 ,1, 陈小飞
,1, 陈小飞 ,2, 阚蕊慈1, 李玉1, 曹慧1, 彭艳伶3, 张斌
,2, 阚蕊慈1, 李玉1, 曹慧1, 彭艳伶3, 张斌 ,1,4
,1,4Molecular Epidemiological Investigation of Porcine Group A Rotavirus in Sichuan from 2017 to 2019
ZHOU Qun ,1, CHEN XiaoFei
,1, CHEN XiaoFei ,2, KAN RuiCi1, LI Yu1, CAO Hui1, PENG YanLing3, ZHANG Bin
,2, KAN RuiCi1, LI Yu1, CAO Hui1, PENG YanLing3, ZHANG Bin ,1,4
,1,4通讯作者:
责任编辑: 林鉴非
收稿日期:2020-04-13接受日期:2020-07-29网络出版日期:2021-03-01
| 基金资助: |
Received:2020-04-13Accepted:2020-07-29Online:2021-03-01
作者简介 About authors
周群,E-mail:
陈小飞,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (3472KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
周群, 陈小飞, 阚蕊慈, 李玉, 曹慧, 彭艳伶, 张斌. 2017-2019年四川地区猪A群轮状病毒的分子流行病学调查[J]. 中国农业科学, 2021, 54(5): 1063-1072 doi:10.3864/j.issn.0578-1752.2021.05.017
ZHOU Qun, CHEN XiaoFei, KAN RuiCi, LI Yu, CAO Hui, PENG YanLing, ZHANG Bin.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】A群轮状病毒(rotavirus A, RVA)为呼肠孤病毒科轮状病毒属成员,是引起婴幼儿及其他多种幼龄动物病毒性腹泻的主要病原体之一[1]。RVA基因组分为11个基因节段,分别编码6种结构蛋白(VP1—VP4,VP6和VP7)和5或6种非结构蛋白(NSP1—NSP5/6),RVA的11个基因之间均易发生重配事件[2]。根据VP6蛋白的抗原性可将RV分类为10个血清群(A—J)[3,4],A—C、E和H群RV已经被证实可感染猪,其中感染人类和动物最常见的为A群轮状病毒[5,6]。RVA感染仔猪呈世界流行,各国猪场中RVA的阳性率为3.3%—67.3%,对世界养猪业产生了严重的经济损失[7]。研究表明猪RVA是人源RVA感染的潜在来源[8,9,10,11],因此了解猪RVA的流行情况和分子特征对RVA的防控以及防止猪RVA传播给人类至关重要。【前人研究进展】VP4和VP7蛋白共同组成RVA的外衣壳,二者都是重要的中和抗原,可以诱导机体产生中和抗体[12]。结构蛋白VP4和VP7分别决定RVA的P型和G型,且G型和P型之间会产生多种组合,不同组合的血清型之间交叉保护性低[2]。在猪RVA中,已经鉴定出了12个G型(G1—G6,G8—G12和G26)和16个P型(P[1]—P[8],P[13],P[19],P[23],P[26],P[27],P[28],P[32],P[34]),其中G3、G4、G5、G9和G11被认为是最常见的G型,通常与P[5]、P[6]、P[7]、P[13]和P[28]组合在一起[13,14,15]。国内的病原学检测结果显示RVA在我国规模化猪场中普遍存在,仔猪腹泻粪便中RVA的阳性率为7.69%—28.76%[16,17,18,19]。近年来中国大陆陆续报道了G11P[13]、G9P[23]和G9P[7]等组合基因型的RVA[20,21,22],然而四川地区RVA的流行情况及遗传多样性资料相对匮乏。【本研究切入点】2017—2019年分别从四川省14个地区40个猪场采集腹泻样本303份,对其进行RVA检测及基因型的鉴定,以期了解四川地区猪群中RVA的感染情况和流行趋势。【拟解决的关键问题】为研究四川地区猪RVA的遗传进化特征提供基础研究资料,预测四川地区猪RVA的流行趋势,从而为猪RVA疫苗的研制提供理论基础。1 材料与方法
试验于2018年3月至2019年12月在西南民族大学生命科学与技术学院动物医学实验室进行。1.1 材料
1.1.1 病料采集 对四川省不同地区、不同时间和不同猪群的腹泻样本进行主动采集,2017—2019年采集了四川省绵阳、泸州、眉山和西昌等14个地区40个猪场共303份仔猪腹泻样本,发病猪均表现为产房哺乳仔猪腹泻、消瘦,腹泻病死率为35%—85%。样本置于-80℃冰箱保存备用。1.1.2 主要试剂与仪器 Trizol试剂(RNAiso Plus)、PrimeScriptTMRT试剂盒、pMD19-T Vector、MarkerⅡ和TB Green Premix ExTaqⅡ购自宝生物工程(大连)有限公司;Quick Taq HS DyeMix购自东洋纺(上海)生物科技有限公司;PCR产物纯化试剂盒和胶回收试剂盒购自OMEGA公司;E. coli DH5α购自北京天根生化科技有限公司;LB培养基购自青岛海博试剂有限公司;高速离心机5804购自Eppendor公司;普通PCR仪、核酸蛋白电泳仪、凝胶成像系统VersaDoc2000均购自Bio-Rad公司。
1.1.3 引物 用西南民族大学生命科学与技术学院动物医学实验室基于RVA VP6基因建立的荧光定量PCR方法检测303份腹泻样本中的RVA;RVA分型引物信息见文献[23—24];用于RVA阳性样本G型和P型全基因扩增的引物信息见文献[23—25]。以上所有引物均由上海生工生物工程技术服务有限公司合成。
1.2 方法
1.2.1 病毒核酸的提取 将适量的粪便样本加入装有400 μL生理盐水的1.5 mL EP管中,根据生产商说明,使用RNAios Plus从400 μL粪便混悬液中提取病毒RNA。反转录按照PrimeScriptTMRT试剂盒说明书进行,反转录产物于-20℃保存。1.2.2 病料样本的检测分型及序列测定 以303份样本的cDNA为模板,使用TB Green Premix ExTaq Ⅱ和Quick Taq HS DyeMix进行RVA的检测和分型,具体方法参照说明书。将PCR扩增产物通过琼脂凝胶核酸电泳及凝胶成像仪进行检测与分析。PCR产物纯化后与pMD19-T载体连接,转化到E. coli DH5α感受态细胞中,筛选阳性重组质粒由上海生工生物有限公司双向测序。对测序结果进行序列拼接,并在NCBI中对拼接的基因组序列进行比对。
1.2.3 RVA VP7和VP4基因型鉴定及序列分析 将测序获得的RVA VP7和VP4基因的核苷酸序列使用RotaC2.0自动基因分析软件进行序列分析以确定相应毒株的基因型;应用Lasergene中MegAlign软件进行同源性分析;并通过MEGA 7.0软件用邻近法构建系统进化树,Bootstrap值选择1 000重复;应用SimPlot和RDP4软件对全基因组序列进行重组分析。
2 结果
2.1 腹泻样本RVA的检测和G/P型分型结果
303份仔猪腹泻的样本RVA检测结果显示:共检出RVA阳性样本98份(32.34%,95% confidence interval(CI)=27.1%—37.9%),40个猪场中有25个猪场检出RVA阳性,检出率为62.5%(95% CI=45.8%—77.3%)。对98份RVA阳性样本分别测定P基因型和G基因型,共得到47株RVA VP4部分基因、30株RVA VP7部分基因、12株RVA VP4全基因组和9株RVA VP7全基因组,GenBank登录号为MT198708—MT198805,将测序获得的RVA VP7和VP4基因的核苷酸序列使用RotaC2.0自动基因分析软件进行序列分析以确定相应毒株的基因型。结果显示:G型存在G9(16/39)、G5(11/39)、G4(9/39)、G3(1/39)和G26(2/39)五种型;P型存在P[13](24/59)、P[23](14/59)、P[6](18/59)和P[1](3/59)4种型。有30份样本成功测定出G/P组合基因型,共包括10种组合,分别为:G9P[23](7/30);G4P[6](5/30);G26P[13](2/30);G9P[13](4/30);G5P[23] (3/30);G4P[23](1/30);G3P[13](1/30);G5P[13](3/30);G4P[13](2/30);G9P[6](2/30),RVA的检测结果及详细分布见表1。Table 1
表1
表1四川地区猪场RVA感染率及基因型的分布
Table 1
| 地区 Region | 猪场个数 Farm | 样本数 Number of samples | RVA阳性数 Positive number | G型 G genotype | P 型 P genotype | G/P组合型 G/P combination genotype |
|---|---|---|---|---|---|---|
| 眉山Meishan | 6 | 62 | 14 | G3,G4,G9 | P[1],P[13],P[23] | G3P[13],G4P[23],G9P[23],G9P[13] |
| 绵阳Mianyang | 5 | 34 | 24 | G5,G9 | P[23] | G9P[23],G5P[23] |
| 泸州Luzhou | 5 | 13 | 3 | G9 | P[1],P[13] | G9P[13] |
| 西昌Xichang | 5 | 25 | 8 | P[1],P[13] | ||
| 成都Chengdu | 4 | 49 | 9 | G9 | P[13],P[23] | G9P[13],G9P[23] |
| 雅安Yaan | 4 | 13 | 3 | G9 | P[13] | G9P[13] |
| 内江Neijiang | 3 | 18 | 2 | G9 | P[6] | G9P[6] |
| 宜宾Yibin | 2 | 7 | 5 | G5 | P[13],P[23] | G5P[23],G5P[13] |
| 乐山Leshan | 1 | 4 | 2 | G4 | P[13] | G4P[13] |
| 遂宁Suining | 1 | 18 | 4 | G9,G26 | P[13],P[23] | G9P[23],G26P[13] |
| 南充Nanchong | 1 | 3 | 3 | G9 | P[13] | G9P[13] |
| 资阳Ziyang | 1 | 18 | 2 | G4 | P[6] | G4P[6] |
| 广元Guangyuan | 1 | 36 | 19 | G4,G5 | P[6],P[13] | G4P[6],G5P[13] |
| 攀枝花Panzhihua | 1 | 3 | 0 | |||
| 共计Total | 40 | 303 | 98 |
新窗口打开|下载CSV
2.2 RVA VP7 基因部分序列遗传进化分析
将本研究中获得的30株RVA VP7部分基因和9株全基因组序列与其他已知G基因型的代表毒株通过MEGA 7.0软件进行遗传进化分析,结果显示(图1):所获得的G3型毒株与四川猪源毒株SCQL-5-3(GenBank No. MG066590)亲缘关系最近;四川地区G4型RVA分为两个不同的分支,来自同一地区4个毒株和RVA/Pig/CHN/SCMS-69与日本猪源毒株GUB71(GenBank No. AB573647)聚为一个分支,另外4株G4型RVA毒株分别来自两个地区,在分支的上端与中国猪源和人源毒株聚为一支;在G5型RVA分支中,来自同一地区的7株毒株聚为一个分支,来自同一地区的4株G5型RVA分别与中国台湾猪源毒株和河北人源RVA毒株聚为一支;在G9型RVA分支中,RVA/Pig/CHN/SCSN-96和RVA/Pig/CHN/SCRS-142虽来自不同地区,但都与中国猪源的毒株HBHP(GenBank No. KX831114)同源性最高为98.3%,并与另外2个地方的5株毒株聚为一支,剩余来自5个地区的7株毒株与湖北猪源毒株(GenBank No. KX831100)同源性最高为95.9%—97.1%,来自内江的2株毒株单独聚为一支;本研究获得的两株来自同一地区的G26型RVA毒株与尼泊尔人源毒株亲缘关系最近。RVA VP7基因同源性分析见表2。图1
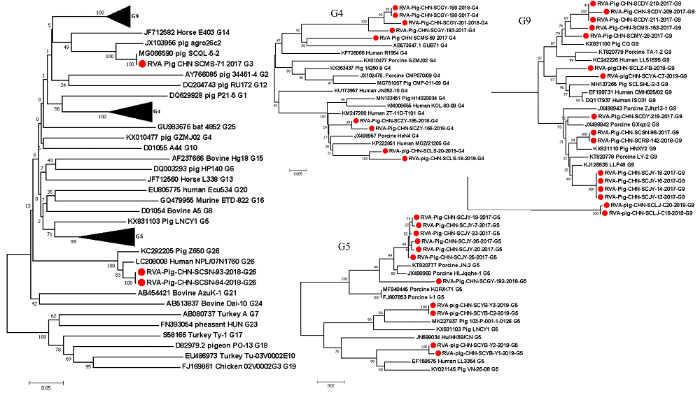 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1基于RVA VP7序列(790 bp)利用邻近法构建的遗传进化树
Fig. 1Phylogenetic tree based on RVA VP7 sequences (790 bp) constructed using the neighbor-joining (NJ) method
●:本研究中的毒株 Strains in this study。下同 The same as below ▲:右侧G9、G4和G5进化树的缩略图 Thumbnails of the right G9, G4 and G5 phylogenetic trees
Table 2
表2
表2RVA VP7 基因同源性分析
Table 2
| 毒株 Strain | 同源性最高的毒株 Strain with highest homology | 国家及宿主 Country and host | 年份 Year | 同源性 Homology |
|---|---|---|---|---|
| SCMS-71 | SCQL-5-2(MG066590) | 中国/猪源China/pig | 2018 | 99.9% |
| SCLS-18,SCLS-20 | MOZ/21205(KP222851) | 莫桑比克/人源Mozambique/human | 2011 | 96.9%,97.4% |
| SCZY-165,SCZY-166 | ZT-11D-T101(KM247280) | 中国/人源China/human | 2011 | 98.9%,98.3% |
| SCMS-69 | GUB71(AB573647) | 日本/猪源Japan/pig | 2006 | 95.6% |
| SCGY-193,SCGY-198,SCGY-199,SCGY-201 | R1954(KF726066) | 中国/人源China/human | 2013 | 95.1% |
| 6株G5型(SCJY),SCGY-192 6 strains of G5 (SCJY) | HLJqqhe-1(JX498960) | 中国/猪源China/pig | 2011 | 97.6%-98.1% |
| SCYB-Y1,SCYB-Y2 | LL3354(EF159575) | 中国/人源China/human | 2000 | 93.9%,94.2% |
| SCYB-C2,SCYB-C3 | 103-P-001-1-0126(MK227837) | 中国/猪源China/pig | 2014 | 93.7% |
| 4株G9型(SCJY) 4 strains of G9 (SCJY) | LLP48(KJ126835) | 中国/猪源China/pig | 2008 | 97.4%-97.6% |
| SCRS-142,SCSN-96 | HBHP(KX831114) | 中国/猪源China/pig | 2016 | 98.3% |
| SCDY-219 | GXqz-2(JX498942) | 中国/猪源China/pig | 2011 | 97.5% |
| SCLZ-FB,SCYA-C7 | ISO31(DQ117937) | 印度/人源India/human | 2008 | 96.2%,96.5% |
| SCMY-28,SCMS-153,SCDY-211, SCDY-210,SCDY-209 | CQ(KX831100) | 中国/猪源China/pig | 2016 | 95.9%-97.1% |
| SCSN-93,SCSN-94 | 07N1760(LC208008) | 尼泊尔/猪源Nepal/pig | 2007 | 96.1% |
| SCLJ-C18,SCLJ-C20 | TA-1-2(KT820779) | 中国/猪源China/pig | 2014 | 85.1% |
新窗口打开|下载CSV
2.3 RVA VP4 基因部分序列遗传进化分析
将本研究中获得的47株RVA VP4部分基因和12株全基因组序列与其他已知P基因型的代表毒株通过MEGA 7.0软件中邻近法构建系统进化树。结果显示(图2):3株P[1]型RVA毒株与人源、牛源和羊源的毒株聚为一个大支;15株来自同一猪场的P[6]型RVA毒株在进化树上端单独聚为一支,其余3株均与斯里兰卡人源毒株R1207(GenBank No. LC389888)的同源性最高为95.7%—98.6%;本研究中的P[13]型毒株RVA/Pig/CHN/SCYB/Y1单独分为一支,其余23株分散在4个亚支中,分支Ⅰ由来自8个地区的11株毒株与四川猪源毒株(GenBank No. MK410284)和人源毒株(GenBank No. MK597985)组成;分支Ⅱ进一步分为2个亚支,分别由RVA/Pig/CHN/SCLZ/FB与日本猪源毒株FGP36(GenBank No. AB573878)和本研究中其余2株毒株与中国猪源毒株LNCY(GenBank No. MF462324)组成;分支Ⅲ由本研究中分别来自3个地区的4株毒株与之前获得的1株四川地区的毒株和1株日本猪源毒株(GenBank No. AB573650)组成;亚支Ⅳ中5株毒株与泰国猪源毒株CMP178(GenBank No. DQ536362)亲缘关系最近。在P[23]型RVA毒株进化树中来自同一个猪场的7个毒株聚为一个小支,位于进化树的最上端,来自2个地区的3株毒株聚为一个小支。RVA/Pig/CHN/SCSN-96和来自同一个地区的2株毒株在进化树的最下端分别聚为一个小支。RVA VP4基因同源性分析见表3。图2
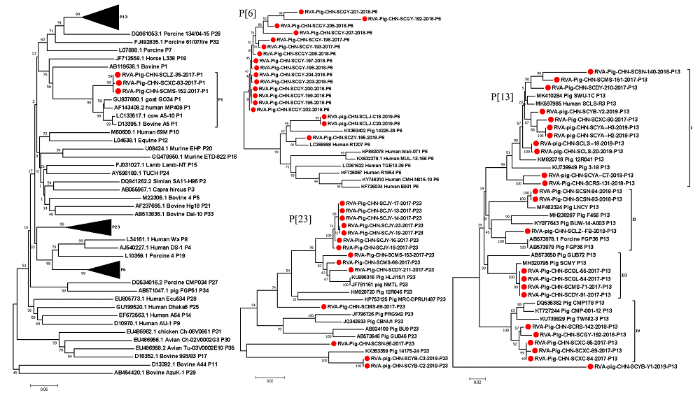 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2基于RVA VP4序列(800 bp)利用邻近法构建的遗传进化树
▲:右侧P[6],P[13]和 P[23]进化树的缩略图
Fig. 2Phylogenetic tree based on RVA VP4 sequences (800 bp) constructed using the neighbor-joining (NJ) method
▲:Thumbnails of the right P[6], P[13] and P[23] phylogenetic trees
Table 3
表3
表3RVA VP4 基因同源性分析
Table 3
| 毒株 Strain | 同源性最高的毒株 Strain with highest homology | 国家及宿主 Country and host | 年份 Year | 同源性 Homology |
|---|---|---|---|---|
| SCMS-152,SCLZ-35,SCXC-83 | A5-10(LC133517) | 泰国/牛源Thailand/cow | 1988 | 93.4% |
| 15株P[6]型(SCGY)15 strains of P[6] (SCGY) | GUB88(AB573872) | 日本/猪源Japan/pig | 2006 | 92.6%-98.6% |
| SCLJ-C18,SCLJ-C20,SCZY-165 | R1207(LC389888) | 斯里兰卡/人源Sri Lanka/human | 2009 | 95.7%-98.6% |
| SCXC-89,SCXC-85,SCXC-84,SCRS-142,SCGY-192 | CMP178(DQ536362) | 泰国/猪源Thailand/pig | 2008 | 94.5%-96.8% |
| SCYB-Y1,SCQL-54,SCQL-55,SCMS-71,SCDY-91 | SCMY(MH320795) | 中国/猪源China/pig | 2018 | 79.1%-98.8% |
| SCLZ-FB | FGP36(AB573878) | 日本/猪源Japan/pig | 2009 | 95.1% |
| SCSN-93,SCSN-94 | LNCY(MF462324) | 中国/猪源China/pig | 2016 | 98.3% |
| SCYA-C7,SCRS-131,SCLS-18,SCLS-20, SCYA-H2,SCYA-H3,SCXC-90,SCYB-Y2 | SWU-1C(MK410284) | 中国/猪源China/pig | 2018 | 92.5%-97.7% |
| SCDY-210,SCSN-140,SCMS-151 | SCLS-R3(MK597985) | 中国/人源China/human | 2018 | 92.6%-97.0% |
| SCYB-C2,SCYB-C3 | 14175-24(KX363359) | 越南/猪源Viet Nam/pig | 2012 | 95.3% |
| SCSN-96 | SC11(MH624176) | 中国/猪源China/pig | 2017 | 97.4% |
| SCMS-153,SCMS-68,SCMS-69,SCDY-211, 7株P[23]型(SCJY)7 strains of P[23] (SCJY) | HLJ/15/1(KU886316) | 中国/猪源China/pig | 2015 | 92.1-96.4% |
新窗口打开|下载CSV
2.4 RVA VP7基因和VP4基因重组分析
应用SimPlot和RDP4软件对12株RVA VP4全基因组和9株RVA VP7全基因组序列进行重组分析。结果显示:RVA/Pig/CHN/SCLJ/C18和RVA/Pig/CHN/ SCLJ/C20的VP7基因,RVA/Pig/CHN/SCYB/Y1和RVA/Pig/CHN/SCYB/Y2的VP4基因均发生了重组。RVA/Pig/CHN/SCLJ/C20和RVA/Pig/CHN/SCLJ/C18(图略)均为猪源G9型YN(GenBank No. KJ466985)与人源G4型KOL-80-09(GenBank No. KM008655)的重组毒株,预测重组值为0.692和0.703,预测重组断点位于253和707 bp处(图3);RVA/Pig/CHN/ SCYB/Y1为猪源P[23]型14150-53(GenBank No. KX363351)与鼠源P[13]型SCLS-M(GenBank No. MK606442)的重组毒株,预测重组值为0.692,预测重组断点位于860 bp(图3);RVA/Pig/CHN/SCYB/Y2为猪源P[23]型14175-24(GenBank No. KX363359)与猪源P[13]型SWU-1C(GenBank No. MK410284)的重组毒株,预测重组值为0.634,预测重组断点位于1 604 bp(图略)。本研究分别对每个毒株重组断点前后区域构建了进化树,结果显示毒株重组断点前后均表现出不一致的遗传进化关系,进一步证实了本研究中毒株的重组事件。图3
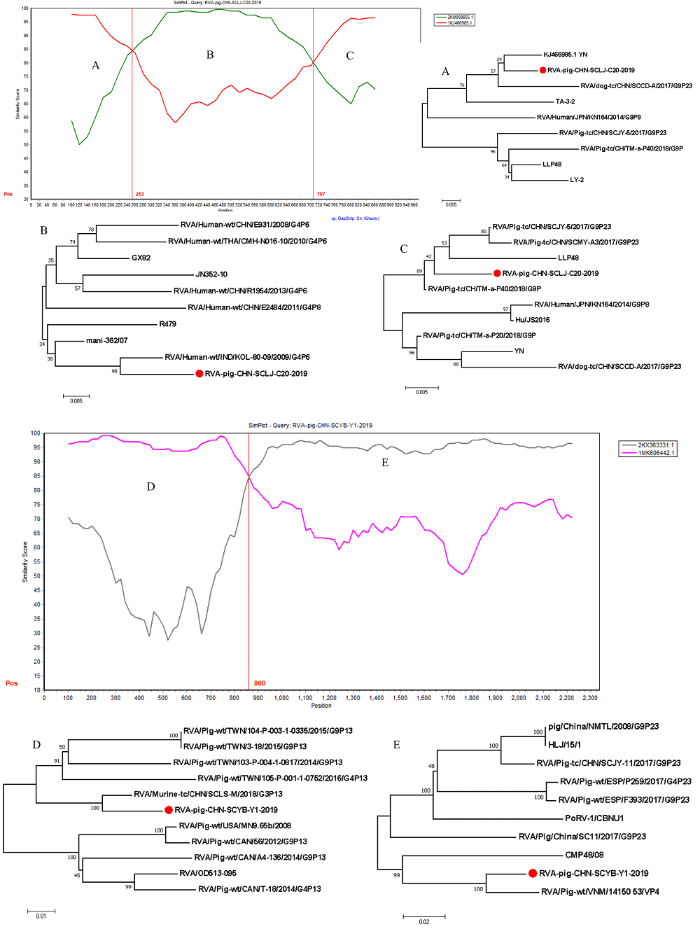 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3毒株RVA/Pig/CHN/SCLJ/C20/2019和RVA/Pig/CHN/SCYB/Y1/2019 VP4的重组序列相似性分析
纵轴表示查询毒株与其他参考毒株核苷酸序列的相似性(%),水平轴表示核苷酸位置。A、B、C为Simplot软件分析毒株RVA/Pig/CHN/ SCLJ/C20/2019 VP7基因重组区域及基于A、B、C重组区域构建的进化树;D、E为Simplot软件分析毒株RVA/Pig/CHN/SCYB/Y1/2019 VP4基因重组区域及基于D、E重组区域构建的进化树
Fig. 3Similarity analysis of recombinant sequence of RVA/Pig/CHN/SCLJ/C20/2019 strain and RVA/Pig/CHN/SCYB/Y1/2019 strain
The vertical axis indicates the similarity (%) of nucleotide sequences between the query strain and other reference strains, the horizontal axis indicates the nucleotide positions. The recombinant region A, B and C of VP7 gene of RVA/Pig/CHN/SCLJ/C20/2019 strain was analyzed by Simplot software, the phylogenetic tree based on recombination region A, B and C. The recombinant region D and E of VP4 gene of RVA/Pig/CHN/SCYB/Y1/2019 strain was analyzed by Simplot software, the phylogenetic tree based on recombination region D and E
3 讨论
RVA是引起婴幼儿以及多种幼龄动物急性肠胃炎的重要病原[26,27,28],对养殖业造成重大的经济损失。国内病原学检测结果显示RVA在我国规模化猪场中普遍存在,仔猪腹泻粪便中RVA的阳性率为7.69%—28.76%[16,17,18,19]。本研究采用荧光定量RT-PCR的方法对2017—2019年四川遂宁、宜宾、资阳、西昌和眉山等14个地区共40个规模化猪场的303份仔猪腹泻粪便样本进行了RVA的检测,结果显示猪场RVA阳性率为62.5%(25/40),RVA个体阳性率为32.34%(98/303),较2014—2016年四川地区RVA的阳性率(7.69%—26.7%)明显增高[16,17]。由于不同猪场的卫生条件和地理位置不同,不同猪场中RVA阳性率区别较大,在四川不同地区的猪场中RVA个体阳性率也有区别,绵阳、宜宾和广元地区的RVA阳性率较高,均超过了50%,而内江和资阳等地的RVA阳性率相对较低,为10%左右。本研究表明RVA在四川地区猪场中感染严重且分布广泛。因此,有必要加强对四川地区RVA的监测,本研究结果对四川地区RVA的防控提供了重要的科学依据。VP7和VP4分别决定RVA的G型和P型,且G型与P型之间会产生不同的组合,不同组合型的RVA毒株诱导的交叉保护性低[2,12]。因此,加强对RVA不同G/P型的监测和研究对了解RVA的遗传进化和疫苗研究非常重要。在本研究中,四川地区RVA中G9型、G5型和G4型为最为流行的G型,分别占分型毒株的41%、28.2%和23%;P[13]型、P[6]型和P[23]型为最为流行的P型,分别占分型毒株的40.7%、30.5%和23.7%,而其中P[13]型RVA最具多样性和复杂性;优势G/P基因型为G9P[23],占分型样本的23.3%,其中眉山地区的组合基因型最为复杂,存在3种不同的基因组合型(G3P[13]、G4P[23]和G9P[23])(表1)。本研究还在四川地区仔猪中检测出了G4P[23]、G4P[13]、G9P[6]、G26P[13]和G5P[23] 5种首次在国内出现的组合基因型。此外,对39株RVA VP7基因片段和59株RVA VP4基因片段同源性分析表明(表2、表3),本研究所获毒株与东南亚地区的猪源和人源毒株具有高度同源性。在30株成功鉴定出G/P组合基因型的毒株中,有13株毒株存在猪RVA与人RVA基因重配现象,推测在我国和东南亚地区猪RVA与人RVA存在交叉传播,且存在大量的基因重配毒株。本研究结果对四川地区猪RVA的防控提供了重要参考,其中首次报道的在国内新出现的RVA组合基因型丰富了国内RVA的资料,有利于对国内RVA的监测。对了解四川地区猪RVA的遗传多样性提供了科学依据,丰富了四川地区RVA的流行病学资料。
研究表明猪RVA与人RVA具有高度的相似性,且常发生重组和重配事件[1,29-30]。近年来,四川地区RVA重组事件陆续被报道[21,31],本研究中获得的12株RVA VP4全基因组和9株RVA VP7全基因组中分别存在2株RVA VP4和2株RVA VP7重组毒株。其中G型重组毒株为G4型和G9型重组,P型重组毒株为P[13]型与P[23]型重组,表明在四川地区RVA感染率升高的同时,RVA流行基因型之间重组频率也正在逐渐增高,提示应加强对四川地区RVA分子流行病学的监测和对猪RVA的综合防控。两株P型重组毒株来自同一猪场,均为猪源毒株与猪源毒株发生重组,但两株毒株预测的重组断点和亲本株存在差异。两株G型重组毒株来自内江某猪场,为人源毒株与猪源毒株重组,但两株毒株预测的重组断点相近,并且亲本株一致。推测两株P型重组毒株所在场内猪群存在多种猪RVA毒株混合感染,而两株G型重组毒株所在场内猪群可能存在跨种间传播,但该场内具体的传播途径还需要根据具体情况进行进一步调查研究。
4 结论
通过对2017—2019年间在四川省不同地区采集的猪腹泻样本进行猪RVA的检测和分型鉴定,揭示了近年来四川省猪A群轮状病毒的主要流行毒株,G9型和P[13]型为分别为优势G基因型和P基因型,G9P[23]为优势组合基因型。总体看来,A群轮状病毒在四川地区猪群中的分布广泛且基因型多样,与人源毒株关系密切,并且个别毒株存在基因重组现象。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/j.vaccine.2017.11.002URLPMID:29274701 [本文引用: 2]

BACKGROUND: Rotavirus is the leading cause of severe acute gastroenteritis (AGE) in children <5years of age in Myanmar. The purpose of this analysis is to report from the sentinel surveillance system for rotavirus gastroenteritis (RVGE), which collects information on the epidemiology and circulating genotypes to assess the disease burden and support vaccine introduction in Myanmar. METHODS: Prospective, active surveillance for RVGE-associated hospitalizations was conducted during 2009 -2014 at Yangon Children's Hospital. Stool samples collected from children <5years of age admitted for AGE were screened for rotavirus antigen by ELISA (ProSpecT Rotavirus, OXOID-UK). G and P genotyping was performed by reverse transcription polymerase chain reaction. RESULTS: Overall, 1860/3724 (49.9%) of stool samples tested positive for rotavirus, ranging from 42-56% of hospitalized AGE cases each year. RVGE was predominant in the 6-11months age group 889/1860 (47.8%) as compared with 12-23months 633/1860 (34.0%), 0-5 months 226/1860 (12.2 %) and 24-59months 112/1860 (6.0%). RVGE occurred in a seasonal cycle with peak occurrence in the cold and dry months (November to February), accounting for 65.3% (1151/1763) among enrolled AGE cases. Vomiting (84.1% Vs 67.9%; P<.01), fever (84.5% Vs 75.6%; P<.01) and dehydration (78% Vs 69%; P<.01) were more frequently observed in RVGE than non-RVGE. Genotyping revealed that G1P[8] was predominant from January to June 2009, G12P[8] was predominant throughout 2009-2012 which was replaced in 2012-2013 by G2P[4] and changed again to G1P[8] in 2013-2014 and G9P[8] in late 2014. CONCLUSIONS: Rotavirus is accounting for approximately half of AGE-associated hospitalizations among children <5years of age in Myanmar. There is immense diversity of rotavirus strains similar to that reported previously for other countries in the region. Information gained from this surveillance system highlights consideration of rotavirus vaccine introduction into this target population.
URL [本文引用: 3]
DOI:10.3201/eid2104.141370URLPMID:25811414 [本文引用: 1]

We identified unusual rotavirus strains in fecal specimens from sheltered dogs in Hungary by viral metagenomics. The novel rotavirus species displayed limited genome sequence homology to representatives of the 8 rotavirus species, A-H, and qualifies as a candidate new rotavirus species that we tentatively named Rotavirus I.
DOI:10.1007/s00705-012-1273-3URL [本文引用: 1]

Indirect immunofluorescence techniques targeting the rotavirus (RV) protein VP6 are used to differentiate RV species. The ICTV recognizes RV species A to E and two tentative species, F and G. A potential new RV species, ADRV-N, has been described. Phylogenetic trees and pairwise identity frequency graphs were constructed with more than 400 available VP6 sequences and seven newly determined VP6 sequences of RVD strains. All RV species were separated into distinct phylogenetic clusters. An amino acid sequence cutoff value of 53% firmly permitted differentiation of RV species, and ADRV-N was tentatively assigned to a novel RV species H (RVH).
URLPMID:23761160 [本文引用: 1]
DOI:10.3390/v9030048URL [本文引用: 1]
URLPMID:31030833 [本文引用: 1]
URLPMID:21801631 [本文引用: 1]
URLPMID:30144568 [本文引用: 1]
DOI:10.1007/s00705-006-0848-2URL [本文引用: 1]

Molecular characterization of two porcine group A rotavirus strains (HP113 and HP140), detected from eastern India, revealed a VP7 closely related to those of human G6P[14] strains, VP4 with a borderline P[13] genotype, and VP6 related to bovine and human SGI strains rather than porcine SGI and/or SGII group A rotaviruses. Both strains had NSP4 and NSP5 of porcine origin. Therefore, to our knowledge, the present study is the first report of detection of group A rotavirus strains with G6P[13] genotype specificities and provides evidence for independent segregation of the VP6- and NSP4-encoding genes in porcine group A rotaviruses.
DOI:10.1016/j.meegid.2017.06.026URL [本文引用: 1]
DOI:10.1128/IAI.39.1.91-99.1983URLPMID:6185436 [本文引用: 2]

Ten monoclones directed to the 42,000-dalton inner structural protein of rotavirus were analyzed. Eight monoclones reacted broadly with antigenic domains common to virtually all mammalian rotaviruses. Two monoclones had specificities similar or identical to previously characterized subgroup specificities. These subgroup monoclones were more efficient in detecting subgroup antigen than either hyperimmune or postinfection antisera. Using the subgroup monoclones, we determined that some animal as well as human rotavirus strains carry subgroup 2 specificity and that epizootic diarrhea of infant mice virus and turkey rotavirus are antigenically distinct from other mammalian rotavirus strains.
DOI:10.1016/j.vetmic.2013.03.020URLPMID:23642647 [本文引用: 1]

Group A rotavirus (RVA) infections cause severe economic losses in intensively reared livestock animals, particularly in herds of swine and cattle. RVA strains are antigenically heterogeneous, and are classified in multiple G and P types defined by the two outer capsid proteins, VP7 and VP4, respectively. This study summarizes published literature on the genetic and antigenic diversity of porcine and bovine RVA strains published over the last 3 decades. The single most prevalent genotype combination among porcine RVA strains was G5P[7], whereas the predominant genotype combination among bovine RVA strains was G6P[5], although spatiotemporal differences in RVA strain distribution were observed. These data provide important baseline data on epidemiologically important RVA strains in swine and cattle and may guide the development of more effective vaccines for veterinary use.
DOI:10.1007/s00705-011-1006-zURL [本文引用: 1]

In April 2008, a nucleotide-sequence-based, complete genome classification system was developed for group A rotaviruses (RVs). This system assigns a specific genotype to each of the 11 genome segments of a particular RV strain according to established nucleotide percent cutoff values. Using this approach, the genome of individual RV strains are given the complete descriptor of Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx. The Rotavirus Classification Working Group (RCWG) was formed by scientists in the field to maintain, evaluate and develop the RV genotype classification system, in particular to aid in the designation of new genotypes. Since its conception, the group has ratified 51 new genotypes: as of April 2011, new genotypes for VP7 (G20-G27), VP4 (P[28]-P[35]), VP6 (I12-I16), VP1 (R5-R9), VP2 (C6-C9), VP3 (M7-M8), NSP1 (A15-A16), NSP2 (N6-N9), NSP3 (T8-T12), NSP4 (E12-E14) and NSP5/6 (H7-H11) have been defined for RV strains recovered from humans, cows, pigs, horses, mice, South American camelids (guanaco), chickens, turkeys, pheasants, bats and a sugar glider. With increasing numbers of complete RV genome sequences becoming available, a standardized RV strain nomenclature system is needed, and the RCWG proposes that individual RV strains are named as follows: RV group/species of origin/country of identification/common name/year of identification/G- and P-type. In collaboration with the National Center for Biotechnology Information (NCBI), the RCWG is also working on developing a RV-specific resource for the deposition of nucleotide sequences. This resource will provide useful information regarding RV strains, including, but not limited to, the individual gene genotypes and epidemiological and clinical information. Together, the proposed nomenclature system and the NCBI RV resource will offer highly useful tools for investigators to search for, retrieve, and analyze the ever-growing volume of RV genomic data.
DOI:10.1111/tbed.12756URLPMID:29148270 [本文引用: 1]

Group A rotaviruses (RVAs) are a major cause of serious intestinal disease in piglets. In this study, a novel pig strain was identified in a stool sample from China. The strain was designated RVA/Pig/China/LNCY/2016/G3P[13] and had a G3-P[13]-I5-R1-C1-M1-A8-N1-T1-E1-H1 genome. The viral protein 7 (VP7) and non-structural protein 4 (NSP4) genes of RVA/Pig/China/LNCY/2016/G3P[13] were closely related to cogent genes of human RVAs, suggesting that a reassortment between pig and human strains had occurred. Recombination analysis showed that RVA/Pig/China/LNCY/2016/G3P[13] is a natural recombinant strain between the P[23] and P[7] RVA strains, and crossover points for recombination were found at nucleotides (nt) 456 and 804 of the VP4 gene. Elucidating the biological characteristics of porcine rotavirus (PoRV) will be helpful for further analyses of the epidemic characteristics of this virus. The results of this study provide valuable information for RVA recombination and evolution and will facilitate future investigations into the molecular pathogenesis of RVAs.
[本文引用: 3]
[本文引用: 3]
[本文引用: 3]
[本文引用: 3]
DOI:10.4149/av_2018_216URLPMID:30160138 [本文引用: 2]

Porcine rotavirus (PoRV) is one of the major causes of neonatal diarrhea in swine worldwide. Multiple serotypes of PoRV have been detected in diarrhea cases of suckling and weaning pigs. To date, the prevalence and molecular characterizations of PoRV circulating in swine in Shandong province of China remains largely unknown. Two hundred and twenty-six feces samples were collected from ten farms showing diarrhea in Shandong. All the samples were tested by RT-PCR for the presence of PoRV, TGEV, or PEDV. The results showed that all farms are positive for PEDV, and 60% and 10% of the farms are positive for PoRV and TGEV respectively. PoRV was detected in 65 out of 226 (28.76%) samples collected from 1-3 months old suckling and weaning pigs, while the positive rates of the TGEV and PEDV were 2.21% and 34.96%, respectively. The present data emphasized that PoRV is an important pathogen causing diarrhea in swine in China. In addition, VP6 and VP7 genes of PoRVs were sequenced and analyzed. Phylogenetical analysis of VP6 showed that all of the five PoRVs belong to group Arotavirus, meanwhile VP7 genes belong to the G3, G5, and G9 genotypes. Moreover, G5 and G9 genotypes are the dominant genotypes. Taken together, co-infections of TGEV, PEDV, and PoRV occur in pig population in Shandong, and the multiple serotypes of PoRVs are circulating in those herds, suggesting the active surveillance and matched vaccine application.
[本文引用: 2]
[本文引用: 2]
URL [本文引用: 1]

业微生物学国家重点实验室,武汉430070;2. 华中农业大学动物医学院,武汉430070)
摘要: 目前国内对猪轮状病毒的研究较少,作者旨在对猪A群轮状病毒进行分离与鉴定,为后续猪轮状病毒致病机理和分子生物学特性研究奠定基础。用胰蛋白酶处理RTPCR检测猪A群轮状病毒病原阳性的临床腹泻粪样,然后接种长成单层的MA104细胞,进行病毒分离传代。再对分离毒株进行常规RTPCR鉴定和电镜观察,并对分离毒株的VP6、VP7和VP8基因进行测序及序列分析。结果成功分离猪A群轮状病毒TMa株,经序列同源性分析发现,与TMa株VP6、VP7和VP8基因同源性最高的毒株分别为中国北京人源LL3354株、印度人源RMC321株和日本野猪源GUB71株,其相似性为96.0%、95.1%和97.0%。该TMa株轮状病毒不同基因表现出与人源轮状病毒和猪源轮状病毒的高度同源性,推测猪A群轮状病毒可能在人畜间传播,发生基因重组现象。
URL [本文引用: 1]

业微生物学国家重点实验室,武汉430070;2. 华中农业大学动物医学院,武汉430070)
摘要: 目前国内对猪轮状病毒的研究较少,作者旨在对猪A群轮状病毒进行分离与鉴定,为后续猪轮状病毒致病机理和分子生物学特性研究奠定基础。用胰蛋白酶处理RTPCR检测猪A群轮状病毒病原阳性的临床腹泻粪样,然后接种长成单层的MA104细胞,进行病毒分离传代。再对分离毒株进行常规RTPCR鉴定和电镜观察,并对分离毒株的VP6、VP7和VP8基因进行测序及序列分析。结果成功分离猪A群轮状病毒TMa株,经序列同源性分析发现,与TMa株VP6、VP7和VP8基因同源性最高的毒株分别为中国北京人源LL3354株、印度人源RMC321株和日本野猪源GUB71株,其相似性为96.0%、95.1%和97.0%。该TMa株轮状病毒不同基因表现出与人源轮状病毒和猪源轮状病毒的高度同源性,推测猪A群轮状病毒可能在人畜间传播,发生基因重组现象。
[本文引用: 2]
[本文引用: 2]
URL [本文引用: 1]
URL
URLPMID:16333070
URLPMID:30796978
URL [本文引用: 1]
URLPMID:31828508 [本文引用: 1]
URL [本文引用: 1]
DOI:10.1016/j.jcv.2007.12.013URLPMID:18304868 [本文引用: 1]

BACKGROUND: We found an unusual human rotavirus, LL36755 of G5P[6] genotype, in a stool sample collected in Lulong County, Hebei Province, China. This is the first detection of rotavirus serotype G5 in Asia. OBJECTIVES: To identify and characterize G5 rotaviruses in 988 stool samples collected from children under 5 years old with acute gastroenteritis. STUDY DESIGN: We analyzed 459 rotavirus-positive samples with RT-PCR using G5 genotype-specific primers. The G5 strains were sequenced. RESULTS: Two additional G5-positive samples (LL3354 and LL4260) were identified. VP7, VP4, VP6 and NSP4 genes of LL3354, LL4260 and LL36755 strains were sequenced. The VP4 sequences formed a group with porcine P[6] strains. The VP6 sequences of strains LL3354 and LL36755 were phylogenetically close to the major clusters of SGI and SGII rotaviruses, respectively. The deduced VP6 protein of strain LL4260 had characteristics of both SGI and SGII strains, but best fit with a cluster of atypical SGI viruses. In addition, based on NSP4 sequences, the three G5 strains belonged to genogroup B and were closest to human strain Wa. CONCLUSION: These results indicate a dynamic interaction of human and porcine rotaviruses and suggest that reassortment could result in the stable introduction and successful spread of porcine gene alleles into human rotaviruses.
URL [本文引用: 1]
URLPMID:30810805 [本文引用: 1]
