 ,, 刘新香, 王元东, 张如养, 王继东, 孙轩, 王夏青北京市农林科学院玉米研究中心/玉米DNA指纹及分子育种北京市重点实验室,北京100097
,, 刘新香, 王元东, 张如养, 王继东, 孙轩, 王夏青北京市农林科学院玉米研究中心/玉米DNA指纹及分子育种北京市重点实验室,北京100097Heterosis and Genetic Recombination Dissection of Maize Key Inbred Line Jing2416
ZHAO JiuRan, LI ChunHui, SONG Wei ,, LIU XinXiang, WANG YuanDong, ZHANG RuYang, WANG JiDong, SUN Xuan, WANG XiaQingMaize Research Center, Beijing Academy of Agriculture and Forestry Sciences/Beijing Key Laboratory of maize DNA Fingerprinting and Molecular Breeding, Beijing 100097
,, LIU XinXiang, WANG YuanDong, ZHANG RuYang, WANG JiDong, SUN Xuan, WANG XiaQingMaize Research Center, Beijing Academy of Agriculture and Forestry Sciences/Beijing Key Laboratory of maize DNA Fingerprinting and Molecular Breeding, Beijing 100097通讯作者:
责任编辑: 李莉
收稿日期:2020-02-15接受日期:2020-03-25网络出版日期:2020-11-16
| 基金资助: |
Received:2020-02-15Accepted:2020-03-25Online:2020-11-16
作者简介 About authors
赵久然,E-mail:
李春辉,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (882KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
赵久然, 李春辉, 宋伟, 刘新香, 王元东, 张如养, 王继东, 孙轩, 王夏青. 玉米骨干自交系京2416杂种优势及遗传重组解析[J]. 中国农业科学, 2020, 53(22): 4527-4536 doi:10.3864/j.issn.0578-1752.2020.22.001
ZHAO JiuRan, LI ChunHui, SONG Wei, LIU XinXiang, WANG YuanDong, ZHANG RuYang, WANG JiDong, SUN Xuan, WANG XiaQing.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】骨干自交系的形成和有效利用对作物遗传育种进程具有重要的推动作用[1]。选自地方品种塘四平头的骨干自交系黄早四[2],开创了中国紧凑型玉米育种新局面,利用黄早四衍生出众多优良自交系,包括昌7-2、Lx9801、京24、京2416等[3,4,5]。其中,优良自交系京2416是在中国玉米籽粒机收重大产业需求背景下育成的,具有耐密植抗倒伏、抗病抗逆性强、适宜区域广、早熟脱水快、自身产量及配合力高等多方面优点,已成为当前中国广泛利用的重要骨干自交系之一。利用京2416组配选育出京农科728、京农科828、NK718、MC121、MC812等21个优良玉米新品种(电子附表1),具有高产优质、多抗广适、易制种等综合优点,已在生产上累计推广300万hm2以上。其中,京农科728突破了黄淮海夏玉米籽粒机收技术瓶颈,成为中国首批通过国家审定的机收籽粒品种,引领夏玉米机收籽粒育种方向[6,7,8]。【前人研究进展】京2416是通过优良黄改群自交系京24和5237遗传重组形成的。重组是基因组进化的重要驱动力之一,可以增强物种的遗传多样性。通过重组实现多态位点重新排列,从而有机会获得新的、更优良等位基因组合,对作物育种具有重大贡献[9]。高通量芯片和测序技术的发展,使重现骨干亲本形成过程中的重组事件成为可能[9,10,11,12]。近年来,对玉米骨干自交系遗传重组进行了大量的研究。LAI等[10]对6个玉米骨干自交系(沈5003、掖478、郑58、昌7-2、178和Mo17)进行了全基因组测序,通过同源传递片段(identity-by-descent,IBD)分析证实自交系8112、沈5003、掖478和郑58之间的关系,重现了郑58形成过程中的重组事件。WU等[12]利用高通量芯片构建了黄改系的精细重组图谱,发现了15个遗传传递保守基因组区域。PAN等[9]利用骨干自交系形成的12个分离群体,解析了染色体上重组频率的分布和变化规律,并鉴定到143个重组热点区域。ZHANG等[11]利用高密度芯片重现了黄早四和14份黄改系的重组事件,确定了每个黄改系的重组事件次数,并通过IBD分析对郑58未知亲本进行推测,结果发现,未知亲本含有自交系丹340的同源染色体片段,使郑58形成过程的重组信息进一步细化。LI等[13]利用黄早四亲代及后代衍生系全基因组测序数据重现了黄早四及黄改系的形成历史,发现862个IBD保守区域,并且超过60%的IBD保守区域中存在选择信号,在这些选择区域中富集了大量的产量性状相关QTL。【本研究切入点】随着近年来优良玉米杂交种的审定与推广,大量优异的自交系得以培育,但对于京2416这一优新自交系并未进行系统地遗传解析。【拟解决的关键问题】本研究通过分析京2416形成过程中的重组事件和黄早四基因组片段传递规律,解析京2416和X群[14,15](由X1132x等杂交种构建的基础材料选育出的优新种质)代表系高配合力的遗传基础,以期为黄改系的遗传改良提供有益参考。1 材料与方法
1.1 试验材料
选用4份优异黄改群自交系(黄早四及其衍生系京2416、京24和5237)为试验材料。其中,京2416为京24和5237同群优系聚合选系。另外选用5份X群代表性自交系京724、京464、DH382、京725和京MC01作为测验种,对4份黄改系进行杂种优势分析。系谱来源见表1。Table 1
表1
表19份自交系及其系谱来源
Table 1
| 序号Code | 自交系Inbred lines | 系谱来源Pedigree |
|---|---|---|
| 1 | 京2416 Jing2416 | 京24×5237 Jing24×5237 |
| 2 | 京24 Jing24 | 早熟302×黄野四 Zaoshu302×Huangyesi |
| 3 | 5237 | 502×丹340 502×Dan340 |
| 4 | 黄早四 Huangzaosi | 塘四平头选系 Selected from Tangsipingtou |
| 5 | 京724 Jing724 | X1132x选系 Selected from X1132x |
| 6 | 京464 Jing464 | X1132x选系 Selected from X1132x |
| 7 | DH382 | X1132x选系 Selected from X1132x |
| 8 | 京725 Jing725 | X1132x选系 Selected from X1132x |
| 9 | 京MC01 JingMC01 | X1132x选系 Selected from X1132x |
新窗口打开|下载CSV
1.2 杂交组合的配制及田间鉴定
采用NCII遗传设计将上述4份黄改系和5份X系组配20个杂交组合,于2016年在黄淮海、东北和华北挑选5个有代表性的种植区(北京市、河北省、河南省、吉林省和辽宁省)进行种植,随机区组设计,2次重复。成熟后每行选取长势一致的5株植株用于单穗粒重测定。1.3 基因组DNA的提取
将9份试验材料种子室温萌发15 d左右,各取10株幼苗的幼嫩叶片混合磨样,参照ROGERS等[16] CTAB法提取玉米基因组DNA。检测DNA纯度浓度,以达到重测序标准。1.4 杂种优势和配合力分析
利用产量相关性状单穗粒重的中亲优势和超高亲优势值评估4份黄改系的杂种优势。中亲优势和超高亲优势的计算方法[17]如下:中亲优势(mid-parent heterosis,%)=(F1-MP)/MP×100
超高亲优势(over-high parent heterosis,%)=(F1-HP)/HP×100
其中,F1、MP和HP分别代表F1各性状平均值、2个亲本各性状平均值和高值亲本各性状平均值。
一般配合力(general combining ability,GCA)和特殊配合力(special combining ability,SCA)的相对效应计算方法[18]如下:
一般配合力:
$\hat{g}_{i}^{'}=\frac{\sum_{i=1}^{n_{1}}(x_{ij}- \bar{x})}{n_{1}} × \frac{1}{\bar{x}} × 100$
$\hat{g}_{i}^{'}=\frac{\sum_{j=1}^{n_{2}}(x_{ij}- \bar{x})}{n_{2}} × \frac{1}{\bar{x}} × 100$
特殊配合力:
$\hat{S}_{ij}^{'}=\frac{x_{ij}-\bar{x}-\hat{g}_{i}-\hat{g}_{j}}{\bar{x}}×100$
$\hat{g}_{i}^{'}$代表P1中第i个亲本的一般配合力相对效应值,$\hat{g}_{j}^{'}$代表P2中第j个亲本的一般配合力相对效应值,
$\hat{S}_{ij}^{'}$代表亲本i和亲本j组合的特殊配合力相对效应值。
n1代表P1中亲本个数,n2代表P2中亲本个数,xij代表亲本i和亲本j组合的平均值,$\bar{x}$代表总平均值。
1.5 全基因组重测序
利用双末端2×150 bp测序方法对供试材料进行基因组重测序。将原始数据进行检测过滤,去除被污染和低质量序列,对过滤后得到的clean reads进行后续分析。使用BWA(version: 0.6.1-r104)软件[19,20]的默认参数将clean reads比对到B73 RefGen_v3参考基因组上[21,22]。为了减少SNP和InDel错误的数量,进一步过滤不匹配reads和非唯一reads,并通过GATK(version 20171018)中的IndelRealigener程序进行矫正。SNP和InDel的calling使用GATK和Picard V1.119软件[23]进行计算。每个样本SNP和InDel的calling使用GATK中的HaplotypeCaller程序包独立计算。使用GATK中的GenotypeGVCFs程序包进行多个样本的vcf文件合并。对所有样品进行进一步过滤,变异calling使用的最小phred值置信阈值为60,mapping质量高于40,mapping质量的秩和检验阈值设置为-12.5,ReadPosRankSum参数设置为-8,每个碱基的测序深度大于2。1.6 主成分分析及系统发生树构建
基于获得的高质量SNP和InDel,使用GCTA V1.26.0软件[24]进行主成分分析(principal component analysis,PCA)。使用Treebest v1.9.2软件[25]计算遗传距离矩阵,并在遗传距离基础上利用邻接法(neighbor-joining method)构建系统发生树。引导值(bootstrap values)经过1 000次计算获得。1.7 IBD片段分析
使用IBDseq[26]软件对黄改系进行IBD片段识别,检测IBD片段的LOD值设为3,IBD片段端部边缘的LOD值设为2.5,滑动窗口内用于检测的标记数量设为4 000,最大等位基因错误率设为0.0001。当京2416和两选系亲本之间都具有IBD片段时,以重组次数最少为原则,并且认定具有较高P值的IBD片段作为最终结果。2 结果
2.1 京2416与X群种质的杂种优势分析
利用京2416、京24、5237和黄早四4份黄改系材料与X群代表系京724、京464、DH382、京725和京MC01的杂交组合F1一年五点的单穗粒重表型数据,计算中亲优势和超高亲优势(表2)。结果表明,黄改系和X系之间的杂种优势普遍存在,但不同杂交组合表现出的杂种优势程度存在一定差异。20个杂交组合的单穗粒重中亲优势,为58.03%—113.93%,平均为85.72%,具有较高的中亲优势。其中京2416和5个X群代表系的中亲优势较为突出,均值为94.00%,明显高于其2个选系亲本和X系的杂交组合。20个杂交组合的单穗粒重超高亲优势为27.65%—84.57%,平均为64.02%,其中,京2416和5个X群代表系的超高亲优势均较高,均值为76.13%,整体水平高于其他3个黄改系和X系的杂交组合。Table 2
表2
表24个黄改系和5个X系之间的杂交组合单穗粒重的杂种优势
Table 2
| 杂种优势 Heterosis | 亲本 Parents | DH382 | 京MC01 JingMC01 | 京464 Jing464 | 京724 Jing724 | 京725 Jing725 |
|---|---|---|---|---|---|---|
| 中亲优势 Mid-parent heterosis | 京2416 Jing2416 | 99.35 | 89.27 | 103.30 | 87.31 | 90.78 |
| 京24 Jing24 | 94.06 | 89.51 | 80.88 | 76.55 | 84.20 | |
| 5237 | 70.95 | 60.91 | 72.39 | 82.32 | 58.03 | |
| 黄早四 Huangzaosi | 113.93 | 100.77 | 106.84 | 82.11 | 70.86 | |
| 超高亲优势 Over-high parent heterosis | 京2416 Jing2416 | 84.48 | 83.87 | 82.46 | 67.25 | 62.60 |
| 京24 Jing24 | 80.02 | 84.57 | 62.71 | 58.00 | 57.33 | |
| 5237 | 63.37 | 60.11 | 59.59 | 67.90 | 38.65 | |
| 黄早四 Huangzaosi | 70.79 | 66.77 | 61.08 | 41.24 | 27.65 |
新窗口打开|下载CSV
进一步分析4个黄改系材料的配合力效应值,表3为4个黄改系亲本单穗粒重的一般配合力相对效应值(relative effect of general combining ability,RGCA)和20个杂交组合特殊配合力相对效应值(relative effect of specific combining ability,RSCA)。结果发现,4份黄改系的RGCA为-6.75%—7.50%,20个杂交组合的RSCA为-6.54%—9.29%。黄改系中京2416的RGCA为7.50%,高于其2个选系亲本京24和5237(2.68%和-3.68%),也高于黄早四(-6.75%)。说明京2416通过2个选系亲本的基因组重组,较其选系亲本和黄早四具有更高的配合力和杂种优势潜力。
2.2 全基因组变异检测及遗传结构分析
为了从基因组层面解析京2416形成的遗传机制,利用双末端2×150 bp测序的方法对9份玉米自交系进行了全基因组重测序(表4),其中4份黄改系数据来源于已发表文献[13],5份X群材料为新测数据。平均深度约为18.32倍基因组,共产生约348.52 Gb的高质量测序数据。平均基因组覆盖度和比对率分别为90.14%和92.32%,共鉴定到31.6 M的SNP和InDel变异信息。利用鉴定到的高质量SNP和InDel对9份玉米自交系进行主成分分析和系统发生树构建。通过主成分分析材料之间的亲缘关系(图1),黄改系和X系之间的遗传关系较远,在4份黄改系中京2416和京24的遗传关系较近,5237与京24的遗传关系较远。通过系统发生树分析9份材料的遗传关系,结果和主成分分析结果保持一致。
Table 3
表3
表34个黄改系亲本单穗粒重一般配合力相对效应值和20个杂交组合特殊配合力相对效应值
Table 3
| 亲本 Parents | RGCA (%) | RSCA (%) | ||||
|---|---|---|---|---|---|---|
| DH382 | 京MC01 JingMC01 | 京464 Jing464 | 京724 Jing724 | 京725 Jing725 | ||
| 京2416 Jing2416 | 7.50 | -1.78 | -2.43 | 2.47 | -2.04 | 3.78 |
| 京24 Jing24 | 2.68 | 0.43 | 2.75 | -5.02 | -3.06 | 4.89 |
| 5237 | -3.43 | -3.15 | -3.15 | -0.85 | 9.29 | -2.13 |
| 黄早四 Huangzaosi | -6.75 | 4.50 | 2.83 | 3.40 | -4.19 | -6.54 |
新窗口打开|下载CSV
Table 4
表4
表49份玉米自交系的测序信息
Table 4
| 材料名称 Sample name | 读长数 Reads (M) | 碱基数量 Bases (Gb) | 读长比对率 Map reads (%) | 比对reads数量 Map reads | 测序深度 Depth | 覆盖度 Coverage (%) |
|---|---|---|---|---|---|---|
| 京2416 Jing2416 | 237.49 | 35.07 | 90.45 | 212725920 | 15.90 | 88.93 |
| 京24 Jing24 | 230.52 | 33.89 | 89.65 | 204550192 | 15.36 | 88.90 |
| 5237 | 148.07 | 22.21 | 90.36 | 133672852 | 13.03 | 91.52 |
| 黄早四 Huangzaosi | 296.63 | 37.08 | 91.03 | 293757182 | 16.81 | 97.24 |
| DH382 | 155.61 | 22.73 | 88.87 | 136266112 | 11.04 | 88.80 |
| 京MC01 JingMC01 | 458.36 | 61.74 | 90.88 | 410270768 | 29.98 | 89.91 |
| 京724 Jing724 | 183.13 | 26.86 | 88.66 | 159907952 | 13.04 | 87.15 |
| 京725 Jing725 | 393.95 | 53.81 | 90.01 | 349216966 | 26.13 | 89.36 |
| 京464 Jing464 | 350.51 | 48.59 | 89.99 | 310638312 | 23.60 | 89.47 |
| 均值 Mean | 281.80 | 38.72 | 92.32 | 257719899 | 18.32 | 90.14 |
新窗口打开|下载CSV
图1
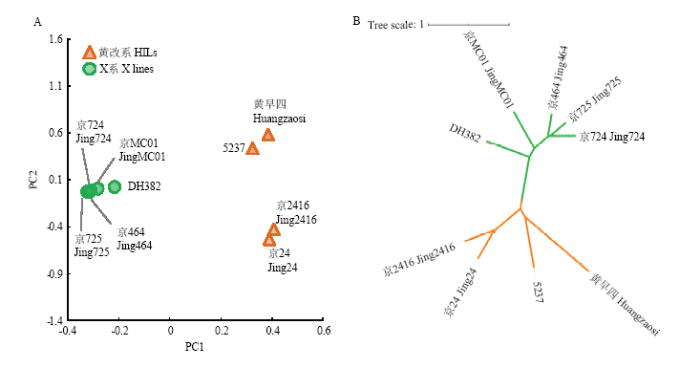 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图19份自交系的主成分分析(A)和系统发生树(B)
Fig. 1PCA analysis (A)and phylogenetic tree (B) of 9 inbred lines
2.3 京2416形成过程中的重组事件
高通量测序为解析骨干亲本形成的遗传基础提供了丰富的数据信息。高密度SNP和InDel分子标记的发掘和已知的系谱关系可以高分辨率地解析骨干自交系京2416形成的遗传基础。京2416的选育过程:首先将京24与5237杂交,再与京24回交1次,构建基础选系群体,按照高大严育种技术(高密度:大于90 000株/hm2;大群体:S1代群体精量点播出苗9 000株以上;严选择:严格选择标准,坚决淘汰生产中可能导致严重减产隐患的不良性状),经8代自交选育,创制出骨干自交系京2416(图2-A)。利用高密度分子标记分析发现,京2416和5237、京24之间的遗传相似度分别为0.765和0.906;5237和京24之间的遗传相似度为0.745,说明两者差异较大;黄早四和京2416、京24、5237三者之间的遗传相似度分别为0.763、0.748和0.796。IBD分析结果表明,京24和5237两者共发生了32次重组事件最终形成了京2416(图2-B)。其中第1、3、4、5和7染色体的重组次数较多,分别为6次、3次、6次、4次和10次重组;第6、9和10染色体重组次数较少,均为1次重组;第2和8染色体没有重组。重组的数量和频率与前人的研究结果类似[10]。京24和5237在京2416基因组中保留的比例分别为80.96%和19.04%,两者对于京2416的遗传贡献差异较大。图2
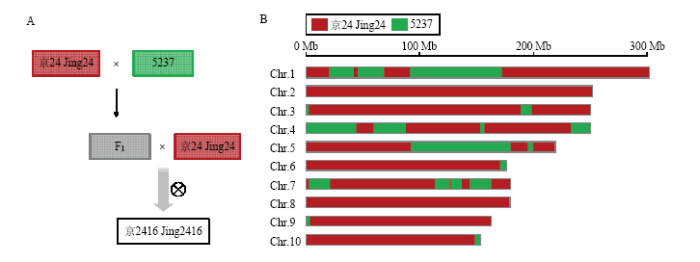 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2京2416及其2个选系亲本的遗传背景
A:京2416的系谱结构;B:京2416形成过程中的重组事件
Fig. 2The genetic background of Jing2416 and the two parents
A: The pedigree of Jing2416; B: The recombination events of Jing2416 formation
2.4 黄早四到京2416基因组片段传递
京2416保留了黄早四传递下来的很多优势性状,例如抗病抗逆性强、适宜区域广、早熟脱水快、自身产量及配合力高等,这些优势性状通过黄早四的IBD基因组片段进行传递。为了确定黄早四传递到京2416的IBD基因组片段,利用IBD方法对黄早四、京2416及其2个选系亲本京24和5237的全基因组重测序数据进行分析。结果表明,在京24和5237中均存在一定数量的、连续的、大片段的IBD区段,并且重组位点基本没有重合,暗示着两者的育种选择具有特异性(图3)。黄早四基因组传递给京24和5237的比例分别为0.3849和0.5464,最终传递给京2416的比例为0.4236。比较黄早四传递给京24和5237的基因组片段,位置分布各具特点,根据LI等[13]黄改系驯化改良研究结果可知,黄改系中重要的IBD传递区域和选择消除区域在2个京2416选系亲本中被各自保留,而京2416从其亲本黄改系中聚合了所有9个重要的IBD保守区域和选择消除区域,例如5237的第4染色体和第5染色体区域,京24的第6染色体区域。说明京2416经过京24和5237的染色体重组,聚合获得了更多的黄改系重要基因组区域(图3)。图3
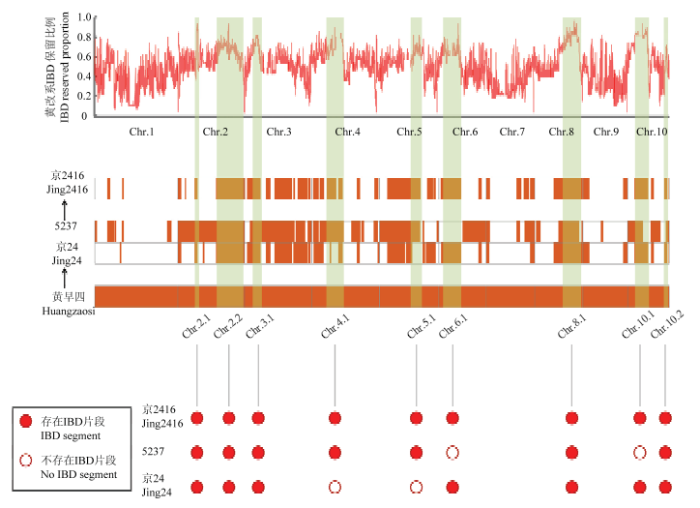 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3京2416形成过程中重要基因组区段的传递
图中“IBD保留比例”数据引自LI等[13]已发表论文
Fig. 3The important genomic segments transferred in the process of Jing2416 formation The data of “IBD reserved proportion” in
3 讨论
京2416是通过京24和5237 2个选系亲本自交系的遗传重组形成的。京24为北京市农林科学院玉米研究中心选育的黄改群骨干自交系,具有较强的抗倒伏能力和良好的早熟性。但一般配合力不高,产量潜力有限,感丝黑穗病和青枯病等主要病害。所组配的杂交种主要适宜京津冀早熟区域,在其他区域表现不突出。为了对京24进行有针对性的改良,通过大量鉴定筛选出符合目标需求的优良自交系5237,5237有一定比例的旅系血缘,具有旅系的优良性状及较好的抗病性和抗逆性。以(京24×5237)×京24构建基础选系群体,通过“高大严”自交系选育方法[27,28],创制出骨干自交系京2416,具有配合力高、株型紧凑、抗倒伏、抗斑病、耐密等多个优良性状,且适应性广,综合抗性好。该系目前已成为国家玉米良种攻关、七大农作物育种、国家玉米产业技术体系等多个重点项目及育种平台研究应用的骨干自交系,为今后中国玉米种质的进一步改良和利用提供了良好材料基础。本研究以产量相关性状单穗粒重为例,对京2416等4份黄改系与5份X群种质进行杂种优势分析。其中京2416的中亲优势(94.00%)明显高于其他2个选系亲本,但略低于黄早四(94.9%),这可能是由于黄早四作为黄改系的原始系,本身的单穗粒重值较小,但其具有高配合力特性,这也是其能够衍生形成中国最重要杂种优势群的主要原因之一。玉米骨干自交系黄早四于20世纪70年代由北京市农林科学院和中国农业科学院作物科学研究所共同选育,具有适应性强、配合力高、株型紧凑、灌浆速度快等多方面优良性状[3,4,5]。黄早四衍生形成的黄改系保持了黄早四的优良性状和高配合力,LI等[13]对黄改系的研究结果表明,黄早四传递给其他黄改系的IBD片段中存在一些重要的基因组区域,这些区域在几乎所有的黄改系基因组区域中均得到了保留,进一步研究发现,超过60%的选择消除信号聚集在IBD保守区域。在育种过程中,人工选择会使基因组产生大量的选择区域,而这些选择区域通常会富集稀有等位基因和优异农艺性状基因[29,30],对于黄改系的研究也证实黄改系的特征性选择区域中富集了与产量相关的基因/QTL[13]。本研究发现,京2416的IBD传递区域中包含了大量的黄改系IBD保守区域和选择消除区域,并且这些IBD区域分别来自不同的选系亲本,同时京2416从其选系亲本中聚合了黄改系几乎所有的重要特征性选择区域,证明高大严及同群优系聚合育种技术的有效性。基于上述选择区域,将进一步挖掘功能位点,开发分子标记进行辅助选择育种,以期在黄改系的遗传改良中发挥更大作用。
4 结论
与其他3份材料(黄改系京24、5237和黄早四)相比,骨干自交系京2416具有更高的超高亲优势和一般配合力。京2416在选育过程中,由2个选系亲本(京24和5237)中聚合了所有的9个重要黄改系特征性选择区域,从分子水平解释了京2416与X群代表系组配表现出更高配合力的遗传基础。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URL [本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1111/nph.13810URLPMID:26720856 [本文引用: 3]

Meiotic recombination is a major driver of genetic diversity, species evolution, and agricultural improvement. Thus, an understanding of the genetic recombination landscape across the maize (Zea mays) genome will provide insight and tools for further study of maize evolution and improvement. Here, we used c. 50 000 single nucleotide polymorphisms to precisely map recombination events in 12 artificial maize segregating populations. We observed substantial variation in the recombination frequency and distribution along the ten maize chromosomes among the 12 populations and identified 143 recombination hot regions. Recombination breakpoints were partitioned into intragenic and intergenic events. Interestingly, an increase in the number of genes containing recombination events was accompanied by a decrease in the number of recombination events per gene. This kept the overall number of intragenic recombination events nearly invariable in a given population, suggesting that the recombination variation observed among populations was largely attributed to intergenic recombination. However, significant associations between intragenic recombination events and variation in gene expression and agronomic traits were observed, suggesting potential roles for intragenic recombination in plant phenotypic diversity. Our results provide a comprehensive view of the maize recombination landscape, and show an association between recombination, gene expression and phenotypic variation, which may enhance crop genetic improvement.
DOI:10.1038/ng.684URLPMID:20972441 [本文引用: 3]

We have resequenced a group of six elite maize inbred lines, including the parents of the most productive commercial hybrid in China. This effort uncovered more than 1,000,000 SNPs, 30,000 indel polymorphisms and 101 low-sequence-diversity chromosomal intervals in the maize genome. We also identified several hundred complete genes that show presence/absence variation among these resequenced lines. We discuss the potential roles of complementation of presence/absence variations and other deleterious mutations in contributing to heterosis. High-density SNP and indel polymorphism markers reported here are expected to be a valuable resource for future genetic studies and the molecular breeding of this important crop.
DOI:10.1007/s00122-018-3072-zURLPMID:29492618 [本文引用: 2]

KEY MESSAGE: Genetic relationships among Chinese maize germplasms reveal historical trends in heterotic patterns from Chinese breeding programs and identify line Dan340 as a potential genome donor for elite inbred line Zheng58. The characterization of the genetic relationships, heterotic patterns and breeding history of lines in maize breeding programs allows breeders to efficiently use maize germplasm for line improvement over time. In this study, 269 temperate inbred lines, most of which have been widely used in Chinese maize breeding programs since the 1970s, were genotyped using the Illumina MaizeSNP50 BeadChip, which contains 56,110 single-nucleotide polymorphisms. The STRUCTURE analysis, cluster analysis and principal coordinate analysis results consistently revealed seven groups, of which five were consistent with known heterotic groups within the Chinese maize germplasm-Domestic Reid, Lancaster, Zi330, Tang SPT and Tem-tropic I (also known as
[本文引用: 2]
DOI:10.1016/j.molp.2019.02.009URLPMID:30807824 [本文引用: 7]

Maize is a globally important crop that was a classic model plant for genetic studies. Here, we report a 2.2 Gb draft genome sequence of an elite maize line, HuangZaoSi (HZS). Hybrids bred from HZS-improved lines (HILs) are planted in more than 60% of maize fields in China. Proteome clustering of six completed sequenced maize genomes show that 638 proteins fall into 264 HZS-specific gene families with the majority of contributions from tandem duplication events. Resequencing and comparative analysis of 40 HZS-related lines reveals the breeding history of HILs. More than 60% of identified selective sweeps were clustered in identity-by-descent conserved regions, and yield-related genes/QTLs were enriched in HZS characteristic selected regions. Furthermore, we demonstrated that HZS-specific family genes were not uniformly distributed in the genome but enriched in improvement/function-related genomic regions. This study provides an important and novel resource for maize genome research and expands our knowledge on the breadth of genomic variation and improvement history of maize.
DOI:10.3864/j.issn.0578-1752.2018.04.003URL [本文引用: 1]

【Objective】 Understanding the genetic diversity and population structure of representative maize accessions are of importance in breeding practice for the guidance and reference. 【Method】A total of 344 maize inbred lines were selected, including American heterotic group, local germplasm, New germplasm used in maize breeding in China in recent years which were broadly representative. These lines were genotyped by 3 072 SNP markers which were developed by Maize Research Center, BAAFS to reveal the genetic diversity and population structure. 【Result】For 3 072 high-quality SNPs, the gene diversity averaged 0.442, ranging from 0.028 to 0.646, and the PIC averaged 0.344, ranging from 0.028 to 0.570. The result of population structure based on a model-based method indicated that these 344 lines could be divided into eight groups, including Lüda red cob, Huangzaosi improved lines, Iodent, Lancaster, P group, Improved Reid group, Reid and X group. The seven groups above were well-known, and the X group was selected from the populations constructed from X1132X. Among the eight groups, the Fst ranged from 0.319 to 0.512, and the genetic distance ranged from 0.229 to 0.514. AMOVA results indicated that 38.6% of the total genetic variation occurred among groups, 58.1% within groups and 3.3% within lines. PCA results showed that X group had higher genetic differentiation with Huangzaosi improved lines and Lancaster, but lower with Iodent. The genetic diversity of subpopulations indicated that with the increase of breeding years, the average of genetic diversity in each subpopulation was decreased, and among them, X group had the highest genetic diversity. Further analysis showed that the genetic diversity of core accessions in American heterotic group and local germplasm were higher decreased compared with that in P group and Improved Reid group. However, the genetic diversity of core accessions in X group was no decreased, which indicated that the core accessions of X group still maintained higher genetic diversity and had potential application in breeding.【Discussion】X group was different from the other seven known groups, which can be defined as an independent group. Furthermore, X group had further genetic relationship with Huangzaosi improved lines which indicated the strong heterosis pattern of "X group × Huangzaosi improved lines" had application potential.
DOI:10.3864/j.issn.0578-1752.2018.04.003URL [本文引用: 1]

【Objective】 Understanding the genetic diversity and population structure of representative maize accessions are of importance in breeding practice for the guidance and reference. 【Method】A total of 344 maize inbred lines were selected, including American heterotic group, local germplasm, New germplasm used in maize breeding in China in recent years which were broadly representative. These lines were genotyped by 3 072 SNP markers which were developed by Maize Research Center, BAAFS to reveal the genetic diversity and population structure. 【Result】For 3 072 high-quality SNPs, the gene diversity averaged 0.442, ranging from 0.028 to 0.646, and the PIC averaged 0.344, ranging from 0.028 to 0.570. The result of population structure based on a model-based method indicated that these 344 lines could be divided into eight groups, including Lüda red cob, Huangzaosi improved lines, Iodent, Lancaster, P group, Improved Reid group, Reid and X group. The seven groups above were well-known, and the X group was selected from the populations constructed from X1132X. Among the eight groups, the Fst ranged from 0.319 to 0.512, and the genetic distance ranged from 0.229 to 0.514. AMOVA results indicated that 38.6% of the total genetic variation occurred among groups, 58.1% within groups and 3.3% within lines. PCA results showed that X group had higher genetic differentiation with Huangzaosi improved lines and Lancaster, but lower with Iodent. The genetic diversity of subpopulations indicated that with the increase of breeding years, the average of genetic diversity in each subpopulation was decreased, and among them, X group had the highest genetic diversity. Further analysis showed that the genetic diversity of core accessions in American heterotic group and local germplasm were higher decreased compared with that in P group and Improved Reid group. However, the genetic diversity of core accessions in X group was no decreased, which indicated that the core accessions of X group still maintained higher genetic diversity and had potential application in breeding.【Discussion】X group was different from the other seven known groups, which can be defined as an independent group. Furthermore, X group had further genetic relationship with Huangzaosi improved lines which indicated the strong heterosis pattern of "X group × Huangzaosi improved lines" had application potential.
[本文引用: 1]
[本文引用: 1]
Plant Mol
DOI:10.12692/ijb/5.1.69-73URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/bioinformatics/btp324URLPMID:19451168 [本文引用: 1]

MOTIVATION: The enormous amount of short reads generated by the new DNA sequencing technologies call for the development of fast and accurate read alignment programs. A first generation of hash table-based methods has been developed, including MAQ, which is accurate, feature rich and fast enough to align short reads from a single individual. However, MAQ does not support gapped alignment for single-end reads, which makes it unsuitable for alignment of longer reads where indels may occur frequently. The speed of MAQ is also a concern when the alignment is scaled up to the resequencing of hundreds of individuals. RESULTS: We implemented Burrows-Wheeler Alignment tool (BWA), a new read alignment package that is based on backward search with Burrows-Wheeler Transform (BWT), to efficiently align short sequencing reads against a large reference sequence such as the human genome, allowing mismatches and gaps. BWA supports both base space reads, e.g. from Illumina sequencing machines, and color space reads from AB SOLiD machines. Evaluations on both simulated and real data suggest that BWA is approximately 10-20x faster than MAQ, while achieving similar accuracy. In addition, BWA outputs alignment in the new standard SAM (Sequence Alignment/Map) format. Variant calling and other downstream analyses after the alignment can be achieved with the open source SAMtools software package. AVAILABILITY: http://maq.sourceforge.net.
DOI:10.1093/bioinformatics/btp352URLPMID:19505943 [本文引用: 1]

SUMMARY: The Sequence Alignment/Map (SAM) format is a generic alignment format for storing read alignments against reference sequences, supporting short and long reads (up to 128 Mbp) produced by different sequencing platforms. It is flexible in style, compact in size, efficient in random access and is the format in which alignments from the 1000 Genomes Project are released. SAMtools implements various utilities for post-processing alignments in the SAM format, such as indexing, variant caller and alignment viewer, and thus provides universal tools for processing read alignments. AVAILABILITY: http://samtools.sourceforge.net.
DOI:10.1126/science.aav7479URLPMID:30523095 [本文引用: 1]
DOI:10.1038/nature22971URLPMID:28605751 [本文引用: 1]

Complete and accurate reference genomes and annotations provide fundamental tools for characterization of genetic and functional variation. These resources facilitate the determination of biological processes and support translation of research findings into improved and sustainable agricultural technologies. Many reference genomes for crop plants have been generated over the past decade, but these genomes are often fragmented and missing complex repeat regions. Here we report the assembly and annotation of a reference genome of maize, a genetic and agricultural model species, using single-molecule real-time sequencing and high-resolution optical mapping. Relative to the previous reference genome, our assembly features a 52-fold increase in contig length and notable improvements in the assembly of intergenic spaces and centromeres. Characterization of the repetitive portion of the genome revealed more than 130,000 intact transposable elements, allowing us to identify transposable element lineage expansions that are unique to maize. Gene annotations were updated using 111,000 full-length transcripts obtained by single-molecule real-time sequencing. In addition, comparative optical mapping of two other inbred maize lines revealed a prevalence of deletions in regions of low gene density and maize lineage-specific genes.
DOI:10.1101/gr.107524.110URLPMID:20644199 [本文引用: 1]

Next-generation DNA sequencing (NGS) projects, such as the 1000 Genomes Project, are already revolutionizing our understanding of genetic variation among individuals. However, the massive data sets generated by NGS--the 1000 Genome pilot alone includes nearly five terabases--make writing feature-rich, efficient, and robust analysis tools difficult for even computationally sophisticated individuals. Indeed, many professionals are limited in the scope and the ease with which they can answer scientific questions by the complexity of accessing and manipulating the data produced by these machines. Here, we discuss our Genome Analysis Toolkit (GATK), a structured programming framework designed to ease the development of efficient and robust analysis tools for next-generation DNA sequencers using the functional programming philosophy of MapReduce. The GATK provides a small but rich set of data access patterns that encompass the majority of analysis tool needs. Separating specific analysis calculations from common data management infrastructure enables us to optimize the GATK framework for correctness, stability, and CPU and memory efficiency and to enable distributed and shared memory parallelization. We highlight the capabilities of the GATK by describing the implementation and application of robust, scale-tolerant tools like coverage calculators and single nucleotide polymorphism (SNP) calling. We conclude that the GATK programming framework enables developers and analysts to quickly and easily write efficient and robust NGS tools, many of which have already been incorporated into large-scale sequencing projects like the 1000 Genomes Project and The Cancer Genome Atlas.
DOI:10.1101/gr.073585.107URLPMID:19029536 [本文引用: 1]

We have developed a comprehensive gene orientated phylogenetic resource, EnsemblCompara GeneTrees, based on a computational pipeline to handle clustering, multiple alignment, and tree generation, including the handling of large gene families. We developed two novel non-sequence-based metrics of gene tree correctness and benchmarked a number of tree methods. The TreeBeST method from TreeFam shows the best performance in our hands. We also compared this phylogenetic approach to clustering approaches for ortholog prediction, showing a large increase in coverage using the phylogenetic approach. All data are made available in a number of formats and will be kept up to date with the Ensembl project.
DOI:10.1016/j.ajhg.2010.11.011URLPMID:21167468 [本文引用: 1]

For most human complex diseases and traits, SNPs identified by genome-wide association studies (GWAS) explain only a small fraction of the heritability. Here we report a user-friendly software tool called genome-wide complex trait analysis (GCTA), which was developed based on a method we recently developed to address the
DOI:10.1016/j.ajhg.2013.09.014URLPMID:24207118 [本文引用: 1]

Existing methods for identity by descent (IBD) segment detection were designed for SNP array data, not sequence data. Sequence data have a much higher density of genetic variants and a different allele frequency distribution, and can have higher genotype error rates. Consequently, best practices for IBD detection in SNP array data do not necessarily carry over to sequence data. We present a method, IBDseq, for detecting IBD segments in sequence data and a method, SEQERR, for estimating genotype error rates at low-frequency variants by using detected IBD. The IBDseq method estimates probabilities of genotypes observed with error for each pair of individuals under IBD and non-IBD models. The ratio of estimated probabilities under the two models gives a LOD score for IBD. We evaluate several IBD detection methods that are fast enough for application to sequence data (IBDseq, Beagle Refined IBD, PLINK, and GERMLINE) under multiple parameter settings, and we show that IBDseq achieves high power and accuracy for IBD detection in sequence data. The SEQERR method estimates genotype error rates by comparing observed and expected rates of pairs of homozygote and heterozygote genotypes at low-frequency variants in IBD segments. We demonstrate the accuracy of SEQERR in simulated data, and we apply the method to estimate genotype error rates in sequence data from the UK10K and 1000 Genomes projects.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/ng.2312URL [本文引用: 1]

The success of modern maize breeding has been demonstrated by remarkable increases in productivity over the last four decades. However, the underlying genetic changes correlated with these gains remain largely unknown. We report here the sequencing of 278 temperate maize inbred lines from different stages of breeding history, including deep resequencing of 4 lines with known pedigree information. The results show that modern breeding has introduced highly dynamic genetic changes into the maize genome. Artificial selection has affected thousands of targets, including genes and non-genic regions, leading to a reduction in nucleotide diversity and an increase in the proportion of rare alleles. Genetic changes during breeding happen rapidly, with extensive variation (SNPs, indels and copy-number variants (CNVs)) occurring, even within identity-by-descent regions. Our genome-wide assessment of genetic changes during modern maize breeding provides new strategies as well as practical targets for future crop breeding and biotechnology.
DOI:10.1038/ng.2309URL [本文引用: 1]

Domestication and plant breeding are ongoing 10,000-year-old evolutionary experiments that have radically altered wild species to meet human needs. Maize has undergone a particularly striking transformation. Researchers have sought for decades to identify the genes underlying maize evolution(1,2), but these efforts have been limited in scope. Here, we report a comprehensive assessment of the evolution of modern maize based on the genome-wide resequencing of 75 wild, landrace and improved maize lines(3). We find evidence of recovery of diversity after domestication, likely introgression from wild relatives, and evidence for stronger selection during domestication than improvement. We identify a number of genes with stronger signals of selection than those previously shown to underlie major morphological changes(4,5). Finally, through transcriptome-wide analysis of gene expression, we find evidence both consistent with removal of cis-acting variation during maize domestication and improvement and suggestive of modern breeding having increased dominance in expression while targeting highly expressed genes.
