 ,*, 耿帅锋
,*, 耿帅锋 ,*中国农业科学院作物科学研究所, 北京 100081
,*中国农业科学院作物科学研究所, 北京 100081Establishment of a CRISPR/Cas9-VQR gene editing system
CHEN Kai*, SUN Guo-Liang*, SONG Gao-Yuan*, LI Ai-Li*, XIE Chuan-Xiao*, MAO Long ,*, GENG Shuai-Feng
,*, GENG Shuai-Feng ,*Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
,*Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China通讯作者:
收稿日期:2018-10-24接受日期:2019-01-19网络出版日期:2019-02-26
| 基金资助: |
Received:2018-10-24Accepted:2019-01-19Online:2019-02-26
| Fund supported: |
作者简介 About authors
陈凯,E-mail:
孙国梁,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (949KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
陈凯, 孙国梁, 宋高原, 李爱丽, 谢传晓, 毛龙, 耿帅锋. 一个CRISPR/Cas9-VQR基因编辑系统的构建[J]. 作物学报, 2019, 45(6): 848-855. doi:10.3724/SP.J.1006.2019.82052
CHEN Kai, SUN Guo-Liang, SONG Gao-Yuan, LI Ai-Li, XIE Chuan-Xiao, MAO Long, GENG Shuai-Feng.
CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeat and Cas9)作为新一代基因组编辑技术, 与ZFN (Zinc Finger Nucleases)、TALEN (Transcriptional Action Like Effector Nucleases)技术相比, 具有操作简便、切割效率高、靶位点多等优点, 在模式植物和作物中得到广泛的应 用[1]。CRISPR/Cas9系统在gRNA的引导之下, 将Cas9带到目的基因上, 利用RuvC和HNH切割DNA双链, 形成双链断裂结构(Double Strand Break), 然后生物体内会在修复DSB过程中引起一些碱基的随机插入或缺失, 从而产生移码突变[2,3]。虽然CRISPR/Cas9相对于其他基因编辑技术具有较高的编辑效率, 但对不同物种或同一物种中不同基因的编辑效率却不相同[4,5]。在拟南芥中, 研究人员以AtFT、AtSPL4、AtBRI1、AtJAZ1、AtADH1、AtFLS2和AtPDS3等为目的基因验证CRISPR/Cas9的功能, 其切割效率为1.10%~84.78%[6,7,8,9,10]。在烟草、水稻、大豆、西红柿、高粱、玉米、棉花、小麦和药用植物丹参中的切割效率分别为1.8%~87.5%[6,11-13]、2.1%~ 100.0%[8,14-21]、14.7%~20.2%[22]、83.56%[23]、33.33%[24]、13.1%[25]、47.6%~81.8%[26]、5.6%[27]和42.3%[28]。
根据现有文献报道, 影响CRISPR/Cas9系统编辑效率的因素, 主要包括Cas9基因密码子的优化、Cas9基因启动子的替换、gRNA启动子的替换和gRNA的靶标数目[29,30,31,32]。在拟南芥、烟草、番茄、大豆、水稻和玉米中, 都有优化Cas9基因密码子的报道, 但只有单子叶植物中的优化能够提高编辑效率[29,31]。利用植物内源组成型启动子, 如ZmUbi、OsUBQ和OsActin1, 启动Cas9基因, 也能获得较高的编辑效率[15,30,33]。在单子叶植物中, 利用植物内源的U6或者U3启动子启动gRNA更有利于基因编辑[31,34]。在拟南芥和玉米中, 串联2个gRNA比单个gRNA的编辑效率高[31,32]。在水稻中, 分别设计1、2、3个gRNA靶向一个基因, 结果表明靶向同一个基因的多个位点时编辑效率提高[35]。另外, CRISPR/Cas9的PAM位点NGG, 也在一定程度上限制了其使用范围。虽然在哺乳动物细胞中CRISPR/ StCas9可识别NNAGAA PAM位点, CRISPR/SaCas9可识别NNGGGT、NNGAAT和NNGAGT PAM位 点[36,37], 但这些CRISPR系统在植物中报道较少。在水稻中, 通过将SpCas9突变, 可以识别NGA、NGAG和NGNG PAM位点, 但是CRISPR/Cas9- VQR识别NGA PAM位点的切割效率仅为2.1%~ 23.4%, 低于原有CRISPR/Cas9系统[38]。通过优化gRNA结构和使用水稻内源强启动子启动gRNA可以提高CRISPR/Cas9-VQR的切割效率, 但对影响编辑效率的其他因素的改造还未见报道[39]。
本研究通过突变SpCas9编码氨基酸、使用玉米Ubi启动子启动Cas9-VQR蛋白、优化SpCas9蛋白、加入核定位信号序列、增加单子叶中保守的 3′ UTR 序列、使用水稻U6 启动子启动gRNA, 构建了一个能够识别NGA PAM位点, 并进行有效切割的CRISPR/Cas9-VQR基因编辑系统, 扩大了CRISPR/ Cas9的使用范围。
1 材料与方法
1.1 植物材料
水稻(Oryza sativa)材料日本晴(Nipponbare) 由中国农业科学院作物科学研究所吴传银老师提供。1.2 实验试剂
DNA 限制性内切酶、PCR扩增酶、T4-DNA连接酶、qRT-PCR荧光酶均购自TaKaRa公司; 反转录试剂盒 SuperScript III Reverse Transcriptase、RNA 提取试剂TRIzol Reagent均购自Invitrogen公司; 卡那霉素、X-gal、IPTG等购自Simga公司。其他化学试剂如氯仿、异丙醇等购自北京化工公司。实验中所用到引物均由生工生物工程(上海)股份有限公司合成。1.3 CRISPR/Cas9密码子改造及优化
CRISPR/Cas9-VQR质粒由实验室和北京唯尚立德生物科技有限公司合作构建, 主要利用PCR定点突变技术, 将SpCas9序列进行突变, 进而改变编码氨基酸, 从而改变CRISPR/Cas9载体识别的PAM位点。同时通过基因合成将哺乳动物里应用的CRISPR/Cas9载体进行植物密码子优化, 并加入核定位序列、增加单子叶中3′ UTR序列、使用玉米Ubi启动Cas9-VQR转录、水稻U6启动gRNA转录, 构建双元表达载体系统。1.4 gRNA靶位点引物设计
以水稻 (Oryza sativa)八氢番茄红素脱氢酶基因(phytoene desaturase, PDS)为靶基因[27], 根据靶位点设计的原则, 在OsPDS基因不同外显子上筛选并设计了4个长度为 23 bp的靶位点序列, 分别命名为 gRNA1、gRNA2、gRNA3和gRNA4 (表1)。Table 1
表1
表1OsPDS-gRNAs序列
Table 1
| 靶位点 Target site | 序列 Targeted sequence (5′-3′) | GC含量 GC content (%) | 外显子位置 Location |
|---|---|---|---|
| OsPDS-gRNA1 | CTTGGAAGGATGAAGATGGAGA | 45 | 3 |
| OsPDS-gRNA2 | CCAGGAGAATTCAGCCGGTTTGA | 55 | 6 |
| OsPDS-gRNA3 | TCAGGAGAAGCATGGTTCTAAGA | 45 | 8 |
| OsPDS-gRNA4 | TCCCGGACTGTGAACCTTGCCGA | 60 | 13 |
新窗口打开|下载CSV
1.5 Cas9/VQR-gRNA体外酶切活性检测
利用Cas9/VQR-gRNA靶点效率检测试剂盒(北京唯尚立德生物科技有限公司)检测4个gRNA对靶位点的切割效率。首先, 参考试剂盒分别合成gRNA1、gRNA2、gRNA3和gRNA4引物(表2); 然后以标准gRNA模板为模板, 分别通过PCR扩增gRNA1、gRNA2、gRNA3和gRNA4及试剂盒提供的标准gRNA (S1和S2), 回收纯化PCR产物作为DNA模板分别进行体外转录, 回收并定量转录gRNA;其次, 通过搭桥PCR的方式, 将gRNA靶位点(包括PAM序列) 构建到PCR产物中, 获得用于酶切的DNA; 最后, 参照试剂盒提供的Cas9/VQR酶切体系, 加入2 μL 10 × Cas9 buffer、1 μL Cas9/VQR酶、50 ng体外转录的gRNA、50 ng酶切的DNA、补DEPC H2O至20 μL, 充分混合后, 37℃反应 30 min, 然后加入3 μL DNA Loading buffer, 混合后65℃ 5 min, 2%琼脂糖凝胶电泳检测分析酶切结果。此外, 每次酶切实验的同时做标准靶位点的gRNA酶切实验, 以标准靶位点酶切实验结果作为阳性对照比较活性。通过与2个阳性参照标准S1和S2估算gRNA的活性。根据SSA活性, 标准S1=3, 标准S2=10, 通过酶切条带的灰度与标准S1和标准 S2对比, 换算出gRNA1 (g1)、gRNA2 (g2)、gRNA3 (g3)、gRNA4 (g4)的活性(若检测gRNA酶切条带的灰度小于标准S1的灰度, 则活性估值小于3; 若检测gRNA酶切条带的灰度大于标准S1的灰度, 且小于标准S2的灰度则活性估值大于3小于10; 若检测gRNA酶切条带的灰度大于标准S2的灰度, 则活性估值大于10)。Table 2
表2
表2本研究所用引物
Table 2
| 引物名称 Primer name | 序列 Sequence (5′-3′) |
|---|---|
| T7-gRNA1-FPg | TAATACGACTCACTATAGCTTGGAAGGATGAAGATGGGTTTTAGAGCTAGAAATAGC |
| T7-gRNA2-FPg | TAATACGACTCACTATAGCCAGGAGAATTCAGCCGGTTGTTTTAGAGCTAGAAATAGC |
| T7-gRNA3-FPg | TAATACGACTCACTATAGTCAGGAGAAGCATGGTTCTAGTTTTAGAGCTAGAAATAGC |
| T7-gRNA4-FPg | TAATACGACTCACTATAGTCCCGGACTGTGAACCTTGCGTTTTAGAGCTAGAAATAGC |
| gRNA-RP | AGCACCGACTCGGTGCCACTT |
| oligo-gRNA1-F | TTGCTTGGAAGGATGAAGATGG |
| oligo-gRNA1-R | AACCCATCTTCATCCTTCCAAG |
| oligo-gRNA2-F | TTGCCAGGAGAATTCAGCCGGTT |
| oligo-gRNA2-R | AACAACCGGCTGAATTCTCCTGG |
| oligo-gRNA4-F | TTGTCCCGGACTGTGAACCTTGC |
| oligo-gRNA4-R | AACGCAAGGTTCACAGTCCGGGA |
| PCR-gRNA1-F | TCAGAGTAAAGCAAAGATTC |
| PCR-gRNA1-R | TCAGGCTCCTACGAGATA |
| PCR-gRNA2-F | GAATTTTCGCTTAGAGGC |
| PCR-gRNA2-R | CCAGTTATTTGAGTTCCATC |
| PCR-gRNA4-F | TGTGCCTGTATGTAACCA |
| PCR-gRNA4-R | GAGCAAACTTCACCTTCT |
新窗口打开|下载CSV
1.6 水稻转化
依据4对gRNA体外酶切活性检测结果, 选取3对分别设计oligo引物(表2), 通过正义链和反义链gRNA的oligo引物的退火反应, 形成oligo二聚体, 并插入CRISPR/Cas9-VQR载体, 转化大肠杆菌, 进而挑选阳性克隆测序, 获得构建好的CRISPR/Cas9- VQR-gRNA载体, 参照Hiei等的方法[40], 通过液氮冻融法将CRISPR/Cas9-VQR-gRNA载体转入农杆菌菌株EHA105感受态细胞中, 涂板挑取单克隆并进行鉴定, 将阳性克隆用于水稻转化。选用日本晴作为转化受体, 将预培养的水稻愈伤组织与农杆菌共培养, 并将共培养后的愈伤组织转移至NB1S1筛选培养基上继续培养, 获得转基因植株。1.7 阳性植株鉴定
采用CTAB 法提取转化水稻叶片全基因组DNA。通过特异性引物PCR, 以提取的转基因材料DNA为模板, 鉴定阳性转基因植株。引物见表2。2 结果与分析
2.1 CRISPR/Cas9 密码子优化
植物体内翻译中对G和C有一定的偏好性, 以水稻为模型, 对野生型SpCas9基因进行密码子优化, 优化后Cas9中碱基G含量由原来的733个增加到1094个, 碱基C由原来的566个增加到931个, GC总含量由31.56%增长到49.20%, 提高了SpCas9 GC碱基的含量(表3)。Table 3
表3
表3Cas9密码子优化结果
Table 3
| 名称 Name | 长度 Length (bp) | G含量 G contents | C含量 C contents | GC百分比 GC percent (%) |
|---|---|---|---|---|
| SpCas9 | 4116 | 733 | 566 | 31.56 |
| Cas9-codon optimized | 4116 | 1094 | 931 | 49.20 |
新窗口打开|下载CSV
2.2 CRISPR/Cas9 PAM位点分布
利用生物信息学技术, 通过对水稻和小麦的CDS区域NGG、NGA、NGC和NGT PAM位点的统计分析, 发现水稻和小麦中NGA PAM位点数量均多于传统NGG PAM位点。水稻中NGA PAM位点比NGG位点多出大约330,000个, 约占NGG位点的12.34%。小麦中NGA PAM位点比NGG多出大约1,300,000个位点, 约占NGG位点的24.78%。因此通过改造SpCas9, 使其能够识别NGA PAM位点, 可以有效增加该基因编辑技术的使用范围, 尤其是在水稻和小麦中(图1)。图1
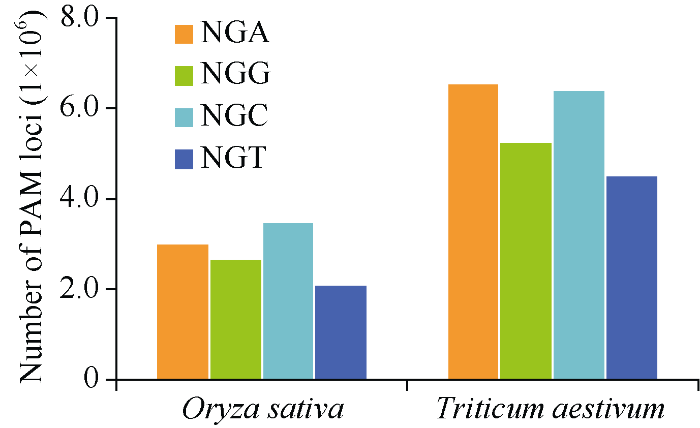 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1水稻和小麦全基因组CDS中PAM位点的数量
Fig. 1Number of PAM loci in whole genome CDS of rice and wheat
2.3 CRISPR/Cas9载体改造
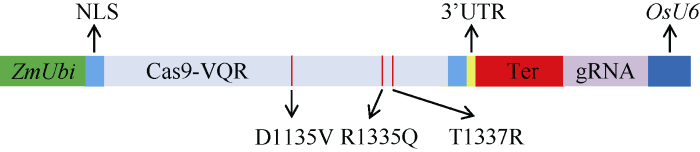
利用PCR定点突变技术, 将传统的SpCas9基因序列进行突变, 改变编码氨基酸: 由1135位的天冬氨酸(D)突变为缬氨酸(V), 由1335位的精氨酸(R)突变为谷胱氨酸(Q), 由1337位的苏氨酸(T)突变为精氨酸(R), 命名为Cas9-VQR (图2)。从而使CRISPR/ Cas9-VQR的PAM识别位点变为NGA。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2CRISPR/Cas9-VQR载体
ZmUbi: 玉米泛素启动子; NLS: 核定位信号; D1135V: 1135位的天冬氨酸(D)突变为缬氨酸(V); R1335Q: 1335位的精氨酸(R)突变为谷胱氨酸(Q); T1337R: 1337位的苏氨酸(T)突变为精氨酸(R); 3′UTR: 3′非翻译区; Ter: 终止子; gRNA: 指导RNA; OsU6: 水稻U6启动子。
Fig. 2Construction of CRISPR/Cas9-VQR vector
ZmUbi: ubiquitin promoter; NLS: nuclear localization signal; D1135V: substituting the 1135D residue with V; R1335Q: substituting the 1335R residue with Q; T1337R: substituting the 1337 T residue with R; 3′ UTR: 3′ untranslated region; Ter: terminator; gRNA: guide RNA; OsU6: OsU6 promoter.
同时, 根据影响基因编辑效率的因素, 对载体进行修饰, 在Cas9-VQR序列两端加上保守的核定位序列, 增加单子叶中保守的3′UTR序列, 使用玉米Ubi启动子启动Cas9-VQR蛋白, 优化的Cas9-VQR蛋白和使用水稻U6启动子表达gRNA (图2)。
2.4 CRISPR/Cas9-VQR 蛋白活性和体外酶切活性检测
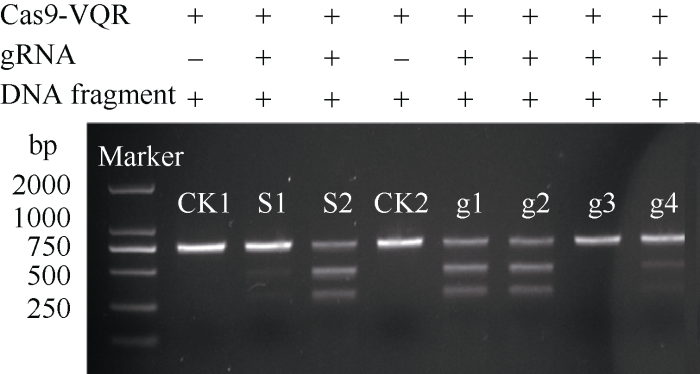
为验证Cas9-VQR的蛋白活性, 我们通过Cas9- VQR蛋白切割标准样(S1, S2)检测, 发现该Cas9- VQR具有酶切活性, 可以有效地切割标准样(S1, S2)(图3), CK1和CK2为未加gRNA的对照。经过SSA luciferase检测活性, 发现S1和S2的SSA活性分别为3和10。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3体外酶切检测活性结果
S1和S2: 标准样品1和2; CK1和CK2: 阴性对照1和2; g1: OsPDS-gRNA1; g2: OsPDS-gRNA2; g3: OsPDS-gRNA3; g4: OsPDS-gRNA4。
Fig. 3Cas9-VQR enzyme digestion activity in vitro
S1 and S2: standard sample 1 and 2; CK1 and CK2: negative control 1and 2; g1: OsPDS-gRNA1; g2: OsPDS-gRNA2; g3: OsPDS- gRNA3; g4: OsPDS-gRNA4.
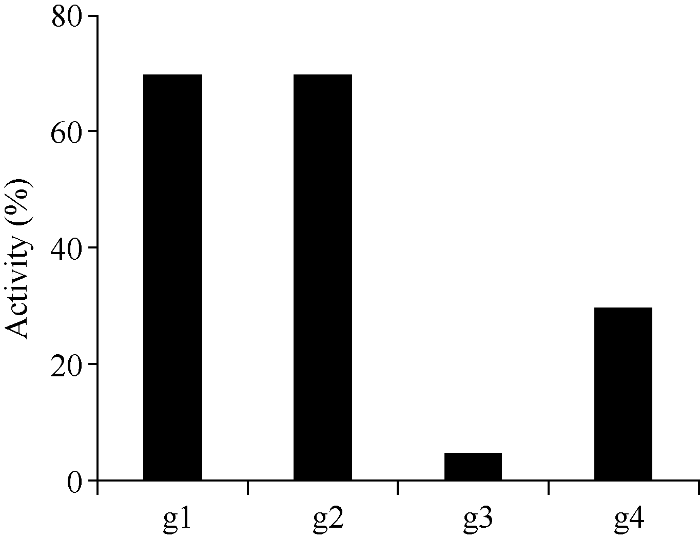
在水稻PDS基因第3、第6、第8和第13外显子上, 分别设计gRNA (表1), 然后分别进行体外酶切活性检测。与对照S1和S2相比, g1、g2、g3、g4靶位点均有活性, 但活性强度有所不同, g1和g2靶位点最好, 基本和阳性对照一致, 而g4靶位点较弱, g3靶位点最差(图3)。根据酶切条带灰度换算gRNA的活性, g1和g2体外酶切活性约为70%, g3体外酶切活性约为5%, g4体外酶切活性约为30% (图4)。
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4OsPDS-gRNA体外活性检测结果
Fig. 4OsPDS-gRNAs activity in vitrog1: OsPDS-gRNA1; g2: OsPDS-gRNA2; g3: OsPDS-gRNA3; g4: OsPDS-gRNA4.
2.5 转化植株的表型观察及突变位点检测
根据4个gRNA的体外酶切活性, 挑选其中3个(gRNA1、gRNA2和gRNA4)插入CRISPR/Cas9- VQR载体, 转化水稻。获得OsPDS-gRNA1表达载体转化再生植株24个, 其中17株阳性植株, 约占70.8%; 获得OsPDS-gRNA2表达载体再生植株45个, 其中阳性植株为40株, 约占88.89%; 获得53株OsPDS-gRNA4水稻再生植株, 其中阳性植株为49株, 约占92.45%。由于OsPDS基因为剂量基因, 所以突变植株因突变发生时期不同而表现不同的表型。若Cas9-VQR切割OsPDS基因是在早期第一个胚胎细胞发育还没分化之前, 就会产生纯合白化苗(图5-A)。若Cas9-VQR切割OsPDS基因是在分化后的细胞中, 则会产生绿色和白色相间表型, 该表型在叶片、叶鞘和穗部都会出现(图5-B, C)。
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5表型观察
A: 苗期; B: 叶片; C: 穗。WT: 野生型; OsPDS-gRNAs: gRNA载体OsPDS-gRNA1、OsPDS-gRNA2和OsPDS-gRNA4。
Fig. 5Phenotype observation
A: seedling stage; B: leaf; C: spike. WT: wild type; OsPDS-gRNAs: gRNA vector of OsPDS-gRNA1, OsPDS-gRNA2, and OsPDS-gRNA4.
通过对不同的靶位点区域片段的测序, 发现OsPDS-gRNA1产生缺失-28 bp到插入+19 bp等不同类型的突变, 共检测到12株突变植株, 突变效率为70.59%, 其中4株为纯合突变, 约占33.33%; OsPDS-gRNA2产生缺失-11 bp到插入+10 bp等不同类型的突变, 共检测到11株突变植株, 突变效率为27.5%, 其中纯合突变植株为5株, 约占45.45%; OsPDS-gRNA4产生缺失-13 bp到插入+9 bp等多种类型的突变, 共检测到20株突变植株, 突变效率为40.8%, 其中7株为纯合植株, 约占35% (图6和表5)。
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6靶位点突变类型
A: OsPDS-gRNA1靶位点的突变类型; B: OsPDS-gRNA2靶位点的突变类型; C: OsPDS-gRNA4靶位点的突变类型。WT: 野生型; L: 不同株系。
Fig. 6Target site mutation
A: target site mutation of OsPDS-gRNA1; B: target site mutation of OsPDS-gRNA2; C: target site mutation of OsPDS-gRNA4. WT: wild type; L: line.
Table 5
表5
表5转基因植株突变体数量统计
Table 5
| 名称 Name | 阳性植株 Transplant | 突变植株 Mutated plant | 突变率 Mutation ratio (%) | 纯合突变体 Homo plant | 杂合突变体 Het plant |
|---|---|---|---|---|---|
| OsPDS-gRNA1 | 17/24 | 12 | 70.5 | 4/12 | 8/12 |
| OsPDS-gRNA2 | 40/45 | 11 | 27.5 | 5/11 | 6/11 |
| OsPDS-gRNA4 | 49/53 | 20 | 40.8 | 7/20 | 13/20 |
新窗口打开|下载CSV
3 讨论
传统的CRISPR/Cas9系统虽然应用广泛, 但受NGG PAM位点的限制。CRISPR/Cas9-VQR能够识别NGA PAM位点, 并进行有效切割, 极大地扩大了CRISPR/Cas9系统的使用范围。本研究通过突变Cas9序列和修饰基因编辑载体, 获得了一个能够识别NGA PAM位点, 并有效切割靶基因的基因编辑系统, 为该编辑系统在NGA PAM位点较多作物中的应用提供了借鉴。在哺乳动物细胞中, 虽然有CRISPR/StCas9识别NNAGAA PAM位点及CRISPR/SaCas9识别NNGGGT、NNGAAT和NNGAGT PAM位点的报道[36,37], 但在植物中却较少。在水稻中, 通过突变SpCas9改造的CRISPR/Cas9-VQR, 其切割效率约为 2.1%~23.4%, 相对较低[38]。通过优化CRISPR/Cas9- VQR的gRNA结构和使用内源强启动子启动gRNA, 提高了CRISPR/Cas9-VQR的切割效率[39], 但并未考虑影响编辑效率的其他影响因素。本研究除了对Cas9蛋白进行改造, 还对影响编辑效率的因素进行修饰, 获得了一个能够识别NGA PAM位点, 并进行有效切割的基因编辑系统。
传统的CRISPR/Cas9系统在水稻的切割突变效率为2.1%~100.0% [8,14-21], 突变效率变化较大。其中使用玉米Ubi作为SpCas9的启动子, 其突变效率为4%~100%[19,21], 而使用35S作为启动子, 目的基因的突变效率为2.1%~75.0%[8,14-18,20], 相对较低。针对相同基因, 例如OsSWEET11/14, 使用玉米Ubi作为启动子, 突变效率为87%~100%, 平均突变效率为93.5%[19], 而使用35S启动子, 突变效率为66.67%[24]。这可能是由于35S启动子在单子叶植物中容易被甲基化修饰[41], 从而影响SpCas9的转录水平。本实验中使用玉米Ubi作为SpCas9-VQR的启动子, 虽然获得46.23%的平均突变效率, 但玉米Ubi启动子的作用还需设置相应的对照进行深入研究。
另外, 我们在载体构建中的修饰, 如对Cas9- VQR进行密码子优化、增加单子叶植物中保守的 3′ UTR 序列和使用 OsU6 启动子启动 gRNA转录, 都会对系统的编辑效率产生影响, 我们将进一步研究这些修饰对编辑效率的影响。
4 结论
建立了有效识别NGA PAM位点的CRISPR/ Cas9-VQR编辑系统, 并扩大了其使用范围, 为后期在水稻等高NGA PAM位点作物中的应用提供了参考。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/j.cj.2015.12.002URL [本文引用: 1]

The CRISPR/Cas9 technology is evolved from a type II bacterial immune system and represents a new generation of targeted genome editing technology that can be applied to nearly all organisms. Site-specific modification is achieved by a single guide RNA (usually about 20 nucleotides) that is complementary to a target gene or locus and is anchored by a protospacer-adjacent motif. Cas9 nuclease then cleaves the targeted DNA to generate double-strand breaks (DSBs), which are subsequently repaired by non-homologous end joining (NHEJ) or homology-directed repair (HDR) mechanisms. NHEJ may introduce indels that cause frame shift mutations and hence the disruption of gene functions. When combined with double or multiplex guide RNA design, NHEJ may also introduce targeted chromosome deletions, whereas HDR can be engineered for target gene correction, gene replacement, and gene knock-in. In this review, we briefly survey the history of the CRISPR/Cas9 system invention and its genome-editing mechanism. We also describe the most recent innovation of the CRISPR/Cas9 technology, particularly the broad applications of modified Cas9 variants, and discuss the potential of this system for targeted genome editing and modification for crop improvement.
DOI:10.1038/nature07992URLPMID:19404259 [本文引用: 1]

Abstract Agricultural biotechnology is limited by the inefficiencies of conventional random mutagenesis and transgenesis. Because targeted genome modification in plants has been intractable, plant trait engineering remains a laborious, time-consuming and unpredictable undertaking. Here we report a broadly applicable, versatile solution to this problem: the use of designed zinc-finger nucleases (ZFNs) that induce a double-stranded break at their target locus. We describe the use of ZFNs to modify endogenous loci in plants of the crop species Zea mays. We show that simultaneous expression of ZFNs and delivery of a simple heterologous donor molecule leads to precise targeted addition of an herbicide-tolerance gene at the intended locus in a significant number of isolated events. ZFN-modified maize plants faithfully transmit these genetic changes to the next generation. Insertional disruption of one target locus, IPK1, results in both herbicide tolerance and the expected alteration of the inositol phosphate profile in developing seeds. ZFNs can be used in any plant species amenable to DNA delivery; our results therefore establish a new strategy for plant genetic manipulation in basic science and agricultural applications.
DOI:10.1111/tpj.12704URLPMID:25327456 [本文引用: 1]

Summary The CRISPR/Cas nuclease is becoming a major tool for targeted mutagenesis in eukaryotes by inducing double-strand breaks (DSBs) at pre-selected genomic sites that are repaired by non-homologous end joining (NHEJ) in an error-prone way. In plants, it could be demonstrated that the Cas9 nuclease is able to induce heritable mutations in Arabidopsis thaliana and rice. Gene targeting (GT) by homologous recombination (HR) can also be induced by DSBs. Using a natural nuclease and marker genes, we previously developed an in planta GT strategy in which both a targeting vector and targeting locus are activated simultaneously via DSB induction during plant development. Here, we demonstrate that this strategy can be used for natural genes by CRISPR/Cas-mediated DSB induction. We were able to integrate a resistance cassette into the ADH1 locus of A.thaliana via HR. Heritable events were identified using a PCR-based genotyping approach, characterised by Southern blotting and confirmed on the sequence level. A major concern is the specificity of the CRISPR/Cas nucleases. Off-target effects might be avoided using two adjacent sgRNA target sequences to guide the Cas9 nickase to each of the two DNA strands, resulting in the formation of a DSB. By amplicon deep sequencing, we demonstrate that this Cas9 paired nickase strategy has a mutagenic potential comparable with that of the nuclease, while the resulting mutations are mostly deletions. We also demonstrate the stable inheritance of such mutations in A.thaliana .
[本文引用: 1]
DOI:10.1111/tpj.12554URLPMID:24836556 [本文引用: 1]

SummaryEngineered nucleases can be used to induce site-specific double-strand breaks (DSBs) in plant genomes. Thus, homologous recombination (HR) can be enhanced and targeted mutagenesis can be achieved by error-prone non-homologous end-joining (NHEJ). Recently, the bacterial CRISPR/Cas9 system was used for DSB induction in plants to promote HR and NHEJ. Cas9 can also be engineered to work as a nickase inducing single-strand breaks (SSBs). Here we show that only the nuclease but not the nickase is an efficient tool for NHEJ-mediated mutagenesis in plants. We demonstrate the stable inheritance of nuclease-induced targeted mutagenesis events in the ADH1 and TT4 genes of Arabidopsis thaliana at frequencies from 2.5 up to 70.0%. Deep sequencing analysis revealed NHEJ-mediated DSB repair in about a third of all reads in T1 plants. In contrast, applying the nickase resulted in the reduction of mutation frequency by at least 740-fold. Nevertheless, the nickase is able to induce HR at similar efficiencies as the nuclease or the homing endonuclease IceI. Two different types of somatic HR mechanisms, recombination between tandemly arranged direct repeats as well as gene conversion using the information on an inverted repeat could be enhanced by the nickase to a similar extent as by DSB-inducing enzymes. Thus, the Cas9 nickase has the potential to become an important tool for genome engineering in plants. It should not only be applicable for HR-mediated gene targeting systems but also by the combined action of two nickases as DSB-inducing agents excluding off-target effects in homologous genomic regions.
[本文引用: 2]
DOI:10.1111/tpj.12554URLPMID:24836556 [本文引用: 1]

SummaryEngineered nucleases can be used to induce site-specific double-strand breaks (DSBs) in plant genomes. Thus, homologous recombination (HR) can be enhanced and targeted mutagenesis can be achieved by error-prone non-homologous end-joining (NHEJ). Recently, the bacterial CRISPR/Cas9 system was used for DSB induction in plants to promote HR and NHEJ. Cas9 can also be engineered to work as a nickase inducing single-strand breaks (SSBs). Here we show that only the nuclease but not the nickase is an efficient tool for NHEJ-mediated mutagenesis in plants. We demonstrate the stable inheritance of nuclease-induced targeted mutagenesis events in the ADH1 and TT4 genes of Arabidopsis thaliana at frequencies from 2.5 up to 70.0%. Deep sequencing analysis revealed NHEJ-mediated DSB repair in about a third of all reads in T1 plants. In contrast, applying the nickase resulted in the reduction of mutation frequency by at least 740-fold. Nevertheless, the nickase is able to induce HR at similar efficiencies as the nuclease or the homing endonuclease IceI. Two different types of somatic HR mechanisms, recombination between tandemly arranged direct repeats as well as gene conversion using the information on an inverted repeat could be enhanced by the nickase to a similar extent as by DSB-inducing enzymes. Thus, the Cas9 nickase has the potential to become an important tool for genome engineering in plants. It should not only be applicable for HR-mediated gene targeting systems but also by the combined action of two nickases as DSB-inducing agents excluding off-target effects in homologous genomic regions.
[本文引用: 4]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.molp.2015.02.011URLPMID:25749112 [本文引用: 1]

Dear Editor,Targeted genome editing in plants will not only facilitate functional genomics studies but also help to discover,expand,and create novel traits of agricultural importance (Pennisi,2010).The most widely used approach for editing plant genomes involves generating targeted double-strand DNA breaks (DSBs)and harnessing the two main DSB repair pathways:imprecise non-homologous end joining and precise homology-directed repair (Voytas,2013).Enzymes that specifically bind the userselected genomic sequences to create DSBs can be generated de novo as synthetic bimodular proteins containing a DNA-binding module,engineered to bind a user-defined sequence,along with a DNA-cleaving module,capable of making DSBs.Several classes of nucleases have been developed with DNA-binding domains engineered to confer sequence specificity,including homing endonucleases,zinc finger nucleases (ZFNs),and transcription activator-like effector nucleases (TALENs).Customization of these genome editing platforms,however,requires protein engineering,a time-consuming and laborintensive process (Puchta and Fauser,2014).Furthermore,delivery of genome engineering reagents into plant cells is a major barrier to the effective use of these technologies for creating novel traits (Baltes et al.,2014).
[本文引用: 1]
DOI:10.1038/nbt.2650URLPMID:23929338 [本文引用: 3]

The article offers information on genome modification of crop plants using a CRISPR-Cas system. It states that genome editing technologies using zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) can also generate genome modifications. Photographs related to genome editing in rice and wheat using an engineered type II CRISPR-Cas system are also presented.
DOI:10.1038/cr.2013.123URLPMID:23999856 [本文引用: 1]

Cell death and differentiation is a monthly research journal focused on the exciting field of programmed cell death and apoptosis. It provides a single accessible source of information for both scientists and clinicians, keeping them up-to-date with advances in the field. It encompasses programmed cell death, cell death induced by toxic agents, differentiation and the interrelation of these with cell proliferation.
DOI:10.1111/pbi.12200URLPMID:24854982

SummaryThe CRISPR/Cas9 system has been demonstrated to efficiently induce targeted gene editing in a variety of organisms including plants. Recent work showed that CRISPR/Cas9-induced gene mutations in Arabidopsis were mostly somatic mutations in the early generation, although some mutations could be stably inherited in later generations. However, it remains unclear whether this system will work similarly in crops such as rice. In this study, we tested in two rice subspecies 11 target genes for their amenability to CRISPR/Cas9-induced editing and determined the patterns, specificity and heritability of the gene modifications. Analysis of the genotypes and frequency of edited genes in the first generation of transformed plants (T0) showed that the CRISPR/Cas9 system was highly efficient in rice, with target genes edited in nearly half of the transformed embryogenic cells before their first cell division. Homozygotes of edited target genes were readily found in T0 plants. The gene mutations were passed to the next generation (T1) following classic Mendelian law, without any detectable new mutation or reversion. Even with extensive searches including whole genome resequencing, we could not find any evidence of large-scale off-targeting in rice for any of the many targets tested in this study. By specifically sequencing the putative off-target sites of a large number of T0 plants, low-frequency mutations were found in only one off-target site where the sequence had 1-bp difference from the intended target. Overall, the data in this study point to the CRISPR/Cas9 system being a powerful tool in crop genome engineering.
DOI:10.1093/mp/sst119URLPMID:23956122

Precise and straightforward methods to edit the plant genome are much needed for functional genomics and crop improvement. Recently, RNA-guided genome editing using bacterial Type II cluster regularly interspaced short palindromic repeats (CRISPR)-associated nuclease (Cas) is emerging as an efficient tool for genome editing in microbial and animal systems. Here, we report the genome editing and targeted gene mutation in plants via the CRISPR–Cas9 system. Three guide RNAs (gRNAs) with a 20–22-nt seed region were designed to pair with distinct rice genomic sites which are followed by the protospacer-adjacent motif (PAM). The engineered gRNAs were shown to direct the Cas9 nuclease for precise cleavage at the desired sites and introduce mutation (insertion or deletion) by error-prone non-homologous end joining DNA repairing. By analyzing the RNA-guided genome-editing events, the mutation efficiency at these target sites was estimated to be 3–8%. In addition, the off-target effect of an engineered gRNA–Cas9 was found on an imperfectly paired genomic site, but it had lower genome-editing efficiency than the perfectly matched site. Further analysis suggests that mismatch position between gRNA seed and target DNA is an important determinant of the gRNA–Cas9 targeting specificity, and specific gRNAs could be designed to target more than 90% of rice genes. Our results demonstrate that the CRISPR–Cas system can be exploited as a powerful tool for gene targeting and precise genome editing in plants. Targeted gene mutation was successfully achieved in rice using the CRISPR–Cas9 system. Experimental analyses of mutation efficiency and off-target effect as well as genome-wide prediction of specific guide RNA seeds suggest that the CRISPR–Cas9 system is a simple and effective tool for plant functional genomics and crop improvement.
[本文引用: 1]
DOI:10.1073/pnas.1420294112URLPMID:25733849 [本文引用: 2]

CRISPR/Cas9 has been widely used for genomic editing in many organisms. Many human diseases are caused by multiple mutations. The CRISPR/Cas9 system provides a potential tool to introduce multiple mutations in a genome. To mimic complicated genomic events in human diseases, such as multiple gene deletions or mutations, two or more small guide RNAs (sgRNA) must be introduced all together. This... [Show full abstract]
DOI:10.1016/j.molp.2015.04.007URLPMID:25917172 [本文引用: 1]

Arobust CRISPR/Cas9 vector system for multiplex genome editing in monocot and dicot plants was developed. Multiple sgRNA expression cassettes can be assembled into the binary CRISPR/Cas9 vectors in one round of cloning. This system can uniformly, efficiently, and simultaneously produce multiple heritable mutations in rice and Arabidopsis by targeting multiple genes or genomic sites via single transformation events.
DOI:10.1093/nar/gku806URLPMID:4176183 [本文引用: 3]

The Cas9/sgRNA of the CRISPR/Cas system has emerged as a robust technology for targeted gene editing in various organisms, including plants, where Cas9/sgRNA-mediated small deletions/insertions at single cleavage sites have been reported in transient and stable transformations, although genetic transmission of edits has been reported only in Arabidopsis and rice. Large chromosomal excision between two remote nuclease-targeted loci has been reported only in a few non-plant species. Here we report in rice Cas9/sgRNA-induced large chromosomal segment deletions, the inheritance of genome edits in multiple generations and construction of a set of facile vectors for high-efficiency, multiplex gene targeting. Four sugar efflux transporter genes were modified in rice at high efficiency; the most efficient system yielding 87-100% editing in T0 transgenic plants, all with di-allelic edits. Furthermore, genetic crosses segregating Cas9/sgRNA transgenes away from edited genes yielded several genome-edited but transgene-free rice plants. We also demonstrated proof-of-efficiency of Cas9/sgRNAs in producing large chromosomal deletions (115-245 kb) involving three different clusters of genes in rice protoplasts and verification of deletions of two clusters in regenerated T0 generation plants. Together, these data demonstrate the power of our Cas9/sgRNA platform for targeted gene/genome editing in rice and other crops, enabling both basic research and agricultural applications.
DOI:10.1038/srep10342URLPMID:26022141 [本文引用: 1]

Abstract Genome editing is a valuable technique for gene function analysis and crop improvement. Over the past two years, the CRISPR-Cas9 system has emerged as a powerful tool for precisely targeted gene editing. In this study, we predicted 11 U6 genes in soybean (Glycine max L.). We then constructed two vectors (pCas9-GmU6-sgRNA and pCas9-AtU6-sgRNA) using the soybean U6-10 and Arabidopsis U6-26 promoters, respectively, to produce synthetic guide RNAs (sgRNAs) for targeted gene mutagenesis. Three genes, Glyma06g14180, Glyma08g02290 and Glyma12g37050, were selected as targets. Mutations of these three genes were detected in soybean protoplasts. The vectors were then transformed into soybean hairy roots by Agrobacterium rhizogenes infection, resulting in efficient target gene editing. Mutation efficiencies ranged from 3.2-9.7% using the pCas9-AtU6-sgRNA vector and 14.7-20.2% with the pCas9-GmU6-sgRNA vector. Biallelic mutations in Glyma06g14180 and Glyma08g02290 were detected in transgenic hairy roots. Off-target activities associated with Glyma06g14180 and Glyma12g37050 were also detected. Off-target activity would improve mutation efficiency for the construction of a saturated gene mutation library in soybean. Targeted mutagenesis using the CRISPR-Cas9 system should advance soybean functional genomic research, especially that of genes involved in the roots and nodules.
DOI:10.1038/srep24765URLPMID:27097775 [本文引用: 1]

Abstract The CRISPR/Cas9 system has successfully been used in various organisms for precise targeted gene editing. Although it has been demonstrated that CRISPR/Cas9 system can induce mutation in tomato plants, the stability of heredity in later generations and mutant specificity induced by the CRISPR/Cas9 system in tomato plants have not yet been elucidated in detail. In this study, two genes, SlPDS and SlPIF4, were used for testing targeted mutagenesis in tomato plants through an Agrobacterium tumefaciens-mediated transformation method. A high mutation frequency was observed in all tested targets in the T0 transgenic tomato plants, with an average frequency of 83.56%. Clear albino phenotypes were observed for the psd mutants. High frequencies of homozygous and biallelic mutants were detected even in T0 plants. The majority of the detected mutations were 1- to 3-nucleotide deletions, followed by 1-bp insertions. The target mutations in the T0 lines were stably transmitted to the T1 and T2 generations, without new modifications or revision. Off-target activities associated with SlPDS and SlPIF4 were also evaluated by sequencing the putative off-target sites, and no clear off-target events were detected. Our results demonstrate that the CRISPR/Cas9 system is an efficient tool for generating stable and heritable modifications in tomato plants.
[本文引用: 2]
DOI:10.1016/j.jgg.2013.12.001URLPMID:24576457 [本文引用: 1]

Transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems have emerged as powerful tools for genome editing in a variety of species. Here, we report, for the first time, targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. We designed five TALENs targeting 4 genes, namely ZmPDS, ZmIPK1A, ZmIPK, ZmMRP4, and obtained targeting efficiencies of up to 23.1% in protoplasts, and about 13.3% to 39.1% of the transgenic plants were somatic mutations. Also, we constructed two gRNAs targeting the ZmIPK gene in maize protoplasts, at frequencies of 16.4% and 19.1%, respectively. In addition, the CRISPR/Cas system induced targeted mutations in Z. mays protoplasts with efficiencies (13.1%) similar to those obtained with TALENs (9.1%). Our results show that both TALENs and the CRISPR/Cas system can be used for genome modification in maize.
DOI:10.1038/srep43902URLPMID:5335549 [本文引用: 1]

The complex allotetraploid genome is one of major challenges in cotton for repressing gene expression. Developing site-specific DNA mutation is the long-term dream for cotton breeding scientists. The clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) system is emerging as a robust biotechnology for targeted-DNA mutation. In this study, two sgRNAs, GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2, were designed in the identical genomic regions ofGhMYB25-like AandGhMYB25-like D, which were encoded by cotton A subgenome and the D subgenome, respectively, was assembled to direct Cas9-mediated allotetraploid cotton genome editing. High proportion (14.2-21.4%) CRISPR/Cas9-induced specific truncation events, either fromGhMYB25-like ADNA site or fromGhMYB25-like DDNA site, were detected in 50% examined transgenic cotton through PCR amplification assay and sequencing analyses. Sequencing results also demonstrated that 100% and 98.8% mutation frequency were occurred on GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2 target site respectively. The off-target effect was evaluated by sequencing two putative off-target sites, which have 3 and 1 mismatched nucleotides with GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2, respectively; all the examined samples were not detected any off-target-caused mutation events. Thus, these results demonstrated that CRISPR/Cas9 is qualified for generating DNA level mutations on allotetraploid cotton genome with high-efficiency and high-specificity.
DOI:10.1038/nprot.2014.157URLPMID:25232936 [本文引用: 2]

Targeted genome editing nucleases, such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), are powerful tools for understanding gene function and for developing valuable new traits in plants. The clustered regularly interspersed short palindromic repeats (CRISPR)/Cas system has recently emerged as an alternative nuclease-based method for efficient and versatile genome engineering. In this system, only the 20-nt targeting sequence within the single-guide RNA (sgRNA) needs to be changed to target different genes. The simplicity of the cloning strategy and the few limitations on potential target sites make the CRISPR/Cas system very appealing. Here we describe a stepwise protocol for the selection of target sites, as well as the design, construction, verification and use of sgRNAs for sequence-specific CRISPR/Cas-mediated mutagenesis and gene targeting in rice and wheat. The CRISPR/Cas system provides a straightforward method for rapid gene targeting within 1-2 weeks in protoplasts, and mutated rice plants can be generated within 13-17 weeks.
[本文引用: 1]
[本文引用: 2]
DOI:10.1093/mp/sst121URLPMID:23963532 [本文引用: 2]
DOI:10.1186/s12870-014-0327-yURLPMID:4262988 [本文引用: 4]

Background To accelerate the application of the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/ CRISPR-associated protein 9) system to a variety of plant species, a toolkit with additional plant selectable markers, more gRNA modules, and easier methods for the assembly of one or more gRNA expression cassettes is required. Results We developed a CRISPR/Cas9 binary vector set based on the pGreen or pCAMBIA backbone, as well as a gRNA (guide RNA) module vector set, as a toolkit for multiplex genome editing in plants. This toolkit requires no restriction enzymes besides BsaI to generate final constructs harboring maize-codon optimized Cas9 and one or more gRNAs with high efficiency in as little as one cloning step. The toolkit was validated using maize protoplasts, transgenic maize lines, and transgenic Arabidopsis lines and was shown to exhibit high efficiency and specificity. More importantly, using this toolkit, targeted mutations of three Arabidopsis genes were detected in transgenic seedlings of the T1 generation. Moreover, the multiple-gene mutations could be inherited by the next generation. Conclusions We developed a toolkit that facilitates transient or stable expression of the CRISPR/Cas9 system in a variety of plant species, which will facilitate plant research, as it enables high efficiency generation of mutants bearing multiple gene mutations.
[本文引用: 2]
[本文引用: 1]
DOI:10.1038/nbt.2969URLPMID:25038773 [本文引用: 1]

Sequence-specific nucleases have been applied to engineer targeted modifications in polyploid genomes, but simultaneous modification of multiple homoeoalleles has not been reported. Here we use transcription activator-like effector nuclease (TALEN) and clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9 (refs. 4,5) technologies in hexaploid bread wheat to introduce targeted mutations in the three homoeoalleles that encode MILDEW-RESISTANCE LOCUS (MLO) proteins. Genetic redundancy has prevented evaluation of whether mutation of all three MLO alleles in bread wheat might confer resistance to powdery mildew, a trait not found in natural populations. We show that TALEN-induced mutation of all three TaMLO homoeologs in the same plant confers heritable broad-spectrum resistance to powdery mildew. We further use CRISPR-Cas9 technology to generate transgenic wheat plants that carry mutations in the TaMLO-A1 allele. We also demonstrate the feasibility of engineering targeted DNA insertion in bread wheat through nonhomologous end joining of the double-strand breaks caused by TALENs. Our findings provide a methodological framework to improve polyploid crops.
DOI:10.1371/journal.pone.0154027URLPMID:27116122 [本文引用: 1]

Rice blast is one of the most destructive diseases affecting rice worldwide. The adoption of host resistance has proven to be the most economical and effective approach to control rice blast. In recent years, sequence-specific nucleases (SSNs) have been demonstrated to be powerful tools for the improvement of crops via gene-specific genome editing, and CRISPR/Cas9 is thought to be the most effective SSN. Here, we report the improvement of rice blast resistance by engineering a CRISPR/Cas9 SSN (C-ERF922) targeting theOsERF922gene in rice. Twenty-one C-ERF922-induced mutant plants (42.0%) were identified from 50 T0transgenic plants. Sanger sequencing revealed that these plants harbored various insertion or deletion (InDel) mutations at the target site. We showed that all of the C-ERF922-induced allele mutations were transmitted to subsequent generations. Mutant plants harboring the desired gene modification but not containing the transferred DNA were obtained by segregation in the T1and T2generations. Six T2homozygous mutant lines were further examined for a blast resistance phenotype and agronomic traits, such as plant height, flag leaf length and width, number of productive panicles, panicle length, number of grains per panicle, seed setting percentage and thousand seed weight. The results revealed that the number of blast lesions formed following pathogen infection was significantly decreased in all 6 mutant lines compared with wild-type plants at both the seedling and tillering stages. Furthermore, there were no significant differences between any of the 6 T2mutant lines and the wild-type plants with regard to the agronomic traits tested. We also simultaneously targeted multiple sites withinOsERF922by using Cas9/Multi-target-sgRNAs (C-ERF922S1S2 and C-ERF922S1S2S3) to obtain plants harboring mutations at two or three sites. Our results indicate that gene modification via CRISPR/Cas9 is a useful approach for enhancing blast resistance in rice.
[本文引用: 2]
[本文引用: 2]
DOI:10.1016/j.molp.2016.03.003URLPMID:26995294 [本文引用: 2]

Dear Editor, The CRISPR/Cas9 system has emerged as a versatile molecular tool for genome editing in various organisms,including plants.In this system,the specificity of Cas9-directed DNA cleavage strictly requires the presence of a chimeric single guide RNA (sgRNA) and a short trinucleotide protospacer adjacent motif (PAM) in the genome (Anders et al.,2014;Sternberg et al.,2014).To date,the Cas9 used in plants has only been shown to recognize PAM sequences in the canonical form NGG (Li et al.,2013;Miao et al.,2013;Shan et al.,2013;Ma et al.,2015b).As such,the range of sequences for genome editing in plants is limited to sites containing an NGG motif.Many attempts have been made to overcome this constraint,and recent work has revealed that Cas9 can be modified to recognize alternative PAM sequences in zebrafish and human cells (Kleinstiver et al.,2015).However,the targeting range limitations of the CRISPR/Cas9 systems in plants have yet to be resolved.The widely cultivated plant rice is not only an important food crop but also a model crop plant because of its relatively small genome and relative ease of transformation.To expand the range of genome editing in rice,we generated two Cas9 variants as reported (Kleinstiver et al.,2015) and investigated their genome editing performance in rice.We demonstrate that Cas9 can be engineered to target sites containing alternative non-canonical PAMs in rice,which significantly broadens the range of genome editing.
DOI:10.1111/pbi.12771URLPMID:28605576 [本文引用: 2]

Abstract Clustered regularly interspaced short palindromic repeats-associated protein 9 (CRISPR-Cas9) is a revolutionary technology that enables efficient genomic modification in many organisms. Currently, the wide use of Streptococcus pyogenes Cas9 (SpCas9) primarily recognises sites harbouring a canonical NGG protospacer adjacent motif (PAM). The newly developed VQR (D1135V/R1335Q/T1337R) variant of Cas9 has been shown to cleave sites containing NGA PAM in rice, which greatly expanded the range of genome editing. However, the low editing efficiency of the VQR variant remains, which limits its wide application in genome editing. In this study, by modifying the single guide RNA (sgRNA) structure and using strong endogenous promoters, we significantly increased the editing efficiency of the VQR variant. The modified CRISPR-Cas9-VQR system provides a robust toolbox for multiplex genome editing at sites containing non-canonical NGA PAM. This article is protected by copyright. All rights reserved. This article is protected by copyright. All rights reserved.
[本文引用: 1]
[本文引用: 1]
