 ,2,3,*, 唐晓丹3, 赵新辉3, 周在为3, 符星学3, 王凯4, 史江伟3, 李艳锋4, 符辰建2,3, 杨远柱
,2,3,*, 唐晓丹3, 赵新辉3, 周在为3, 符星学3, 王凯4, 史江伟3, 李艳锋4, 符辰建2,3, 杨远柱 ,1,2,3,4,*
,1,2,3,4,*Construction of tms5 Mutants in Rice Based on CRISPR/Cas9 Technology
HUANG Zhong-Ming1, ZHOU Yan-Biao ,2,3,*, TANG Xiao-Dan3, ZHAO Xin-Hui3, ZHOU Zai-Wei3, FU Xing-Xue3, WANG Kai4, SHI Jiang-Wei3, LI Yan-Feng4, FU Chen-Jian2,3, YANG Yuan-Zhu
,2,3,*, TANG Xiao-Dan3, ZHAO Xin-Hui3, ZHOU Zai-Wei3, FU Xing-Xue3, WANG Kai4, SHI Jiang-Wei3, LI Yan-Feng4, FU Chen-Jian2,3, YANG Yuan-Zhu ,1,2,3,4,*
,1,2,3,4,*通讯作者:
第一联系人:
收稿日期:2017-10-25接受日期:2018-03-26网络出版日期:2018-06-12
| 基金资助: |
Received:2017-10-25Accepted:2018-03-26Online:2018-06-12
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (2976KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
黄忠明, 周延彪, 唐晓丹, 赵新辉, 周在为, 符星学, 王凯, 史江伟, 李艳锋, 符辰建, 杨远柱. 基于CRISPR/Cas9技术的水稻温敏不育基因tms5突变体的构建[J]. 作物学报, 2018, 44(6): 844-851. doi:10.3724/SP.J.1006.2018.00844
HUANG Zhong-Ming, ZHOU Yan-Biao, TANG Xiao-Dan, ZHAO Xin-Hui, ZHOU Zai-Wei, FU Xing-Xue, WANG Kai, SHI Jiang-Wei, LI Yan-Feng, FU Chen-Jian, YANG Yuan-Zhu.
水稻是我国最重要的粮食作物之一, 针对紧缺的自然资源和有限的耕地面积, 粮食的供需变得越来越不平衡, 培育高产稳产的优质作物品种是确保粮食安全的根本途径。杂交水稻的产量比常规水稻提高20%~30%, 因此发展杂交水稻是解决粮食短缺问题的重要方法[1]。
杂交水稻的育种方法主要包括两系法和三系法。三系即保持系、不育系和恢复系, 主要是通过细胞质雄性不育系和恢复系杂交产生杂交种子, 利用保持系和不育系杂交繁殖细胞质雄性不育系。两系即不育系和恢复系, 主要是利用光/温敏雄性核不育系与恢复系杂交产生杂交种子。光/温敏不育系在长日/高温条件下花粉败育, 而在短日/低温条件下能恢复花粉育性, 故可一系两用。光/温敏雄性核不育系的育性由核基因控制, 无恢保关系限制, 配组自由, 选出优良组合机率高[2]。因此, 光/温敏不育系在杂交水稻育种中潜力巨大, 被广泛应用。杂交水稻的品种选育, 从三系发展到两系, 如何进一步突破产量潜力, 实现高产与优质高效结合, 是目前的迫切需求。2014年, Zhou等[3]克隆了安农S-1和株1S的温敏不育基因TMS5, 首次揭示了水稻温敏雄性不育的分子机制。序列分析结果显示, 安农S-1和株1S中TMS5编码区第71位碱基均由C突变成A, 形成了一个提前的终止密码子, 导致TMS5蛋白不能正确合成。
培育优良的杂交水稻组合, 首要要实现不育系的突破。常规杂交转育是培育两系不育系较常用的方法。但常规杂交育种有选育周期漫长和一些优良性状会丢失等缺点, 越来越难以实现日益提高的育种目标。目前, CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR- associated protein 9)基因编辑技术可对作物内源基因进行定向改造, 并且具有成本低廉、技术门槛低、操作简单、试验周期短等优点, 为作物遗传改良提供了新的途径[4,5,6]。CRISPR/Cas9 系统主要包括 Cas9核酸酶和 sgRNA 两个重要组分。这两个组分形成复合体, 切割与sgRNA上的间隔序列(spacers)互补的基因组DNA, 引起DNA双链断裂, 并通过体内的非同源末端连接(non-homologous end joining, NHEJ)进行修复形成碱基缺失、插入、替换等突变类型[7,8]。2013年, Shan等[9]利用CRISPR/Cas9技术对水稻OsBADH2、OsPDS基因进行编辑, 获得osbadh2及ospds突变体, 可以用于水稻香米株系和耐冷性株系的创建。同年, 冯争艳等[10]对水稻ROC5、OsWaxy基因进行突变, 可以用于水稻卷叶育种及糯性育种。2016 年, 中国科学院遗传与发育生物学研究所高彩霞研究组和李家洋研究组合作利用在修复途径中占主导地位的非同源末端连接(NHEJ)修复方式在水稻中建立了基于 CRISPR/Cas9 技术的基因替换以及基因定点插入体系, 并通过碱基替换策略获得抗草甘膦的水稻材料[11]。CRISPR/Cas9 基因编辑技术的发展使其在水稻遗传改良中的应用越来越广泛[12,13]。本研究利用CRISPR/Cas9技术定点编辑温敏不育基因TMS5, 获得了粳稻F197的tms5突变体, 并对其不育起点温度进行了研究, 为利用TMS5基因创制新型的温敏不育新种质提供了材料基础。
1 材料与方法
1.1 靶位点设计
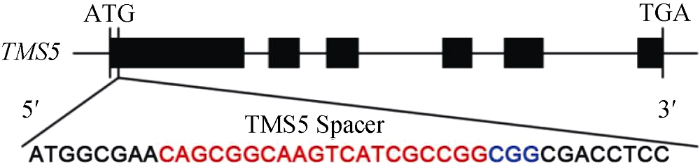
设计TMS5 (LOC_Os02g12290)靶点序列参照Xie等的方法[14]。在TMS5第1外显子上, 靠近编码蛋白质的N’端, 设计长度为20 bp的靶位点序列, PAM序列为CGG (图1)。靶点序列正义链和反义链的5°端分别添加TGGC和AAAC, 使其与中间载体18T-Cas9经Bbs I酶切后形成黏性互补末端。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1TMS5靶点位置
红色的字母为靶点序列; 蓝色的字母为PAM序列。
Fig. 1Schematic diagram of the targeted site in TMS5
The red letters are the target genome sequences; The blue letters are the protospacer adjacent motif (PAM) sequences.
1.2 CRISPR/Cas9表达载体的构建
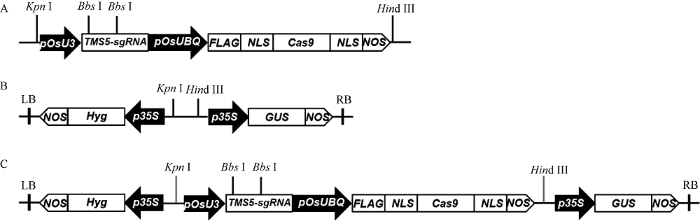
利用合成的靶点序列引物Target-1 (表1), 经过98℃处理30 s, 再室温放置5 min制成互补双链DNA, 用于载体构建。用Bbs I酶切18T-Cas9载体、酶切体系包括18T-Cas9载体2 μg、10×FastDigest buffer 2 μL、Bbs I 0.1 μL (10 U μL-1), 加ddH2O至20 μL。对酶切后的18T-Cas9质粒进行胶回收, 然后用T4 DNA连接酶将TMS5的接头与回收后的18T-Cas9载体连接。连接产物转化大肠杆菌DH5a, 用18T-Cas9载体上的引物M13F与靶点序列引物Target-1R进行菌液PCR检测, 挑选阳性菌株送公司测序, 筛选正确的重组中间载体18T-Cas9-TMS5- gRNA, 然后分别用Kpn I和Hind III酶切植物表达载体pCAMBIA1301和重组的中间载体, 回收酶切产物, 再通过T4 DNA连接酶连接, 构建pCAMBIA1301-Cas9-TMS5-gRNA表达载体, 再通过酶切鉴定确认植物表达载体构建成功后转化农杆菌EHA105, 载体构建的具体流程见图2。Table 1
表1
表1本研究所用的引物
Table 1
| 引物名称 Primer name | 引物序列 Primer sequence (5°-3°) |
|---|---|
| Target-1F | TGGCGAACAGCGGCAAGTCATCGC |
| Target -1R | AAACGCGATGACTTGCCGCTGTTC |
| M13F | GTAAAACGACGGCCAG |
| GUS-JC-F | CGTCCGTCCTGTAGAAACCC |
| GUS-JC-R | GTGCGGATTCACCACTTGC |
| TMS5-CX-F | TCCAACGCATAGCAGTAGTCG |
| TMS5-CX-R | TGCCATCGTATCTCCGGTAAA |
新窗口打开|下载CSV
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2CRISPR/Cas9-TMS5载体构建示意图
A: 18T-Cas9-TMS5-gRNA重组载体结构示意图; B: pCAMBIA1301载体的T-DNA片段; C: pCAMBIA1301-Cas9-TMS5-gRNA重组载体结构示意图。LB: 左边界; RB: 右边界。
Fig. 2Schematic diagram of the CRISPR/Cas9-TMS5 vector construction
A: schematic diagram of the recombinant vector of 18T-Cas9-TMS5-gRNA; B: T-DNA fragment of pCAMBIA1301 vector; C: schematic diagram of the pCAMBIA1301-Cas9-TMS5-gRNA construct. LB: left border; RB: right border.
1.3 转基因阳性植株的获得
将构建好的阳性农杆菌EHA105转化粳稻F197的愈伤组织, 用潮霉素筛选获得T0代植株。提取T0代植株的DNA, 用载体上的特异引物GUS-JC-F/ GUS-JC-R (表1)进行PCR扩增, 扩增产物大小为718 bp, 能扩增出目的大小片段的植株为阳性转基因植株。同时, 因为pCAMBIA1301载体上有GUS报告基因, 因此可以通过GUS组织化学染色[15]的方法快速鉴定阳性转基因植株。1.4 测序检测靶位点
为了检测靶位点的突变情况, 在TMS5的靶位点两端分别设计测序引物TMS5-CX-F和TMS5- CX-R (表1), 扩增产物大小为974 bp。将测序引物扩增的目的片段送测序公司测序。同时, 以野生型F197的基因组DNA为模板, 将测序引物扩增目的片段的测序结果作为对照序列。1.5 T1代无选择标记基因突变株的获得
对T1代植株的叶片进行GUS组织化学染色, 不能染上蓝色的为无选择标记基因的植株, 以野生型叶片的染色为对照。再以无选择标记基因植株的基因组为模板用TMS5-JC-F和TMS5-JC-R引物扩增, 将目的条带测序, 根据测序结果分析TMS5是否有突变。1.6 碘-碘化钾染色检测花粉育性
对照F197和T1代无转基因元件且TMS5功能缺失的突变株在主茎幼穗分化到六期的时候进行人工控光温处理, 分别用24℃、28℃和32℃处理14 d (12 h光照/12 h黑夜)后, 在自然条件下生长, 用碘-碘化钾染色[16]观察除主茎外的其他茎节上花粉的育性, 并调查成熟期F197和tms5突变体的结实率。1.7 大田农艺性状调查
参照Zhou等的方法[17], 以F197为对照, 于2017年5月至10月在湖南长沙自然条件下调查tms5突变体的田间农艺性状。5月25日播种, 8月11日至20日为tms5突变体幼穗敏感期, 这期间日平均气温为27.8℃。在成熟期选择F197和tms5突变体各20株, 分别考察株高、千粒重、有效穗、结实率和单株产量, 采用t检验统计分析数据。2 结果与分析
2.1 CRISPR/Cas9表达载体的构建
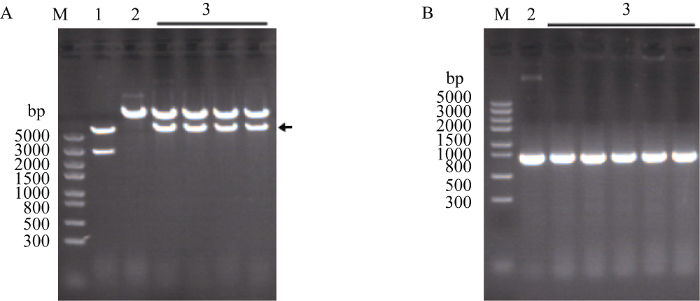
为了获得粳稻F197的TMS5基因敲除的突变体, 根据CRISPR/Cas9技术原理, 结合生物信息学网站http://www.genome.arizona.edu/crispr/, 在TMS5第1外显子区域设计20 bp的靶点序列Target-1 (表1和图1)。退火形成的双链互补靶序列与中间载体18T-Cas9连接形成重组载体18T-Cas9-Rice-TMS5- gRNA (图2-A)。重组载体经Kpn I和Hind III双酶切, 将CRISPR/Cas9元件连接到植物表达载体pCAMBIA1301中形成重组载体pCAMBIA1301- Cas9-TMS5-gRNA (图2-B, C)。通过酶切鉴定, 说明pCAMBIA1301-Cas9-TMS5-gRNA表达载体构建成功(图3-A)。将构建成功的重组载体通过电击转化农杆菌EHA105, 再用pCAMBIA1301载体上的报告基因GUS的检测引物GUS-JC-F/GUS-JC-R (表1)进行菌液PCR, 扩增产物大小为718 bp (图3-B), 能扩增出目的片段大小的菌液为阳性菌株, 将阳性的农杆菌菌株用于转化粳稻F197。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3酶切和PCR鉴定pCAMBIA1301-Cas9-TMS5-gRNA载体
A: Kpn I和Hind III酶切鉴定pCAMBIA1301-Cas9-TMS5-gRNA载体。箭头所示为CRISPR/Cas9系统元件的片段; B: 利用GUS-JC-F和GUS-JC-R引物PCR鉴定农杆菌中的pCAMBIA1301-Cas9-TMS5-gRNA载体。M: DL2000; 1: 18T-Cas9-TMS5-gRNA; 2: pCAMBIA1301; 3: pCAMBIA1301-Cas9-TMS5-gRNA。
Fig. 3Identification of the pCAMBIA1301-Cas9-TMS5-gRNA plasmid by restriction enzyme digestion and PCR
A: identification of the pCAMBIA1301-Cas9-TMS5-gRNA plasmid digested with Kpn I and Hind III; B: identification of the pCAMBIA1301-Cas9-TMS5-gRNA plasmid in Agrobacterium tumefaciens by PCR with primers of GUS-JC-F and GUS-JC-R. M: DL2000; 1: 18T-Cas9-TMS5-gRNA; 2: pCAMBIA1301; 3: pCAMBIA1301-Cas9-TMS5-gRNA.
2.2 转基因植株的鉴定及突变位点测序分析
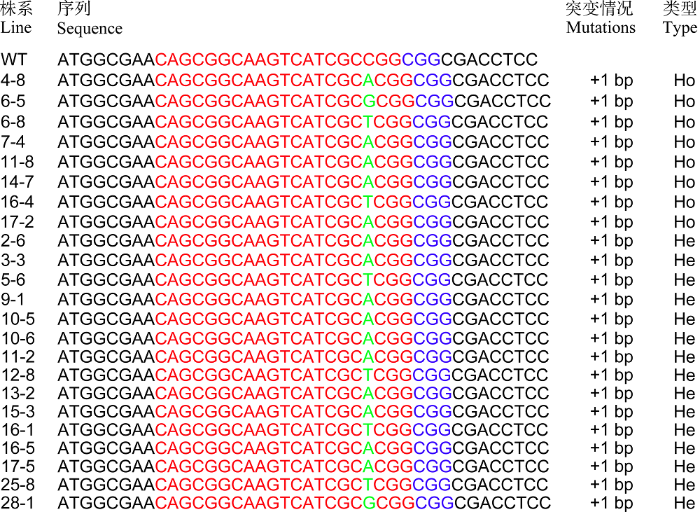
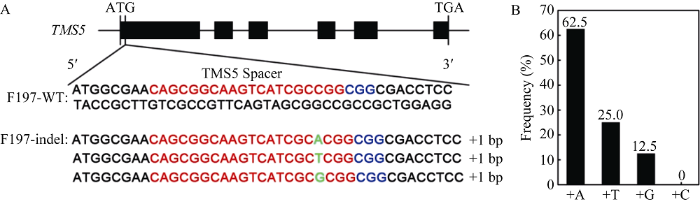
采用CTAB法提取T0代水稻植株叶片的基因组DNA, 用GUS报告基因的特异引物GUS-JC-F/GUS- JC-R进行PCR, 扩增目的片段大小为718 bp的植株为阳性转基因植株(图4-A)。另外, 对T0代植株进行GUS组织化学染色可以快速地鉴定转基因植株, 能染上蓝色的为阳性转基因植株(图4-B)。在F197中共获得57株组培苗, 其中有36株阳性转基因植株。为了分析T0代阳性转基因植株TMS5的突变情况, 在TMS5靶点附近设计测序引物TMS5-CX-F/ TMS5-CX-R。PCR测序结果表明, 在36株阳性转基因植株中, 检测到23株TMS5发生突变, 其中纯合突变有8株, 纯合突变率为34.78%, 其余15株的靶序列突变位点附近出现双峰。将出现双峰的测序文件通过在线工具DSDecode (http://skl.scau.edu.cn/ dsdecode/)自动解码[18], 结果显示均为杂合突变(附图)。纯合突变主要来自PAM前第3和第4碱基之间有单碱基A、T和G的插入, 分别占62.5%、25.0%和12.5%, 而没有检测到单碱基C的插入(图5)。附图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图1靶位点突变序列分析
红色的字母为靶点序列; 蓝色的字母为PAM序列; 绿色的字母为插入的碱基; +表示插入; WT表示野生型; Ho表示纯合突变; He表示杂合突变。
Supplementary Fig. 1Analysis of mutation sequences of target
The red letters are the target genome sequences; the blue letters are PAM; the green letters are the insert base; +: Insertion; WT: Wild-type; Ho: homozygous mutation; He: heterozygous mutation.
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4转基因植株的鉴定
A: PCR检测阳性转基因植株; B: GUS组织化学染色检测阳性转基因植株。M: DL2000; +: 阳性对照; 1-18: 转基因植株。
Fig. 4Identification of the transgenic plants
A: identification of the positive transgenic plants by PCR; B: identification of the positive transgenic plants by GUS histochemical staining. M: DL2000; +: positive control; 1-18: transgenic plants.
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5T0代转基因植株TMS5纯合突变类型和频率
A: 纯合突变类型序列比对; B: 纯合突变类型不同碱基插入的频率。红色字母为靶点序列; 蓝色字母为PAM序列; 绿色的字母为插入的碱基。
Fig. 5Mutation types and frequencies at the TMS5 loci in T0 transgenic plants
A: Sequence alignment of homozygous mutation types; B: Frequency of different base inserts in homozygous mutation types. The red letters are the target genome sequences; The blue letters are PAM; The green letters are the insert base.
2.3 T1代无选择标记基因突变株的获得及突变株不育起点温度的鉴定
将上述单碱基A或T或G插入的纯合突变单株自交, 结实后得到T1代种子, 将其播种成苗后进行GUS染色, 没有染上蓝色的为不含转基因成分的单株, 然后提取非转基因植株叶片的基因组DNA进行PCR测序, 分别得到单碱基A或T或G插入的不含转基因成分的单株, 编号分别为tms5-1、tms5-2、tms5-3用于不育起点温度的鉴定。当野生型(WT)和tms5突变体主茎的幼穗处于六期的时候, 将WT和tms5突变体置于24℃、28℃和32℃的恒温培养箱分别培养14 d, 然后对除主茎外的其他茎节上的花粉进行镜检, 并考察成熟期结实率。结果表明, 在24℃培养下, WT和tms5突变体的花粉都是可育的, 在成熟期WT和tms5突变体的结实率没有显著变化; 28℃培养下, tms5突变体的花粉大部分是不育的, 而WT的花粉是可育的, 在成熟期tms5突变体的结实率显著低于WT的; 32℃培养下, tms5突变体的花粉是完全不育的, 而野生型的花粉是可育的, 在成熟期tms5突变体的结实率为0 (图6)。因此, 28℃是突变株花粉育性的转换温度。
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6野生型和tms5突变体在不同温度处理条件下的花粉育性和结实率
数据表示平均值±方差。**表示tms5突变体与WT之间存在显著差异(P < 0.01)。
Fig. 6Pollen fertility and seed setting rate of wild-type (WT) and tms5 mutant at different temperatures treatments
Data are presented as average values ± standard error of the mean. **, significant difference between tms5 mutant and WT (P < 0.01).
2.4 T1代植株农艺性状分析
为了研究F197的TMS5基因缺失后对植株农艺性状的影响。我们调查了野生型和tms5突变体的株高、千粒重、结实率、有效穗和单株重。结果表明tms5突变体的结实率和单株重比野生型明显降低(P<0.01), 而株高、千粒重、有效穗与野生型相比均无显著差异(表2)。Table 2
表2
表2野生型(WT)和tms5突变体的农艺性状
Table 2
| 株系 Line | 株高 Plant height (cm) | 千粒重 1000-grain weight (g) | 有效穗 Panicle number per plant | 结实率 Seed setting rate (%) | 单株产量 Grain yield per plant (g) |
|---|---|---|---|---|---|
| WT | 78.96±3.56 | 20.82±0.97 | 9.3±1.87 | 72.21±3.57 | 17.30±1.09 |
| tms5 | 78.57±3.87 | 21.20±0.88 | 9.85±1.79 | 61.60±3.43** | 15.22±1.05** |
新窗口打开|下载CSV
3 讨论
CRISPR/Cas9基因编辑技术是一种新兴的基因组定点编辑技术, 具有操作简单、编辑效率高、成本低廉等特点。由于插入位置与作用位点分离, 在转基因植物的后代中通过自交等手段可将转基因成分从基因组中分离出去, 从而最小范围地特异性地改变植物基因组。因此, CRISPR/Cas9基因组编辑技术可以帮助育种工作者更加快速、准确地定向改造作物品种。目前该技术已经在许多作物中广泛应用, 如水稻[19,20]、玉米[21,22]、小麦[23,24]、大豆[25,26]等。2014年, Zhou等[3]克隆了安农S-1和株1S的温敏不育基因TMS5, 首次揭示了水稻温敏雄性不育的分子机制。本研究为探讨TMS5基因功能缺失在粳稻中的效应, 利用CRISPR/Cas9基因编辑技术在粳稻品种F197中敲除TMS5基因, 成功获得了功能缺失的tms5突变体, 并在T1代tms5功能缺失的非转基因材料中考察了花粉育性的起点温度和农艺性状。CRISPR/Cas9系统中的Cas9核酸酶以PAM上游第3碱基处为切割中心进行突变[27]。其中, 超过一半的突变为单碱基插入, 其中又以A 或T插入为主[28,29]。本研究中, TMS5基因的纯合突变发生在PAM上游第3碱基和第4碱基之间, 并且是单碱基A或T或G的插入, 其中单碱基A插入占62.5%, 单碱基T插入占25%, 单碱基G插入占12.5%。TMS5基因的突变情况与前人的研究结果类似。
研究显示我国25个主要使用的两用光/温敏核不育系中有24个均含有 TMS5基因, 表明TMS5基因是控制我国温敏型不育系的主要不育基因[1]。Zhou等[3]利用CRISPR/Cas9技术对粳稻品种中花11、GAZ和籼稻品种粤晶丝苗、泰丰B、五山丝苗、ReB、珍汕97B的TMS5基因进行敲除, 并分别获得了这些品种的TMS5功能缺失的突变体。粳稻品种中花11和GAZ的tms5突变体在28℃培养下花粉表现为完全不育, 而籼稻品种粤晶丝苗、泰丰B、五山丝苗、ReB、珍汕97B的tms5突变体分别在24℃、26℃、26℃、26℃、28℃培养下花粉表现为完全不育[18]。同时, 水稻广亲和温敏不育系籼稻株1S的不育起点温度为22.6℃[30,31]。因此, 可以看出籼稻品种的TMS5功能缺失突变体的不育起点温度较粳稻品种低, 这种结果可能是粳稻和籼稻的遗传背景不同造成的, 同时也说明TMS5基因功能缺失与不育起点温度无关。本研究中, 粳稻F197的tms5突变体花粉在24℃培养下完全可育, 28℃培养下大部分不育, 32℃培养下完全不育(图6)。因此, 28℃可能是tms5突变体花粉育性的转换温度。
产量是作物最重要的性状之一, 由多个产量相关性状共同决定, 并且易受外界环境的影响。产量的决定因素主要有千粒重、结实率和有效穗数[32]。TMS5编码一个短版的RNAase Z同源蛋白RNAase ZS1, 通过对UbL40mRNA的加工调控水稻温敏雄性不育[1,3]。本试验中粳稻品种F197的tms5突变体由于花粉育性降低, 导致其结实率比野生型的低, 但是tms5突变体的株高、千粒重和有效穗数与野生型相比没有明显变化。
4 结论
利用CRISPR/Cas9技术获得了敲除温敏不育基因TMS5的粳稻品种F197的突变体, 为培育水稻温敏雄性核不育系提供了重要材料基础。28℃是tms5突变体的育性转换温度, 并且在32℃表现为完全不育, 这为敲除TMS5的粳稻品种不育起点温度的研究提供了重要参考。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 3]
[本文引用: 3]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/ncomms5884URLPMID:25208476 [本文引用: 4]

Thermosensitive genic male-sterile (TGMS) lines, which are male-sterile at restrictive (high) temperatures but male-fertile at permissive (low) temperatures, have been widely used in breeding two-line hybrid rice (Oryza sativa L.). Here we find that mutation of thermosensitive genic male sterile 5 (tms5) in rice causes the TGMS trait through a loss of RNase Z(S1) function. We show that RNase Z(S1) processes the mRNAs of three ubiquitin fusion ribosomal protein L40 (UbL40) genes into multiple fragments in vitro and in vivo. In tms5 mutants, high temperature results in increased levels of UbL40 mRNAs. Overaccumulation of UbL40 mRNAs causes defective pollen production and male sterility. Our results uncover a novel mechanism of RNase Z(S1)-mediated UbL40 mRNA regulation and shows that loss of this regulation produces TGMS in rice, a finding with potential applications in hybrid crop breeding.
DOI:10.1016/j.copbio.2014.11.007URL [本文引用: 1]
DOI:10.1016/j.tibtech.2014.11.008URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/nature10886URL [本文引用: 1]
DOI:10.1146/annurev-genet-110410-132435URL [本文引用: 1]
DOI:10.1038/nbt.2650URLPMID:23929338 [本文引用: 1]

The article offers information on genome modification of crop plants using a CRISPR-Cas system. It states that genome editing technologies using zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) can also generate genome modifications. Photographs related to genome editing in rice and wheat using an engineered type II CRISPR-Cas system are also presented.
DOI:10.1038/nbt.2501URLPMID:23360964 [本文引用: 1]

In bacteria, foreign nucleic acids are silenced by clustered, regularly interspaced, short palindromic repeats (CRISPR)--CRISPR-associated (Cas) systems. Bacterial type II CRISPR systems have been adapted to create guide RNAs that direct site-specific DNA cleavage by the Cas9 endonuclease in cultured cells. Here we show that the CRISPR-Cas system functions in vivo to induce targeted genetic modifications in zebrafish embryos with efficiencies similar to those obtained using zinc finger nucleases and transcription activator-like effector nucleases.
DOI:10.1038/nplants.2016.139URLPMID:27618611 [本文引用: 1]

Abstract Sequence-specific nucleases have been exploited to create targeted gene knockouts in various plants(1), but replacing a fragment and even obtaining gene insertions at specific loci in plant genomes remain a serious challenge. Here, we report efficient intron-mediated site-specific gene replacement and insertion approaches that generate mutations using the non-homologous end joining (NHEJ) pathway using the clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9) system. Using a pair of single guide RNAs (sgRNAs) targeting adjacent introns and a donor DNA template including the same pair of sgRNA sites, we achieved gene replacements in the rice endogenous gene 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) at a frequency of 2.0%. We also obtained targeted gene insertions at a frequency of 2.2% using a sgRNA targeting one intron and a donor DNA template including the same sgRNA site. Rice plants harbouring the OsEPSPS gene with the intended substitutions were glyphosate-resistant. Furthermore, the site-specific gene replacements and insertions were faithfully transmitted to the next generation. These newly developed approaches can be generally used to replace targeted gene fragments and to insert exogenous DNA sequences into specific genomic sites in rice and other plants.
DOI:10.1016/j.molp.2016.12.001URLPMID:27940306 [本文引用: 1]
DOI:10.1016/j.molp.2016.11.013URLPMID:27932049 [本文引用: 1]
DOI:10.1093/mp/ssu009URLPMID:24482433 [本文引用: 1]

Dear Editor, RNA-guided genome editing (RGE) using the Streptococcus pyogenes CRISPR-Cas9 system (Jinek et al.,2012;Cong et al.,2013;Mali et al.,2013b) is emerging as a simple and highly efficient tool for genome editing in many organisms.The Cas9 nuclease can be programmed by dual or single guide RNA (gRNA) to cut target DNA at specific sites,thereby introducing precise mutations by error-prone non-homologousend-joining repairing or by incorporating foreign DNAs via homologous recombination between target site and donor DNA.The gRNA-Cas9 complex recognizes targets based on the complementarity between one strand of targeted DNA (referred as protospacer) and the 5'-end leading sequence of gRNA (referred to as gRNA spacer) that is approximately 20 base pairs (bp) long (Figure 1A).Besides gRNA-DNA pairing,a protospacer-adjacent motif (PAM) following the paired region in the DNA is also required for Cas9 cleavage.Recent studies reveal that Cas9 could cut the PAM-containing DNA sites that imperfectly match gRNA spacer sequences,resulting in genome editing at undesired positions.
DOI:10.1007/s00299-009-0706-2URLPMID:19455339 [本文引用: 1]

Traditional transformation methods are complex and time consuming. It is generally difficult to transform indica rice varieties using traditional transformation methods due to their poor regeneration. In this contribution, a simple method was developed for the transformation of indica rice. In this method, the mature embryos of soaked seeds were pierced by a needle, and then soaked in the Agrobacterium inoculum under vacuum infiltration. The inoculated seeds germinated and grew to maturation ( T 0 ) under nonsterile conditions. The herbicide or antibiotic analysis and molecular analysis were conducted on T 0 plants. The results showed that although the efficiency of transformation was about 6.0%, it was easier to transform indica rice using the proposed method, and the transformation process was significantly shortened. The success of transformation was further confirmed by the genetic and molecular analyses of T 1 transformants.
DOI:10.1073/pnas.1213041110URL [本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.molp.2015.05.009URL [本文引用: 2]
DOI:10.1038/srep37395URL [本文引用: 1]
DOI:10.3389/fpls.2016.00377URLPMID:27066031 [本文引用: 1]

Abstract Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated (Cas) systems have been successfully used as efficient tools for genome editing in a variety of species. We used the CRISPR/Cas9 system to mutate the Gn1a (Os01g0197700), DEP1 (Os09g0441900), GS3 (Os03g0407400), and IPA1 (Os08g0509600) genes of rice cultivar Zhonghua 11, genes which have been reported to function as regulators of grain number, panicle architecture, grain size and plant architecture, respectively. Analysis of the phenotypes and frequencies of edited genes in the first generation of transformed plants (T0) showed that the CRISPR/Cas9 system was highly efficient in inducing targeted gene editing, with the desired genes being edited in 42.5% (Gn1a), 67.5% (DEP1), 57.5% (GS3), and 27.5% (IPA1) of the transformed plants. The T2 generation of the gn1a, dep1, and gs3 mutants featured enhanced grain number, dense erect panicles, and larger grain size, respectively. Furthermore, semi-dwarf, and grain with long awn, phenotypes were observed in dep1 and gs3 mutants, respectively. The ipa1 mutants showed two contrasting phenotypes, having either fewer tillers or more tillers, depending on the changes induced in the OsmiR156 target region. In addition, we found that mutants with deletions occurred more frequently than previous reports had indicated and that off-targeting had taken place in highly similar target sequences. These results proved that multiple regulators of important traits can be modified in a single cultivar by CRISPR/Cas9, and thus facilitate the dissection of complex gene regulatory networks in the same genomic background and the stacking of important traits in cultivated varieties.
DOI:10.1038/ncomms13274URLPMID:27848933 [本文引用: 1]

Targeted DNA double-strand breaks have been shown to significantly increase the frequency and precision of genome editing. In the past two decades, several double-strand break technologies have been developed. CRISPR–Cas9 has quickly become the technology of choice for genome editing due to its simplicity, efficiency and versatility. Currently, genome editing in plants primarily relies on delivering double-strand break reagents in the form of DNA vectors. Here we report biolistic delivery of pre-assembled Cas9–gRNA ribonucleoproteins into maize embryo cells and regeneration of plants with both mutated and edited alleles. Using this method of delivery, we also demonstrate DNA- and selectable marker-free gene mutagenesis in maize and recovery of plants with mutated alleles at high frequencies. These results open new opportunities to accelerate breeding practices in a wide variety of crop species. Genome editing in plants typically requires the expression of Cas9 and guide RNA from stably transformed plasmid DNA. Here, the authors show that successful editing can be achieved after delivery of the Cas9-guide RNA complex as a ribonucleoprotein to maize embryos via biolistics.
DOI:10.1016/j.jgg.2015.10.002URL [本文引用: 1]
DOI:10.1038/nprot.2014.157URL [本文引用: 1]
DOI:10.1038/nbt.2969URL [本文引用: 1]
DOI:10.1371/journal.pone.0136064URL [本文引用: 1]
DOI:10.1186/s12896-015-0131-2URL [本文引用: 1]
DOI:10.1126/science.1225829URL [本文引用: 1]
DOI:10.1016/j.molp.2015.04.007URL [本文引用: 1]
DOI:10.1111/pbi.2014.12.issue-6URL [本文引用: 1]
DOI:10.3969/j.issn.1006-8082.2007.06.005URL [本文引用: 1]

简要介绍了水稻温敏核不育基因株1S的发现及两用核不育系株1S、陆1KS的选育过程,阐述 了株1S、陆1KS的特征特性及利用株1S、陆18S选育超级杂交早稻取得的重要进展,提出选育细胞核具有部分粳稻血缘、细胞质保持完全籼型的广亲和温敏 核不育系,是实现部分利用籼粳亚种间杂种优势、选育南方广适性超级杂交早稻的有效途径之一。
DOI:10.3969/j.issn.1006-8082.2007.06.005URL [本文引用: 1]

简要介绍了水稻温敏核不育基因株1S的发现及两用核不育系株1S、陆1KS的选育过程,阐述 了株1S、陆1KS的特征特性及利用株1S、陆18S选育超级杂交早稻取得的重要进展,提出选育细胞核具有部分粳稻血缘、细胞质保持完全籼型的广亲和温敏 核不育系,是实现部分利用籼粳亚种间杂种优势、选育南方广适性超级杂交早稻的有效途径之一。
DOI:10.3321/j.issn:0496-3490.2003.06.023URL [本文引用: 1]

培矮64S、810S、株1S、陆18S、729S、139S、 康201S、179S、香125S等是一类高温条件下不育、低温条件下可育的高温敏型核不育水稻,它们的临界不育温度为23~24℃左右;go543S是 一类高温条件下可育、低温条件下不育的低温敏感核不育水稻,其临界不育温度为29.5℃.分别在自然长日(光长14.2~13.7 h)、高温(日均气温29.0~34.1℃)条件下和短日(光长13.0~12.4 h)、较低气温(日均气温23.9~28.1℃)条件下对处于育性敏感期的温敏核不育水稻进行1~6 d的冷水(21.5℃/20.5℃)处理,研究不同温敏核不育水稻育性对低温持续时间的敏感性差异.结果表明:株1S和陆18S经6 d低温处理表现为稳定不育,对低温反应钝感,杂交制种最为安全;729S、139S、康201S在长日、高温阶段经5 d处理表现为不育,在短日、较低气温阶段经4 d处理开始转为可育,对低温反应较钝感;培矮64S和810S在长日、高温阶段经4 d处理,在短日、较低气温阶段经2 d处理开始转为可育,对低温反应较敏感;179S、香125S在长日、高温阶段经3 d处理,在短日、较低气温阶段经2 d处理就会出现可染花粉和自交结实,对低温反应敏感,杂交制种风险大;短暂低温和高温都会引起go543S的育性发生变化,因此必须选择温度稳定的特殊生 态条件下才能进行自交繁殖和杂交制种.另外,各个材料中不同单株之间育性对低温的敏感性存在差异.
DOI:10.3321/j.issn:0496-3490.2003.06.023URL [本文引用: 1]

培矮64S、810S、株1S、陆18S、729S、139S、 康201S、179S、香125S等是一类高温条件下不育、低温条件下可育的高温敏型核不育水稻,它们的临界不育温度为23~24℃左右;go543S是 一类高温条件下可育、低温条件下不育的低温敏感核不育水稻,其临界不育温度为29.5℃.分别在自然长日(光长14.2~13.7 h)、高温(日均气温29.0~34.1℃)条件下和短日(光长13.0~12.4 h)、较低气温(日均气温23.9~28.1℃)条件下对处于育性敏感期的温敏核不育水稻进行1~6 d的冷水(21.5℃/20.5℃)处理,研究不同温敏核不育水稻育性对低温持续时间的敏感性差异.结果表明:株1S和陆18S经6 d低温处理表现为稳定不育,对低温反应钝感,杂交制种最为安全;729S、139S、康201S在长日、高温阶段经5 d处理表现为不育,在短日、较低气温阶段经4 d处理开始转为可育,对低温反应较钝感;培矮64S和810S在长日、高温阶段经4 d处理,在短日、较低气温阶段经2 d处理开始转为可育,对低温反应较敏感;179S、香125S在长日、高温阶段经3 d处理,在短日、较低气温阶段经2 d处理就会出现可染花粉和自交结实,对低温反应敏感,杂交制种风险大;短暂低温和高温都会引起go543S的育性发生变化,因此必须选择温度稳定的特殊生 态条件下才能进行自交繁殖和杂交制种.另外,各个材料中不同单株之间育性对低温的敏感性存在差异.
DOI:10.1146/annurev-arplant-042809-112209URLPMID:20192739 [本文引用: 1]

Abstract Grain yield in rice is a complex trait multiplicatively determined by its three component traits: number of panicles, number of grains per panicle, and grain weight; all of which are typical quantitative traits. The developments in genome mapping, sequencing, and functional genomic research have provided powerful tools for investigating the genetic and molecular bases of these quantitative traits. Dissection of the genetic bases of the yield traits based on molecular marker linkage maps resolved hundreds of quantitative trait loci (QTLs) for these traits. Mutant analyses and map-based cloning of QTLs have identified a large number of genes required for the basic processes underlying the initiation and development of tillers and panicles, as well as genes controlling numbers and sizes of grains and panicles. Molecular characterization of these genes has greatly advanced the mechanistic understanding of the regulation of these rice yield traits. These findings have significant implications in crop genetic improvement.
