦幼苗的生长。综上所述, 藜麦GRF基因具有明显的组织表达特异性, 在种子中的表达量相对较高, 其次是花序和根, 在其他组织中的表达量相对较低, 揭示它们在藜麦不同组织中可能具有不同的生物学功能。图5
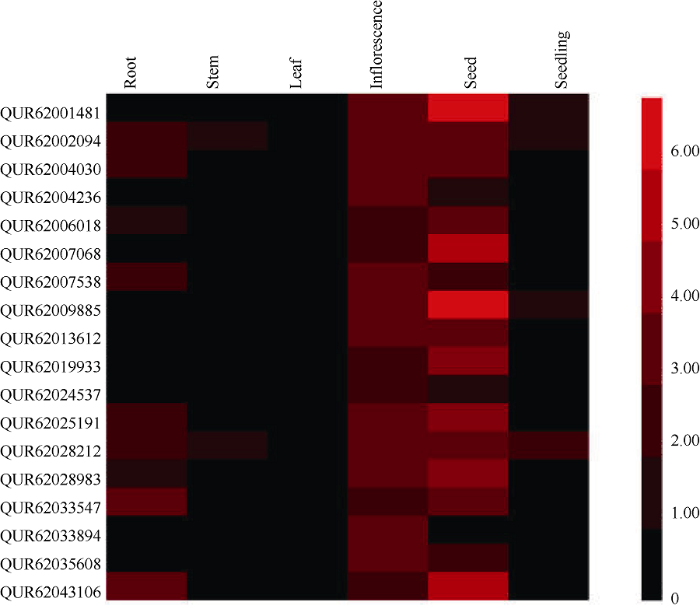 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5GRF基因在藜麦不同组织中的表达模式分析
Fig. 5Expression profile analysis of GRF genes in different tissues of quinoa
为了进一步确定GRF基因在藜麦不同组织器官中具有明显的表达特异性, 从表达图谱中挑选6个GRF基因并对其进行实时荧光定量PCR分析以确定其相应的表达模式。由图6可知, AUR62001481在种子中的表达量显著高于其他组织, 其次是花序, 在根、茎、叶和幼苗中几乎不表达; AUR62002094和AUR62028212有着相同的组织表达模式, 在种子中的表达量相对较高, 其次是幼苗和花序, 在根、茎、叶中的表达量相对较低; AUR62009885和AUR62043106有着相似的组织表达模式, 在根中的表达量最高, 其次是幼苗, 在其他组织中表达量相对较低甚至不表达; AUR62033894在花序中的表达量最高, 其次是根和幼苗但表达量均相对较低, 在茎、叶和种子中几乎不表达。定量PCR分析结果与上述表达图谱结果基本一致。
图6
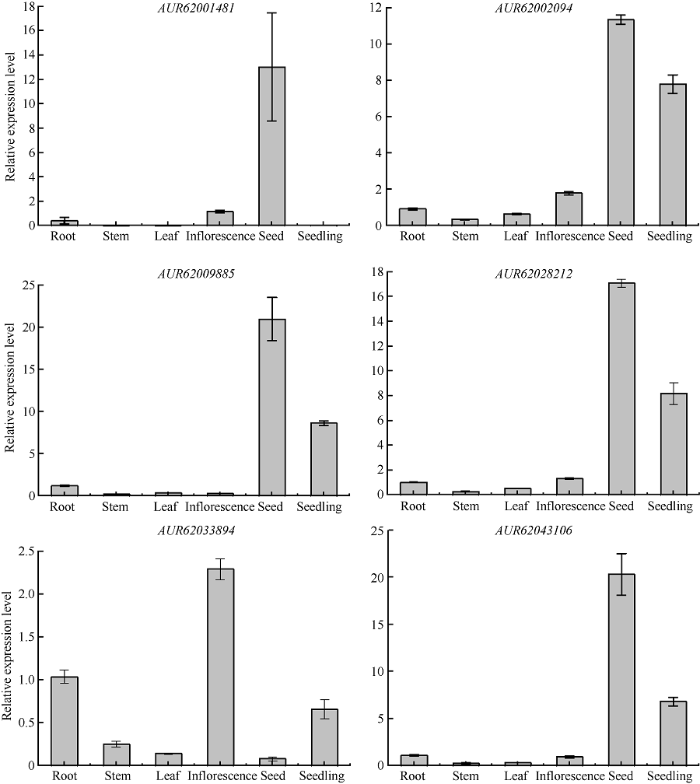 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图66个GRF基因在藜麦不同组织的表达
Fig. 6Expression level of six GRF genes in different tissues of quinoa
3 讨论
随着基因组测序技术的逐渐完善和分子生物学研究的不断深入, 大量植物全基因组测序已经完成并陆续公布了其基因组数据库, 这为鉴定植物基因家族、挖掘功能基因及研究基因功能等提供了极为有利的条件。GRF转录因子是植物特有的一类转录因子, 在植物的生长发育过程中发挥着重要的调控作用。目前, GRF基因家族在拟南芥[8]、玉米[39]、水稻[13]、番茄[40]、大白菜[5]、大豆[41]等多种植物中已有研究。藜麦作为未来最具潜力的农作物之一, 富含极高的营养价值, 但关于其GRF基因家族的研究至今仍缺乏报道。关于模式植物拟南芥和水稻GRF基因功能的研究较为深入。AtGRF5基因通过调控叶绿体的分裂来增加叶片面积和叶绿素含量, 从而增强植物的光合作用以促进植物生长发育[42]; AtGRF7是多种渗透胁迫响应基因的抑制因子, 通过与脱水反应元件结合蛋白DREB2A的启动子区域结合, 从而抑制该基因及其下游一系列渗透应激反应基因的表达, 可在正常生长条件下防止植物生长抑制[43]。过表达OsGRF6、OsGRF7、OsGRF8、OsGRF9的转基因水稻植株均增强稻瘟病的抗性, 但各自生长变化不同, OsGRF7过表达导致植株生长缺陷, 而OsGRF6、OsGRF8、OsGRF9过表达则导致水稻产量性状发生较好变化[44]。研究表明, OsGRF4能够调控2种细胞分裂素脱氢酶前体基因CKX1和CKX5的表达, 导致细胞分裂素水平升高, 从而增加穗长度, 此外, OsGRF4过量表达可增加籽粒硬度[45], OsGRF4与miR396c、OsGIF1互作, 从而调控水稻籽粒大小[46], 对水稻高产育种和机械化收获具有重要意义。本研究利用生物信息学手段, 从藜麦全基因组数据库中共鉴定出18个GRF基因家族成员, 比拟南芥(9个)和水稻(12个)成员数目更多, 这可能是由于异源四倍体植物藜麦在进化过程中发生了全基因组复制和串联复制而导致GRF基因数目增加[47], 基因扩张同时也增强了藜麦对外界复杂多变环境的适应能力。藜麦GRF转录因子的蛋白长度、分子量、等电点等基本特性存在较大差异; 由于内含子的不断插入使其基因结构也发生一定程度的变化, 每个成员含有1~4个内含子和2~5个外显子, 部分GRF基因含有非翻译区。氨基酸序列分析表明, 大部分成员同时具有QLQ和WRC两个保守结构域, 少部分成员只具有其中一种结构域, 其中, QLQ结构域的基序为QX3LX2Q, WRC结构域的基序为CX9CX10CX2H, WRC结构域的保守性比QLQ结构域更高; QLQ和WRC结构域中这些氨基酸保守区域可能通过与其他因子相互结合或相互作用, 从而影响藜麦GRF蛋白的转录激活与抑制。对藜麦、拟南芥与水稻GRF基因家族构建系统进化树, 可将其分为4个亚家族, 其中藜麦和拟南芥的GRF广泛分布于每个亚家族, 水稻的GRF基因只存在于Class I和Class II, 表明GRF基因的进化在双子叶植物和单子叶植物中存在差异, 藜麦与拟南芥GRF转录因子的亲缘关系比水稻更为密切。
藜麦GRF基因主要在特定的组织和器官中表达, 可能在这些组织或器官的生长发育中发挥着重要的作用。本研究发现, AUR62001481、AUR62002094、AUR62009885、AUR62028212和AUR62043106在藜麦种子中的表达量均显著高于其他检测组织, 说明这些基因主要参与了藜麦种子的生长发育, 因此, 提高这些基因的表达水平可能有利于增加藜麦的产量。同源性越高的基因, 其功能越相似。例如, AtGRF8与拟南芥花的发育有关[48], AUR62013612和AUR62004236均与AtGRF8的同源性较高, 2个基因在藜麦花序中的表达量又相对较高, 说明AUR62013612和AUR62004236可能参与了藜麦花序的发育过程。水稻的OsGRF4基因与籽粒大小和籽粒硬度有关[45,46], 其在藜麦中同源性较高的基因为AUR62002094和AUR62028212, 二者在种子中的表达量均显著高于其他组织, 说明这2个基因可能与藜麦产量性状有关。在后续的研究中还需要利用转基因技术或基因沉默技术对这些基因的功能和作用机制进一步验证。
4 结论
通过对藜麦GRF转录因子家族全基因组鉴定, 共获得18个成员, 分属4个不同亚家族, 具有典型的QLQ或WRC保守结构域, 基因表达具有明显的组织表达特异性, 在种子和花序中表达量较高, 与藜麦籽实产量紧密相关。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/S1360-1385(98)01302-8URL [本文引用: 1]
DOI:10.1007/s11105-014-0835-9URL [本文引用: 1]
DOI:10.1093/jxb/erv349URL [本文引用: 1]
[本文引用: 1]
URL [本文引用: 2]
DOI:10.1111/tpj.12567URL [本文引用: 1]

The growth-regulating factors (GRFs) are plant-specific transcription factors. They form complexes with GRF-interacting factors (GIFs), a small family of transcriptional co-activators. In Arabidopsis thaliana, seven out of the nine GRFs are controlled by microRNA miR396. Analysis of Arabidopsis plants carrying a GRF3 allele insensitive to miR396 revealed a strong boost in the number of cells in leaves, which was further enhanced synergistically by an additional increase of GIF1 levels. Genetic experiments revealed that GRF3 can still increase cell number in gif1 mutants, albeit to a much lesser extent. Genome-wide transcript profiling indicated that the simultaneous increase of GRF3 and GIF1 levels causes additional effects in gene expression compared to either of the transgenes alone. We observed that GIF1 interacts in vivo with GRF3, as well as with chromatin-remodeling complexes, providing a mechanistic explanation for the synergistic activities of a GRF3-GIF1 complex. Interestingly, we found that, in addition to the leaf size, the GRF system also affects the organ longevity. Genetic and molecular analysis revealed that the functions of GRFs in leaf growth and senescence can be uncoupled, demonstrating that the miR396-GRF-GIF network impinges on different stages of leaf development. Our results integrate the post-transcriptional control of the GRF transcription factors with the progression of leaf development.
DOI:10.1016/j.molp.2015.01.013URL [本文引用: 1]
DOI:10.1046/j.1365-313X.2003.01862.xURL [本文引用: 4]
DOI:10.1105/tpc.15.00452URL [本文引用: 1]
DOI:10.1073/pnas.0405450101URL [本文引用: 1]
DOI:10.1104/pp.122.3.695URL [本文引用: 2]
[本文引用: 1]
DOI:10.1093/pcp/pch098URL [本文引用: 2]
DOI:10.1242/dev.043067URL [本文引用: 2]
DOI:10.1111/tpj.2005.43.issue-1URL [本文引用: 1]
DOI:10.1007/BF03031127URL [本文引用: 1]

Previously, we identified aGROWTH-REGULATING FACTOR gene family, comprising nine members, which encodes putative transcription factors inArabidopsis thaliana. Thegrf1 grf2 grf3 triple mutants produced partially fused cotyledons and developed small leaves due to a reduction in cell numbers. To understand the functional role of another member of this gene family,GRF4, we have now identified agrf4 null mutant and constructed a quadruple mutant by crossing it to thegrf triple mutant. The quadruple mutant has much smaller leaves than its parental mutants, with this reduced size again due to fewer cells, interestingly, the quadruple mutant displays not only a much stronger fusion of cotyledons but also the phenotype of theshoot meristemless mutant. The aberrant cotyledons of the quadruple mutants result from a fusion of cotyledon primordia during embryogenesis. These results suggest that GRF4 is required for both leaf cell proliferation and the embryonic development of cotyledons and the shoot apical meristem.
DOI:10.1111/ppl.2009.136.issue-2URL [本文引用: 1]
DOI:10.1080/15592324.2016.1184809URL [本文引用: 1]
DOI:10.4161/15592324.2014.988071URL [本文引用: 1]
DOI:10.1007/s12374-009-9061-7URL [本文引用: 1]

MicroRNAs (miRNAs) are single-stranded, noncoding small RNAs that usually function as posttranscriptional negative regulators by base pairing to target genes. They are pivotal to plant development. MiR396 is conserved among plant species and is predicted to target GRF (growth-regulating factor) genes in Arabidopsis. Here, overexpression of ath-miR396 in tobacco reduced the levels of three NtGRF-like genes containing an miR396 match site. Furthermore, its elevated expression resulted in a small, narrow leaf phenotype similar to that found with the Arabidopsis grf1grf2grf3 triple mutant. We also demonstrated that 35S:MIR396a transgenic plants were defective in the four whorls of floral organs. These results provide a link between the miR396-mediated regulatory pathway of NtGRF-like gene expression and the developmental processes for leaves and flowers in tobacco.
DOI:10.1104/pp.113.225144URL [本文引用: 1]

The precise control of gene regulation, and hence, correct spatiotemporal tissue patterning, is crucial for plant development. Plant microRNAs can constrain the expression of their target genes at posttranscriptional levels. Recently, microRNA396 (miR396) has been characterized to regulate leaf development by mediating cleavage of its GROWTH-REGULATING FACTOR (GRF) targets. miR396 is also preferentially expressed in flowers. However, its function in flower development is unclear. In addition to narrow leaves, pistils with a single carpel were also observed in miR396 overexpression plants. The dramatically reduced expression levels of miR396 targets (GRF1, GRF2, GRF3, GRF4, GRF7, GRF8, and GRF9) caused pistil abnormalities, because the miR396-resistant version of GRF was able to rescue miR396-overexpressing plants. Both GRF and GRF-INTERACTING FACTOR (GIF) genes are highly expressed in developing pistils, and their expression patterns are negatively correlated with that of miR396. GRF interacted with GIF to form the GRF/GIF complex in plant cell nucleus. miR396 suppressed the expression of GRF genes, resulting in reduction of GRF/GIF complex. gif single mutant displayed normal pistils, whereas gif triple mutant gif1/gif2/gif3 produced abnormal pistils, which was a phenocopy of 35S:MIR396a/grf5 plants. GRF and GIF function as cotranscription factors, and both are required for pistil development. Our analyses reveal an important role for miR396 in controlling carpel number and pistil development via regulation of the GRF/GIF complex.
DOI:10.1111/plb.2013.15.issue-5URL [本文引用: 1]
DOI:10.1371/journal.pone.0026231URL [本文引用: 1]
DOI:10.1093/jxb/ers066URL [本文引用: 1]

Seed yield and oil content are two important agricultural characteristics in oil crop breeding, and a lot of functional gene research is being concentrated on increasing these factors. In this study, by differential gene expression analyses between rapeseed lines (zy036 and 51070) which exhibit different levels of seed oil production, BnGRF2 (Brassica napus growth-regulating factor 2-like gene) was identified in the high oil-producing line zy036. To elucidate the possible roles of BnGRF2 in seed oil production, the cDNA sequences of the rapeseed GRF2 gene were isolated. The Blastn result showed that rapeseed contained BnGRF2a/2b which were located in the A genome (A1 and A3) and C genome (Cl and C6), respectively, and the dominantly expressed gene BnGRF2a was chosen for transgenic research. Analysis of 35S-BnGRF2a transgenic Arabidopsis showed that overexpressed BnGRF2a resulted in an increase in seed oil production of >50%. Moreover, BnGRF2a also induced a >20% enlargement in extended leaves and >40% improvement in photosynthetic efficiency because of an increase in the chlorophyll content. Furthermore, transcriptome analyses indicated that some genes associated with cell proliferation, photosynthesis, and oil synthesis were up-regulated, which revealed that cell number and plant photosynthesis contributed to the increased seed weight and oil content. Because of less efficient self-fertilization induced by the longer pistil in the 35S-BnGRF2a transgenic line, Napin-BnGRF2a transgenic lines were further used to identify the function of BnGRF2, and the results showed that seed oil production also could increase >40% compared with the wild-type control. The results suggest that improvement to economically important characteristics in oil crops may be achieved by manipulation of the GRF2 expression level.
DOI:10.1104/pp.109.141838URL [本文引用: 1]
[本文引用: 1]
DOI:10.1105/tpc.113.115907URL [本文引用: 1]

The transcriptional coactivator ANGUSTIFOLIA3 (AN3) stimulates cell proliferation during Arabidopsis thaliana leaf development, but the molecular mechanism is largely unknown. Here, we show that inducible nuclear localization of AN3 during initial leaf growth results in differential expression of important transcriptional regulators, including GROWTH REGULATING FACTORs (GRFs). Chromatin purification further revealed the presence of AN3 at the loci of GRF5, GRF6, CYTOKININ RESPONSE FACTOR2, CONSTANS-LIKE5 (COL5), HECATE1 (HEC1), and ARABIDOPSIS RESPONSE REGULATOR4 (ARR4). Tandem affinity purification of protein complexes using AN3 as bait identified plant SWITCH/SUCROSE NONFERMENTING (SWI/SNF) chromatin remodeling complexes formed around the ATPases BRAHMA (BRM) or SPLAYED. Moreover, SWI/SNF ASSOCIATED PROTEIN 73B (SWP73B) is recruited by AN3 to the promoters of GRF5, GRF3, COL5, and ARR4, and both SWP73B and BRM occupy the HEC1 promoter. Furthermore, we show that AN3 and BRM genetically interact. The data indicate that AN3 associates with chromatin remodelers to regulate transcription. In addition, modification of SWI3C expression levels increases leaf size, underlining the importance of chromatin dynamics for growth regulation. Our results place the SWI/SNF-AN3 module as a major player at the transition from cell proliferation to cell differentiation in a developing leaf.
DOI:10.1111/jipb.12220URL [本文引用: 1]

It has long been thought that growth-regulating factors (GRFs) gene family members act as transcriptional activators to play important roles in multiple plant developmental processes. However, the recent characterization of Arabidopsis GRF7 showed that it functions as a transcriptional repressor of osmotic stress-responsive genes. This highlights the complex and diverse mechanisms by which different GRF members use to take action. In this study, the maize (Zea mays L.) GRF10 was functionally characterized to improve this concept. The deduced ZmGRF10 protein retains the N-terminal QLQ and WRC domains, the characteristic regions as protein-interacting and DNA-binding domains, respectively. However, it lacks nearly the entire C-terminal domain, the regions executing transactivation activity. Consistently, ZmGRF10 protein maintains the ability to interact with GRF-interacting factors (GIFs) proteins, but lacks transactivation activity. Overexpression of ZmGRF10 in maize led to a reduction in leaf size and plant height through decreasing cell proliferation, whereas the yield-related traits were not affected. Transcriptome analysis revealed that multiple biological pathways were affected by ZmGRF10 overexpression, including a few transcriptional regulatory genes, which have been demonstrated to have important roles in controlling plant growth and development. We propose that ZmGRF10 aids in fine-tuning the homeostasis of the GRF-GIF complex in the regulation of cell proliferation.
Wu L, Zhang D, Xue M, Qian J, He Y, Wang S (2014) Overexpression of the maize GRF10, an endogenous truncated GRF protein, leads to reduction in leaf size and plant height. J Integr Plant Biol 56:1053–1063. doi: 10.1111/jipb.12220
DOI:10.1105/tpc.15.00269URL [本文引用: 1]
DOI:10.1111/jipb.12220URL [本文引用: 1]

It has long been thought that growth-regulating factors (GRFs) gene family members act as transcriptional activators to play important roles in multiple plant developmental processes. However, the recent characterization of Arabidopsis GRF7 showed that it functions as a transcriptional repressor of osmotic stress-responsive genes. This highlights the complex and diverse mechanisms by which different GRF members use to take action. In this study, the maize (Zea mays L.) GRF10 was functionally characterized to improve this concept. The deduced ZmGRF10 protein retains the N-terminal QLQ and WRC domains, the characteristic regions as protein-interacting and DNA-binding domains, respectively. However, it lacks nearly the entire C-terminal domain, the regions executing transactivation activity. Consistently, ZmGRF10 protein maintains the ability to interact with GRF-interacting factors (GIFs) proteins, but lacks transactivation activity. Overexpression of ZmGRF10 in maize led to a reduction in leaf size and plant height through decreasing cell proliferation, whereas the yield-related traits were not affected. Transcriptome analysis revealed that multiple biological pathways were affected by ZmGRF10 overexpression, including a few transcriptional regulatory genes, which have been demonstrated to have important roles in controlling plant growth and development. We propose that ZmGRF10 aids in fine-tuning the homeostasis of the GRF-GIF complex in the regulation of cell proliferation.
Wu L, Zhang D, Xue M, Qian J, He Y, Wang S (2014) Overexpression of the maize GRF10, an endogenous truncated GRF protein, leads to reduction in leaf size and plant height. J Integr Plant Biol 56:1053–1063. doi: 10.1111/jipb.12220
DOI:10.1371/journal.pgen.1007484URL [本文引用: 1]
DOI:10.1016/j.foodchem.2006.01.016URL [本文引用: 1]
DOI:10.1080/0963748031000084106URL [本文引用: 1]
DOI:10.1111/j.1439-037X.2011.00473.xURL [本文引用: 1]

Drought and salinity are the two major factors limiting crop growth and production in arid and semi-arid regions. The separate and combined effects of salinity and progressive drought in quinoa (Chenopodium quinoa Willd.) were studied in a greenhouse experiment. Stomatal conductance (gs), leaf water potential (Psi(1)), shoot and root abscisic acid concentration ([ABA]) and transpiration rate were measured in full irrigation (FI; around 95 % of water holding capacity (WHC)) and progressive drought (PD) treatments using the irrigation water with five salinity levels (0, 10, 20, 30 and 40 dS m(-1)); the treatments are referred to as FI(0), FI(10), FI(20), FI(30), FI(40); PD(0), PD(10), PD(20), PD(30), PD(40), respectively. The measurements were carried out over 9 days of continuous drought. The results showed that increasing salinity levels decreased the total soil water potential (Psi(T)) and consequently decreased g(s) and Psi(1) values in both FI and PD. During the drought period, the xylem [ABA] extracted from the shoots increased faster than that extracted from the roots. A reduction in Psi(T), caused by salinity and soil drying, reduced transpiration and increased apparent root resistance (R) to water uptake, especially in PD(0) and PD(40) during the last days of the drought period. The reasons for the increase in apparent root resistance are discussed. At the end of the drought period, the minimum value of relative available soil water (RAW) was reached in PD(0). Under non-saline conditions, Psi(1) decreased sharply when RAW reached 0.42 or lower, but under the saline conditions of PD(10) and PD(20), the threshold values of RAW were 0.67 and 0.96, respectively. In conclusion, due to the additive effect of osmotic and matric potential during soil drying on soil water availability, quinoa should be re-irrigated at higher RAW in salt-affected soils, i.e. before the soil water content reaches the critical threshold level causing the drop in Psi(1) resulting in stomatal closure.
DOI:10.1016/j.eja.2004.01.003URL [本文引用: 1]

Frost is one of the principal limiting factors for agricultural production in the high Andean region. One of the most important grain crops in that region, quinoa (Chenopodium quinoa Willd.), is generally less affected by frost than most other crop species, but little is known about its specific mechanisms for frost resistance. This study was undertaken to help understand quinoa’s response to various intensities and durations of frost under different levels of relative humidity (RH). The effect of frost on seed yield and plant death rate was studied, and content of soluble sugars, proteins, and free proline, was analyzed, in order to develop criteria for the selection of cultivars with improved resistance to frost. On the basis of greenhouse and phytotron experiments, it was concluded that at the two-leaf stage, cultivars from the altiplano of Peru, 3800 m above sea level, tolerated −8 °C for 4 h, whereas a cultivar from the Andean valleys tolerated the same temperature for only 2 h. At −4 °C, plant death rate increased from 25% at high relative humidity to 56% at low RH After a frost treatment of −4 °C applied at the two-leaf stage, final seed yield was reduced by 9% compared to control plants not exposed to frost. For the same treatment applied at the 12-leaf and flowering stages, yield reductions were 51 and 66%, respectively, indicating that frost for 2 h or more during anthesis caused significant damage to the plants. In general, an increased level of soluble sugars implied a greater tolerance to frost, resulting in higher yields.
DOI:10.1093/jxb/erq257URL [本文引用: 1]

Ionic and osmotic relations in quinoa (Chenopodium quinoa Willd.) were studied by exposing plants to six salinity levels (0-500 mM NaCl range) for 70 d. Salt stress was administered either by pre-mixing of the calculated amount of NaCl with the potting mix before seeds were planted or by the gradual increase of NaCl levels in the irrigation water. For both methods, the optimal plant growth and biomass was achieved between 100 mM and 200 mM NaCl, suggesting that quinoa possess a very efficient system to adjust osmotically for abrupt increases in NaCl stress. Up to 95% of osmotic adjustment in old leaves and between 80% and 85% of osmotic adjustment in young leaves was achieved by means of accumulation of inorganic ions (Na(+), K(+), and Cl(-)) at these NaCl levels, whilst the contribution of organic osmolytes was very limited. Consistently higher K(+) and lower Na(+) levels were found in young, as compared with old leaves, for all salinity treatments. The shoot sap K(+) progressively increased with increased salinity in old leaves; this is interpreted as evidence for the important role of free K(+) in leaf osmotic adjustment under saline conditions. A 5-fold increase in salinity level (from 100 mM to 500 mM) resulted in only a 50% increase in the sap Na(+) content, suggesting either a very strict control of xylem Na(+) loading or an efficient Na(+) removal from leaves. A very strong correlation between NaCl-induced K(+) and H(+) fluxes was observed in quinoa root, suggesting that a rapid NaCl-induced activation of H(+)-ATPase is needed to restore otherwise depolarized membrane potential and prevent further K(+) leak from the cytosol. Taken together, this work emphasizes the role of inorganic ions for osmotic adjustment in halophytes and calls for more in-depth studies of the mechanisms of vacuolar Na(+) sequestration, control of Na(+) and K(+) xylem loading, and their transport to the shoot.
DOI:10.1038/nature21370URL [本文引用: 1]
URL [本文引用: 1]
DOI:10.1016/j.plantsci.2008.08.002URL [本文引用: 1]
URL [本文引用: 1]
DOI:10.1093/jxb/erz022URL [本文引用: 1]
DOI:10.1104/pp.114.256180URL [本文引用: 1]
DOI:10.1105/tpc.112.100933URL [本文引用: 1]

Arabidopsis thaliana DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) functions as a transcriptional activator that increases tolerance to osmotic and heat stresses; however, its expression also leads to growth retardation and reduced reproduction. To avoid these adverse effects, the expression of DREB2A is predicted to be tightly regulated. We identified a short promoter region of DREB2A that represses its expression under nonstress conditions. Yeast one-hybrid screening for interacting factors identified GROWTH-REGULATING FACTOR7 (GRF7). GRF7 bound to the DREB2A promoter and repressed its expression. In both artificial miRNA-silenced lines and a T-DNA insertion line of GRF7, DREB2A transcription was increased compared with the wild type under nonstress conditions. A previously undiscovered cis-element, GRF7-targeting cis-element (TGTCAGG), was identified as a target sequence of GRF7 in the short promoter region of DREB2A via electrophoretic mobility shift assays. Microarray analysis of GRF7 knockout plants showed that a large number of the upregulated genes in the mutant plants were also responsive to osmotic stress and/or abscisic acid. These results suggest that GRF7 functions as a repressor of a broad range of osmotic stress-responsive genes to prevent growth inhibition under normal conditions.
[本文引用: 1]
DOI:10.1111/jipb.12473URL [本文引用: 2]

Traits such as grain shape, panicle length and seed shattering, play important roles in grain yield and harvest. In this study, the cloning and functional analysis of PANICLE TRAITS 2 (PT2), a novel gene from the Indica rice Chuandali (CDL), is reported. PT2 is synonymous with Growth-Regulating Factor 4 (OsGRF4), which encodes a growth-regulating factor that positively regulates grain shape and panicle length and negatively regulates seed shattering. Higher expression of OsGRF4 is correlated with larger grain, longer panicle and lower seed shattering. A unique OsGRF4 mutation, which occurs at the OsmiRNA396 target site of OsGRF4, seems to be associated with high levels of OsGRF4 expression, and results in phenotypic difference. Further research showed that OsGRF4 regulated two cytokinin dehydrogenase precursor genes (CKX5 and CKX1) resulting in increased cytokinin levels, which might affect the panicle traits. High storage capacity and moderate seed shattering of OsGRF4 may be useful in high-yield breeding and mechanized harvesting of rice. Our findings provide additional insight into the molecular basis of panicle growth.
DOI:10.1111/pbi.2016.14.issue-11URL [本文引用: 2]
[本文引用: 1]
DOI:10.1104/pp.15.00307URL [本文引用: 1]
