 ,*广州大学生命科学学院, 广东广州 510006
,*广州大学生命科学学院, 广东广州 510006Creation of high oleic acid soybean mutation plants by CRISPR/Cas9
HOU Zhi-Hong, WU Yan, CHENG Qun, DONG Li-Dong, LU Si-Jia, NAN Hai-Yang, GAN Zhuo-Ran, LIU Bao-Hui ,*School of Life Sciences, Guangzhou University, Guangzhou 510006, Guangdong, China
,*School of Life Sciences, Guangzhou University, Guangzhou 510006, Guangdong, China通讯作者:
收稿日期:2018-11-21接受日期:2019-01-19网络出版日期:2019-03-16
| 基金资助: |
Received:2018-11-21Accepted:2019-01-19Online:2019-03-16
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (3352KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
侯智红, 吴艳, 程群, 董利东, 芦思佳, 南海洋, 甘卓然, 刘宝辉. 利用CRISPR/Cas9技术创制大豆高油酸突变系[J]. 作物学报, 2019, 45(6): 839-847. doi:10.3724/SP.J.1006.2019.84157
HOU Zhi-Hong, WU Yan, CHENG Qun, DONG Li-Dong, LU Si-Jia, NAN Hai-Yang, GAN Zhuo-Ran, LIU Bao-Hui.
大豆(Glycine max L. Merrill)起源于中国, 是世界上重要的油料和经济作物之一, 也是人类植物油和植物性蛋白的主要来源。随着生活水平的提高以及饮食结构的改善, 人们对优质大豆油的需求日益增长, 培育高品质大豆成为大豆育种的重要目标之一。大豆油约占我国植物食用油消费的40%, 大豆脂肪酸的组分及其配比决定大豆油脂的品质[1], 其主要由棕榈酸(16:0)、硬脂酸(18:0)、油酸(18:1Δ9)、亚油酸(18:2Δ9,12)和亚麻酸(18:3Δ9,12,15) 5种脂肪酸组成, 分别约占大豆脂肪酸总量的10%、4%、18%、55%和13% [2,3]。其中, 硬脂酸、棕榈酸为饱和脂肪酸; 油酸、亚油酸、亚麻酸为不饱和脂肪酸[4]。饱和脂肪酸不容易被人体消化吸收, 易引起肥胖症及脑血管疾病[5]。亚油酸、亚麻酸是多不饱和脂肪酸, 稳定性差, 在高温加工时易氧化, 使营养价值降低, 影响油的品质, 在工业上的加氢反应也易产生对人体有害的反式脂肪酸[6]。而油酸作为一种单不饱和脂肪酸, 性质稳定, 抗氧化作用强, 在人体的脂类代谢中能降低有害胆固醇, 保持有益胆固醇, 从而减缓动脉粥样硬化, 有效预防心血管疾病发生的几率[7,8]。因此, 提高油酸的相对含量, 增加油酸/亚油酸的比值, 已成为大豆品质改良育种的重要目标之一。
所有生物的脂肪酸合成途径基本类似, 但是, 动物和酵母的脂肪酸合成主要在细胞质中进行, 而植物脂肪酸合成主要发生在质体中和内质网上[9]。在脂肪酸合成途径中, Δ12-脂肪酸脱饱和酶(delta- twelve fatty acid desaturase 2 enzyme, FAD2)是催化油酸转化为亚油酸的关键酶[10]。目前, 拟南芥[11]、棉花[12]、向日葵[13]、大豆[14]等植物中的FAD2基因已经被克隆。大豆中存在2个FAD2基因, 分别为FAD2-1和FAD2-2, 是2个非等位基因。其中, FAD2-1基因在种子中特异性表达, 是决定种子油酸含量的主要基因[15]。基于稳定性及磷酸化作用不同, FAD2-1基因又分为FAD2-1A和FAD2-1B两种[14]。抑制或过表达FAD2-1A和FAD2-1B基因, 可以显著提高或降低油酸/亚油酸比例[16,17,18]。Li等[16]研究发现, 酵母细胞中不含有亚油酸, 而通过在酵母细胞中过表达GmFAD2-1B基因后, 亚油酸的含量显著提高, 说明GmFAD2-1B基因是亚油酸代谢途径中的关键基因。Pham等[17,18]分别获得FAD2-1A突变体与FAD2-1B突变体, 以及FAD2-1A/FAD2-1B双突变体材料, 并发现无论是单突变还是双突变体中, 大豆的油酸含量均显著提高。Wang等[19]利用RNAi技术, 获得干涉FAD2基因转基因植株, 并发现转基因大豆油酸含量高达71.5%~81.9%。Zhang等[20]和Yang等[21]分别通过RNAi技术, 获得油酸含量为51.71%和27.38%~80.42%的干涉FAD2-1B基因转基因大豆。Haun等[22]利用人工核酶技术定点突变大豆FAD2-1A 和FAD2-1B 基因, 获得油酸含量高达80%的转基因大豆。因此, FAD2-1A与FAD2-1B在决定大豆种子油酸含量中起着重要作用, 是调控油酸含量的2个主要基因[15]。
CRISPR/Cas9系统是近几年开发的一种准确、便捷、高效率的生物基因组编辑方法。通过向导RNA的介导和Cas9蛋白的切割来实现对靶基因的定点编辑[23]。目前, CRISPR/Cas9系统已经成功应用在拟南芥[24]、玉米[25]、小麦[26]、水稻[27,28]、大豆[29]等植物中。Jacobs等[29]第一次利用 CRISPR/ Cas9技术在大豆中实现了定点突变。Cai等[30]利用CRISPR/Cas9技术原理选取3个不同靶位点, 首次应用农杆菌介导的大豆子叶节遗传转化体系, 成功创制出稳定遗传的大豆突变体材料。该技术应用在大豆基因功能研究中起到重要作用, 然而, 应用在大豆育种上的报道较少。本研究利用CRISPR/Cas9技术, 定点编辑了控制油酸含量的关键基因FAD2-1A (Glyma10g42470), 并获得纯合FAD2-1A突变体, 对大豆分子育种和提高大豆品质具有重要意义, 为进一步提高大豆品质奠定了重要的材料基础。
1 材料与方法
1.1 大豆材料、CRISPR/Cas9及gRNA载体
大豆材料华夏3号(简称H3)由华南农业大学和广西壮族自治区农业科学院合作选育, 以桂早1号为母本, 巴西13号为父本经有性杂交和系谱选育而成。该品种田间抗病性较强, 抗倒性强, 耐旱性好, 对酸性低磷土壤有很好的耐性且高产、优质、抗大豆病毒病, 适宜在广东、广西、海南、福建中南部、江西和湖南南部地区夏播种植。pYLgRNA载体是由pUC18载体为骨架改造而成, 大小约3 kb (图1-A)。pYLCRISPR/Cas9-DB载体大小约15.5 kb, 含有一个35S启动子驱动的Bar基因、35S启动子驱动的Cas9基因、终止子、ccdB大肠杆菌致死基因(图1-B)。Cas9载体pYLCRISPR/ Cas9-DB及gRNA载体pYLgRNA-AtU3d/AtU3b/ AtU6-1由华南农业大学刘耀光研究院馈赠[31]。
图1
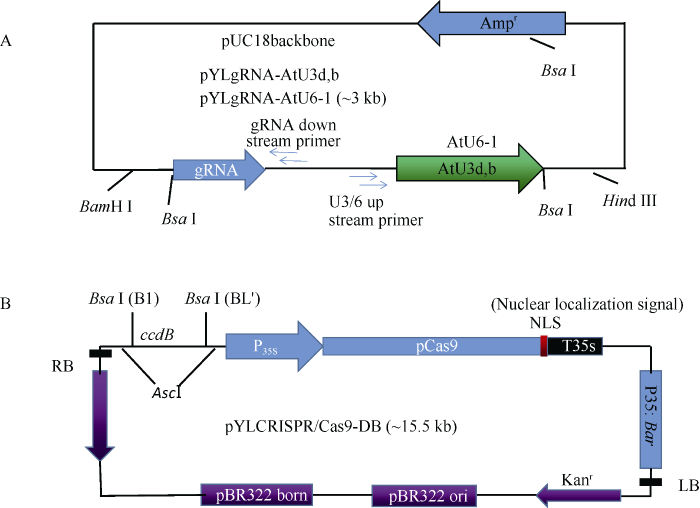 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1pYLgRNA及pYLCRISPR/Cas9-DB质粒图
Fig. 1Maps of pYLgRNA and pYLCRISPR/Cas9-DB vectors
1.2 gRNA靶点选择及其寡核苷酸链引物的设计
根据大豆基因组中GmFAD2-1A基因的外显子序列, 利用CRISPRdirect网站(http://crispr.dbcls.jp/)设计3个长度为20 bp的靶位点序列(图2), 分别为FAD-AtU3d-T1-F1/FAD-AtU3d-T1-R1、FAD-AtU3b- T2-F2/FAD-AtU3b-T2-R2、FAD-AtU6-1-T3-F3/FAD- AtU6-1-T3-R3 (表1)。同时设计CRISPR鉴定引物FAD2-1A-CasF-Test/FAD2-1A-CasR-Test (表1), 所有寡核苷酸链引物均由睿博测序公司合成。图2
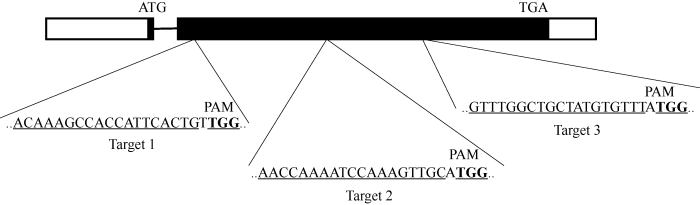 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图23个靶点分别在GmFAD2-1A基因内的位置
左边和右边白色方框代
Fig. 2Positions of three targets in the GmFAD2-1A gene locus
The white boxes on the left and the right represent 5' and 3' noncoding regions; the black box stands for CDS; the solid line represents the intron.
Table 1
表1
表1引物序列
Table 1
| 引物名称 Primer name | 寡核苷酸序列 Oligonucleotide sequence (5'-3') |
|---|---|
| FAD-AtU3d-T1-F1 | gtcACAAAGCCACCATTCACTGT |
| FAD-AtU3d-T1-R1 | aaacACAGTGAATGGTGGCTTTG |
| FAD-AtU3b-T2-F2 | gtcAACCAAAATCCAAAGTTGCA |
| FAD-AtU3b-T2-R2 | aaacTGCAACTTTGGATTTTGGT |
| FAD-AtU6-1-T3-F3 | attGTTTGGCTGCTATGTGTTTA |
| FAD-AtU6-1-T3-R3 | aaacTAAACACATAGCAGCCAAA |
| FAD2-1A-CasF-Test | GAGGGATTGTAGTTCTGTTG |
| FAD2-1A-CasR-Test | CTATGGCCCATTGGTTGCTC |
| U-F | CTCCGTTTTACCTGTGGAATCG |
| gRNA-R | CGGAGGAAAATTCCATCCAC |
| B1’ | TTCAGAggtctcTctcgCACTGGAATCGGCAGCAAAGG |
| B2 | AGCGTGggtctcGtcagGGTCCATCCACTCCAAGCTC |
| B2’ | TTCAGAggtctcTctgaCACTGGAATCGGCAGCAAAGG |
| B3 | AGCGTGggtctcGtcttGGTCCATCCACTCCAAGCTC |
| B3’ | TTCAGAggtctcTaagaCACTGGAATCGGCAGCAAAGG |
| BL | AGCGTGggtctcGaccgGGTCCATCCACTCCAAGCTC |
| Bar-F | TGCCAGTTCCCGTGCTTGAA |
| Bar-R | CTGCACCATCGTCAACCACTA |
新窗口打开|下载CSV
1.3 三靶点pYLCRISPR/Cas9-FAD2-1A-gRNA载体的构建
参照刘耀光研究员提供的实验指南, 方法如下: (1)靶点接头制备。将接头正反向引物用TE buffer稀释到1 μmol L-1, 90℃ 30 s后移至室温冷却完成退火。(2) gRNA载体酶切与连接反应。配制10 μL反应体系, 包括1 μL接头、10 ng gRNA质粒、5 U Bsa I、35 U T4 DNA ligase、1 μL 10×buffer for T4 DNA ligase, PCR反应程序为37℃ 5 min, 20℃ 5 min, 5个循环。(3)扩增gDNA表达盒。第一轮扩增取1 μL连接产物为模板, 适量高保真PCR酶, 通用引物U-F/gRNA-R (表1)各0.2 μmol L-1。PCR反应程序为95℃ 15 s, 55℃ 15 s, 68℃ 10 s, 10个循环; 95℃ 15 s, 60℃ 15 s, 68℃ 10 s, 17~20个循环。各取4 μL电泳检查。第二轮扩增取第一轮PCR产物1 μL稀释20倍, 取1 μL为模板, 各表达盒20~50 μL PCR, 分别利用引物B1’/B2、B2’/B3、B3’/BL (表1)对gDNA进行扩增。95℃ 10 s, 60℃ 15 s, 68℃ 30 s, 18~25个循环。各取2 μL电泳检查产物长度是否符合。(4)产物纯化、酶切和连接。将所有靶点gDNA PCR产物参照全式金公司胶回收试剂盒混合后回收, 取约20~70 ng胶回收产物, 加入约40~60 ng未切割的pYLCRISPR/Cas9质粒, 在15 μL反应用5~10 U Bsa I 37℃酶切10 min, 然后加1.5 μL 10×buffer for T4 DNA ligase和35 U T4 DNA ligase, 用变温循环酶切连接, 37℃ 5 min, 10℃ 5 min, 20℃ 5 min, 10~15个循环。(5)转化及质粒测序。连接产物转化大肠杆菌DH5α感受态细胞后, 挑取单克隆接种培养, 抽提质粒后利用Asc I酶切鉴定, 并挑取酶切验证正确的克隆送睿博公司测序。将验证正确的重组质粒转化农杆菌EHA101以备大豆遗传转化。pYLCRISPR/ Cas9-FAD2-1A-gRNA载体构建如图3所示。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3pYLCRISPR/Cas9-FAD2-1A-gRNA载体构建示意图
Fig. 3Schematic diagram of the pYLCRISPR/Cas9-FAD2-1A-gRNA vector construction
1.4 农杆菌介导大豆遗传转化
采用农杆菌介导的大豆子叶节遗传转化法构建大豆转基因材料[32], 选用大豆品种H3为受体材料, 实验中用到的培养基见表2。利用氯气对大豆表面灭菌, 在无菌超净工作台中, 将大豆种子接种于萌发培养基上, 16 h/8 h (光/暗), 温度为25℃, 湿度为60%的培养室培养1~3 d后, 在子叶节处用手术刀片创造伤口, 用农杆菌侵染液(OD650 nm ≈ 0.6), 在28℃的摇床中, 侵染30 min。随后, 将子叶放于表面附有灭菌滤纸的共培养培养基中, 25℃, 16 h/8 h光暗条件下共培养4 d, 转移B5芽诱导培养基诱导不定芽。4周后, 不定芽经芽伸长培养基培养, 培养条件为25℃, 16 h/8 h光暗周期。待其伸长大约达到5~7 cm时, 再将其转入生根培养基生根, 根系健壮后移到土里, 获得转基因植株。Table 2
表2
表2大豆遗传转化中使用的培养基及成分
Table 2
| 萌发培养基 Germination medium | 共培养培养基 Co-cultivation medium | 诱导培养基 Shoot induction medium | 伸长培养基 Shoot elongation medium | 生根培养基 Rooting medium | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MS合成盐 MS salt mixture | — | — | — | 1× | 1× | |||||
| B5合成盐 B5 salt mixture | 1× | 1/10× | 1× | — | — | |||||
| 2-(4-吗啉)乙磺酸 MES (g L-1) | — | 4.2 | 0.6 | 0.6 | — | |||||
| 6-苄基腺嘌呤 6-BAP (mg L-1) | — | 3.2 | 1.6 | — | — | |||||
| 赤霉素 GA3 (mg L-1) | — | — | — | 0.5 | — | |||||
| 羧苄青霉素Car (mg L-1) | — | — | 50 | 50 | — | |||||
| 二硫苏糖醇 DTT (mg L-1) | — | 150 | — | — | — | |||||
| 替卡西林 Tic (mg L-1) | — | — | 100 | 100 | 25 | |||||
| 头孢霉素 Cef (mg L-1) | — | — | 75 | 75 | 25 | |||||
| L-天冬酰胺 L-Asp (mg L-1) | — | — | — | 50 | — | |||||
| 谷氨酰胺 Glu (mg L-1) | — | — | — | 50 | — | |||||
| 草铵膦 Glufosinate (mg L-1) | — | — | 250 | — | — | |||||
| 萌发培养基 Germination medium | 共培养培养基 Co-cultivation medium | 诱导培养基 Shoot induction medium | 伸长培养基 Shoot elongation medium | 生根培养基 Rooting medium | ||||||
| 吲哚乙酸 IAA (mg L-1) | — | — | — | 0.1 | — | |||||
| 玉米素 ZR (mg L-1) | — | — | — | 1 | — | |||||
| 吲哚丁酸 IBA (mg L-1) | — | — | — | — | 1 | |||||
| 蔗糖 Sucrose (g L-1) | 20 | 30 | 30 | 30 | 20 | |||||
| 琼脂 Agar (g L-1) | 12 | 6.5 | 8.5 | 9 | 12 | |||||
| pH | 5.8 | 5.4 | 5.7 | 5.6 | 5.8 | |||||
新窗口打开|下载CSV
1.5 转基因大豆T1代植株抗草铵膦筛选、PCR鉴定及测序
将获得的转基因植株T0代种子以及对照H3种子, 种植于16 h/8 h (光照/黑暗)、湿度为60%的温室中, 当T1代植株长出第1片三出复叶时, 选择长势一致的嫩叶, 用毛笔蘸取浓度为150 mg L-1的草铵膦溶液, 对其中1/3叶片进行均匀的涂抹, 初步筛选抗草铵膦的植株。参照康为世纪公司的新型植物基因组DNA提取试剂盒(NuClean Plant Genomic DNA Kit)说明书, 提取转基因大豆和对照H3的基因组DNA, 并以DNA为模板, 用Bar-F/R引物(表1)进行PCR扩增, 检测出阳性植株。以筛选出的阳性植株DNA为模板, 用FAD2-1A-CasF-Test/FAD2-1A- CasR-Test靶点鉴定引物(表1)进行PCR扩增, 并将PCR产物送至测序公司测序。1.6 品质分析及农艺性状调查
分别选取100粒T3代纯合GmFAD2-1A突变体大豆种子和对照大豆品种华夏3号, 委托东北农业大学农学院, 采用气相色谱法检测脂肪酸各组分的含量[33], 福斯谷物分析仪(INFRATEC1241)检测总脂肪(%)及总蛋白含量(%), 试验进行3次生物学重复。农艺性状包括株高、主茎节数、单枝分枝数、叶形、花色、种皮色、种脐色和生育期。2 结果与分析
2.1 pYLCRISPR/Cas9-FAD2-1A-gRNA载体的构建
根据CRISPR/Cas9技术的原理, 构建pYLCRISPR/Cas9-FAD2-1A-gRNA表达载体, 将pYLCRISPR/Cas9-FAD2-1A-gRNA转化大肠杆菌后, 经菌落PCR鉴定(图4-A) 3~7号为阳性单克隆, 挑取3号单克隆接种培养, 抽提质粒后利用Asc I进行酶切鉴定, 如图4-B所示, 切出的目的片段大小约为1.5 kb左右, 表明FAD2-1A的3个靶点gRNA表达盒顺利插入到pYLCRISPR/Cas9载体上。为进一步确定靶位点的准确性, 通过设计特异性引物对构建载体上的各靶点测序, 得到完全一致的序列, 表明表达载体构建成功, 可用于后续试验。图4
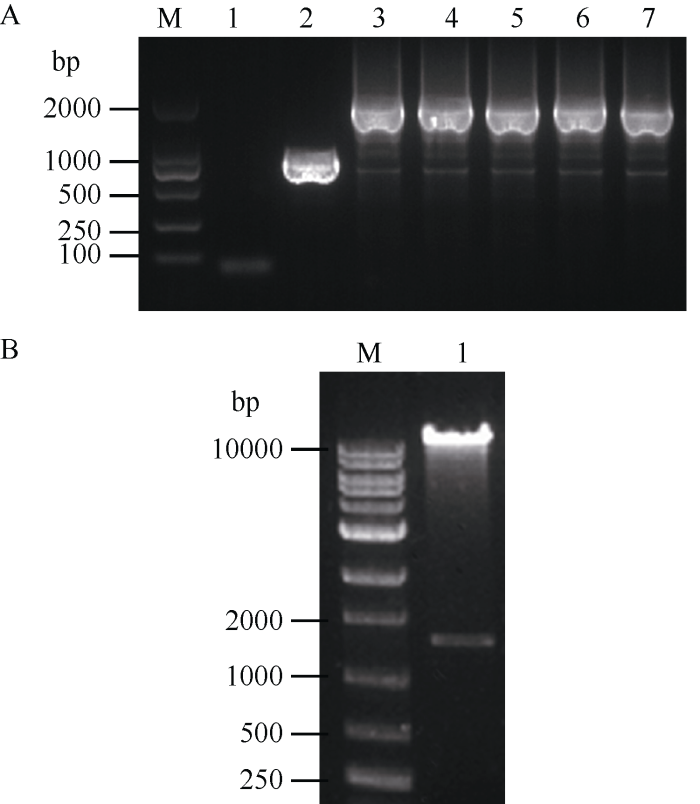 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4重组质粒鉴定
A: 菌落PCR检测电泳; M: DNA marker (DM2000); 1: H2O空白对照; 2: pYLCRISPR/Cas9-DB质粒; 3~7: FAD2-1A基因敲除阳性单菌落。B: Asc I酶切鉴定pYLCRISPR/Cas9-FAD2-1A-gRNA载体; M: 1 kb DNA ladder marker; 1: pYLCRISPR/Cas9-FAD2-1A- gRNA。
Fig. 4Identification of recombinant plasmids
A: Electrophoresis of PCR detection of clony; M: DNA marker (DM2000); 1: H2O blank control; 2: pYLCRISPR/Cas9-DB plasmid; 3-7: FAD2-1A gene knockout positive single clones. B: Identification of the pYLCRISPR/Cas9-FAD2-1A-gRNA plasmid digested with Asc I; M: 1 kb DNA ladder marker; 1: pYLCRISPR/Cas9- FAD2-1A-gRNA.
2.2 转基因植株的获得
用制备好的含有pYLCRISPR/Cas9-FAD2-1A- gRNA载体的农杆菌侵染液侵染大豆的子叶节, 经萌发、暗培养、恢复、筛选培养、伸长培养和生根培养后移入无菌土中保温保湿培养(图5-A)。涂抹草铵膦后, T0代转基因大豆对草铵膦产生抗性, 叶片表面无明显变化(图5-B-a), 而未转化成功的大豆, 则叶片表面发黄, 干枯(图5-B-b)。选取T0代涂抹草铵膦后无明显反应的3棵植株, 扩繁种子, 共获得5粒T0代种子。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5转化过程及叶片涂抹草铵膦筛选结果
A: 农杆菌介导的子叶节法转化大豆; 1: 无菌苗的萌发; 2: 共培养; 3: 丛生芽的诱导; 4: 丛生芽的伸长; 5: 诱导生根; 6: 移栽土里。 B: T0转基因大豆植株草铵膦涂抹鉴定; a: 阳性转基因植株; b: 阴性转基因植株。
Fig. 5Conversion process and the results of leaf painting
A: transformation soybean cotyledonine by agrobacterium; 1: germination of sterile seedlings; 2: co-cultivation; 3: shoot induction; 4: shoot elongation; 5: rooting of the resistant shoot; 6: seedling acclimatization. B: identification of the herbicide resistance of the leaves of T0 transgenic soybean plants using glufosinate; a: positive transgenic plant; b: negative transgenic plant.
2.3 GmFAD2-1A大豆突变体的鉴定
提取T1代转基因大豆叶片的总DNA, 以CRISPR/Cas9载体上含有的外源标记Bar基因引物对转基因大豆植株进行PCR扩增, 检测结果显示, T1代5株转基因植株均为阳性植株(图6-A)。利用靶点筛选引物FAD2-1A-CasF-Test/FAD2-1A-CasR-Test (表1), 对T1代阳性植株进行PCR扩增, 连接T载体后测序发现, 仅编号为4的植株发生编辑(以下称GmFAD2- 1A mutant), 该突变体仅在靶点1发生编辑(图6-B): 在靶点位置由碱基CATT替换成ACGA, AC替换成CT, 插入4个碱基GAAC; 并且在靶点位置后由碱基GCC替换成TTT, 插入1个T碱基。该靶点碱基突变导致FAD2-1A的蛋白移码突变(图6-C)。图6
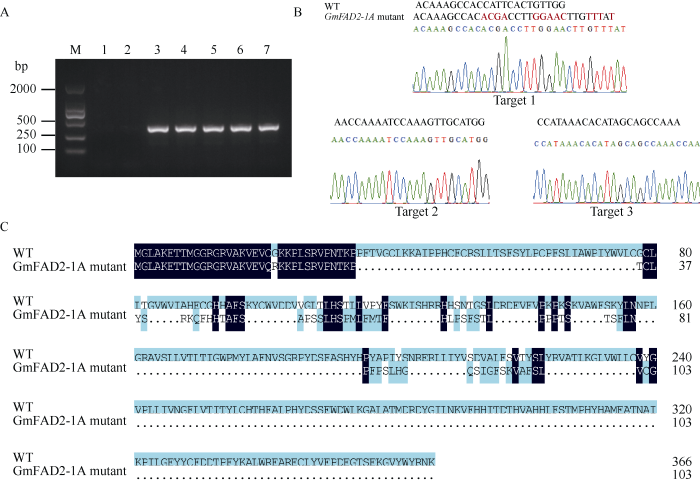 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6GmFAD2-1A突变体鉴定
A: T1代转基因大豆植株的PCR检测; M: DNA marker (DM2000); 1: H2O 空白对照; 2: H3野生型; 3~7: 转基因大豆草铵膦抗性阳性苗。B: GmFAD2-1A突变体与野生型序列比对分析; C: GmFAD2-1A突变体与野生型蛋白序列比对结果。
Fig. 6Identification of GmFAD2-1A mutant
A: PCR detection of T1 transgenic soybean; M: DNA marker (DM2000); 1: H2O blank control; 2: WT H3; 3-7: positive seedling of glufosinate tolerence of transgenic soybean. B: sequence alignment of GmFAD2-1A mutant compared to the WT line; C: sequence alignment of GmFAD2-1A mutant and wild-type protein.
2.4 GmFAD2-1A大豆突变体品质分析及农艺性状调查
GmFAD2-1A突变体大豆株系在株高、主茎节数、单枝分枝数、叶形、花色、种皮色、种脐色、生育期等方面与对照品种H3没有明显差异(表3和图7-A), 表明GmFAD2-1A 基因的突变对转基因大豆的农艺性状表型没有影响。GmFAD2-1A突变体的油酸含量为23%, 极显著高于对照品种H3 (20%), 而亚油酸、棕榈酸和硬脂酸极显著降低, 亚麻酸没有显著变化(图7-B), 表明GmFAD2-1A基因是调控大豆种子油酸含量的关键基因。转基因大豆的蛋白和脂肪含量分别为40.61%和20.50%, 与对照品种H3无显著差异(表3), 表明利用CRISPR/Cas9技术对控制油酸含量的GmFAD2-1A基因定点编辑没有显著影响转基因大豆种子中总蛋白和总脂肪的积累。Table 3
表3
表3转基因大豆表型性状
Table 3
| 农艺性状 Agronomic trait | 华夏3号 Huaxia 3 | GmFAD2-1A突变体 GmFAD2-1A mutant |
|---|---|---|
| 株高 Plant height (cm) | 58.53±1.47 | 57.60±0.54 |
| 主茎节数 Main stem number | 11±0 | 11±0 |
| 单株分枝数 Branching number per plant | 4±0 | 4±0 |
| 叶形 Leaf shape | 椭圆形 Oval | 椭圆形 Oval |
| 花色 Flower color | 白色 White | 白色 White |
| 种皮色 Seed coat color | 黄色 Yellow | 黄色 Yellow |
| 种脐色 Hilum color | 浅褐色 Pale brown | 浅褐色 Pale brown |
| 生育期 Growth period (d) | 107±0 | 107±0 |
| 蛋白 Protein (%) | 40.47±0.54 | 40.61±0.12 |
| 油脂 Oil (%) | 20.06±0.12 | 20.5±0.16 |
新窗口打开|下载CSV
图7
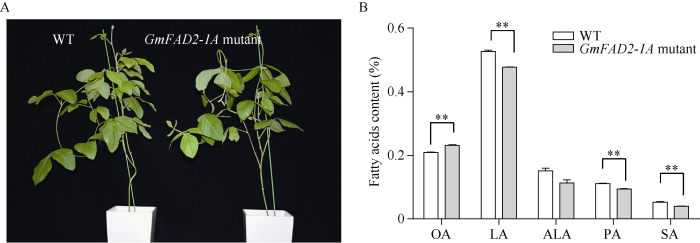 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7GmFAD2-1A突变体表型及其脂肪酸组分含量
OA: 油酸; LA: 亚油酸; ALA: 亚麻酸; PA: 棕榈酸; SA: 硬脂酸。
Fig. 7GmFAD2-1A mutants phenotype and the fatty acids content
OA: oleic acid; LA: linoleic acid; ALA: linolenic acid; PA: palmitic acid; SA: stearic acid.
3 讨论
随着人口的增长和生活水平的提高, 中国对大豆产品的品质需求不断增加, 因此改善大豆品质, 提高大豆营养成分是大豆育种的主要目标之一。油酸含量高的油非常有益于健康, 油酸可以降低人体血液中有害的低密度脂蛋白, 而对有益的高密度脂蛋白没有影响[34,35]。油酸含量高的油中多不饱和脂肪酸含量低, 在加工过程不需要加氢除去多不饱和脂肪酸, 从而有效避免产生对人体有害的反式脂肪酸73%)因其油酸含量高而被认为是一种有益人体健康的食用油。但由于国内不能生产橄榄, 橄榄油全部进口于地中海国家, 价格昂贵, 普通百姓难以消费。因此高油酸大豆油国产化, 具有价格竞争优势, 可替代橄榄油。大量研究表明, 抑制大豆种子中控制油酸转化为亚油酸的关键酶基因FAD2-1的表达, 可以有效提高大豆种子中的油酸含量[16,17,18,19,20,21,22]。但目前却没有可以大量推广到市场上的高油酸大豆。本研究利用CRISPR/Cas9技术定点编辑大豆FAD2-1A基因, 获得了油酸含量达23%的纯合突变体大豆, 其油酸含量极显著高于对照H3, 该结果与前人研究基本相同。不同的是, 本研究利用CRISPR/Cas9技术获得稳定遗传, 且无外源基因骨架的FAD2-1A突变体大豆植株。本实验选择的大豆受体品种H3为广东、广西、海南、福建中南部等低纬度地区大面积推广种植的大豆品种, 前景广阔。同时本研究所获得的大豆突变体材料与受体材料有着相同的分枝性好, 株型健壮, 具有丰产性突出、综合抗性好的优点。
近年来, CRISPR/Cas9基因编辑系统的发展越来越完善, 与前两代基因编辑技术ZFNs和TALENs相比, 用RNA替代组装蛋白, 实现与靶基因的特异性识别与结合, 使得该技术构建简单灵活[23]。因此, CRISPR/Cas9系统作为一种新兴的快速、稳定、廉价的基因编辑技术, 未来将会更加广泛应用于育种。
目前农杆菌介导大豆遗传转化体系中的外植体有很多, 但子叶节再生体系应用最为广泛和成熟[37]。自从该系统首次由Cheng等[38]报道以后的几十年间, 众多科研工作者对该系统进行了改良和优化[37,39]。这个体系具有取材不受时间限制、周期短、再生植株遗传稳定等优点, 其缺点也很明显, 如再生苗嵌合体多, 筛选困难, 对培养条件和操作技术要求较高。虽然大豆的遗传转化效率已经提高了很多, 但相对于水稻、玉米等其他作物, 大豆的转化效率仍然较低, 因此建立高效、稳定的遗传转化技术仍然是大豆研究者亟需解决的问题。
4 结论
利用CRISPR/Cas9技术, 获得适合低纬度种植的高油酸大豆新种质GmFAD2-1A突变体。与受体品种华夏3号(H3)相比, GmFAD2-1A突变体的油酸含量由20%提高到23%, 达到极显著水平, 同时, GmFAD2-1A突变体农艺性状与华夏3号没有显著差异。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1006/mben.2001.0204URLPMID:11800570 [本文引用: 1]

Fatty acids are the most abundant form of reduced carbon chains available from nature and have diverse uses ranging from food to industrial feedstocks. Plants represent a significant renewable source of fatty acids because many species accumulate them in the form of triacylglycerol as major storage components in seeds. With the advent of plant transformation technology, metabolic engineering of oilseed fatty acids has become possible and transgenic plant oils represent some of the first successes in design of modified plant products. Directed gene down-regulation strategies have enabled the specific tailoring of common fatty acids in several oilseed crops. In addition, transfer of novel fatty acid biosynthetic genes from noncommercial plants has allowed the production of novel oil compositions in oilseed crops. These and future endeavors aim to produce seeds higher in oil content as well as new oils that are more stable, are healthier for humans, and can serve as a renewable source of industrial commodities. Large-scale new industrial uses of engineered plant oils are on the horizon but will require a better understanding of factors that limit the accumulation of unusual fatty acid structures in seeds.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]

Modern biotechnology has and will continue to play a valuable role in public and private soybean breeding programs. Tools provided by biotechnology will not replace soybean breeding in the public and private sectors but rather will help provide new discoveries and improve upon the overall efficiency of soybean improvement. Public soybean breeders conduct basic plant breeding research in breeding methodology and germplasm development. In addition, public breeders are involved in educating future plant breeders with the latest biotechnological advances in molecular genetics/biology and laboratory techniques as well as conventional plant breeding practices. The majority of variety development is conducted by private soybean breeders. Biotechnology can be used in virtually every facet of plant breeding activities by both public and private soybean breeders.
DOI:10.1111/j.1445-5994.1992.tb00480.xURLPMID:1449442 [本文引用: 1]

Music goes back a very long way in human experience. Music therapy is now used in many disparate areas-from coronary care units to rehabilitation after a stroke. But its widespread adoption has a poor scientific evidence base, founded more on enthusiasm than on proper evaluation in any controlled way. This has led to a lack of clarity about whether any particular type of music is superior, or... [Show full abstract]
DOI:10.1136/bmj.306.6893.1648URLPMID:8518602 [本文引用: 1]

To investigate the level of risk of death from coronary heart disease above which cholesterol lowering treatment produces net benefits. Meta-analysis of results of randomised controlled trials of cholesterol lowering treatments. Published and unpublished data from all identified randomised controlled trials of cholesterol lowering treatments with six months or more follow up and with at least one death were included in the meta-analysis. The analyses were stratified by the rate of death from coronary heart disease in the control arms of the trials. Death from all causes, from coronary heart disease, and from causes other than coronary heart disease. In the pooled analysis, net benefit in terms of total mortality from cholesterol lowering was seen only for trials including patients at very high initial risk of coronary heart disease (odds ratio 0.74; 95% confidence interval 0.60 to 0.92). In a medium risk group no net effect was seen, and in the low risk group there were adverse treatment effects (1.22; 1.06 to 1.42). In a weighted regression analysis a significant (p < 0.001) trend of increasing benefit with increasing initial risk of coronary heart disease was shown. Raised mortality from causes other than coronary heart disease was seen in trials of drug treatment (1.21; 1.05 to 1.39) but not in the trials of non-drug treatments (1.02; 0.88 to 1.19). Cumulative meta-analysis showed that these results seem to have been stable as new trials appeared. Currently evaluated cholesterol lowering drugs seem to produce mortality benefits in only a small proportion of patients at very high risk of death from coronary heart disease. Population cholesterol screening could waste resources and even result in net harm in substantial groups of patients. Overall risk of coronary heart disease should be the main focus of clinical guidelines, and a cautious approach to the use of cholesterol lowering drugs should be advocated. Future trials should aim to clarify the level of risk above which treatment is of net benefit.
DOI:10.1073/pnas.76.3.1194URLPMID:286305 [本文引用: 1]

This communication demonstrates that all de novo fatty acid biosynthesis in spinach leaf cells requires acyl carrier protein (ACP) and occurs specifically in the chloroplasts. Antibodies raised to purified spinach ACP inhibited at least 98% of malonyl CoA-dependent fatty acid synthesis by spinach leaf homogenates. Therefore, the presence of ACP in a compartment of the spinach leaf cell would serve as a marker for de novo fatty acid biosynthesis. A radioimmunoassay capable of detecting 10-15mol (10-11g) of spinach ACP was developed to measure the levels of ACP in leaf cell components isolated by sucrose gradient centrifugation of a gentle lysate of spinach leaf protoplasts. All of the ACP of the leaf cell could be attributed to the chloroplast. Less than 1% of the ACP associated with chloroplasts resulted from binding of free ACP to chloroplasts. Of interest, ACP from Escherichia coli, soybean, and sunflower showed only partial crossreactivity with spinach ACP by the radioimmunoassay. These results strongly suggest that, in the leaf cell, chloroplasts are the sole site for the de novo synthesis of C16and C18fatty acids. These fatty acids are then transported into the cytoplasm for further modification and are either inserted into extrachloroplastic membrane lipids or returned to the chloroplast for insertion into lamellar membrane lipids.
DOI:10.2307/2657130URLPMID:11159130 [本文引用: 1]

The FAD2-1 microsomal desaturase gene contains a large intron (\sim1133 bp [base pairs]) in the 5' untranslated region that may participate in gene regulation and, in Gossypium, is evolving at an evolutionary rate useful for elucidating recently diverged lineages. FAD2-1 is single copy in diploid Gossypium species, and two orthologs are present in the allotetraploid species. Among the diploid species, the D-genome FAD2-1 introns have accumulated substitutions 1.4-1.8 times faster than the A-genome introns. In the tetraploids, the difference between the D-subgenome introns and their A-subgenome orthologs is even greater. The substitution rate of the intron in the D-genome diploid G. gossypioides more closely approximates that of the A genome than other D genome species, highlighting its unique evolutionary history. However, phylogenetic analyses support G. raimondii as the closest living relative of the D-subgenome donor. The Australian K-genome species diverged 8-16 million years ago into two clades. One clade comprises the sporadically distributed, erect to suberect coastal species; a second clade comprises the more widely spread, prostrate, inland species. A comparison of published gene trees to the FAD2-1 intron topology suggests that G. bickii arose from an early divergence, but that it carries a G. australe-like rDNA captured via a previously undetected hybridization event.
DOI:10.1105/tpc.6.1.147URLPMID:7907506 [本文引用: 1]

The polyunsaturated fatty acids linoleate and alpha-linolenate are important membrane components and are the essential fatty acids of human nutrition. The major enzyme responsible for the synthesis of these compounds is the plant oleate desaturase of the endoplasmic reticulum, and its activity is controlled in Arabidopsis by the fatty acid desaturation 2 (fad2) locus. A fad2 allele was identified in a population of Arabidopsis in which mutations had been created by T-DNA insertions. Genomic DNA flanking the T-DNA was cloned by plasmid rescue and used to isolate cDNA and genomic clones of FAD2. A cDNA containing the entire FAD2 coding sequence was expressed in fad2 mutant plants and shown to complement the mutant fatty acid phenotype. The deduced amino acid sequence from the cDNA showed homology to other plant desaturases, and this confirmed that FAD2 is the structural gene for the desaturase. Gel blot analyses of FAD2 mRNA levels showed that the gene is expressed throughout the plant and suggest that transcript levels are in excess of the amount needed to account for oleate desaturation. Sequence analysis identified histidine-rich motifs that could contribute to an iron binding site in the cytoplasmic domain of the protein. Such a position would facilitate interaction between the desaturase and cytochrome b5, which is the direct source of electrons for the desaturation reaction, but would limit interaction of the active site with the fatty acyl substrate.
DOI:10.1016/j.plaphy.2008.12.024URLPMID:19217793 [本文引用: 1]

A cotton ( Gossypium hirsutum L.) genomic clone encompassing a 17.9-kb DNA fragment was found to contain a delta-12 fatty acid desaturase gene (designated FAD2-4). The FAD2-4 open reading frame has 1,155 bp and is uninterrupted, encoding a conceptual FAD2-4 polypeptide of 384 amino acids that has 98% identity with the cotton FAD2-3 polypeptide. The FAD2-4 gene has a single intron of 2,780 bp in its 5′-untranslated region (5′-UTR). The 3′-flanking regions and 5′-UTR introns in the FAD2-4 and FAD2-3 genes are quite different, indicating that the genes might be paralogs in the cotton genome. Reverse transcriptase (RT)-PCR analysis indicated that the FAD2-4 and FAD2-3 genes were expressed in all tissues examined, including seeds, seedling tissues, young and mature leaves, roots, stems, developing flower buds, and ovule fibers. These constitutive patterns of expression were notably different from that of the FAD2-1 gene, which was restricted to seeds and developing flower buds, or to the expression of a newly-identified FAD2-2 gene isoform, which was barely detectable in roots, hypocotyls, stems, and fibers, but was expressed in all other tissues. The FAD2-4 coding region was expressed in yeast and shown to encode a functional delta-12 desaturase, converting oleic acid (C18:1) to linoleic acid (C18:2) in recombinant yeast cells. In addition, both the FAD2-4 and the FAD2-3 genes were transferred into the Arabidopsis thaliana fad2-1 mutant background where they effectively restored wild type fatty acid composition and growth characteristics. Finally, the cotton FAD2-4 green fluorescent protein (GFP) fusion polypeptide appeared to be localized in the endomembrane system of transgenic Arabidopsis plants in the complemented fad2-1 mutant background, supporting a functional ER location for the cotton FAD2-4 polypeptide in this heterologous plant system. Thus, a new functional member of the FAD2 gene family in cotton has been characterized, indicating a complex regulation of membrane lipid desaturation in this important fiber/oilseed crop.
DOI:10.2135/cropsci1998.0011183X003800050022xURL [本文引用: 1]

The seed oils of wild-type sunflower (Helianthus annuus L.) germplasm typically contain 140 to 350 g [kg.sup.-1] oleic acid (18:1). Sunflower germplasm with 800 to 950 g [kg.sup.-1] 18:1 (high oleic germplasm) has been developed with a dominant mutation (01) originating from the cultivar `Pervenets'. We speculated that this mutation reduced the activity or expression of a [Delta]-12 oleate desaturase (OLD) gene. This was tested by cloning OLD genes expressed in developing seeds and comparing nucleotide sequences, OLD transcript concentrations, and OLD restriction fragment lengths among wild-type (Iow oleic) and backcross-derived mutant (high oleic) lines fixed for ol or Ol alleles, respectively. An Arabidopsis thaliana L. OLD (At OLD) cDNA probe was used to isolate a full-length cDNA clone (OLD-7) from a developing seed cDNA library of sunflower. OLD-7 was strongly expressed in developing seeds, hut was not expressed in the other tissues sampled. The nucleotide sequences for OLD-7 ORFs from partially isogenic Iow and high oleic acid lines (HA89 and HA341) were 100% identical. High oleic lines had substantially lower OLD-7 transcript concentrations than Iow oleic lines. Restriction fragment analyses showed that OLD-7 is duplicated and rearranged in mutant lines and linked to the Ol locus; thus, increased oleic acid content seems to be caused by DNA changes affecting OLD-7 cis regulatory sequences. OLD-7 RFLPs distinguish between Ol locus genotypes and can be used to accelerate the development of high oleic lines in sunflower.
DOI:10.1016/j.jplph.2006.08.007URLPMID:17141918 [本文引用: 2]

In plants, the endoplasmic reticulum (ER)-associated oleate desaturase (FAD2) is the key enzyme responsible for the production of linoleic acid in non-photosynthetic tissues. In soybean three FAD2-like genes have been reported including two seed-specific genes, FAD2-1A and FAD2-1B, and a house-keeping gene FAD2-2. In this study, we isolated a novel gene encoding FAD2 isoform, designated as FAD2-3. The deduced amino acid sequences of the FAD2-3 displayed the typical three histidine boxes characteristic of all membrane-bound desaturases, and possessed a C-terminal signal for ER retention. Phylogenetic analysis showed that FAD2-3 is grouped within plant house-keeping FAD2 sequences. Yeast cells transformed with a plasmid construct containing the FAD2-3 coding region accumulated a considerable amount of linoleic acid (18:2), normally not present in wild-type yeast cells, suggesting that the isolated gene encodes a functional FAD2 enzyme. Semi-quantitative RT-PCR and in silico analysis showed that FAD2-3 gene is constitutively expressed in both vegetative tissues and developing seeds. In soybean leaves, the level of linolenic acid (18:3) increases with the decrease of linoleic aicd (18:2) under cold treatment. However, no significant change of transcript levels of FAD2-2 and FAD2-3 genes was detected. These results indicated that the altered polyunsaturated fatty acid levels in leaves treated with cold stress have no direct correlation with the expression of these two microsomal oleate desaturase genes.
DOI:10.1104/pp.110.1.311URLPMID:8587990 [本文引用: 2]

The polyunsaturated fatty acid content is one of the major factors influencing the quality of vegetable oils. Edible oils rich in monounsaturated fatty acid provide improved oil stability, flavor, and nutrition for human and animal consumption. In plants, the microsomal desaturase-catalyzed pathway is the primary route of production of polyunsaturated lipids. We report the isolation of two different cDNA sequences, FAD2-1 and FAD2-2, encoding microsomal desaturase in soybeans and the characterization of their developmental and temperature regulation. The FAD2-1 gene is strongly expressed in developing seeds, whereas the FAD2-2 gene is constitutively expressed in both vegetative tissues and developing seeds. Thus, the FAD2-2 gene-encoded desaturase appears to be responsible for production of polyunsaturated fatty acids within membrane lipids in both vegetative tissues and developing seeds. The seed-specifically expressed FAD2-1 gene is likely to play a major role in controlling conversion of oleic acid to linoleic acid within storage lipids during seed development. In both soybean seed and leaf tissues, linoleic acid and linolenic acid levels gradually increase as temperature decreases. However, the levels of transcripts for FAD2-1, FAD2-2, and the plastidial desaturase gene (FAD 6) do not increase at low temperature. These results suggest that the elevated polyunsaturated fatty acid levels in developing soybean seeds grown at low temperature are not due to the enhanced expression of desaturase genes.
DOI:10.1080/10425170701207208URLPMID:18300159 [本文引用: 3]

Abstract In plants, the endoplasmic reticulum (ER)-associated oleate desaturase (FAD2) is the key enzyme responsible for the production of linoleic acid in non-photosynthetic tissues. In this study, we report the characterization of a seed-specific isoform of microsomal omega-6 fatty acid desaturase gene (FAD2-1B) sharing high sequence similarity with FAD2-1 from soybean. Several potential promoter elements including seed-specific motifs are found in the 5'-flanking region of FAD2-1B gene. The ORF of FAD2-1B is 1161 bp long and encodes a protein of 387 amino acids. This deduced protein holds three histidine boxes and four putative membrane-spanning helices, and possesses a signal for endoplasmic reticulum retention at C-terminal. Yeast cells transformed with the plasmid construct containing soybean FAD2-1B accumulate an appreciable amount of linoleic acid (18:2), normally not present in wild-type yeast cells, indicating that the cloned gene encodes a functional FAD2 enzyme. Both semi-quantitative RT-PCR and in silico analysis show that FAD2-1B gene is specifically expressed in developing seeds of soybean.
URLPMID:20828382 [本文引用: 3]

pAbstract/p pBackground/p pThe alteration of fatty acid profiles in soybean [itGlycine max /it(L.) Merr.] to improve soybean oil quality is an important and evolving theme in soybean research to meet nutritional needs and industrial criteria in the modern market. Soybean oil with elevated oleic acid is desirable because this monounsaturated fatty acid improves the nutrition and oxidative stability of the oil. Commodity soybean oil typically contains 20% oleic acid and the target for high oleic acid soybean oil is approximately 80% of the oil; previous conventional plant breeding research to raise the oleic acid level to just 50-60% of the oil was hindered by the genetic complexity and environmental instability of the trait. The objective of this work was to create the high oleic acid trait in soybeans by identifying and combining mutations in two delta-twelve fatty acid desaturase genes, itFAD2-1A /itand itFAD2-1B/it./p pResults/p pThree polymorphisms found in the itFAD2-1B /italleles of two soybean lines resulted in missense mutations. For each of the two soybean lines, there was one unique amino acid change within a highly conserved region of the protein. The mutant itFAD2-1B /italleles were associated with an increase in oleic acid levels, although the itFAD2-1B /itmutant alleles alone were not capable of producing a high oleic acid phenotype. When existing itFAD2-1A /itmutations were combined with the novel mutant itFAD2-1B /italleles, a high oleic acid phenotype was recovered only for those lines which were homozygous for both of the mutant alleles./p pConclusions/p pWe were able to produce conventional soybean lines with 80% oleic acid in the oil in two different ways, each requiring the contribution of only two genes. The high oleic acid soybean germplasm developed contained a desirable fatty acid profile, and it was stable in two production environments. The presumed causative sequence polymorphisms in the itFAD2-1B /italleles were developed into highly efficient molecular markers for tracking the mutant alleles. The resources described here for the creation of high oleic acid soybeans provide a framework to efficiently develop soybean varieties to meet changing market demands./p
[本文引用: 3]
DOI:10.1007/s00299-008-0535-8URLPMID:18347801 [本文引用: 2]

An efficient system of gene transformation is necessary for soybean [ Glycine max (L.) Merrill] functional genomics and gene modification by using RNA interference ( RNAi ) technology. To establish such system, we improved the conditions of tissue culture and transformation for increasing the frequency of adventitious shoots and decreasing the browning and necrosis of hypocotyls. Adding N 6 -benzylaminopurine (BAP) and silver nitrate in culture medium enhanced the shoot formation on hypocotyls. BAP increased the frequency of the hypocotyls containing adventitious shoots, while silver nitrate increased the number of shoots on the hypocotyls. As a result, the number of adventitious shoots on hypocotyls cultured in medium containing both BAP and silver nitrate was 5-fold higher than the controls. Adding antioxidants in co-cultivation medium resulted in a significant decrease in occurrence of browning and necrosis of hypocotyls and increase in levels of -Glucuronidase (GUS) gene expression. Histochemical assays showed that the apical meristem of hypocotyls was the arget tissue for Agrobacterium tumefaciens transformation of soybean. Gene silencing of functional gene by using RNAi technology was carried out under above conditions. A silencing construct containing an inverted-repeat fragment of the GmFAD2 gene was introduced into soybean by using the A. tumefaciens -mediated transformation. Several lines with high oleic acid were obtained, in which mean oleic acid content ranged from 71.5 to 81.9%. Our study demonstrates that this transgenic approach could be efficiently used to improve soybean quality and productivity through functional genomics.
DOI:10.1155/2014/921950URLPMID:4147191 [本文引用: 2]

The Delta-12 oleate desaturase gene (FAD2-1), which converts oleic acid into linoleic acid, is the key enzyme determining the fatty acid composition of seed oil. In this study, we inhibited the expression of endogenous Delta-12 oleate desaturase GmFad2-1b gene by using antisense RNA in soybean Williams 82. By employing the soybean cotyledonary-node method, a part of the cDNA of soybean GmFad2-1b 801???bp was cloned for the construction of a pCAMBIA3300 vector under the soybean seed promoter BCSP. Leaf painting, LibertyLink strip, PCR, Southern blot, qRT-PCR, and fatty acid analysis were used to detect the insertion and expression of GmFad2-1b in the transgenic soybean lines. The results indicate that the metabolically engineered plants exhibited a significant increase in oleic acid (up to 51.71%) and a reduction in palmitic acid (to <3%) in their seed oil content. No structural differences were observed between the fatty acids of the transgenic and the nontransgenic oil extracts.
[本文引用: 2]
[本文引用: 2]
DOI:10.1111/pbi.12201URLPMID:24851712 [本文引用: 2]

Summary Soybean oil is high in polyunsaturated fats and is often partially hydrogenated to increase its shelf life and improve oxidative stability. The trans-fatty acids produced through hydrogenation pose a health threat. Soybean lines that are low in polyunsaturated fats were generated by introducing mutations in two fatty acid desaturase 2 genes ( FAD2-1A and FAD2-1B ), which in the seed convert the monounsaturated fat, oleic acid, to the polyunsaturated fat, linoleic acid. Transcription activator-like effector nucleases (TALENs) were engineered to recognize and cleave conserved DNA sequences in both genes. In four of 19 transgenic soybean lines expressing the TALENs, mutations in FAD2-1A and FAD2-1B were observed in DNA extracted from leaf tissue; three of the four lines transmitted heritable FAD2 -1 mutations to the next generation. The fatty acid profile of the seed was dramatically changed in plants homozygous for mutations in both FAD2-1A and FAD2-1B : oleic acid increased from 20% to 80% and linoleic acid decreased from 50% to under 4%. Further, mutant plants were identified that lacked the TALEN transgene and only carried the targeted mutations. The ability to create a valuable trait in a single generation through targeted modification of a gene family demonstrates the power of TALENs for genome engineering and crop improvement.
[本文引用: 2]
DOI:10.1038/cr.2013.114URLPMID:23958582 [本文引用: 1]

In the past few years, the development of sequence-specific DNA nucleases has progressed rapidly and such nucleases have shown their power in generating efficient targeted mutagenesis and other genome editing applications. For zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), an engineered array of sequence-specific DNA binding domains are fused with the DNA nuclease Fok1. These nucleases have been successful in genome modifications by generating double strand breaks (DSBs), which are then repaired through non-homologous end joining (NHEJ) or homologous recombination (HR) in different species, including mouse, tobacco and rice. Recently, another breakthrough technology for genome editing, the CRISPR/Cas system, was developed. CRISPR (clustered regulatory interspaced short palindromic repeats) loci are variable short spacers separated by short repeats, which are transcribed into non-coding RNAs. The non-coding RNAs form a functional complex with CRISPR-associated (Cas) proteins and guide the complex to cleave complementary invading DNA. After the initial development of a programmable CRISPR/Cas system, it has been rapidly applied to achieve efficient genome editing in human cell lines, zebrafish and mouse. However, there is still no successful application in plants reported.
DOI:10.1016/j.jgg.2013.12.001URLPMID:24576457 [本文引用: 1]

Transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems have emerged as powerful tools for genome editing in a variety of species. Here, we report, for the first time, targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. We designed five TALENs targeting 4 genes, namely ZmPDS, ZmIPK1A, ZmIPK, ZmMRP4, and obtained targeting efficiencies of up to 23.1% in protoplasts, and about 13.3% to 39.1% of the transgenic plants were somatic mutations. Also, we constructed two gRNAs targeting the ZmIPK gene in maize protoplasts, at frequencies of 16.4% and 19.1%, respectively. In addition, the CRISPR/Cas system induced targeted mutations in Z. mays protoplasts with efficiencies (13.1%) similar to those obtained with TALENs (9.1%). Our results show that both TALENs and the CRISPR/Cas system can be used for genome modification in maize.
DOI:10.1038/nbt.2969URLPMID:25038773 [本文引用: 1]

Sequence-specific nucleases have been applied to engineer targeted modifications in polyploid genomes, but simultaneous modification of multiple homoeoalleles has not been reported. Here we use transcription activator-like effector nuclease (TALEN) and clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9 (refs. 4,5) technologies in hexaploid bread wheat to introduce targeted mutations in the three homoeoalleles that encode MILDEW-RESISTANCE LOCUS (MLO) proteins. Genetic redundancy has prevented evaluation of whether mutation of all three MLO alleles in bread wheat might confer resistance to powdery mildew, a trait not found in natural populations. We show that TALEN-induced mutation of all three TaMLO homoeologs in the same plant confers heritable broad-spectrum resistance to powdery mildew. We further use CRISPR-Cas9 technology to generate transgenic wheat plants that carry mutations in the TaMLO-A1 allele. We also demonstrate the feasibility of engineering targeted DNA insertion in bread wheat through nonhomologous end joining of the double-strand breaks caused by TALENs. Our findings provide a methodological framework to improve polyploid crops.
DOI:10.1038/nbt.2650URLPMID:23929338 [本文引用: 1]

The article offers information on genome modification of crop plants using a CRISPR-Cas system. It states that genome editing technologies using zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) can also generate genome modifications. Photographs related to genome editing in rice and wheat using an engineered type II CRISPR-Cas system are also presented.
[本文引用: 1]
[本文引用: 1]
URLPMID:4365529 [本文引用: 2]

Background The ability to selectively alter genomic DNA sequences in vivo is a powerful tool for basic and applied research. The CRISPR/Cas9 system precisely mutates DNA sequences in a number of organisms. Here, the CRISPR/Cas9 system is shown to be effective in soybean by knocking-out a green fluorescent protein (GFP) transgene and modifying nine endogenous loci. Results Targeted DNA mutations were detected in 95% of 88 hairy-root transgenic events analyzed. Bi-allelic mutations were detected in events transformed with eight of the nine targeting vectors. Small deletions were the most common type of mutation produced, although SNPs and short insertions were also observed. Homoeologous genes were successfully targeted singly and together, demonstrating that CRISPR/Cas9 can both selectively, and generally, target members of gene families. Somatic embryo cultures were also modified to enable the production of plants with heritable mutations, with the frequency of DNA modifications increasing with culture time. A novel cloning strategy and vector system based on In-Fusion?? cloning was developed to simplify the production of CRISPR/Cas9 targeting vectors, which should be applicable for targeting any gene in any organism. Conclusions The CRISPR/Cas9 is a simple, efficient, and highly specific genome editing tool in soybean. Although some vectors are more efficient than others, it is possible to edit duplicated genes relatively easily. The vectors and methods developed here will be useful for the application of CRISPR/Cas9 to soybean and other plant species.
DOI:10.1111/pbi.12758URLPMID:28509421 [本文引用: 1]

Abstract Flowering is an indication of the transition from vegetative growth to reproductive growth and has considerable effects on the life cycle of soybean (Glycine max). In this study, we employed the CRISPR/Cas9 system to specifically induce targeted mutagenesis of GmFT2a, an integrator in the photoperiod flowering pathway in soybean. The soybean cultivar Jack was transformed with three sgRNA/Cas9 vectors targeting different sites of endogenous GmFT2a via Agrobacterium tumefaciens-mediated transformation. Site-directed mutations were observed at all targeted sites by DNA sequencing analysis. T1 generation soybean plants homozygous for null alleles of GmFT2a frameshift mutated by a 1-bp insertion or short deletion exhibited late flowering under natural conditions (summer) in Beijing, China (N39 58', E116 20'). We also found that the targeted mutagenesis was stably heritable in the following T2 generation, and the homozygous GmFT2a mutants exhibited late flowering under both long-day and short-day conditions. We identified some "transgene-clean" soybean plants that were homozygous for null alleles of endogenous GmFT2a and without any transgenic element from the T1 and T2 generations. These "transgene-clean" mutants of GmFT2a may provide materials for more in-depth research of GmFT2a functions and the molecular mechanism of photoperiod responses in soybean. They will also contribute to soybean breeding and regional introduction. This article is protected by copyright. All rights reserved. This article is protected by copyright. All rights reserved.
DOI:10.1016/j.molp.2015.04.007URLPMID:25917172 [本文引用: 1]

A robust CRISPR/Cas9 vector system for multiplex genome editing in monocot and dicot plants was developed. Multiple sgRNA expression cassettes can be assembled into the binary CRISPR/Cas9 vectors in one round of cloning. This system can uniformly, efficiently, and simultaneously produce multiple heritable mutations in rice and Arabidopsis by targeting multiple genes or genomic sites via single transformation events.
DOI:10.1385/1-59745-130-4:385URLPMID:16988361 [本文引用: 1]

Agrobacterium tumefaciens-mediated transformation of soybeans has been steadily improved since its development in 1988. Soybean transformation is now possible in a range of genotypes from different maturity groups using different explants as sources of regenerable cells, various selectable marker genes and selective agents, and different A. tumefaciens strains. The cotyledonary-node method has been extensively investigated and across a number of laboratories yields on average greater than 1% transformation efficiency (one Southern-positive, independent event per 100 cotyledonary-node explants). Continued improvements in the cotyledonary-node method concomitant with further increases in transformation efficiency will enhance broader adoption of this already productive transformation method for use in crop improvement and functional genomics research efforts.
[本文引用: 1]
DOI:10.1007/s11745-998-0231-9URLPMID:9625595 [本文引用: 1]

The effects of dietary monounsaturated fatty acid (MUFA) and polyunsaturated fatty acid+MUFA/saturated fatty acid (PUFA+MUFA/SFA) ratio on plasma and liver lipid concentrations were studied. In experiment I, when rats were fed with 40% fat (energy%, PUFA/SFA ratio 1.0) and 1% (w/w) cholesterol (C) diets for 21 d, a large amount of MUFA (28.1 energy%, PUFA+MUFA/SFA=5.7) in the diet was found to increase the plasma total C, triacylglycerol (TAG), and phospholipid (PL) as compared with the low-MUFA diet (7.0 energy%, PUFA+MUFA/SFA=1.4). The plasma very low density lipoprotein (VLDL)-C, VLDL-TAG, VLDL-PL, and low density lipoprotein (LDL)-C increased significantly in the high-MUFA diet group, but high density lipoprotein (HDL)-C did not change significantly. The high-MUFA diet resulted in greater accumulation of liver C but lesser accumulation of TAG. In experiment II, when dietary SFA was fixed at a certain level (13.2 energy%; PUFA+MUFA/SFA=2.0), rats given a larger amount of MUFA (23.1 energy%; PUFA/MUFA=0.2; MUFA/SFA=1.8) showed higher plasma and liver C levels than did the low-MUFA diet (7.7 energy%; PUFA/MUFA=2.5; MUFA/SFA=0.6). When PUFA was fixed at a certain level (24.4 energy%), there was not a significant difference in the plasma C level between the high-and low-MUFA dietary groups (PUFA+MUFA/SFA=4.8 and 8.4), but the higher PUFA+MUFA/SFA diet, which was high in MUFA/SFA ratio, significantly decreased the plasma HDL-C and TAG levels. However, when MUFA content was fixed at a certain level (16.4 energy%), no significant difference was observed between the two groups with different PUFA/SFA ratios of 0.2 and 4.1, but liver C level was raised in the higher PUFA/SFA diet. It appears that the PUFA/SFA ratio alone is unsuitable to predict the change of plasma C level, because a large amount of dietary MUFA may lead to an increase of plasma and liver lipids in rats. It seems that the prerequisites for keeping low plasma and liver C are (i) low MUFA/SFA ratio, (ii) high PUFA/MUFA ratio, and (iii) PUFA+MUFA/SFA ratio not to exceed 2.
DOI:10.1016/S0735-1097(98)00681-0URLPMID:10091835 [本文引用: 1]

Ingestion of a meal rich in fat previously used for deep frying in a commercial fast food restaurant resulted in impaired arterial endothelial function. These findings suggest that intake of degradation products of heated fat contribute to endothelial dysfunction.
DOI:10.1002/1438-9312(200009)102:8/9<587::aid-ejlt587>3.0.co;2-#URL

Results of studies investigating the toxicity of components of thermoxidized fats and oils are discussed. Aspects considered include: demonstration by early studies that overheated fats were toxic if fed to animals; evaluation of the results of long-term feeding trials showing that fats and oils produced from deep fat fryers under conditions of good commercial practice have no detrimental effects on the health of animals (consideration of the possible adverse effects of the total polar materials fraction when fed in high doses); use of cell culture and enzymic techniques to study resorption and metabolism of lipid components; and conclusions following use of new analytical techniques to isolate, quantify and assess the toxicity of well defined fractions from heated fats (mono- and diglycerides and free fatty acids being harmless; dimeric and polymeric triglycerides having very low resorption rates and thus also being harmless; and oxidized triglyceride monomers that are saponified and degraded to oxidized monomeric and dimeric acids, oxidized cyclic fatty acids and other polar compounds that are potentially harmful).
[本文引用: 2]
DOI:10.1016/0304-4211(80)90084-XURL [本文引用: 1]

Stimulation of multiple shoot-bud formation of soybeans in culture has been accomplished using conditioned cotyledonary node segments. Germination of soybean seeds in the presence of 10–50 μM BAP stimulated varying numbers of buds to form at the cotyledonary node region. Culturing cotyledonary node segments excised from such conditioned seedlings on a medium containing 5–50 μM BAP stimulated multiple shoot-bud formation. Continual bud multiplication was obtained by periodic subculture of the organized tissue mass onto fresh media containing the same BAP concentration. Bud growth was stimulated by transferring cultures from a high BAP concentration of 10–50 μM to a low BAP concentration of 1 μM. Soybean plants were produced by rooting tissue culture shoots on a basal medium without auxin.
DOI:10.1007/s00425-002-0922-2URLPMID:12624759 [本文引用: 1]

The efficiency of soybean [Glycine max (L.) Merrill] transformation was significantly increased from an average of 0.7% to 16.4% by combining strategies to enhance Agrobacterium tumefaciens-mediated T-DNA delivery into cotyledonary-node cells with the development of a rapid, efficient selection protocol based on hygromycin B. Wounded cotyledonary-node explants were inoculated with A. tumefaciens carrying either a standard-binary or super-binary plasmid and co-cultivated in the presence of mixtures of the thiol compounds, L-cysteine, dithiothreitol, and sodium thiosulfate. Transformed shoots began elongating only 8 weeks after co-cultivation. Southern analysis confirmed integration of the T-DNA into genomic DNA and revealed no correlation between the complexity of the integration pattern and thiol treatment applied at co-cultivation. All T0 plants were fertile and the majority of the lines transmitted the glucuronidase (GUS) phenotype in 3:1 or 15:1 ratios to their progenies.
