 ,, 尚鹏祥
,, 尚鹏祥 ,, 李景远, 丁新伦, 吴祖建, 张洁
,, 李景远, 丁新伦, 吴祖建, 张洁 ,福建农林大学植物保护学院植物病毒研究所/闽台作物有害生物生态防控国家重点实验室/福建省植物病毒学重点实验室,福州 350002
,福建农林大学植物保护学院植物病毒研究所/闽台作物有害生物生态防控国家重点实验室/福建省植物病毒学重点实验室,福州 350002Effects of Proteins Encoded by “C4 ORFs” of Cotton Leaf Curl Multan Virus on Viral Pathogenicity
ZHENG XinShi ,, SHANG PengXiang
,, SHANG PengXiang ,, LI JingYuan, DING XinLun, WU ZuJian, ZHANG Jie
,, LI JingYuan, DING XinLun, WU ZuJian, ZHANG Jie ,Institute of Plant Virology, College of Plant Protection, Fujian Agriculture and Forestry University/State Key Laboratory for Ecological Pest Control of Fujian and Taiwan Crops/Key Laboratory of Plant Virology of Fujian Province, Fuzhou 350002
,Institute of Plant Virology, College of Plant Protection, Fujian Agriculture and Forestry University/State Key Laboratory for Ecological Pest Control of Fujian and Taiwan Crops/Key Laboratory of Plant Virology of Fujian Province, Fuzhou 350002通讯作者:
责任编辑: 岳梅
收稿日期:2020-08-18接受日期:2020-09-28网络出版日期:2021-05-16
| 基金资助: |
Received:2020-08-18Accepted:2020-09-28Online:2021-05-16
作者简介 About authors
郑信诗,E-mail:
尚鹏祥,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (3457KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
郑信诗, 尚鹏祥, 李景远, 丁新伦, 吴祖建, 张洁. 木尔坦棉花曲叶病毒“C4 ORF”编码蛋白对病毒致病性的影响[J]. 中国农业科学, 2021, 54(10): 2095-2104 doi:10.3864/j.issn.0578-1752.2021.10.006
ZHENG XinShi, SHANG PengXiang, LI JingYuan, DING XinLun, WU ZuJian, ZHANG Jie.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】双生病毒科(Geminiviridae)病毒是一类植物单链环状DNA病毒,世界上的热带、亚热带等地区均有分布,为典型的双联体颗粒形态,大小约22 nm×38 nm[1,2]。双生病毒的寄主范围广阔,能够侵染多种单子叶及双子叶植物,由烟粉虱、叶蝉等昆虫介体以持久性方式进行传播,对世界各地的农作物造成重大经济损失[3,4,5]。菜豆金黄花叶病毒属(Begomovirus)是双生病毒科最大的属,从基因结构上分成了单组分begomovirus与双组分begomovirus[1]。木尔坦棉花曲叶病毒(cotton leaf curl Multan virus,CLCuMuV)是单组分begomovirus,基因组仅含DNA-A,伴随一个β卫星分子(betasatellite)[6,7]。自2006年在我国发现由CLCuMuV侵染引起的朱槿曲叶病以来[7],该病毒已经在我国江苏、福建、海南、广西、广东等省(自治区)多个地区发生,可侵染棉花、黄秋葵、扶桑及垂花悬铃花等多种寄主植物[8,9,10,11,12,13]。CLCuMuV可以随昆虫介体烟粉虱及带病苗木传播,对我国的棉花产区造成潜在威胁[7,12]。【前人研究进展】CLCuMuV基因组的DNA-A大约2.7—2.8 kb,含6个ORF,2个在病毒链方向,编码V1和V2蛋白,4个在互补链方向,编码C1、C2、C3和C4蛋白[6,7,8,9,10,11,12,13]。CLCuMuV伴随的β卫星分子,仅编码一个βC1蛋白,β卫星依赖DNA-A组分进行复制[11-12,14-16]。双生病毒C4编码11.3 kD蛋白,是一个症状决定因子,可编码复制或转录调控因子,影响植物正常细胞发育,引起叶脉肿大等症状,也具有沉默抑制子活性[17,18,19,20]。2018年,ISMAYIL等发现CLCuMuV C4通过抑制S-腺苷甲硫氨酸合成酶(SAMS)活性来抑制转录水平基因沉默(TGS)和转录后水平基因沉默(PTGS),从而增强植物中CLCuMuV的侵染[21]。【本研究切入点】通过对NCBI登录的多个CLCuMuV分离物的序列进行分析,发现不同CLCuMuV C4开放阅读框附近可编码不同大小的ORF,而NCBI数据库登录的C4蛋白大小也不尽相同,甚至利用NCBI ORF Finder在线分析软件在部分CLCuMuV分离物中找不到目前公认的最小ORF,鉴于此,本研究对CLCuMuV的多个假定的“C4 ORF”编码的蛋白进行分析。【拟解决的关键问题】分析C4附近不同ORF对病毒致病性的影响,为C4开放阅读框的确切定位提供依据。1 材料与方法
试验于2018—2019年在福建农林大学植物病毒研究所完成。1.1 材料及主要试剂
CLCuMuV感病植株总DNA(CLCuMuV DNA-A的序列登录号为JX861210,DNA-β序列登录号为JX861211)、PVX载体、pDONR221载体、pEarleyGate- 101载体、pBINPLUS载体、农杆菌(Agrobacterium)GV3101感受态、大肠杆菌(Escherichia coli)Trans1-T1 Phage Resistant感受态、YFP-C4-S载体、DNA-β侵染性克隆重组质粒农杆菌、CLCuMuV C4蛋白多克隆抗体、本氏烟(Nicotiana benthamiana)种子均由本实验室保存。同源重组酶试剂盒(ClonExpressTM Ultra One Step Cloning Kit)和高保真酶试剂盒(PhantaTM Super- Fidelity DNA Polymerase)均购自南京诺唯赞生物科技有限公司,Gateway LR/BP Clonase II Enzyme Mix试剂盒购自Invitrogen公司。
本研究所用到的引物序列见表1。
Table 1
表1
表1本研究中所用引物序列
Table 1
| 引物用途及名称 Application and name of primer | 引物序列a Primer sequence (5′- 3′) | 引物在病毒基因组中的位置b Position of primers in viral genome |
|---|---|---|
| PVX异源表达For PVX heterologous expression | ||
| C4-L-F | cagctagcatcgattcccgggATGATTTCTCAAATTACGATAATGC | 2694-2670 |
| C4-M-F | cagctagcatcgattcccgggATGCCATTTGGAGACACTAT | 2673-2654 |
| C4-S-F | cagctagcatcgattcccgggATGGGAGCCCTCATCTCCAT | 2430-2411 |
| C4-R | cttatcggcggtcgacccgggCTAGTTCCTTAATGACTCTAAGAGC | 2128-2152 |
| PVX-F | CCCTATCAAGCTTATCGGCGGTC | -- |
| PVX-R | CATTGCCGATCTCAAGCCACTCTC | -- |
| 亚细胞定位For subcellular localization | ||
| C4-L-221-F | ggggacaagtttgtacaaaaaagcaggcttcATGATTTCTCAAATTACGATAATG | 2694-2671 |
| C4-221-R | ggggaccactttgtacaagaaagctgggtcGTTCCTTAATGACTCTAAGAGCC | 2131-2153 |
| M13-F | TGTAAAACGACGGCCAGT | -- |
| M13-R | CAGGAAACAGCTATGACC | -- |
| Gateway-F | GGGCTGGCAAGCCACGTTTGGTG | -- |
| Gateway-R | CCGGGAGCTGCATGTGTCAGAGG | -- |
| 侵染性克隆For infectious clones | ||
| CLCuMuV-L-F | acgccaagctggcgcgccaagcttGATCCATTGTTAAACGAATTTCCTGATACG | 123-152 |
| CLCuMuV-L-R | TTTAACAATGGATCCCACATgtttgaatttgaaacttagtgcgca | 136-117 |
| CLCuMuV-R-F | ATGTGGGATCCATTGTTAAACGAATTTCC | 117-145 |
| CLCuMuV-R-R | agaattcgagctcggtacccgggcacatGTTTGAATTTGAAACTTAGTGCGCA | 116-92 |
| pBIN-F | GTATGTTGTGTGGAATTGTGAGCGGATAAC | -- |
| pBIN-R | CACTGGCCGTCGTTTTACAACGTCGTGAC | -- |
| C4L-i2o-R-pBIN-F | TCACATTTCAAAATCCTCACGCTCCAAAAAG | 2692-2719 |
| C4L-i2o-L-pBIN-R | GGAGCGTGAGGATTTTGAAATGTGATCTCAAATTACGATAATGCCATTTG | 2713-2692, 2688-2664 |
| C4S-G2O-R-pBIN-F | ACATCTTCGTGTAACTCTCTGCAGAC | 2430-2452 |
| C4S-G2O-L-pBIN-R | TCTGCAGAGAGTTACACGAAGATGTGAGCCCTCATCTCCATGTGCTCATC | 2451-2428, 2424-2402 |
| CLCuMuV-RBamH1-pBIN-F | TTCAAATTCAAACATGTGGGATCCATTGTTAAACGAATTTCCTG | 104-147 |
| CLCuMuV-CPprobe-F | GCAGCCCATACACCAGCCGTGTTGC | 347-371 |
| CLCuMuV-CPprobe-R | GTTCCTTACTCGCATATTGACCACC | 862-838 |
| CLCuMuV-BETAprobe-F | GTCCCACTGCTGGTATTGACTTGATTTG | 37-64 |
| CLCuMuV-BETAprobe-R | CCATGAAATAAGCAGATATGACGAGGAGC | 567-539 |
新窗口打开|下载CSV
1.2 载体构建
1.2.1 PVX载体构建 以实验室保存CLCuMuV感病植株总DNA为模板,根据NCBI登录的CLCuMuV DNA-A序列设计CLCuMuV C4 L、M、S的ORF特异性扩增引物(表1)。PCR反应条件为95℃预变性5 min,95℃变性30 s,50℃退火35 s,72℃延伸30 s,30个循环后72℃继续延伸5 min。将反应产物在1%琼脂糖凝胶中电泳,目的条带经由DNA凝胶纯化试剂盒回收纯化。通过同源重组的方法,将回收纯化的片段与线性化PVX载体(Sma I 37℃ 酶切70 min)进行连接。热激法转化大肠杆菌后利用引物PVX-F/PVX-R(表1)筛选阳性克隆,即获得最终植物表达载体PVX-C4-L、PVX-C4-M、PVX-C4-S。测序结果正确的提取质粒电击法转化农杆菌GV3101感受态细胞,PCR检测阳性克隆。1.2.2 亚细胞定位载体构建 利用设计的特异性引物(表1)对所需片段进行PCR扩增并回收。利用Gateway系统BP反应构建中间载体,将回收的片段与pDONR221载体重组,转化大肠杆菌。利用引物M13-F/M13-R(表1)筛选阳性克隆并进行测序验证,然后提取质粒,Mlu I酶切中间载体并回收大片段,通过LR反应将其构建至终载体pEarleyGate-101并转化大肠杆菌,利用引物Gateway-F/Gateway-R(表1)鉴定阳性克隆,即获得最终植物表达载体YFP-C4-L。将终载体用电击法转化农杆菌GV3101,PCR检测阳性克隆。
1.2.3 侵染性克隆载体构建 根据CLCuMuV基因组的序列,设计引物CLCuMuV-R-F/R与CLCuMuV- L-F/R(表1)分别扩增其全长基因组和缺失基因组,延伸时间为170 s,扩增得到均为2 700 bp左右条带,PCR产物进行回收纯化;将pBINPLUS空载用限制性内切酶Hind III和Sma I线性化,通过同源重组,将回收纯化的目的条带与回收纯化的线性化pBINPLUS克隆载体连接,获得重组质粒;将重组质粒转到大肠杆菌中,利用引物pBIN-F/pBIN-R(表1)进行阳性克隆检测,测序结果正确的克隆进行质粒提取,即为侵染性克隆重组质粒CLCuMuV。将终载体用电击法转化农杆菌GV3101,PCR检测阳性克隆。
1.2.4 侵染性克隆突变载体构建 获得的重组质粒CLCuMuV为模板,用限制性内切酶BamH I和Sma I将未缺失基因组切下,PCR产物回收纯化后作为模板,利用引物C4L-i2o-R-pBIN-F/CLCuMuV-R-R、C4L-i2o-R- pBIN-R/CLCuMuV-R-F、C4S-G2O-R-pBIN-F/CLCuMuV- R-R、C4S-G2O-L-pBIN-R/CLCuMuV-R-F(表1)分别扩增片段并回收,回收片段命名为C4-L-1、C4-L-2、C4-S-1、C4-S-2;利用引物CLCuMuV-R-R/CLCuMuV- R-F(表1)将C4-L-1和C4-L-2、C4-S-1和C4-S-2分别进行同源重组,构建C4-ΔL和C4-ΔS的突变片段,将PCR产物回收纯化;用限制性内切酶BamH I和Sma I对CLCuMuV侵染性克隆载体进行酶切,将其回收纯化;通过同源重组,使用两组引物CLCuMuV- RBamH1-pBIN-F/C4L-i2o-R-pBIN-R、CLCuMuV- RBamH1-pBIN-F/C4S-G2O-L-pBIN-R分别将C4-ΔL和C4-ΔS与线性化的CLCuMuV侵染性克隆载体连接;重组质粒转到大肠杆菌中,利用引物pBIN-F/ pBIN-R(表1)进行阳性克隆检测,测序结果正确的克隆进行质粒提取,即为突变体侵染性克隆重组质粒CLCuMuV-ΔL、CLCuMuV-ΔS。其中CLCuMuV-ΔL和CLCuMuV-ΔS分别将C4-L和C4-S ORF第2位氨基酸突变为终止密码子。将终载体用电击法转化农杆菌GV3101,PCR检测阳性克隆。
1.3 农杆菌介导的植物接种
将新鲜培养的阳性克隆农杆菌菌液离心去除上清后,加入新鲜配置的烟草浸润缓冲液(10 mmol·L-1 MgCl2、10 mmol·L-1 MES pH 5.7、200 μmol·L-1 AS),调整其OD600≈1.0,室温下静置3—4 h后接种长至5—6片叶的本氏烟。接种后的植株培养于26℃、14 h光照/10 h黑暗的光照培养箱。侵染性克隆接种试验中需将重组质粒农杆菌分别与DNA-β重组质粒农杆菌等比例混合接种本氏烟。1.4 激光共聚焦显微镜观察
浸润48 h后,剪取浸润区叶片让其叶背朝上放置在载玻片上,滴上清水后盖上盖玻片,要注意避免产生气泡,在激光共聚焦显微镜(Leica TCS SP5 Ⅱ)下观察并拍照。1.5 蛋白检测
取0.3 g接种后的新叶叶片组织,液氮研磨后加900 μL蛋白抽提液(50 mmol·L-1 Tris-HCl pH 8.0、150 mmol·L-1 NaCl、0.2% Tween-20、2% β-巯基乙醇、蛋白酶抑制剂1片/10 mL),离心后吸上清液至新的2 mL EP管,-20℃保存。取10 μL总蛋白与2.5 μL蛋白Loading混匀,100℃水浴锅内煮15—20 min,12 000×g离心5 min后置于12% SDS-PAEG电泳,同时电泳两块胶。将一块凝胶泡在考马斯亮蓝染色液中置于水平摇床染色1—2 h,染色结束后加水煮胶进行脱色。将另外一块凝胶通过电转法转印到PVDF膜上。Western blot检测方法参考文献[22]进行。其中一抗使用1:1 000稀释的CLCuMuV C4蛋白多克隆抗体,二抗使用1:5 000稀释的HRP标记的羊抗鼠IgG。最后将PVDF膜置于检测反应液中避光孵育1 min,于自动化学发光凝胶成像分析仪(Amershm Imager600)中曝光10 s显色。1.6 DNA检测
植物总DNA的提取采用CTAB法[23]。Southern blot检测采用Roche DIG High Prime Lab/Detection Kit,详细步骤参见其说明书。2 结果
2.1 CLCuMuV“C4 ORF”编码蛋白的致病性
2.1.1 PVX表达载体的构建 通过序列分析,对CLCuMuV C4开放阅读框附近的3个ORF(图1-A)分别设计了引物,其中片段之间重复序列完全一致,也即起始密码子均位于同一个大的开放阅读框,并且共用同一个终止密码子。PCR结果显示分别扩增到C4-L(567 bp)、C4-M(546 bp)和C4-S(303 bp)的片段(图1-B)。利用同源重组将该3个目的片段与PVX线性化处理后的载体连接,获得重组载体PVX-C4-L、PVX-C4-M与PVX-C4-S。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1不同“C4 ORF”在病毒基因组上的可能分布及其PCR扩增
A:不同大小“C4 ORF”在CLCuMuV基因组上的位置示意图Position schematic diagram of “C4 ORFs” on the CLCuMuV genome。B:PCR扩增产物The amplified production of PCR。 M:DNA分子量标准 Trans2K Plus DNA Marker;1:C4-L的扩增Amplification of C4-L;2:C4-M的扩增Amplification of C4-M;3:C4-S的扩增Amplification of C4-S
Fig. 1Possible distribution and PCR amplification of different “C4 ORFs” in viral genome
2.1.2 PVX介导的3个蛋白在本氏烟表达对本氏烟症状影响比较 重组载体转化农杆菌后分别接种本氏烟,结果发现接种17 d后PVX-C4-L、PVX-C4-M与PVX-C4-S接种的植株略高于PVX空载接种对照,叶片卷曲,并且叶柄有伸长的现象,其症状严重程度由强至弱依次为PVX-C4-S、PVX-C4-L、PVX-C4-M,接种25 d后PVX-C4-S叶片卷曲症状更加明显,值得注意的是PVX-C4-S叶片多为上卷,而PVX-C4-L和PVX-C4-M叶片下卷(图2-A),测量叶柄长后发现,PVX-C4-S、PVX-C4-L接种组的长度明显长于PVX- C4-M及对照组(P<0.01)(图2-B)。结果表明C4-S ORF编码的蛋白致病性最强,C4-L ORF次之,而C4-M ORF编码蛋白的致病性非常弱。
图2
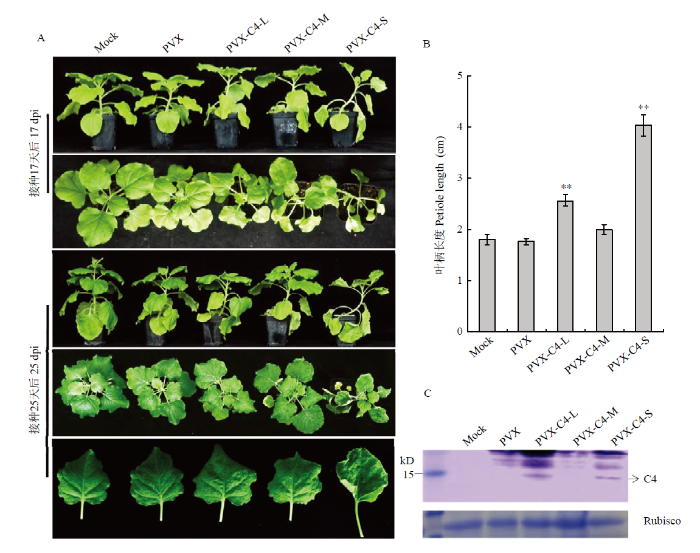 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2PVX载体介导的CLCuMuV 3个“C4 ORF”接种本氏烟植株
A:本氏烟症状Symptoms of N. benthamiana plants。B:接种25 d后本氏烟叶柄长度Petiole length of N. benthamiana plants 25 days post inoculation (dpi);数据为平均值±标准误,柱上**表示在P<0.01水平差异显著All values are shown as the mean±SE, ** on the column indicates significant difference at P<0.01 level。C:Western blot检测接种25 d后本氏烟中的C4蛋白Western blot analysis of the C4 protein accumulation in N. benthamiana plants at 25 dpi;Rubisco:核酮糖-1,5-二磷酸羧化酶 Ribulose-1,5-bisphosphate carboxylase
Fig. 2N. benthamiana plants inoculated by three “C4 ORFs” of CLCuMuV using the PVX vector
2.1.3 PVX介导C4蛋白在本氏烟植株中表达的检测 为验证C4蛋白在PVX异源载体中的表达,提取接种25 d后烟草新叶总蛋白进行Western blot检测,结果发现PVX-C4-L与PVX-C4-S接种的烟草叶片中均检测到C4蛋白的表达,遗憾的是虽然经过多次重复,PVX-C4-M没有检测到特异蛋白的积累(图2-C)。
2.2 CLCuMuV“C4 ORF”编码蛋白的亚细胞定位
为分析C4-L和C4-S的亚细胞定位情况,将重组表达载体YFP-C4-L和YFP-C4-S分别利用农杆菌介导的植物接种法浸润本氏烟叶片,48 h后,剪取本氏烟叶片表皮细胞于激光共聚焦显微镜下观察。结果发现YFP-C4-L主要定位在叶绿体,YFP-C4-S则是定位在细胞膜或细胞质周边,并且在细胞膜上可以观察到点状聚集体结构(图3)。图3
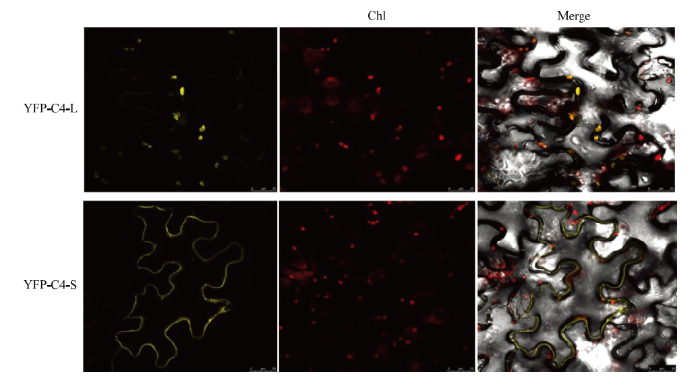 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3CLCuMuV C4在本氏烟叶片表皮细胞中的亚细胞定位
Chl:叶绿体Chloroplast;Merge:合并
Fig. 3Subcellular localization of CLCuMuV C4 in the epidermal cells of N. benthamiana leaves
2.3 CLCuMuV及其突变体侵染性克隆接种
2.3.1 CLCuMuV及其突变体侵染性克隆接种本氏烟 为分析不同“C4 ORF”对病毒诱导寄主症状的影响,将CLCuMuV、CLCuMuV-ΔL、CLCuMuV-ΔS分别与DNA-β侵染性克隆等比例混合接种本氏烟,以未转化质粒的农杆菌接种本氏烟为阴性对照(Mock)。结果发现接种7 d后CLCuMuV、CLCuMuV-ΔL接种的植株新叶即有向下卷曲的现象(图片未示出);接种14 d 和21 d后,叶片出现明显的皱缩并且增生、叶柄和茎秆扭曲,植株的高度明显矮于Mock和CLCuMuV-ΔS,CLCuMuV-ΔS与Mock相同,没有出现症状(图4-A)。结果说明病毒对寄主症状的诱导过程中,C4-L不是必需,C4-S起到重要作用。图4
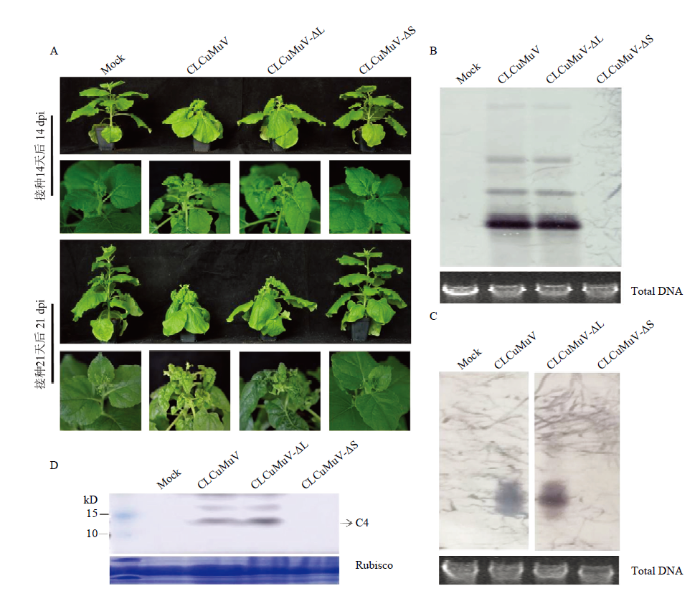 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4CLCuMuV及其突变体侵染性克隆接种本氏烟植株
A:本氏烟症状Symptoms of N. benthamiana plants。B:接种21 d后本氏烟中DNA-A的Southern blot检测Southern blot analysis of the DNA-A accumulation in N. benthamiana plants at 21 dpi。C:接种21 d后本氏烟中DNA-β的Southern blot检测Southern blot analysis of the DNA-β accumulation in N. benthamiana plants at 21 dpi;Total DNA:总DNA。D:接种14 d后本氏烟中C4蛋白的Western blot检测Western blot analysis of the C4 protein accumulation in N. benthamiana plants at 14 dpi;Rubisco:核酮糖-1,5-二磷酸羧化酶 Ribulose-1,5-bisphosphate carboxylase
Fig. 4N. benthamiana plants inoculated by infectious clones of CLCuMuV and the mutants
2.3.2 接种本氏烟植株的病毒DNA检测 为检测病毒基因组在接种烟草叶片中的复制积累情况,利用CLCuMuV的CP基因设计探针,对接种21 d后提取的叶片总DNA进行Southern blot检测,结果发现CLCuMuV、CLCuMuV-ΔL接种的烟草叶片中均能检测到病毒DNA-A的积累,而CLCuMuV-ΔS以及Mock没有检测到病毒DNA-A的积累(图4-B)。同时,利用DNA-β组分设计探针,对接种21 d后提取的叶片总DNA进行Southern blot检测,结果发现CLCuMuV、CLCuMuV-ΔL接种的烟草叶片中均能检测到病毒DNA-β的积累,而CLCuMuV-ΔS以及Mock均未检测到病毒DNA-β的积累(图4-C)。结果说明病毒对寄主症状的诱导与病毒基因组的积累成正比。
2.3.3 接种本氏烟植株的C4蛋白检测 为检测病毒C4蛋白在接种烟草叶片中的表达情况,提取接种14 d叶片总蛋白,以C4蛋白多克隆抗体为一抗进行Western blot检测。结果发现CLCuMuV和CLCuMuV-ΔL接种的烟草叶片中检测到了C4蛋白,且蛋白积累量基本一致,而CLCuMuV-ΔS以及Mock没有检测到C4蛋白的积累。值得注意的是,CLCuMuV接种的烟草蛋白检测条带跟CLCuMuV-ΔL十分一致,即C4蛋白抗体未能够在CLCuMuV接种的烟草中检测到C4-L蛋白的表达(图4-D)。结果说明病毒在侵染寄主过程中或许并不编码C4-L蛋白。
3 讨论
利用MEGA软件对国内外50多种CLCuMuV分离物的全基因组进行系统发育关系分析,结果发现印度、巴基斯坦、泰国、菲律宾等地的分离物亲缘关系较近,聚类为一个大分支,而所有来自中国的分离物聚类为一个大分支。值得注意的是,通过编码ORF序列分析,发现来自中国的CLCuMuV C4 ORF均为567 nt,国外绝大部分为303 nt以及个别546 nt,而NCBI登录的C4蛋白的大小也不尽相同。本研究利用PVX异源表达载体发现PVX-C4-L和PVX-C4-S具有较强致病性,PVX-C4-M的致病性可忽略不计。更进一步利用侵染性克隆接种试验发现C4-S ORF对病毒的侵染不可或缺。然而Southern blot在CLCuMuV-ΔS突变体上检测不到病毒DNA-A和DNA-β组分,推测可能的原因之一是C4-S ORF的缺失使病毒基因组DNA-A的复制受到了影响,而β卫星需要依赖DNA-A组分进行复制[12,14-16,24-25],所以β卫星的复制也受到了影响;再者也可能是C4-S ORF的缺失影响病毒的运动,导致病毒无法正常运动而不能进行系统侵染,这与已报道的C4蛋白参与病毒的运动[26,27,28]一致。亚细胞定位被认为与蛋白质所发挥的功能有关,亚细胞定位的信息将有助于推断蛋白质功能,进而研究其作用机制。番茄黄化曲叶病毒(tomato yellow leaf curl virus,TYLCV)C4定位在细胞周边并与胞间连丝互作[28],在病毒运动中介导细胞间的转运。同样,云南番茄曲叶病毒(tomato leaf curl Yunnan virus,TLCYnV)C4蛋白也被证实定位在细胞膜,通过增强CyclinD1.1的稳定性,抑制NbSKη介导的磷酸化而诱导细胞分裂[29]。甘薯卷叶病毒(sweet potato leaf curl virus,SPLCV)的C4蛋白主要与质膜中的AtBIN2相互作用,激活AtBES1/AtBZR1成分,从而使它们可以进入细胞核并激活BR反应基因的转录[30]。这些说明C4蛋白的膜定位与其功能的发挥存在密切的联系。本研究结合之前的研究[31]发现YFP-C4-S蛋白主要定位在细胞膜上或细胞质周边,并且在细胞膜上可以观察到点状聚集体结构,推测其与胞间连丝相关,与前人的研究结果一致。而YFP-C4-L主要定位在叶绿体,结合病毒侵染寄主后C4蛋白的Western blot检测结果,或许暗示了其不作为CLCuMuV致病ORF而存在。
4 结论
CLCuMuV的3个假定的“C4 ORF”编码蛋白中,C4-S对CLCuMuV的侵染必不可少,而C4-L和C4-M对CLCuMuV的侵染不是必需。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 2]
DOI:10.1007/s00705-002-0957-5URL [本文引用: 1]
DOI:10.1126/science.286.5446.1835URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1046/j.1364-3703.2003.00188.xURL [本文引用: 2]
[本文引用: 4]
[本文引用: 4]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 3]
[本文引用: 3]
[本文引用: 5]
[本文引用: 5]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
DOI:10.1016/j.virusres.2007.04.002URL
DOI:10.1007/s00705-003-0149-yURL [本文引用: 2]
DOI:10.1046/j.1365-313X.1997.11061273.xURL [本文引用: 1]
DOI:10.1094/MPMI-04-12-0094-RURL [本文引用: 1]
DOI:10.1371/journal.ppat.1008829URL [本文引用: 1]
DOI:10.1006/viro.1994.1606URL [本文引用: 1]
DOI:10.1371/journal.ppat.1007282URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/BF02914055URL [本文引用: 1]
DOI:10.3390/v9100280URL [本文引用: 1]
DOI:10.1007/s11262-019-01716-5URL [本文引用: 1]
DOI:10.1016/j.molp.2018.12.006URL [本文引用: 1]
DOI:10.1099/vir.0.83049-0URL [本文引用: 1]
DOI:10.1006/viro.2001.1194URL [本文引用: 2]
DOI:10.1371/journal.ppat.1006789URL [本文引用: 1]
DOI:10.3389/fpls.2017.01689URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
