闫慧贞, 徐邹洋, 陈文杰, 王璐, 刘倩, 高岭岭, 侯丹迪, 张德民, 王凯


宁波大学海洋学院, 浙江 宁波 315832
收稿日期:2020-10-31;修回日期:2020-12-27;网络出版日期:2021-01-11
基金项目:国家自然科学基金(41977192)
*通信作者:王凯, E-mail: wangkai@nbu.edu.cn.
摘要:[目的] 海洋浮游古菌是生物地球化学循环的关键驱动者,但其在近岸海域的水平空间分布特征还未被充分了解。本研究以与陆地紧密相连的环梅山岛海域为例研究浮游古菌在海陆过渡带的水平分布模式。[方法] 利用16S rRNA基因扩增子测序,以期从优势类群分布、群落组成变化和物种共现模式3个层面揭示梅山湾潟湖区和临近海域春季浮游古菌的空间分布特征。[结果] 该海域浮游古菌在原核生物群落中的相对丰度为0.6%-26.5%,向海侧古菌丰度显著高于潟湖区。浮游古菌群落由奇古菌门Marine Group(MG)I和广古菌门MGII主导,MGI的物种组成较为单一,而MGII的系统发育多样性较高。古菌群落的空间分布受同质扩散、环境选择和非主导过程(包括生态漂变)的共同塑造,其中环境选择主要由悬浮颗粒物、硝酸盐、溶解氧、水温和铵盐驱动。通过网络分析发现MGI与红杆菌科细菌呈广泛的负相关,MGII则普遍与SAR11、SAR116和SAR86等异养细菌类群呈正相关。[结论] 本研究初步揭示了环梅山岛海域春季浮游古菌群落的空间分布特征及其调控因子,拓展了对古菌在海陆过渡带分布规律的认识。
关键词:浮游古菌近岸海域空间分布共现网络
Spatial distribution of planktonic archaeal community in the coastal waters around Meishan Island in spring
Huizhen Yan, Zouyang Xu, Wenjie Chen, Lu Wang, Qian Liu, Lingling Gao, Dandi Hou, Demin Zhang, Kai Wang


School of Marine Science, Ningbo University, Ningbo 315832, Zhejiang Province, China
Received: 31 October 2020; Revised: 27 December 2020; Published online: 11 January 2021
*Corresponding author: E-mail: wangkai@nbu.edu.cn.
Foundation item: Supported by the National Natural Science Foundation of China (41977192)
Abstract: [Objective] Marine planktonic archaea are the key drivers of biogeochemical cycling, but the knowledge about their horizontal spatial distribution characteristics in the coastal waters has not been well understood. In this study, we used the Meishan Island (closely connected to the land) as an example to study the horizontal distribution patterns of planktonic archaea in the transition zone between land and sea. [Methods] 16S rRNA gene amplicon sequencing was used to reveal the spatial distribution characteristics of planktonic archaea in terms of the distribution of dominant taxa, community composition variation, and the co-occurrence pattern of taxa in the lagoon (called Meishan Bay) and adjacent seawaters around Meishan Island in spring. [Results] The results showed that relative abundance of planktonic archaea in the whole prokaryotic community ranged from 0.6% to 26.5%, and the archaeal abundance in seaward waters was significantly higher than that in the lagoon. Planktonic archaeal communities were dominated by Thaumarchaeota Marine Group (MG) I and Euryarchaeota MGII. The composition of MGI taxa was relatively simple, while MGII taxa had relatively high phylogenetic diversity. The spatial distribution of archaeal communities was shaped by homogenizing dispersal, environmental selection, and undominated processes (including ecological drift), and environmental selection was mainly driven by suspended particle, nitrate, dissolved oxygen, water temperature and ammonium. The network analysis showed that MGI taxa commonly had negative correlation with Rhodobacteraceae taxa, while MGII taxa commonly had positive correlation with heterotrophic bacteria affiliated to the clades SAR11, SAR116, and SAR86. [Conclusion] This study preliminary revealed the spatial distribution characteristics and drivers of planktonic archaeal communities in the coastal waters around Meishan Island in the spring, expanding the understanding of the distribution patterns of archaea in the transitional zone between the land and ocean.
Keywords: planktonic archaeacoastal watersspatial distributionco-occurrence network
自1992年浮游古菌首次在近岸海域中温条件下被发现后[1-2],便由于其在海洋中的普遍性和优势地位受到广泛关注。海洋浮游古菌是生物地球化学循环的关键驱动者,也是微食物网的重要组成部分,主要包含4大类群:奇古菌门(Thaumarchaeota)的Marine Group (MG) I以及广古菌门(Euryarchaeota)的MGII、MGⅢ和MGIV。MGI是氨氧化菌,对海洋氮循环至关重要[3],MGII和MGⅢ则具有降解复杂有机质的潜能[4-5]。海洋浮游古菌在开放海域的垂直空间分布格局已被广泛了解[6-8]。MGI通常在表层海水中丰度较低,在真光层底部丰度达到峰值;MGII则在表层海水占据优势,其丰度随水深增加而降低;MGⅢ整体丰度较低并且主要局限于深水层(已发现一些表层类群),而MGIV目前被认为仅局限于深海[8]。
近年来,古菌在表层海水中的水平空间分布特征也逐步受到关注。例如,Wang等[9]在浙江北部海域区域尺度(regional scale)下发现表层海水中MGI在原核生物群落中的相对丰度从杭州湾到渔山列岛呈现从高到低的梯度分布,而MGII则呈现斑块化分布,在三门湾等区域呈热点分布。MGI往往在近岸表层海水古菌群落中占主导地位,而MGII则在开放海域占优势[10]。由此可见,浮游古菌在近岸海域的分布格局沿区域环境梯度变化。然而,对海陆过渡带局域尺度(local scale)下浮游古菌空间分布特征及其影响因子的认识仍较少。
海洋浮游古菌群落组成受当地环境条件和空间因子的共同调控。例如,沿水柱深度形成的理化条件差异(如光照强度、水温、营养盐等)塑造了古菌群落垂直分布的格局[7-8]。在近岸海域水平空间,例如浙北海域表层海水古菌群落组成的空间变化主要由盐度、溶解氧、pH等基础理化因子以及营养盐驱动[9],这些因子也显著影响了珠江口、九龙江口等河口的浮游古菌群落组成[11-12]。综上,盐度、营养盐和溶解氧可能是塑造浮游古菌群落组成水平空间格局的核心环境因子。但是,海陆过渡带局域尺度下影响浮游古菌群落组成的关键因子仍未被充分了解。
尽管很多影响海洋浮游古菌群落分布的环境因子已被揭示,仍有相当高比例的群落变异无法被解释[7, 10],对海洋浮游古菌群落构建机制的了解则尤为缺乏[8, 13-14]。Vellend[15]提出生态群落构建受4个高级过程调控:选择(selection)、扩散(dispersal)、漂变(drift)和成种(speciation)。生态位差异导致的确定性(deterministic)过程(包括异质选择(heterogeneous selection)和同质选择(homogeneous selection))和随机性(stochastic)过程(包括扩散限制(dispersal limitation)、同质扩散(homogenizing dispersal)和漂变(drift)等)在群落构建中同时发挥作用,解析浮游古菌群落构建机制的关键在于量化这些生态过程的相对重要性。近年来,基于系统发育和物种周转(turnover)零假设模型的微生物群落构建过程判别法[16]已被用于解析不同生境中微生物群落的构建机制[9, 17-20]。Wu等[21]和Logares等[22]均发现选择过程在海洋浮游细菌/原核生物群落构建中的重要性高于扩散限制,但真核微生物群落构建则受细胞尺寸和研究区域尺度的影响。Wang等[9]应用该方法发现随机过程主导了近岸区域尺度下表层海水中浮游古菌群落的构建。然而,对海陆过渡带局域尺度下浮游古菌群落构建机制的了解仍很有限。
一些基于相关性网络的研究已发现古菌与细菌、真核微生物间存在广泛的潜在互作关系[9, 23-25],表明微生物相互作用对维持海洋中浮游古菌群落分布格局可能很重要[7, 26-27]。例如,MGI古菌是海洋中最主要的氨氧化菌,常与硝酸化细菌共现,这可能与它们协作完成硝化作用有关[7, 9, 28]。另外,有研究报道MGI与SAR11、SAR86等异养细菌也存在广泛的共现关系[29]。宏基因组证据表明真光层中的MGII古菌含变形菌视紫红质(proteorhodopsin,PR)基因[30],具有捕捉光能并代谢有机质的潜能,PR基因在光能异养细菌中也广泛存在[29, 31-32],有研究发现MGII与光能异养细菌存在普遍共现性,表明这些类群可能占据相似的生态位,共同驱动海洋碳循环[8, 33]。陆源物质输入导致海陆过渡带在局域尺度下即可能存在复杂环境变化,在这类水域中古菌与细菌的共现/互作关系还未被充分了解。
梅山岛作为宁波-舟山港的重要枢纽之一,与陆地联系紧密,其向陆侧的梅山湾两端有大坝拦截,形成近岸人工潟湖[34];向海侧位于象山港咽喉处,受频繁水体交换的影响。研究该海区局域尺度下浮游古菌群落的空间变化特征不仅有助于拓展对古菌在海陆过渡带分布规律的认识,而且能为揭示近年来趋于密集的开发建设活动对环梅山岛邻近海域的微生态效应提供背景数据。本研究选取梅山湾潟湖区和向海侧的8个站位采集表层海水样品,基于古菌-细菌16S rRNA基因扩增子测序在原核生物群落背景下表征古菌群落,旨在揭示:(1) 环梅山岛海域浮游古菌优势类群的空间分布规律;(2) 浮游古菌群落组成的空间变化、构建过程及调控因子;(3) 浮游古菌与细菌的跨域(cross-domain)共现模式。以期从优势类群分布、群落组成变化和物种共现模式3个层面揭示该海域浮游古菌的空间分布特征。
1 材料和方法 1.1 站位设计、样品采集与海水理化因子测定 本研究在梅山湾潟湖区(Lagoon)设置2个站位(S1–S2),在向海侧(seaward)设置6个站位(S3–S8;表 1)。于2019年春季(5月14日)采集各站位表层海水(0–1 m)样品,每个站位采集5个生物学重复,利用YSI6000多参数水质分析仪(YSI,美国)原位测定水温、pH、盐度和溶解氧(dissolved oxygen,DO)。用0.2 μm孔径的聚碳酸酯膜(Millipore,美国)过滤约500 mL水样收集微生物。使用AutoAnalyzer 3流动注射仪(Seal,德国)测定水样硝酸盐、亚硝酸盐、铵盐和磷酸盐的浓度。参照海洋监测规范(GB 17378.4-2007),用重量法测定悬浮颗粒物(suspended particle,SP)含量。
表 1. 采样站位表 Table 1. Information of sampling stations
| Stations ID | Zones | Longitude/E | Latitude/N |
| S1 | Lagoon | 121°56′56′′ | 29°47′12′′ |
| S2 | Lagoon | 121°56′47′′ | 29°46′50′′ |
| S3 | Seaward | 121°55′22′′ | 29°44′29′′ |
| S4 | Seaward | 121°58′50′′ | 29°45′22′′ |
| S5 | Seaward | 122°0′29′′ | 29°45′31′′ |
| S6 | Seaward | 122°1′4′′ | 29°46′48′′ |
| S7 | Seaward | 122°2′13′′ | 29°48′5′′ |
| S8 | Seaward | 122°2′14′′ | 29°49′15′′ |
表选项
1.2 DNA提取、16S rRNA基因扩增和Illumina测序 用PowerSoil DNA提取试剂盒(MOBIO,美国)提取滤膜上的总DNA,用NanoDrop ND-2000核酸定量仪(Thermo,美国)测定其浓度和纯度,于–80 ℃冰箱保存备用。采用地球微生物组计划推荐的古菌-细菌通用引物515F-Y (5′-GTGYCA GCMGCCGCGGTAA-3′)和926R (5′-CCGYCAA TTYMTTTRAGTTT-3′)扩增原核生物16S rRNA基因V4–V5可变区(http://www.earthmicrobiome.org/protocols-and-standards/16s/),该引物对古菌和细菌均有较高的覆盖度[35]。各个样本取等量的10 ng经纯化的DNA模板在30 μL PCR反应体系中扩增,设3个重复,条件如下:98 ℃ 1 min;98 ℃ 10 s,50 ℃ 30 s,72 ℃ 30 s,30个循环;72 ℃ 5 min。将每个样本的扩增产物混合,用Qiagen Gel Extraction Kit试剂盒(Qiagen,德国)纯化PCR产物,并使用TruSeq DNA PCR-Free Sample Preparation Kit试剂盒(Illumina,美国)构建文库,随后用Illumina Nova PE250平台测序。序列数据存储在NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/bioproject/),登录号为PRJNA673086。
1.3 序列处理 用FLASH对双末端序列进行拼接[36],随后用QIIME 1.9.1对拼接好的序列进行后续处理[37],使用split_libraries_fastq.py脚本控制序列质量。使用UCHIME识别序列中的嵌合体[38],去除嵌合体后,用pick_de_novo_otus.py脚本聚类分类操作单元(operational taxonomic units,OTUs;序列相似性 > 99%)[39]。去除叶绿体、线粒体、未分配到细菌/古菌界的OTUs以及在总序列中占比低于0.0001%的OTUs后[40],所有样品(n=40)获得1705147条高质量原核生物序列,把原核生物OTU表中的每个样本标准化至25500条序列后,计算古菌优势类群在原核生物群落中的相对丰度。从未标准化的原核生物OTU表里提取古菌OTU表,共获得了299792条古菌序列,包含688个OTUs,采用cumulative sum scaling (CSS)算法对古菌OTU表进行标准化[41],随后进行下游分析。用PyNAST把古菌OTUs的代表序列与SILVA 132数据库中的模板进行对齐和过滤[42],随后用FastTree构建古菌的系统发育树[43]。
1.4 基本统计分析 用R包“ggplot2”绘制环境因子箱体图。在SPSS中计算MGI/MGII古菌相对丰度与环境因子的Spearman相关性。在MEGA中用最大似然法结合参考序列构建MGI/MGII优势OTUs (平均相对丰度前20)的系统发育树,并用R包“pheatmap”绘制热图表征其相对丰度的空间变化。在PRIMER-E v5中用相似性分析(analysis of similarity,ANOSIM)检验各采样站位古菌群落组成差异。用R包“vegan”进行主坐标约束排序分析(constrained analysis of principal coordinates,CAP)来表征古菌群落组成空间变化及其关键驱动因子。
1.5 基于零模型量化古菌群落构建过程 运用Stegen[16]零模型方法的前提是OTU间生态位差异具有系统发育信号。为了检验系统发育信号,首先计算古菌OTUs针对各环境因子的最适值(optimum),然后计算成对OTUs之间所有环境因子最适值的欧氏距离以表征OTU间的生态位差异。最后,用R包“vegan”中的‘mantel.correlog’函数生成Mantel相关图以评估古菌OTUs生态位差异与系统发育距离之间的相关性,从而评判系统发育信号的显著性和分布区间。
如果在较短的系统发育距离内检测到显著的系统发育信号,则可使用β平均种间最近距离(β-mean-nearest taxon distance,βMNTD)推断群落构建过程。首先用R包“picante”中的“comdistnt”函数计算成对样本的丰度加权βMNTD与经999次随机打乱OTUs在系统发育树上的位置获得的零分布(βMNTDnull),根据成对样本之间实测βMNTD值与其零分布的差异程度,即βNTI (β-nearest taxon index)值推断异质选择(heterogeneous selection;βNTI > +2)和同质选择(homogeneous selection;βNTI < –2)过程。当|βNTI|≤2时,根据基于Bray-Curtis非相似性的Raup-Crick指数(RCbray)实测值与其零分布的差异程度,推断扩散限制(dispersal limitation;RCbray > +0.95)、同质扩散(homogenizing dispersal;RCbray < –0.95)和包含生态漂变(ecological drift)的非主导过程(undominated;|RCbray|≤0.95)。最后,根据βNTI和RCbray值统计各生态过程的比例。
1.6 网络分析 用R包“igraph”构建优势古菌-细菌类群的跨域共现网络。为了降低组成效应和网络复杂度,取平均相对丰度前20的MGI/MGII OTUs和前50的细菌OTUs用于网络计算。随后,用Gephi软件对共现网络进行模块分析,并对Spearman相关性|ρ| > 0.6且P < 0.01的节点和边进行可视化,节点的度(degree)用来表示与其连接的边数,模块化系数(modularity)用来量化网络可以被分解为较小组分的程度。随后,用Cytoscape软件分别进一步聚焦呈现MGI或MGII古菌与细菌OTUs的共现关系。
2 结果和分析 2.1 水体环境因子的空间变化 梅山湾潟湖区内的水温显著高于向海侧,但湾内的盐度、溶解氧、铵盐、硝酸盐和悬浮颗粒物总体上呈现低于向海侧的趋势(图 1)。测定的大多数水体环境因子与经度或纬度显著相关(表 2),如悬浮颗粒物沿纬度梯度呈升高趋势;而亚硝酸盐、磷酸盐和pH无明显空间变化规律(图 1)。整体上,环梅山岛海域水体环境因子存在空间异质性,向海侧海水的营养水平较潟湖区高。
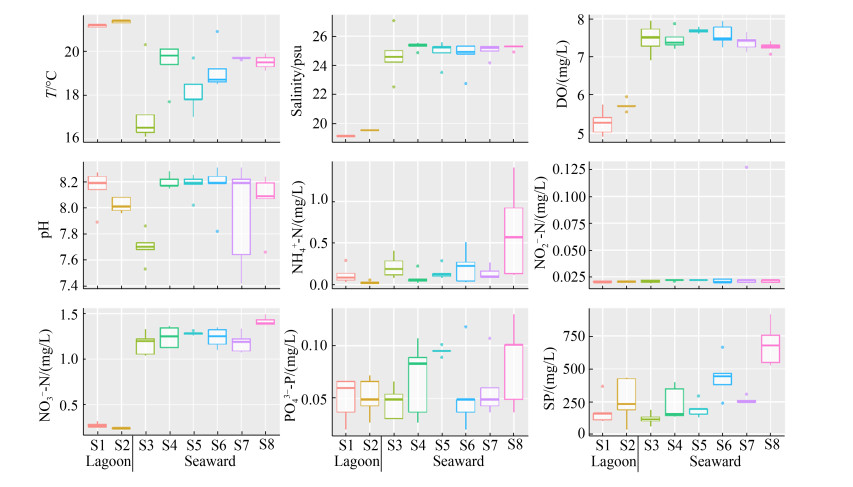 |
| 图 1 海水环境因子空间变化箱体图 Figure 1 Box plots illustrating spatial variability of seawater environmental factors [n=5, except for ammonium in station S3 (n=4)]. |
| 图选项 |
表 2. 海水环境因子与采样站位经纬度的Spearman相关性 Table 2. Spearman's rank correlations between seawater environmental factors and geographic coordinates of sampling stations
| Factors | Latitude | Longitude | |||
| ρ | P | ρ | P | ||
| SP | 0.531** | < 0.001 | 0.672** | < 0.001 | |
| NO3–-N | 0.073 | 0.655 | 0.635** | < 0.001 | |
| DO | –0.436** | 0.005 | 0.225 | 0.162 | |
| Temperature | 0.396* | 0.011 | –0.180 | 0.266 | |
| Salinity | –0.174 | 0.282 | 0.427** | 0.006 | |
| NH4+-N | 0.182 | 0.276 | 0.411** | 0.011 | |
| pH | 0.117 | 0.472 | 0.314* | 0.049 | |
| PO43–-P | 0.087 | 0.652 | 0.269 | 0.093 | |
| NO2–-N | –0.049 | 0.637 | 0.235 | 0.145 | |
| *: P < 0.05, **: P < 0.01. | |||||
表选项
2.2 古菌优势类群的空间分布 环梅山岛海域浮游古菌优势类群是MGI奇古菌(在原核生物群落中的平均相对丰度为14.6%)和MGII广古菌(1.8%) (图 2),向海侧的MGI、MGII相对丰度均高于潟湖区。MGI的相对丰度主要与溶解氧、盐度和硝酸盐显著正相关(P < 0.001),与水温显著负相关(P < 0.001);MGII的相对丰度则主要与溶解氧和盐度显著正相关(P < 0.001),与水温显著负相关(P < 0.001) (表 3)。环梅山岛海域MGI优势OTUs (相对丰度排名前20)均属于表层类群(shallow marine group),其中一个OTU (MGI_1)在MGI总序列中占据绝对优势(75.5%–93.4%) (图 3-A),表明该海域MGI古菌系统发育多样性较低。MGII优势OTUs则分别属于MGII.A和MGII.B两个亚群,系统发育多样性较高。在相对丰度排名前三位的MGII OTUs (均属于MGII.B)中,MGII_1和MGII_3几乎仅分布在向海侧,而MGII_2则在整个海域均匀分布(图 3-B)。
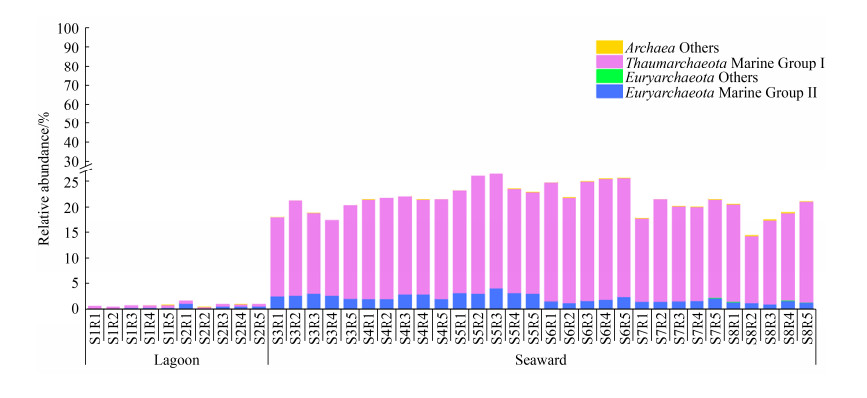 |
| 图 2 浮游古菌优势类群在原核生物群落中的相对丰度 Figure 2 Relative abundance of dominant archaeal groups in the prokaryotic community. |
| 图选项 |
表 3. MGI/MGII古菌相对丰度与环境因子的Spearman相关性 Table 3. Spearman's rank correlations between the relative abundance of MGI/MGII archaea and environmental factors
| Factors | Thaumarchaeota MGI | Euryarchaeota MGII | |||
| ρ | P | ρ | P | ||
| DO | 0.690** | < 0.001 | 0.744** | < 0.001 | |
| Temperature | –0.611** | < 0.001 | –0.701** | < 0.001 | |
| Salinity | 0.574** | < 0.001 | 0.559** | < 0.001 | |
| NO3–-N | 0.572** | < 0.001 | 0.406** | 0.009 | |
| pH | 0.421** | 0.007 | 0.037 | 0.821 | |
| NO2–-N | 0.296 | 0.063 | 0.282 | 0.078 | |
| NH4+-N | 0.279 | 0.085 | 0.198 | 0.227 | |
| SP | 0.234 | 0.147 | –0.263 | 0.101 | |
| PO43–-P | 0.131 | 0.420 | 0.206 | 0.202 | |
| *: P < 0.05, **: P < 0.01. | |||||
表选项
 |
| 图 3 原核生物群落中平均相对丰度排名前20的MGI (A)和MGII (B)古菌优势OTUs的16S rRNA基因V4–V5区的最大似然系统发育树和相对丰度热图 Figure 3 Maximum-likelihood phylogenetic tree based on the V4–V5 region of 16S rRNA gene of top 20 MGI (A) and MGII (B) OTUs in average relative abundance in prokaryotic communities and heatmap of relative abundances of these OTUs in the prokaryotic community. Shallow and deep marine groups are designated similarly as previously reported[7, 44]. Marine Group A and Group B are designated similarly as previously reported[7, 45]. |
| 图选项 |
2.3 古菌群落组成的空间变化及其驱动因子 主坐标约束排序和相似性分析结果表明,潟湖区与向海侧的浮游古菌群落组成差异显著(ANOSIM R=0.490,P=0.001),向海侧6个站位古菌群落组成沿纬度变化,但部分相邻站位如S3与S4,S4与S5,S6与S7,S7与S8之间无显著差异(图 4和表 4)。两个采样区之间的古菌群落组成变化主要由硝酸盐、溶解氧和水温的差异驱动,而向海侧古菌群落组成沿纬度的变化则主要受悬浮颗粒物驱动。
 |
| 图 4 主坐标约束排序分析表征古菌群落组成的空间变化及其关键驱动因子 Figure 4 Constrained analysis of principal coordinates (CAP) bi-plot showing spatial variability of archaeal community composition and the key environmental drivers. |
| 图选项 |
表 4. 基于Bray-Curtis的相似性分析检验采样站位间古菌群落差异(999次置换检验) Table 4. Analysis of similarity based on Bray-Curtis dissimilarity for testing the differences in archaeal communities between sampling stations (with 999 permutations)
| Station | S1 | S2 | S3 | S4 | S5 | S6 | S7 |
| S2 | 0.200 | ||||||
| S3 | 1.000** | 1.000** | |||||
| S4 | 1.000** | 1.000** | 0.084 | ||||
| S5 | 1.000** | 0.996** | –0.048 | –0.116 | |||
| S6 | 1.000** | 1.000** | 0.672** | 0.516** | 0.568** | ||
| S7 | 1.000** | 1.000** | 0.348** | 0.256* | 0.428* | 0.140 | |
| S8 | 1.000** | 1.000** | 0.660** | 0.572* | 0.672** | 0.324* | 0.100 |
| Data are R values, *: P < 0.05, **:P < 0.01. | |||||||
表选项
2.4 古菌群落构建过程 Mantel检验发现,古菌OTUs生态位差异在较近系统发育距离上具有显著的系统发育信号(图 5-A),表明亲缘关系较近物种的生态位差异与其系统发育距离显著相关,满足利用系统发育零模型推断群落构建过程的前提。总体上,古菌群落构建受同质扩散(主导51.3%的群落周转)、包含生态漂变的非主导过程(34.9%)和环境选择(13.8%)的共同调控,但几乎不受扩散限制的影响(图 5-B)。
 |
| 图 5 古菌OTUs生态位差异的系统发育信号检验(A)和古菌群落构建过程的比例(B) Figure 5 Phylogenetic signal test for ecological niche differences between archaeal OTUs (A) and the proportion of archaeal community assembly processes (B). |
| 图选项 |
2.5 古菌-细菌共现网络 环梅山岛海域原核生物优势类群共现网络的模块化程度较高(模块化系数0.758),主要含6个模块,模块Ⅰ-Ⅳ包含了93.2%的节点(图 6-A)。模块Ⅰ以MGI为核心,模块Ⅳ以MGII为核心,模块Ⅱ和Ⅲ由细菌OTUs主导(图 6-A,B),模块Ⅰ和Ⅳ间有31条边,模块Ⅰ和Ⅱ间有145条边,模块Ⅳ和Ⅲ间有109条边,表明两个古菌模块间的共现关系较简单,而古菌与细菌模块间的共现关系较复杂。同时,各古菌/细菌模块内部均存在较紧密的联系。总之,古菌-细菌跨域共现关系较MGI与MGII古菌间的共现关系更复杂。在以MGI为核心的子网络中,MGI OTUs与红杆菌科(Rhodobacteraceae)、生丝单胞菌科(Hyphomonadaceae)、黄杆菌目(Flavobacteriales)、微杆菌科(Microbacteriaceae)、交替单胞菌科(Alteromonadaceae)、岸生菌科(Litoricolaceae)的OTUs呈广泛的负相关,而与SAR11、SAR116、SAR86和Actinomarinaceae的OTUs呈普遍正相关(图 6-C)。在以MGII为核心的子网络中,MGII与SAR11、SAR116、红螺旋菌目(Rhodospirillales)、SAR86、Actinomarinaceae、SAR406的OTUs呈广泛的正相关,而与部分红杆菌科、黄杆菌科(Flavobacteriaceae)的OTUs呈显著负相关(图 6-D)。
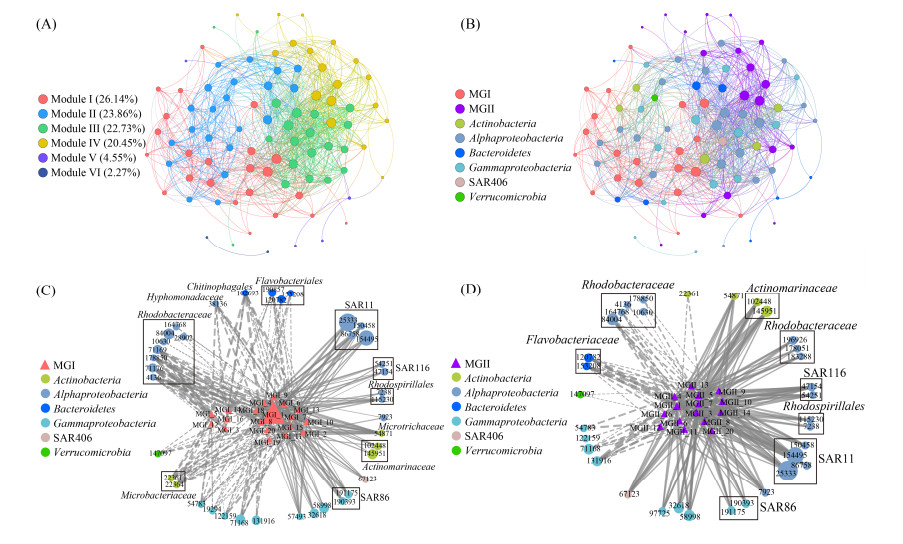 |
| 图 6 古菌-细菌共现网络 Figure 6 Archaeal-bacterial co-occurrence network. The co-occurrence network colored with modules (A) and taxonomy (B). The size of nodes reflects their degree and the edges present significant correlations (Spearman's |ρ| > 0.6, P < 0.01). The value in parentheses represents the percentage of the node number of each module in the total node number (A). The sub-networks of cross-domain co-occurrence between MGI (C)/MGII (D) and bacterial OTUs. The size of the nodes reflects their average relative abundance; the solid and dashed lines present positive and negative correlations, respectively; and the thickness of the lines indicates correlation strength. |
| 图选项 |
3 讨论 3.1 浮游古菌优势类群的多样性和空间变化 海洋浮游古菌主要类群的垂直空间分布特征呈现较固定的模式[7-8, 46],而近岸海域表层浮游古菌群落组成由MGI和MGII间的权衡决定,这往往取决于多种环境因子的共同作用[9, 11]。Liu等[10]发现MGI倾向于分布在近岸区,MGII趋于分布在开放海域。作为海洋环境中最重要的氨氧化微生物,MGI古菌的丰度通常在富营养水域(尤其是氨氮或无机氮浓度高的区域)呈升高趋势[13]。Wang等[9]发现富营养化程度较高的浙北近岸海域表层水中MGI在原核生物群落中的相对丰度总体上高于MGII。而在寡营养的南海东北部,从表层到深层水体中MGII丰度均显著高于MGI[47]。本研究聚焦海陆过渡带水域,其整体营养盐水平较高(按可溶性无机氮划分属于第IV类海水(GB 3838-2002)),这可能是该海域MGI相比MGII占压倒性优势的原因。浮游古菌常被报道在夏季和冬季勃发[9-10, 48-49],其在秋季的勃发也偶有报道[50]。此外,春季浮游植物爆发时也常常伴随异养MGII古菌的勃发[26]。本研究在春季未发现浮游植物爆发的情况下,依然观察到向海测表层水中较高的古菌相对丰度,表明浮游古菌在海陆过渡带的普遍性。
本研究发现MGI和MGII的相对丰度均呈现向海侧显著高于潟湖区的规律,这可能与两个区域中多种环境因子的差异有关。其中,向海侧水体中溶解氧、硝酸盐均高于潟湖区。我们此前在浙北海域的研究发现溶解氧与MGI和MGII在原核群落中的相对丰度均呈显著负相关[9],这与本研究的结果相反。MGI既可生长在富氧环境中,又能在低氧条件下保持较强活力[11],呈现出较广的氧生态位宽度。有研究发现MGI分离株SCM1和PS0的生长速率与氧浓度正相关[51],表明氧浓度的升高可能导致MGI丰度升高。另一方面,MGII被发现能在低氧区富集[52]。本研究中所有水体均处于好氧状态,这些研究结果的不一致性也反映出好氧条件下氧对MGI和MGII的微妙调控。已有较多研究表明盐度是影响浮游古菌分布的关键因子,比如,Liu等[11]发现珠江口盐度梯度下,MGI和MGII分别在低盐和高盐水体中占据主导地位。浙北近岸区域尺度下MGI和MGII的相对丰度分别与盐度呈负相关和正相关[9],这些研究都表明MGI和MGII对盐度具有不同的响应模式。而本研究发现MGI和MGII均与盐度呈正相关,反映出在相对较弱的梯度下(19.1–27.05 psu),MGI和MGII对盐度变化的响应可能呈现一致性,也反映出在局域尺度下研究浮游古菌分布特征和影响因子的必要性。此外,我们发现在硝酸盐水平较高的向海侧水体中古菌的丰度更高。MGI古菌趋向于分布在营养水平较高的海域,其丰度往往和含氮营养盐密切相关。多项研究报道,即使在MGI丰度与氨氮等营养盐无显著相关时,其与硝酸盐均呈正相关[9, 13],这与本研究的结果一致。此外,MGI古菌对光照较为敏感,向海侧水体的悬浮颗粒物含量(平均322.5 mg/L)总体上高于潟湖区(平均221.1 mg/L),减弱了光的穿透强度,可能缓解光照对MGI的抑制作用,为MGI提供更合适的生长条件。虽然MGII古菌尚未获得纯培养,但基于宏基因组组装基因组的证据表明海洋真光层中的MGII是(光能)异养菌,具有代谢多种有机质的功能潜力[33]。一些研究还发现MGII表现出颗粒附着的生长策略[53]。因此,本研究中向海侧较高的悬浮颗粒物含量可能是导致MGII相对丰度较高的原因之一。
通过对MGI和MGII优势OTUs系统发育关系和空间分布特征的分析,我们发现MGI优势类群的系统发育多样性很低,全部站位由一个OTU (MGI_1)主导。而MGII优势类群的系统发育多样性较高,主导类群存在明显的生态位分区(niche partitioning)。有研究发现相比于开放海域,MGII在河口拥有更高的多样性[11, 54]。这可能是由于海陆过渡带是陆源有机物入海的通道,水体中有机质多样性较高,从而为MGII多样化类群的共存和生长提供了条件。本研究还发现向海侧的MGII优势类群更为丰富,这一方面可能是由于悬浮颗粒物含量高,从而提供了更丰富的有机质和生态位;另一方面也可能与梅山岛向海侧包含港口作业区和航道有关,这些人类活动可能排放更多样的有机物,从而使向海侧水体中的有机质组成更为复杂,有利于MGII的多样化。
3.2 浮游古菌群落组成的空间变化及其构建机制 除了相对丰度的差异,梅山湾潟湖区和向海侧浮游古菌群落组成也不同,向海侧不同站位间也存在明显的空间变化,悬浮颗粒物、硝酸盐、溶解氧、水温和铵盐等是驱动古菌群落空间变化的主要环境因子。此前在浙江北部[9]和美国南加州[7]海域的研究也发现盐度、硝酸盐和(或)溶解氧是驱动古菌群落空间变化的关键因子。近年来,人们对微生物地理分布模式的研究焦点逐步从驱动因子转移到生态过程[9, 18, 50]。目前,对于海洋浮游古菌群落构建过程的了解还很有限,仅有个别工作基于零假设模型发现近岸海域古菌群落构建受随机过程主导[9, 50],这与受确定性过程主导的细菌群落构建机制有明显区别[50]。本研究也发现环梅山岛海域浮游古菌群落的空间变化主要受控于随机过程。一些研究发现细胞大小和休眠能力差异是导致细菌和真核微生物的群落构建机制差异的主要原因[21-22]。而古菌和细菌同属原核生物,有相似的细胞大小和结构,两者构建机制的差异可能是由于相对丰度(群落规模)和生态位宽度的差异导致[50]。本研究中古菌的相对丰度远低于细菌,群落规模越小越容易受到生态漂变(个体的随机出生和死亡)的影响,这也部分解释了生态漂变在环梅山岛海域浮游古菌群落构建中的重要贡献。研究区域的环境梯度强度对古菌群落构建过程的权重也有重要影响,在浙江近岸海域的系列研究发现除了非主导过程(主要为生态漂变)外,浙北海域的古菌群落构建主要受控于异质选择过程,而浙南海域古菌群落则主要受控于同质扩散过程,这与浙北海域相对浙南海域具有较强的环境梯度相对应[9, 50]。本研究中,虽然潟湖区与向海侧环境因子存在差异,但在局域尺度下整体环境梯度较弱,从CAP分析也看出,虽然古菌群落存在空间变化,但是环境因子对变异的解释度较低(尤其是CAP第二轴),主要解释了两个采样区间的差异。另一方面,环梅山岛海域空间跨度仅为约15 km,向海侧位于象山港咽喉处,水体交换情况良好,为微生物的扩散提供了便利条件。上述原因能很好地解释环梅山岛海域浮游古菌群落空间构建受同质扩散主导且环境选择作用较弱。
3.3 浮游古菌与细菌的潜在相互作用 除随机过程和非生物环境因子选择外,生物相互作用(例如竞争、寄生和互利共生等)在微生物群落塑造中也起到重要作用。多项研究报道在较小的时空尺度,生物互作对微生物群落组成变化的影响可能超过非生物因子[26, 55-56]。许多研究发现古菌与细菌和真核微生物间存在广泛的跨域共现关系[7, 26],可能在微生物互作网络中处于枢纽位置,发挥重要的生态功能[57]。本研究通过共现网络发现古菌和细菌间可能存在广泛的潜在互作,但MGI与MGII间的共现关系有限,这可能是由MGI和MGII的不同营养类型和生存策略导致。
本研究中,MGI与红杆菌科、黄杆菌科等异养细菌类群呈现广泛的负相关,Wang等[9]在浙北海域区域尺度下也发现类似现象,但有研究在蒙特利湾(Monterey Bay)发现MGI虽与一些黄杆菌科类群呈负相关却很少与红杆菌科类群共现[58],而在美国南加州SPOT站表层水中却观察到MGI与红杆菌科和黄杆菌科类群呈正相关[7],这种差异提示我们海区环境条件的差异可能影响特定微生物类群的共现模式。红杆菌科和黄杆菌科作为海洋中重要的异养菌[59-60],在有机质丰富的近岸海域可快速增殖,进而可能抑制代时较长的化能自养菌MGI[61];另一方面,有研究发现一些分离自近岸海域的MGI菌株的生长需要有机物[62],这表明有机质可能介导MGI古菌与异养细菌间的互作,从而使其共现模式更为复杂。许多研究发现在全球多种海洋环境中MGI古菌与SAR11类群呈广泛正相关[7, 9, 29, 58],与本研究结果一致。这种普遍规律可能与这2种类群多样化(diversification)程度较高[63-64],均占据较宽的生态位有关。MGII目前尚未获得纯培养菌株,因此对其生理生化特性的了解很少。有研究在深圳河流-海湾系统中发现浮游古菌-细菌共现网络中多个MGII OTUs为关键(keystone)类群,提示MGII可能在海陆过渡带中发挥重要生态功能[25]。宏基因组学证据表明真光层MGII与SAR11、SAR116和SAR86等细菌类群相似,均含有视紫红质编码基因,具有利用光能代谢有机质的潜力[33]。与本研究结果一致,多项研究发现MGII与上述细菌OTUs普遍呈正相关[7, 29],这可能由它们相似的生存策略决定。但是,也有一些工作发现近岸海域中MGII与某些SAR11和SAR86类群呈负相关[9, 26],表明MGII古菌与光能异养细菌类群间也可能存在复杂的互作机制,这些异养古菌和细菌间的潜在互作模式及其调控因子有待后续研究的跟进。在环梅山岛海域,这些潜在互作关系可能在古菌群落构建(选择作用)中有一定的贡献,验证和探究这些古菌-细菌共现关系的过程和机制是后续研究的重要方向。
4 结论 本研究从优势类群分布、群落组成变化和物种共现模式3个层面初步探明了环梅山岛海域春季浮游古菌群落空间分布特征及其调控因子,得出以下主要结论。
(1) 环梅山岛海域春季浮游古菌群落由MGI和MGII主导,向海侧古菌在原核生物群落中的相对丰度显著高于潟湖区。
(2) 浮游古菌群落构建由随机过程(尤其是同质扩散)主导,环境选择作用相对较小,悬浮颗粒物、硝酸盐、溶解氧、水温和铵盐是古菌群落组成空间变化的主要环境驱动因子。
(3) MGI和MGII古菌间的共现关系较少,但两类古菌均与细菌存在复杂的共现关系。
References
| [1] | Fuhrman JA, McCallum K, Davis AA. Novel major archaebacterial group from marine plankton. Nature, 1992, 356(6365): 148-149. DOI:10.1038/356148a0 |
| [2] | DeLong EF. Archaea in coastal marine environments. Proceedings of the National Academy of Sciences of the United States of America, 1992, 89(12): 5685-5689. DOI:10.1073/pnas.89.12.5685 |
| [3] | K?nneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature, 2005, 437(7058): 543-546. DOI:10.1038/nature03911 |
| [4] | Orsi WD, Smith JM, Wilcox HM, Swalwell JE, Carini P, Worden AZ, Santoro AE. Ecophysiology of uncultivated marine euryarchaea is linked to particulate organic matter. The ISME Journal, 2015, 9(8): 1747-1763. DOI:10.1038/ismej.2014.260 |
| [5] | Haro-Moreno JM, Rodriguez-Valera F, López-García P, Moreira D, Martin-Cuadrado AB. New insights into marine group Ⅲ Euryarchaeota, from dark to light. The ISME Journal, 2017, 11(5): 1102-1117. DOI:10.1038/ismej.2016.188 |
| [6] | Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature, 2001, 409(6819): 507-510. DOI:10.1038/35054051 |
| [7] | Parada AE, Fuhrman JA. Marine archaeal dynamics and interactions with the microbial community over 5 years from surface to seafloor. The ISME Journal, 2017, 11(11): 2510-2525. DOI:10.1038/ismej.2017.104 |
| [8] | Santoro AE, Richter RA, Dupont CL. Planktonic marine archaea. Annual Review of Marine Science, 2019, 11: 131-158. DOI:10.1146/annurev-marine-121916-063141 |
| [9] | Wang K, Hu HJ, Yan HZ, Hou DD, Wang YT, Dong PS, Zhang DM. Archaeal biogeography and interactions with microbial community across complex subtropical coastal waters. Molecular Ecology, 2019, 28(12): 3101-3118. DOI:10.1111/mec.15105 |
| [10] | Liu Q, Tolar BB, Ross MJ, Cheek JB, Sweeney CM, Wallsgrove NJ, Popp BN, Hollibaugh JT. Light and temperature control the seasonal distribution of thaumarchaeota in the South Atlantic bight. The ISME Journal, 2018, 12(6): 1473-1485. DOI:10.1038/s41396-018-0066-4 |
| [11] | Liu JW, Yu SL, Zhao MX, He BY, Zhang XH. Shifts in archaeaplankton community structure along ecological gradients of Pearl Estuary. FEMS Microbiology Ecology, 2014, 90(2): 424-435. |
| [12] | Hu AY, Hou LY, Yu CP. Biogeography of planktonic and benthic archaeal communities in a subtropical eutrophic estuary of China. Microbial Ecology, 2015, 70(2): 322-335. DOI:10.1007/s00248-015-0597-4 |
| [13] | Herfort L, Schouten S, Abbas B, Veldhuis MJW, Coolen MJL, Wuchter C, Boon JP, Herndl GJ, Sinninghe Damsté JS. Variations in spatial and temporal distribution of archaea in the North Sea in relation to environmental variables. FEMS Microbiology Ecology, 2007, 62(3): 242-257. DOI:10.1111/j.1574-6941.2007.00397.x |
| [14] | Zhou JZ, Ning DL. Stochastic community assembly: does it matter in microbial ecology?. Microbiology and Molecular Biology Reviews, 2017, 81(4): e00002-17. |
| [15] | Vellend M, Agrawal A. Conceptual synthesis in community ecology. The Quarterly Review of Biology, 2010, 85(2): 183-206. DOI:10.1086/652373 |
| [16] | Stegen JC, Lin XJ, Fredrickson JK, Chen XY, Kennedy DW, Murray CJ, Rockhold ML, Konopka A. Quantifying community assembly processes and identifying features that impose them. The ISME Journal, 2013, 7(11): 2069-2079. DOI:10.1038/ismej.2013.93 |
| [17] | Tripathi BM, Stegen JC, Kim M, Dong K, Adams JM, Lee YK. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. The ISME Journal, 2018, 12(4): 1072-1083. DOI:10.1038/s41396-018-0082-4 |
| [18] | Liu JW, Zhu SQ, Liu XY, Yao P, Ge TT, Zhang XH. Spatiotemporal dynamics of the archaeal community in coastal sediments: assembly process and co-occurrence relationship. The ISME Journal, 2020, 14(6): 1463-1478. DOI:10.1038/s41396-020-0621-7 |
| [19] | Wang JJ, Shen J, Wu YC, Tu C, Soininen J, Stegen JC, He JZ, Liu XQ, Zhang L, Zhang EL. Phylogenetic beta diversity in bacterial assemblages across ecosystems: deterministic versus stochastic processes. The ISME Journal, 2013, 7(7): 1310-1321. DOI:10.1038/ismej.2013.30 |
| [20] | Yan QY, Stegen JC, Yu YH, Deng Y, Li XH, Wu S, Dai LL, Zhang X, Li JJ, Wang C, Ni JJ, Li XM, Hu HJ, Xiao FS, Feng WS, Ning DL, He ZL, Van Nostrand JD, Wu LY, Zhou JZ. Nearly a decade-long repeatable seasonal diversity patterns of bacterioplankton communities in the eutrophic Lake Donghu (Wuhan, China). Molecular Ecology, 2017, 26(14): 3839-3850. DOI:10.1111/mec.14151 |
| [21] | Wu WX, Lu HP, Sastri A, Yeh YC, Gong GC, Chou WC, Hsieh CH. Contrasting the relative importance of species sorting and dispersal limitation in shaping marine bacterial versus protist communities. The ISME Journal, 2018, 12(2): 485-494. DOI:10.1038/ismej.2017.183 |
| [22] | Logares R, Tesson SVM, Canb?ck B, Pontarp M, Hedlund K, Rengefors K. Contrasting prevalence of selection and drift in the community structuring of bacteria and microbial eukaryotes. Environmental Microbiology, 2018, 20(6): 2231-2240. DOI:10.1111/1462-2920.14265 |
| [23] | R?ttjers L, Faust K. From hairballs to hypotheses-biological insights from microbial networks. FEMS Microbiol Reviews, 2018, 42(6): 761-780. DOI:10.1093/femsre/fuy030 |
| [24] | Cui YS, Chun SJ, Baek SS, Baek SH, Kim PJ, Son M, Cho KH, Ahn CY, Oh HM. Unique microbial module regulates the harmful algal bloom (Cochlodinium polykrikoides) and shifts the microbial community along the Southern Coast of Korea. Science of the Total Environment, 2020, 721: 137725. DOI:10.1016/j.scitotenv.2020.137725 |
| [25] | Wang YM, Pan J, Yang J, Zhou ZC, Pan YP, Li M. Patterns and processes of free-living and particle-associated bacterioplankton and archaeaplankton communities in a subtropical river-bay system in South China. Limnology and Oceanography, 2020, 65(S1): S161-S179. |
| [26] | Needham DM, Fuhrman JA. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nature Microbiology, 2016, 1(4): 16005. DOI:10.1038/nmicrobiol.2016.5 |
| [27] | Fortunato CS, Herfort L, Zuber P, Baptista AM, Crump BC. Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. The ISME Journal, 2012, 6(3): 554-563. DOI:10.1038/ismej.2011.135 |
| [28] | Kuypers MMM, Marchant HK, Kartal B. The microbial nitrogen-cycling network. Nature Reviews Microbiology, 2018, 16(5): 263-276. DOI:10.1038/nrmicro.2018.9 |
| [29] | Beman JM, Steele JA, Fuhrman JA. Co-occurrence patterns for abundant marine archaeal and bacterial lineages in the deep chlorophyll maximum of coastal California. The ISME Journal, 2011, 5(7): 1077-1085. DOI:10.1038/ismej.2010.204 |
| [30] | Frigaard NU, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic bacteria and archaea. Nature, 2006, 439(7078): 847-850. DOI:10.1038/nature04435 |
| [31] | Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science, 2000, 289(5486): 1902-1906. DOI:10.1126/science.289.5486.1902 |
| [32] | Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature, 2001, 411(6839): 786-789. DOI:10.1038/35081051 |
| [33] | Zhang CL, Xie W, Martin-Cuadrado AB, Rodriguez-Valera F. Marine group Ⅱ archaea, potentially important players in the global ocean carbon cycle. Frontiers in Microbiology, 2015, 6: 1108. |
| [34] | Sun LM, Wang WD, Gao F, Cao CL, Tang JF. Distribution and influencing factors of plankton in an artificial lagoon in Ningbo. Asian Journal of Ecotoxicology, 2018, 13(4): 60-67. (in Chinese) 孙丽梅, 汪文东, 高凤, 曹昌丽, 唐剑锋. 宁波市某人工泻湖浮游生物变化特征及影响因素分析. 生态毒理学报, 2018, 13(4): 60-67. |
| [35] | Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environmental Microbiology, 2016, 18(5): 1403-1414. DOI:10.1111/1462-2920.13023 |
| [36] | Mago? T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 2011, 27(21): 2957-2963. DOI:10.1093/bioinformatics/btr507 |
| [37] | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pe?a AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 2010, 7(5): 335-336. DOI:10.1038/nmeth.f.303 |
| [38] | Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 2011, 27(16): 2194-2200. DOI:10.1093/bioinformatics/btr381 |
| [39] | Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 2010, 26(19): 2460-2461. DOI:10.1093/bioinformatics/btq461 |
| [40] | Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from illumina amplicon sequencing. Nature Methods, 2013, 10(1): 57-59. DOI:10.1038/nmeth.2276 |
| [41] | Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nature Methods, 2013, 10(12): 1200-1202. DOI:10.1038/nmeth.2658 |
| [42] | Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics, 2010, 26(2): 266-267. DOI:10.1093/bioinformatics/btp636 |
| [43] | Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE, 2010, 5(3): e9490. DOI:10.1371/journal.pone.0009490 |
| [44] | Swan BK, Chaffin MD, Martinez-Garcia M, Morrison HG, Field EK, Poulton NJ, Masland ED, Harris CC, Sczyrba A, Chain PS, Koren S, Woyke T, Stepanauskas R. Genomic and metabolic diversity of marine group Ⅰ Thaumarchaeota in the mesopelagic of two subtropical gyres. PLoS ONE, 2014, 9(4): e95380. DOI:10.1371/journal.pone.0095380 |
| [45] | Martin-Cuadrado AB, Garcia-Heredia I, Moltó AG, López-úbeda R, Kimes N, López-García P, Moreira D, Rodriguez-Valera F. A new class of marine Euryarchaeota group Ⅱ from the Mediterranean deep chlorophyll maximum. The ISME Journal, 2015, 9(7): 1619-1634. DOI:10.1038/ismej.2014.249 |
| [46] | Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, Djahanschiri B, Zeller G, Mende DR, Alberti A, Cornejo-Castillo FM, Costea PI, Cruaud C, d'Ovidio F, Engelen S, Ferrera I, Gasol JM, Guidi L, Hildebrand F, Kokoszka F, Lepoivre C, Lima-Mendez G, Poulain J, Poulos BT, Royo-Llonch M, Sarmento H, Vieira-Silva S, Dimier C, Picheral M, Searson S, Kandels-Lewis S, Bowler C, de Vargas C, Gorsky G, Grimsley N, Hingamp P, Iudicone D, Jaillon O, Not F, Ogata H, Pesant S, Speich S, Stemmann L, Sullivan MB, Weissenbach J, Wincker P, Karsenti E, Raes J, Acinas SG, Bork P. Structure and function of the global ocean microbiome. Science, 2015, 348(6237): 1261359. DOI:10.1126/science.1261359 |
| [47] | Liu HD, Zhang CL, Yang CY, Chen SZ, Cao ZW, Zhang ZW, Tian JW. Marine group Ⅱ dominates planktonic archaea in water column of the Northeastern South China Sea. Frontiers in Microbiology, 2017, 8: 1098. DOI:10.3389/fmicb.2017.01098 |
| [48] | Galand PE, Gutiérrez-Provecho C, Massana R, Gasol JM, Casamayor EO. Inter-annual recurrence of archaeal assemblages in the coastal NW Mediterranean Sea (Blanes Bay microbial observatory). Limnology and Oceanography, 2010, 55(5): 2117-2125. DOI:10.4319/lo.2010.55.5.2117 |
| [49] | Church MJ, DeLong EF, Ducklow HW, Karner MB, Preston CM, Karl DM. Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnology and Oceanography, 2003, 48(5): 1893-1902. DOI:10.4319/lo.2003.48.5.1893 |
| [50] | Wang K, Yan HZ, Peng X, Hu HJ, Zhang HJ, Hou DD, Chen W, Qian P, Liu JF, Cai JB, Chai XL, Zhang DM. Community assembly of bacteria and archaea in coastal waters governed by contrasting mechanisms: a seasonal perspective. Molecular Ecology, 2020, 29(19): 3762-3776. DOI:10.1111/mec.15600 |
| [51] | Qin W, Meinhardt KA, Moffett JW, Devol AH, Virginia Armbrust E, Ingalls AE, Stahl DA. Influence of oxygen availability on the activities of ammonia-oxidizing archaea. Environmental Microbiology Reports, 2017, 9(3): 250-256. DOI:10.1111/1758-2229.12525 |
| [52] | Gillies LE, Thrash JC, deRada S, Rabalais NN, Mason OU. Archaeal enrichment in the hypoxic zone in the northern Gulf of Mexico. Environmental Microbiology, 2015, 17(10): 3847-3856. DOI:10.1111/1462-2920.12853 |
| [53] | Liu HD, Yan RM, Zhang CL. Advances in marine group Ⅱ archaea research. Acta Microbiologica Sinica, 2020, 60(9): 1834-1851. (in Chinese) 刘浩东, 闫荣曼, 张传伦. 海洋浮游古菌MGII的研究进展. 微生物学报, 2020, 60(9): 1834-1851. |
| [54] | Lai DX, Xie W, Fan L, Tao JC, Hu AY, Wang P, Zhu YQ, Zhang CL. The spatial patterning of marine group Ⅱ archaea in Chinese coastal and esturine waters. Bulletin of National Natural Science Foundation of China, 2018, 32(5): 459-470. (in Chinese) 赖登训, 谢伟, 范陆, 陶建昌, 胡安谊, 王鹏, 朱元清, 张传伦. MGII古菌在中国近海和河口的空间分布特征. 中国科学基金, 2018, 32(5): 459-470. |
| [55] | Fortunato CS, Eiler A, Herfort L, Needoba JA, Peterson TD, Crump BC. Determining indicator taxa across spatial and seasonal gradients in the Columbia River coastal margin. The ISME Journal, 2013, 7(10): 1899-1911. DOI:10.1038/ismej.2013.79 |
| [56] | Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I, Somerfield P, Fuhrman JA, Field D. Defining seasonal marine microbial community dynamics. The ISME Journal, 2012, 6(2): 298-308. DOI:10.1038/ismej.2011.107 |
| [57] | Ma B, Wang HZ, Dsouza M, Lou J, He Y, Dai ZM, Brookes PC, Xu JM, Gilbert JA. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. The ISME Journal, 2016, 10(8): 1891-1901. DOI:10.1038/ismej.2015.261 |
| [58] | Reji L, Tolar BB, Smith JM, Chavez FP, Francis CA. Differential co-occurrence relationships shaping ecotype diversification within Thaumarchaeota populations in the coastal ocean water column. The ISME Journal, 2019, 13(5): 1144-1158. DOI:10.1038/s41396-018-0311-x |
| [59] | Garrity GM, Bell JA, Lilburn T, Family I. Rhodobacteraceae fam. nov//Brenner DJ, Krieg NR, Staley JT, Garrity GM. Bergey's manual of systematic bacteriology, vol 2, 2nd edn, The Proteobacteria, Part C. The alpha-, beta-, delta-, and Epsilonproteobacteria. New York: Springer, 2005: 161-228. |
| [60] | Alderkamp AC, Sintes E, Herndl GJ. Abundance and activity of major groups of prokaryotic plankton in the coastal North Sea during spring and summer. Aquatic Microbial Ecology, 2006, 45(3): 237-246. |
| [61] | Qin W, Heal KR, Ramdasi R, Kobelt JN, Martens-Habbena W, Bertagnolli AD, Amin SA, Walker CB, Urakawa H, Konneke M, Devol AH, Moffett JW, Armbrust EV, Jensen GJ, Ingalls AE, Stahl DA. Nitrosopumilus maritimus gen. nov., sp. nov., Nitrosopumilus cobalaminigenes sp. nov., Nitrosopumilus oxyclinae sp. nov., and Nitrosopumilus ureiphilus sp. nov., four marine ammonia-oxidizing archaea of the phylum Thaumarchaeota. International Journal of Systematic and Evolutionary Microbiology, 2017, 67(12): 5067-5079. DOI:10.1099/ijsem.0.002416 |
| [62] | Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH, Ingalls AE, Moffett JW, Armbrust EV, Stahl DA. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(34): 12504-12509. DOI:10.1073/pnas.1324115111 |
| [63] | Qin W, Zheng Y, Zhao F, Wang YL, Urakawa H, Martens-Habbena W, Liu HD, Huang XW, Zhang XX, Nakagawa T, Mende DR, Bollmann A, Wang BZ, Zhang Y, Amin SA, Nielsen JL, Mori K, Takahashi R, Virginia Armbrust E, Winkler MKH, DeLong EF, Li M, Lee PH, Zhou JZ, Zhang CL, Zhang T, Stahl DA, Ingalls AE. Alternative strategies of nutrient acquisition and energy conservation map to the biogeography of marine ammonia-oxidizing archaea. The ISME Journal, 2020, 14(10): 2595-2609. DOI:10.1038/s41396-020-0710-7 |
| [64] | Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ, Rappé MS. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio, 2012, 3(5): e00252-12. |
