 ,, 董亚辉
,, 董亚辉 ,, 任真真, 王志勇, 苏慧慧, 库丽霞, 陈彦惠
,, 任真真, 王志勇, 苏慧慧, 库丽霞, 陈彦惠 ,*河南农业大学农学院/省部共建小麦玉米作物学国家重点实验室,郑州 450046
,*河南农业大学农学院/省部共建小麦玉米作物学国家重点实验室,郑州 450046Over-expression of ZmIBH1-1 to Improve Drought Resistance in Maize Seedlings
ZHU FangFang ,, DONG YaHui
,, DONG YaHui ,, REN ZhenZhen, WANG ZhiYong, SU HuiHui, KU LiXia, CHEN YanHui
,, REN ZhenZhen, WANG ZhiYong, SU HuiHui, KU LiXia, CHEN YanHui ,*College of Agronomy, Henan Agricultural University/National Key Laboratory of Wheat and Maize Crop Science, Zhengzhou 450046
,*College of Agronomy, Henan Agricultural University/National Key Laboratory of Wheat and Maize Crop Science, Zhengzhou 450046通讯作者:
责任编辑: 李莉
收稿日期:2021-04-25接受日期:2021-06-16
| 基金资助: |
Received:2021-04-25Accepted:2021-06-16
作者简介 About authors
联系方式:朱芳芳,E-mail:
董亚辉,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2129KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
朱芳芳, 董亚辉, 任真真, 王志勇, 苏慧慧, 库丽霞, 陈彦惠. 过表达ZmIBH1-1提高玉米苗期抗旱性. 中国农业科学, 2021, 54(21): 4500-4513 doi:10.3864/j.issn.0578-1752.2021.21.002
ZHU FangFang, DONG YaHui, REN ZhenZhen, WANG ZhiYong, SU HuiHui, KU LiXia, CHEN YanHui.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】玉米是世界第一大粮食作物,其产量占全球总谷物产量的37.2%[1]。玉米产量的提高对保障国家粮食安全至关重要。干旱、盐碱、矿物质缺乏等非生物胁迫对玉米正常的生长发育会造成极大的影响,进而导致玉米产量的损失。其中,干旱是制约玉米产量提高的重要因素之一。因此,挖掘玉米抗旱耐旱的关键基因,从分子水平上揭示玉米抗旱机制,为培育抗旱耐旱玉米新品种提供理论依据,对保障粮食产量稳定具有十分重要的意义。【前人研究进展】为了适应复杂多变的自然环境,在漫长的进化过程中,植物自身形成了一种特异的适应机制,植物在应对干旱等逆境时,会启动相应的功能基因,调节其生理生化水平,以适应不良环境[2,3]。已有的研究表明,植物中bHLH、NAC、WRKY、bZIP、NF-Y、MYB等转录因子家族的基因通过不同的调控路径,在植物抵御逆境胁迫反应中发挥着重要的作用[4,5,6,7,8,9,10]。在干旱等胁迫条件下,多个玉米转录因子家族基因的过量表达可以提高植物的抗旱耐旱特性。例如,玉米苗期过表达ZmNAC111可以提高玉米水分利用率并诱导干旱响应基因的表达,进而提高玉米干旱耐受性[4];ZmNAC55的表达受干旱、高盐等胁迫条件诱导,过表达ZmNAC55可以增强拟南芥干旱耐受性[5]。ZmWRKY106作为干旱胁迫和高温胁迫的正调控因子,通过调控ABA信号路径相关基因,过表达ZmWRKY106提高了拟南芥中超氧化物歧化酶(superoxide dismutase,SOD)、过氧化物酶(peroxidase,POD)和过氧化氢酶(catalase,CAT)活性,降低了活性氧(reactive oxygen species,ROS)含量,转基因拟南芥表现出耐旱性和耐热性[6]。过表达ZmWRKY40提高干旱胁迫下转基因拟南芥中POD和CAT的活性,降低了ROS积累,通过调控胁迫相关基因提高转基因拟南芥的抗旱性[7]。植物bZIP转录因子调节多种功能,包括植物发育、胁迫反应和信号传导等过程,过表达ZmbZIP72的拟南芥比野生型的电解质渗漏低,水分流失慢,干旱和盐耐受性增强[8]。干旱处理24 h后,地上部ZmbZIP71的表达量升高5.49倍,推测该基因可能参与了玉米干旱胁迫响应[9]。干旱和盐胁迫诱导ZmMYB3R的表达,拟南芥过表达该基因呈现生长性能增强,存活率高,CAT、POD和SOD活性升高,ZmMYB3R增强了转基因拟南芥对干旱和盐胁迫的耐受性[10]。除去转录因子,还有与干旱胁迫响应相关的基因的报道。例如,干旱处理后,过表达ZmPTPN转基因玉米成活率显著提高,而ZmPTPN敲除突变体对干旱表现出更加敏感,说明ZmPTPN增强了玉米干旱耐受性[11]。ZHANG等[12]发现过表达ZmTIP1的转基因玉米根毛长度增加,对水分亏缺的耐性提升。DING等[13]发现ZmGRXCC14的遗传变异与玉米苗期抗旱性显著关联。玉米gl6突变体表皮蜡质减少、角质层渗透性增加,降低了幼苗抗旱性[14]。ZmPIP1;1通过诱导胁迫响应基因和提高ROS清除酶活性,可能在干旱和盐胁迫耐受中起作用[15]。WANG等[16]鉴定到一个玉米Ⅰ类SUMO结合酶基因(ZmSCE1d),该基因在干旱胁迫下表达上调,拟南芥中过表达该基因则增强其抗旱性。在干旱条件下,过表达ZmASR3的转基因株系通过提高SOD和CAT的活性,增加气孔关闭,减少ROS的积累正向调控植物耐旱性[17]。bHLH家族是仅次于MYB家族的第二大家族[18],在植物抗旱中发挥着重要作用。AmDEL与VvbHLH1编码典型的bHLH转录因子,可以促进转基因拟南芥体内类黄酮生物合成、脱落酸(abscisic acid,ABA)信号通路、脯氨酸生物合成及ROS清除酶等基因表达上调,增强拟南芥耐旱性[19,20]。胡杨基因PebHLH35被干旱诱导后表达上调,主要通过调节气孔密度和大小、光合作用及生长发育等从而提高其抗旱性[21]。茶树在干旱胁迫下有39个bHLH转录因子表达上调[22]。拟南芥AtbHLH122是耐旱性、耐盐性等多个逆境胁迫信号的调节因子[23]。在水稻中,OsbHLH148高量表达显著提高了水稻的抗旱性[24]。ZmPTF1是一种磷酸饥饿诱导的bHLH转录因子,过表达ZmPTF1株系改善玉米根系发育,增加ABA含量,激活ABA-、CBF4-、ATAF2-和NAC30介导的胁迫响应,提高玉米的耐旱性[25]。【本研究切入点】ZmIBH1-1编码一个bHLH型转录因子,在叶夹角形成发育中负向调控玉米叶夹角的大小[26]。然而,关于IBH1-1是否可以提高植物的耐旱性,目前尚未报道。【拟解决的关键问题】本研究以B104(WT)及B104为背景的过表达ZmIBH1-1转基因(ZmIBH1-1-OE)株系为材料,对玉米幼苗进行干旱胁迫处理,结合RNA-Seq和DAP-Seq数据分析,解析ZmIBH1-1响应干旱胁迫的分子机制,为挖掘玉米抗旱基因及选育抗旱型玉米新品种奠定理论基础。1 材料与方法
试验于2018年6月—2020年10月在河南农业大学农学院/省部共建小麦玉米作物学国家重点实验室完成。1.1 试验材料
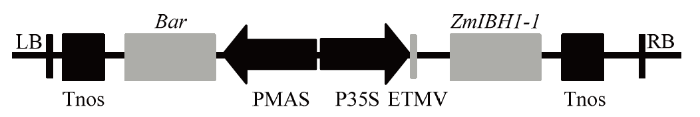
玉米遗传转化所用农杆菌(Agrobacterirum tumefaciens)菌株EHA105购自河南三瑞生物科技有限公司;pFGC5941植物表达载体购自BioVector NTCC典型培养物保藏中心;过表达ZmIBH1-1重组质粒由河南农业大学省部共建小麦玉米作物学国家重点实验室陈彦惠教授课题组构建,T-DNA区结构如图1所示;玉米遗传转化由北京博美兴奥科技有限公司完成;T1—T4自交纯合及阳性转基因植株鉴定由河南农业大学省部共建小麦玉米作物学国家重点实验室陈彦惠教授课题组完成。过表达ZmIBH1-1转基因株系命名为ZmIBH1-1-OE。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1过表达ZmIBH1-1重组质粒的T-DNA区示意图
LB:T-DNA的左边重复序列;Tnos:终止子;Bar:除草剂筛选基因;PMAS:MAS启动子;P35S:35S启动子;ETMV:TMV增强子;RB:T-DNA的右边重复序列
Fig. 1Schematic diagram of the T-DNA region of the ZmIBH1-1 overexpression recombinant plasmid
LB: Left border repeat of T-DNA; Tnos: Terminator; Bar: Herbicide screening gene; PMAS: MAS promoter; P35S: 35S promoter; ETMV: TMV enhancer; RB: Right border repeat of T-DNA
1.2 转基因植株的检测
采用除草剂抗性筛选、标记和目的基因PCR鉴定和目的基因的荧光定量PCR表达分析3种方法对转基因植株进行检测鉴定。除草剂抗性筛选:利用WT和ZmIBH1-1-OE为材料,从植株3片叶开始,每隔3—5 d向叶片喷施草铵膦(300 mg·L-1),共喷施3次,处理一周后观察抗性表型,初步筛选转基因阳性植株。标记和目的基因PCR鉴定:以提取的植株叶片DNA为模板,利用标记基因Bar特异引物(Bar-F和Bar-R)和ZmIBH1-1特异引物(ZmIBH-F和ZmIBH-R)进行扩增检测,根据扩增条带鉴定出转基因阳性植株,所用引物序列见表1。目的基因的荧光定量PCR表达分析:用Trizol法提取玉米第4片完全展开叶的RNA,按PrimeScripTM RT reagent Kit(TaKaRa)的操作步骤进行RNA反转录,用ZmIBH1-1定量引物(qIBH-F和qIBH-R)和内参引物(Tublin-F和Tublin-R)进行荧光定量PCR检测,采用2-ΔΔCt法进行定量分析[27]检测阳性植株。每个样品3次生物学重复,3次技术重复,引物序列见表1。将转基因不同世代经过PCR检测到的阳性单株移栽到大田,在开花期进行人工套袋自交获得种子。Table 1
表1
表1试验所用引物
Table 1
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|
| Bar-F | CATCGAGACAAGCACGGTC |
| Bar-R | AAACCCACGTCATGCCAGTT |
| ZmIBH-F | CAATTACATTTACAATTACCATGGTCATGGCCAGGAAGAGGAC |
| ZmIBH-R | CTCTCTAGACTCACCTAGGATCCTCATTGGGCGGAGAAG |
| qIBH-F | AGGAACCACCGCCAAACC |
| qIBH-R | GCHTCTCCGCAGCAGGAC |
| Tublin-F | CCGCTATCTCCGTCGC |
| Tublin-R | GTTCTTGGATGGCGGTCG |
新窗口打开|下载CSV
1.3 干旱胁迫下WT和ZmIBH1-1-OE的表型鉴定
干旱胁迫处理:将ZmIBH1-1-OE和WT玉米籽粒置于10%的H2O2中浸泡洗涤20 min,无菌水洗涤3次,每次5 min。消毒后的种子放在无菌湿润的发芽纸中,28℃黑暗培养2 d,挑选发芽均一的籽粒种植蛭石中,在(30±2)℃恒温培养室中培养(12 h黑暗/12 h光照)。玉米幼苗生长至2叶1心转入Hoagland营养液,4叶1心时采用添加20% PEG-6000的Hoagland营养液进行干旱处理,同时设置对照(Hoagland营养液),试验重复3次。耐旱表型鉴定:选取对照和干旱处理的WT和ZmIBH1-1-OE植株进行存活率统计和相对含水量测定。
相对含水量(relative water content,RWC)的测定:干旱处理24 h后取新鲜叶片擦干净后称重(fresh weight,FM),将叶片放入水中5—6 h,使叶片吸水达到饱和状态后,取出叶片并擦干叶片表面水分后再称重(saturation weight,TM),再将叶片放入烘箱,105℃杀青30 min,然后在80℃环境下烘至恒重,称重(dry weight,DM)。叶片相对含水量RWC(%)=(FM-DM)/(TM-DM)×100。
1.4 生理生化指标的测定
干旱处理24 h后取处理组和对照组的植株叶片,用于生理生化指标的测定,每个指标3次生物学重复。用韩赞平[28]方法进行SOD、POD、CAT活性测定;用叶绿素用丙酮-乙醇混合液的萃取方法[29]进行叶绿素含量(chlorophyll contents,Cht)和类胡萝卜素含量(carotenoids,Car)测定,称取0.2 g新鲜玉米叶片,加入80%预冷的丙酮,研磨成匀浆,于6 000 r/min离心15 min,抽取上清液测定其在665、649和470 nm波长下的吸光值,试验在黑暗环境中进行。计算公式为:叶绿素a浓度(mg·L-1)Ca=13.95A665-6.8A649
叶绿素b浓度(mg·L-1)Cb=24.96 A649-7.32A665
叶绿素浓度(mg·L-1)Cht=Ca+Cb
类胡萝卜素浓度(mg·L-1)Car=(1000A470-2.05Ca- 114.8Cb)/248
采用考马斯亮蓝染色法[30],对可溶性蛋白(soluble protein,SP)含量进行测定,称取0.1 g新鲜玉米叶片,采用Tris-HCL缓冲液(pH6.8)进行可溶性蛋白研磨提取,通过测定595 nm处吸光值测定可溶性蛋白含量。
1.5 数据分析
应用Microsoft Excel 2010进行数据分析和作图。1.6 转录组测序及分析
以正常、干旱处理的WT和ZmIBH1-1-OE植株的叶片(2个生物学重复)为材料,委托武汉希望组生物科技有限公司完成RNA提取和文库构建,然后通过MGI-T7平台对文库进行PE150测序,共构建8个cDNA文库。使用在线软件Trim Galore(www.bioinformatics.babraham.ac.uk/projects/trim_galore/)处理测序产出的原始数据,去除含有接头、poly-N及低质量reads后获得高质量clean reads,并对Q20、Q30、GC含量以及重复序列进行统计;将上述高质量的clean reads与玉米基因组B73_V4(AGPV4版本)进行比对,通过FPKM(Fragments per Kilobase per Millon Mapped Fragments)对基因表达量进行标准化,以FPKM>1为表达标准,随后根据正常与干旱处理的WT和ZmIBH1-1-OE的比较,用DEseq(|log2 FC(fold change)|≥1和P-value/FDR<0.05)确定DEGs。利用Uniprot、Swissprot、COG、NR、GO和KEGG等数据库对DEGs进行功能注释,利用在线工具WEGO(1.7 ZmIBH1-1对其直接作用的下游靶基因的调控分析
为了明确ZmIBH1-1在玉米发育过程中的调控网络,CAO等[26]采用DAP-seq技术证明了ZmIBH1-1蛋白通过绑定4个Cis-elements(NNCAAGTNG、CANGTN、CTTCGNN和GGNGGAGA)直接作用于启动子区域的靶基因有1 188个,结合RNA-Seq数据明确了调控玉米叶夹角发育的靶基因。为了进一步明确ZmIBH1-1响应干旱胁迫所涉及的调控路径,首先用VLOOKUP对干旱胁迫下的RNA-Seq分析获得的差异表达基因和ZmIBH1-1蛋白DAP-seq获得的1 188个靶基因[26]进行交集分析,初步确定干旱胁迫响应的候选靶基因。然后用基因组可视化软件IGV(integrative genomics viewer)分析ZmIBH1-1蛋白结合候选靶基因的位置。采用Dual-Luciferase试验进一步验证ZmIBH1-1与靶基因的调控关系。将靶基因的启动子(含有ZmIBH1-1的结合位点)克隆到pGreenII0800-luc载体上(含有报告基因);将基因ZmIBH1-1克隆到pCAMBIA1300载体上。将上述重组载体分别转入农杆菌GV3101中,采用3种组合(融合有靶基因启动子的双荧光素酶报告载体、过表达转录因子载体以及融合有靶基因启动子的双荧光素酶报告载体、双荧光素酶报告载体(空载))侵染烟草(N. benthamiana)叶片。培养(14 h光照/10 h黑暗)2 d后,提取烟草叶片蛋白(Cat#E1910,Promega),使用Glomax®20/20生物/化学发光检测仪(Cat#E5311,Promega)测定萤火虫荧光素酶(firefly luciferase,LUC)和海肾荧光素酶(renilla luciferase,REN),每个样品3个生物学重复。
2 结果
2.1 ZmIBH1-1-OE转基因玉米阳性植株的鉴定
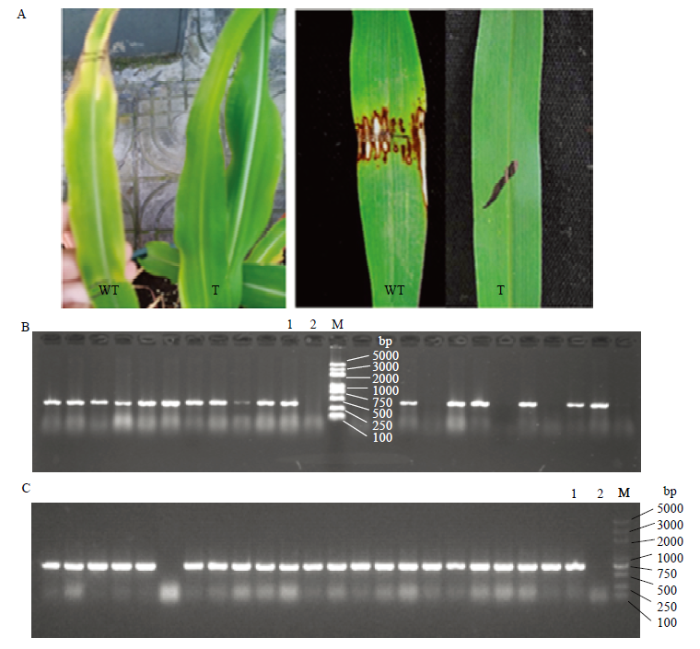
通过玉米遗传转化共获得12个转化事件。T3代时,每个转化事件种植5个株系,每个株系种植10个单株,共600个单株,进行鉴定(图2)。结果表明,T3代株系中约80%具有筛选标记基因Bar和目的基因ZmIBH1-1。标记基因和目的基因同时被检测到的植株有458个,其中,来自于2个(3、8)转化事件的3个株系,共30个单株全部为阳性。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2部分T3转基因玉米植株的鉴定结果
A:喷洒或涂抹除草剂草胺膦的表型鉴定;B:Bar引物PCR检测;C:IBH1-1引物PCR检测。WT:野生型单株;T:转基因单株;1:阳性对照(质粒DNA);2:空白对照;M:DL5000 DNA Marker;其余泳道代表转基因植株
Fig. 2Identification of some T3 transgenic maize plants
A: Phenotypic identification of herbicide glyphosate sprayed or applied; B: PCR detection of Bar; C: PCR detection of IBH1-1. WT: Wild-type plant; T: Transgenic ZmIBH1-1-OE plant; 1: Positive control (plasmid DNA); 2: Blank; M: DL5000 DNA Marker; The rest represent transgenic plants
对T3代12个独立事件的转基因株系在4叶期进行qRT-PCR检测,结果表明,ZmIBH1-1-OE株系中的ZmIBH1-1表达量显著高于WT,且在3、8转化事件中该基因的表达量最高(图3)。结合抗除草剂鉴定和PCR检测结果,说明这两个转化事件已经纯合。转化事件3和8套袋自交获得的T4代转基因株系用于后续试验研究。
图3
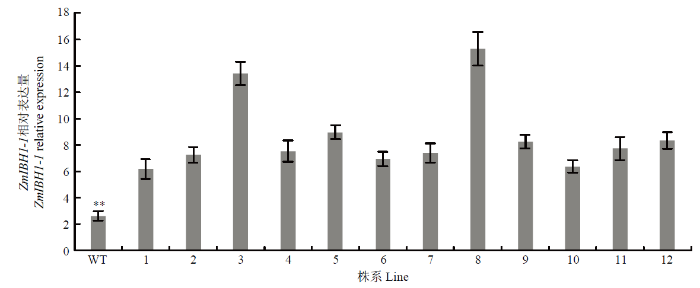 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图312个转化事件株系中ZmIBH1-1的表达量测定
WT:野生型;1—12:12个转化事件的ZmIBH1-1-OE株系(n=3,±SD,**P<0.01)
Fig. 3Expression of ZmIBH1-1 in 12 transformation events lines
WT: Wild-type plant; 1-12: 12 transformation events of ZmIBH1-1-OE lines (n=3, ±SD, **P<0.01)
2.2 过表达ZmIBH1-1增强玉米耐旱性
ZmIBH1-1-OE和WT植株模拟干旱处理后发现,ZmIBH1-1-OE株系比WT植株抗旱性强(图4-A)。具体表现为:干旱胁迫下,ZmIBH1-1-OE株系的存活率显著高于WT(图4-B),ZmIBH1-1-OE株系的叶片相对含水量显著高于WT(图4-C),ZmIBH1-1-OE株系的SOD、CAT及POD酶活性均显著高于WT(图4-D),类胡萝卜素、总叶绿素、可溶性蛋白含量也均显著高于WT(图4-E)。以上结果说明过表达ZmIBH1-1增强玉米的耐旱性。图4
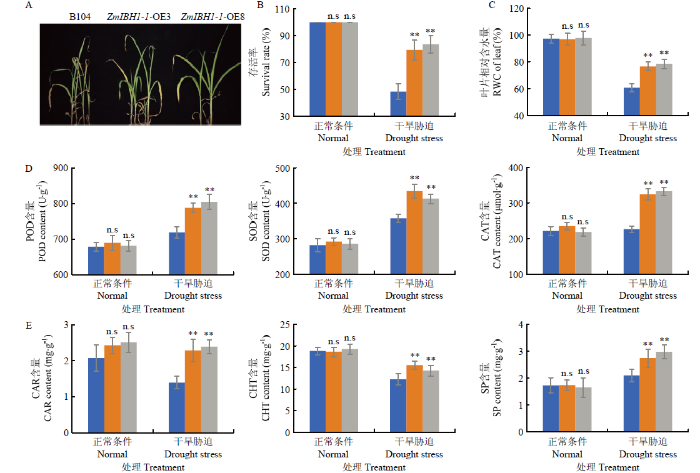 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4ZmIBH1-1-OE和野生型株系B104在干旱迫下的表型及各项生理指标变化
A:B104和ZmIBH1-1-OE株系在PEG6000胁迫下的表型;B:B104和ZmIBH1-1-OE株系在PEG6000胁迫下的存活率(n=3,±SD,**P<0.01,n.s不显著);C:B104和ZmIBH1-1-OE株系在PEG6000胁迫下的叶片平均含水量(n=3,±SD,**P<0.01,n.s不显著);D:B104和ZmIBH1-1-OE株系在PEG6000胁迫下的POD、SOD、CAT酶活性测定(n=3,±SD,**P<0.01,n.s不显著);E:B104和ZmIBH1-1-OE株系在PEG6000胁迫下的总叶绿素(Cht)、类胡萝卜素(Car)和可溶性蛋白(SP)含量的变化测定(n=3,±SD,**P<0.01,n.s不显著)
Fig. 4Phenotype and physiological changes of ZmIBH1-1-OE and wild type B104 lines under drought stress
A: The phenotype of B104 and ZmIBH1-1-OE lines under PEG6000 stress; B: Mean survival rate of B104 and ZmIBH1-1-OE lines under PEG6000 stress (n=3, ±SD, **P<0.01, n.s not significant); C: Mean relative water contents (RWCs) of B104 and ZmIBH1-1-OE lines under PEG6000 stress (n=3, ±SD, **P<0.01, n.s not significant); D: Determination of POD, SOD and CAT enzyme activities of B104 and ZmIBH1-1-OE strains under PEG6000 stress (n=3, ±SD, **P< 0.01, n.s not significant); E: Determination of the contents of Cht, Car and SP of B104 and ZmIBH1-1-OE strains under PEG6000 stress (n=3, ±SD, **P<0.01, n.s not significant)
2.3 干旱胁迫下ZmIBH1-1-OE和WT叶片转录组分析
为明确ZmIBH1-1-OE植株在干旱胁迫下转录水平变化,对正常和干旱胁迫下WT和ZmIBH1-1-OE株系的幼苗进行转录组测序。去除低质量reads后,8个样品均有超过91%的clean reads。将clean reads与玉米B73基因组参考序列V4进行比对,8个样品均有超过88%的unique reads(表2);PCA分析结果显示不同样品间存在差异,同一样品不同生物学重复聚集(图5-A);相关性分析表明,生物学重复间相关系数高(图5-B)。以上结果表明转录组数据稳定可靠,可用于后续分析。Table 2
表2
表2RNA-Seq数据reads数总结
Table 2
| 样品 Sample | 正常条件Normal | 干旱胁迫Drought stress | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 原始reads Raw reads | 过滤后的reads Clean reads | 过滤后的 reads所占比 Reads keep rate (%) | 比对上的 reads Mapped reads | 比对上的 reads占比 Mapped reads rate (%) | 原始reads Raw reads | 过滤后的 reads Clean reads | 过滤后的 reads所占比 Reads keep rate (%) | 比对上的reads Mapped reads | 比对上的 reads占比 Mapped reads rate (%) | |
| WT-1 | 5407756 | 5032404 | 93.05 | 4492503 | 89.27 | 3870308 | 3532833 | 91.28 | 3128759 | 88.56 |
| WT-2 | 7232533 | 6586333 | 91.06 | 5987305 | 90.90 | 4119004 | 3813323 | 92.57 | 3482561 | 91.32 |
| OE-1 | 8523635 | 7780152 | 91.27 | 6893250 | 88.60 | 6322179 | 5776743 | 91.37 | 5367210 | 92.91 |
| OE-2 | 7647821 | 6977539 | 91.23 | 6289035 | 90.13 | 6994016 | 6490634 | 92.80 | 5901352 | 90.92 |
新窗口打开|下载CSV
图5
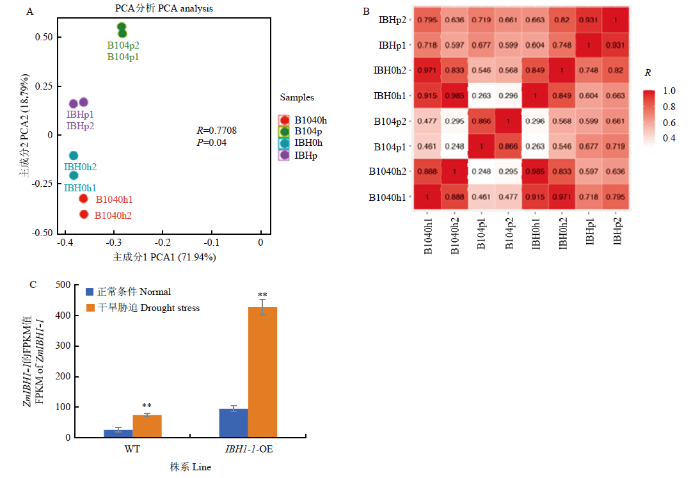 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5B104与ZmIBH1-1-OE株系在干旱胁迫和正常条件下的转录组数据分析
A:转录组数据PCA分析。p代表PEG处理,0 h代表未处理,1和2代
Fig. 5RNA-Seq analysis of B104 and ZmIBH1-1-OE lines under normal and drought conditions
A: PCA analysis of RNA-Seq. p represents PEG treatment, 0 h represents normal conditions, 1 and 2 represents two biological replicates respectively; B: Correlation analysis of RNA-Seq; C: The expression of ZmIBH1-1 in B104 and ZmIBH1-1-OE lines under normal/drought stress (n=2, ±SD, **P<0.01)
转录组分析结果表明,干旱胁迫下ZmIBH1-1的表达量显著高于正常条件,且在ZmIBH1-1-OE株系中表达量显著高于WT(图5-C);WT与ZmIBH1-1-OE株系在干旱胁迫下有1 214个基因表达差异显著。将1 214个差异表达基因进行GO注释分析(电子附图1-A),这些基因主要涉及到生物过程、细胞组分和分子功能。KEGG代谢通路分析结果表明,差异表达基因主要参与植物激素信号传导、新陈代谢等路径(电子附图1-C)。1 214个差异表达基因中还包括Ca+通道蛋白及NAC、WRKY、MYB等类型转录因子(电子附图1-B和电子附表1)。
2.4 ZmIBH1-1蛋白直接作用的干旱胁迫响应的靶基因分析
为进一步解析ZmIBH1-1蛋白直接作用的干旱响应靶基因,对RNA-Seq差异表达基因和DAP-Seq分析得到的ZmIBH1-1蛋白的靶基因[26]进行分析,初步确定11个ZmIBH1-1可能直接调控的与抗旱相关的候选靶基因(电子附表2),包括2个钙信号相关基因(ZmCa-M:Zm00001d025340;ZmAGD12:Zm00001d051676)、3个半胱氨酸代谢相关基因(ZmSYCO:Zm00001d034736;ZmCYS:Zm00001d049110;ZmCYSB:Zm00001d038173)、1个bHLH转录因子(ZmbHLH54:Zm00001d011847)、1个应激响应蛋白(ZmCLPB3:Zm00001d047302)、1个谷胱甘肽转移酶(ZmGlu-r1:Zm00001d036951)、1个氧化还原过程蛋白(ZmP450-99A2:Zm00001d029519)和2个乙烯响应因子(ZmERF-107:Zm00001d017466;ZmEIN3:Zm00001d028974)。ZmIBH1-1的DAP-Seq结果显示ZmIBH1-1在靶基因启动子区域存在peaks富集(图6),说明ZmIBH1-1可以直接结合靶基因启动子。图6
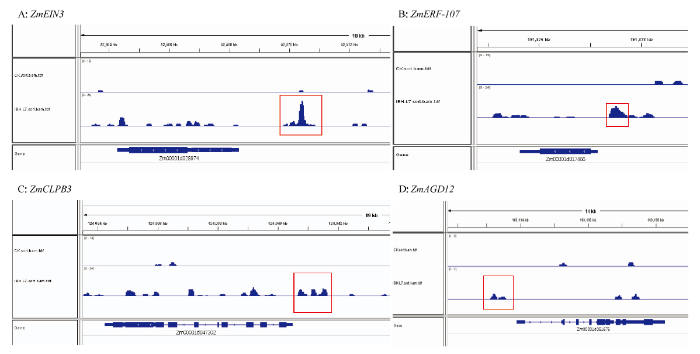 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6IGV展示ZmIBH1-1在部分靶基因启动子区域存在peaks富集
A—D分别代表ZmIBH1-1分别结合ZmEIN3、ZmERF-107、ZmCLPB3和ZmAGD12的启动子区域。红框代表ZmIBH1-1结合峰所在位置
Fig. 6IGV showed that ZmIBH1-1 had peaks enrichment in the promoter region of some target genes
A-D represents that ZmIBH1-1 can bind to the promoter region of target genes ZmEIN3, ZmERF-107, ZmCLPB3, and ZmAGD12, respectively. The red box represents the location of ZmIBH1-1 binding peaks
随后采用Dual-Luciferase试验进一步验证ZmIBH1-1对候选靶基因的调控作用(图7-A)。结果表明,ZmIBH1-1分别显著增强由ZmCa-M、ZmSYCO、ZmbHLH54、ZmGlu-r1、ZmCLPB3和ZmP450-99A2的启动子启动的报告基因LUC的表达(图7-B),而显著降低由ZmAGD12、ZmCYS、ZmCYSB、ZmERF-107和ZmEIN3的启动子启动的报告基因LUC的表达(图7-C);RNA-Seq结果表明,在干旱胁迫下,ZmCa-M、ZmSYCO、ZmbHLH54、ZmGlu-r1、ZmCLPB3和ZmP450-99A2在ZmIBH1-1-OE株系中的表达量显著高于WT,而ZmZmAGD12、ZmCYS、ZmCYSB、ZmERF-107和ZmEIN3在ZmIBH1-1-OE株系中的表达量显著低于WT。以上结果表明ZmIBH1-1可以直接调控靶基因的表达。
图7
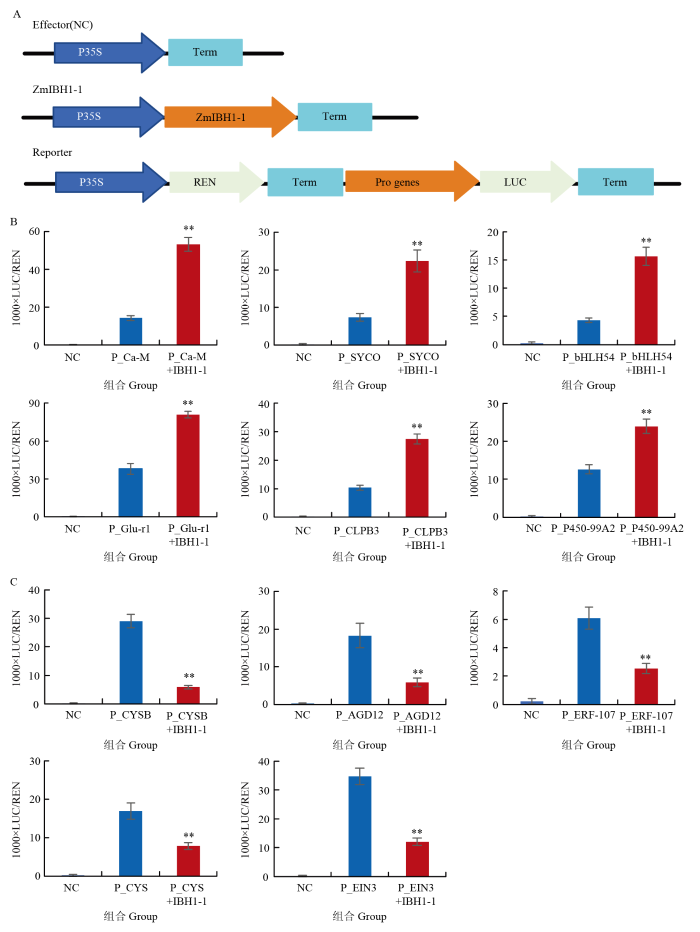 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7ZmIBH1-1对靶基因的调控
A:35S启动子启动海肾荧光素酶(REN)作为内参,基因启动子启动报告基因萤火虫荧光素酶(LUC)基因,ZmIBH1-1作为效应因子,空载体为对照,在本氏烟草中瞬时表达。B—C:LUC/REN的比值代表启动子的活性,每个试验重复3次(n=3,±SD,**P<0.01)。NC:阴性对照
Fig. 7Regulation of target genes by ZmIBH1-1
A: The 35S: REN-Pro gene LUC reporter constructs were transiently expressed in N.benthamiana leaves together with control vector or 35S:ZmIBH1-1 effector, respectively. The expression level of REN was used an internal control; B-C: The LUC/REN ratio represents the relative activity of promoters. Data are values of three independent experiments. Significant differences from the corresponding control values (n=3, ±SD, **P<0.01). NC: Negative control
3 讨论
本研究结果表明,过表达ZmIBH1-1可以增强玉米的耐旱性,ZmIBH1-1蛋白直接结合11个靶基因的启动子调控靶基因的表达,其中包括2个乙烯响应基因ZmEIN3和ZmERF-107,因此推测ZmIBH1-1可能通过乙烯信号通路来响应干旱胁迫。植物在受到干旱胁迫时植物体内的内源乙烯含量会有很大程度的提升[32],过量的乙烯会诱发植物细胞的衰老和程序性死亡[33,34]。在玫瑰中RhEIN3促进RhGAI1表达,而RhGAI1可以显著抑制乙烯受体蛋白基因RhETR的表达[35],ETRs结合乙烯,减少或抑制乙烯信号转导诱发的细胞死亡[36]。ZmEIN3作为EIN3-like转录因子,是乙烯信号通路中的重要转录因子[37]。本研究结果表明,在干旱胁迫下,ZmEIN3的表达被ZmIBH1-1显著抑制。说明ZmIBH1-1可能通过直接抑制ZmEIN3的表达,间接促进玉米中乙烯受体蛋白基因的表达,从而减少或抑制乙烯诱发的细胞程序性死亡,提高玉米的抗旱性。在逆境胁迫下,植物通过转录因子调控相关基因的表达提高植物体抵御逆境胁迫的能力[4,5,6,7,8,9,10]。ERF转录因子在响应逆境胁迫中起到重要作用[38,39]。ERF家族成员通过特异结合基因启动子GCC-box,从而参与乙烯应答及非生物胁迫[38]。水稻中,过表达ERF转录因子基因SNORKEL1(SK1)和SK2可以调节内源乙烯等激素合成的功能,以抵抗淹水胁迫[39]。本研究中ZmIBH1-1蛋白通过抑制乙烯响应因子基因ZmERF-107的表达参与玉米对干旱胁迫响应的调节。钙离子作为第二信使,在植物应对非生物胁迫时(盐、干旱、低温等)发挥着重要作用[40,41]。干旱缺水引起细胞质内钙离子浓度变化从而激活钙依赖性蛋白激酶(calmodulin-dependent protein kinases,CPKs)信号导致ABA释放,ABA浓度的积累导致气孔关闭减少水分损失[42,43]。本研究中,在干旱胁迫下,ZmIBH1-1-OE植株的RWC显著高于WT。RNA-Seq分析发现多个与钙信号相关的基因在WT和ZmIBH1-1-OE株系中存在差异表达,其中,ZmIBH1-1直接作用于ZmCa-M和ZmAGD12的启动子调控其表达,间接调控钙依赖性蛋白激酶Zm00001d014773以及钙调素蛋白Zm00001d040323和Zm00001d028948。说明ZmIBH1-1可能通过调控钙信号相关基因的表达使气孔关闭减少水分蒸腾,从而提高玉米ZmIBH1-1- OE植株的抗旱性。
已有的研究表明,植物中bHLH、NAC、WRKY、bZIP、NF-Y、MYB等转录因子家族的基因通过不同的调控路径,在植物抵御逆境胁迫反应中发挥着重要的作用[4,5,6,7,8,9,10]。MAO等[4]克隆了基因ZmNAC111,该基因过表达明显提高了玉米苗期的抗旱性和水分利用效率,并诱导应答抗旱基因在干旱胁迫下表达上调。REN等[44]对NAC型转录因子ZmNST3展开了研究,敲除ZmNST3导致玉米的耐旱耐盐能力降低。MYB蛋白是植物抵御各种逆境的重要转录因子。如AtMYB60、AtMYB12、ATMYB75、OsMYB3R-2、CmMYB2、TaMYB22等都能通过不同的方式和路径提高植物的抗旱性[45,46,47,48,49]。干旱和盐胁迫可以诱导ZmMYB3R的表达,拟南芥过表达该基因,CAT、POD和SOD活性升高,增强了转基因拟南芥对干旱和盐胁迫的耐受性[10]。植物特有转录因子WRKY在植物非生物胁迫响应过程中发挥重要作用[50]。在拟南芥中过表达ZmWRKY106、ZmWRKY40,通过提高SOD、POD和CAT活性降低ROS含量,进而提高转基因拟南芥的耐旱性和耐热性[6,7]。本研究中,转录组分析表明,在干旱胁迫下,WT与ZmIBH1-1-OE株系的差异表达基因中包括8个NAC、8个WRKY、11个MYB类型转录因子(电子附图1-B和电子附表1)。说明ZmIBH1-1可能通过间接调控NAC、MYB和WRKY转录因子参与干旱胁迫响应。
4 结论
过表达ZmIBH1-1可以增强玉米的耐旱性;ZmIBH1-1可以直接结合11个靶基因的启动子区域,调控11个靶基因的表达;ZmIBH1-1通过直接调控乙烯信号通路中的基因ZmERF-107和ZmEIN3的表达提高玉米的耐旱性;ZmIBH1-1通过直接调控钙信号相关基因ZmCa-M和ZmAGD12增强玉米的耐旱性;ZmIBH1-1可能通过间接调控NAC、WRKY、MYB等转录因子响应干旱胁迫。(责任编辑 李莉)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1080/07352680590910410URL [本文引用: 1]
DOI:10.1038/ncomms9326URL [本文引用: 5]
[本文引用: 4]
DOI:10.3390/ijms19103046URL [本文引用: 5]
DOI:10.3390/ijms19092580URL [本文引用: 5]
DOI:10.1007/s00425-011-1496-7URL [本文引用: 4]
[本文引用: 4]
[本文引用: 4]
DOI:10.1016/j.plaphy.2019.02.010URL [本文引用: 5]
DOI:10.1016/j.molp.2020.02.005URL [本文引用: 1]
DOI:10.1111/pbi.v18.5URL [本文引用: 1]
DOI:10.3390/genes10080610URL [本文引用: 1]
DOI:10.1093/jxb/erz131URL [本文引用: 1]
[本文引用: 1]
DOI:10.3390/ijms21010029URL [本文引用: 1]
DOI:10.3390/ijms20092278URL [本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00425-016-2489-3URL [本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.bbrc.2014.05.139URL [本文引用: 1]
DOI:10.1007/s10142-018-0608-xURL [本文引用: 1]
DOI:10.1111/nph.2014.201.issue-4URL [本文引用: 1]
[本文引用: 1]
DOI:10.1093/jxb/erz307URL [本文引用: 1]
DOI:10.1093/jxb/eraa052URL [本文引用: 4]
DOI:10.1006/meth.2001.1262URL [本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/nar/gkl031URL [本文引用: 1]
DOI:10.1007/s00344-006-0124-4URL [本文引用: 1]
[本文引用: 1]
DOI:10.1046/j.1365-313X.1995.8040595.xURL [本文引用: 1]
DOI:10.1093/jxb/ert296URL [本文引用: 1]
DOI:10.1016/S0092-8674(00)81425-7URL [本文引用: 1]
DOI:10.1093/mp/ssr042URL [本文引用: 1]
[本文引用: 2]
DOI:10.1038/nature08258URL [本文引用: 2]
[本文引用: 1]
DOI:10.1105/tpc.111.084988URL [本文引用: 1]
DOI:10.1016/j.jplph.2015.05.020URL [本文引用: 1]
DOI:10.1105/tpc.15.00144URL [本文引用: 1]
DOI:10.1111/pce.v43.9URL [本文引用: 1]
DOI:10.1007/s11103-011-9796-7URL [本文引用: 1]
DOI:10.1111/tpj.2014.77.issue-3URL [本文引用: 1]
DOI:10.1104/pp.106.094532URL [本文引用: 1]
DOI:10.1007/s12033-011-9451-1URL [本文引用: 1]
DOI:10.1007/s11033-012-1550-yURL [本文引用: 1]
[本文引用: 1]
