 ,1,2, 郭勇2, 邱丽娟1,2
,1,2, 郭勇2, 邱丽娟1,2Establishment and Application of Multiple PCR Detection System for Glyphosate-Tolerant Gene EPSPS/GAT in Soybean
WEN Jing ,1,2, GUO Yong2, QIU LiJuan1,2
,1,2, GUO Yong2, QIU LiJuan1,2责任编辑: 李莉
收稿日期:2020-01-3接受日期:2020-03-7网络出版日期:2020-10-16
| 基金资助: |
Received:2020-01-3Accepted:2020-03-7Online:2020-10-16
作者简介 About authors
文静,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2908KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
文静, 郭勇, 邱丽娟. 耐草甘膦转EPSPS/GAT大豆多重PCR检测体系的建立及应用[J]. 中国农业科学, 2020, 53(20): 4127-4136 doi:10.3864/j.issn.0578-1752.2020.20.003
WEN Jing, GUO Yong, QIU LiJuan.
0 引言
【研究意义】自1994年第一个商业化的转基因植物(FlavrSavr tomato)被批准上市以来,植物基因工程技术在现代农业中得到了广泛的应用[1]。目前已有24个国家/地区种植了转基因作物,42个国家/地区进口转基因作物用于粮食、饲料和加工。全球转基因作物的种植面积从1996年的170万hm2增加到2018年的1.917亿hm2,增长了113倍,而转基因大豆作为种植面积最大的转基因作物,种植面积达到全球转基因作物种植面积的50%[2]。随着转基因作物产业化和商业化程度的不断扩大,全球对转基因食品安全、环境风险和伦理问题的争议加剧[3],越来越多的国家和地区要求对转基因食品和成分进行标识[4]。部分国家要求转基因生物(genetically modified organism,GMO)含量需要低于一定阈值水平的限制。例如,欧盟将非转基因背景下的转基因物质的阈值设定为0.9%[5]。因此,标准化的转基因成分检测和鉴定方法是转基因发展和商业化的关键步骤,也在转基因生物的安全评估和风险管理中发挥了重要作用[6]。现有的转基因检测方法大多基于聚合酶链反应(polymerase chain reaction,PCR),并由于其具有通用性、敏感性、特异性和高通量的特点,可用于复杂转基因成分分析,包括筛查外源调控元件和目的基因,并对特定的转基因成分进行鉴定[7]。多重PCR技术是在同一个PCR反应体系中加入多对特异性引物,实现对多个靶标的扩增,更能节省转基因生物检测中时间和成本[8,9]。【前人研究进展】吕山花等[10]应用PCR检测方法,以大豆凝集素(lectin)基因为内参,检测了4个抗草甘膦转基因大豆品种及草甘膦敏感大豆品种豫豆12的外源CaMV35S启动子、NOS终止子和CP4-EPSPS成分。目前,中国允许进口的转基因大豆有16种(具体包括A2704-12、CV127、MON87701、MON87769、GTS40-3-2、MON87708、305423、A5547-127、FG72、MON89788、SYHT0H2、MON87701×MON89788、DAS-44406-6、305423× GTS40-3-2、MON87705和356043,其中MON87701× MON89788和305423×GTS40-3-2为复合性状转基因大豆),农业部针对这些转基因大豆品系发布的检测方法以核酸检测为主,但是其方法均为单重PCR检测。针对转基因大豆建立系统的、高通量的由“公共引物”介导的多重PCR转基因大豆检测体系,可大幅降低检测成本、提高检测效率,为转基因安全评价提供新的检测技术和方法[11]。多重PCR检测体系并不是多个单重PCR体系检测体系的简单叠加,它需要对PCR体系和程序进行反复试验、优化和验证,保证多重PCR具有足够的特异性、灵敏性和适用性,最终实现多个目标片段的同时扩增,因此,与普通PCR相比,难度更大[12]。近年来,国内外有诸多****对其进行研究,并建立了许多关于通用元件、基因特异性和转化事件特异性的多重PCR检测体系[8,13-16]。尹全等[17]以抗草甘膦大豆GTS40-3-2的通用元件作为检测参数,建立了大豆内源基因lectin、CaMV35S启动子、NOS终止子和CP4-EPSPS的四重PCR检测体系。AO等[18]根据外源基因的特异性建立了适用于转基因大豆、玉米、水稻中CP4-EPSPS、Cry1A(b)、BAR、PAT检测的多重PCR方法。KIM等[19]针对RRS、GTS40-3-2、A2704-12、DP356043-5、MON89788、A5547-127和DP305423-1 6个商业化转基因大豆特异性事件,建立了包含内参基因的七重PCR检测体系。付伟等[20]以转基因大豆DAS44406-6品系为研究对象,利用Multiplex PCR技术与其他技术相结合,建立了多重实时荧光PCR检测体系,检测灵敏度为0.01%。【本研究切入点】目前,大多数转基因大豆的多重PCR技术都是针对通用元件和基因特异性进行检测,或者针对多个商业化特异性转化事件进行筛选,而单一转化事件特异性鉴定鲜见报道[21]。在转基因生物检测中,转化事件特异性检测的目标片段是外源基因和宿主DNA之间的连接区域,故比单纯基因特异性检测和通用元件检测具有更高的特异性[22,23]。【拟解决的关键问题】本研究基于转基因成分检测技术及要求,以转基因大豆ZH10-6为研究对象,针对内参基因、外源基因和侧翼序列设计5对特异性引物,构建了转EPSPS/GAT大豆ZH10-6多重PCR反应体系,并用本研究建立的多重PCR检测体系检测了ZH10-6的11个衍生品系,为中国抗草甘膦转基因大豆新品种目标基因鉴定提供信息和方法。1 材料与方法
1.1 试验材料
转基因大豆ZH10-6、受体中黄10及11个ZH10-6衍生品系均为中国农业科学院作物科学研究所大豆基因资源挖掘与利用实验室保存样品。1.2 样品基因组DNA的提取
参照植物基因组提取试剂盒说明书提取样品DNA,用紫外分光光度计测量DNA样品质量及浓度,并用1×TE缓冲液将DNA稀释至50 ng·μL-1。1.3 引物的设计及筛选
使用软件Primer Premier 5.0进行引物设计,设计时遵从多重PCR检测引物设计与筛选的原则,根据抗草甘膦大豆ZH10-6和受体中黄10的分子特征,设计大豆内源参考基因(Actin)、外源基因(G2- EPSPS和GAT)以及侧翼序列(G2EPSPS-2/ZH10P2和ZH10P1/GAT-2)的特异性引物(表1)。将各个检测基因的上、下游引物使用灭菌的超纯水稀释为10 μmol·L-1。Table 1
表1
表1多重PCR标记及其相关信息
Table 1
| 基因/标记名称 Gene/Marker name | 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 产物大小 Fragment size (bp) |
|---|---|---|---|
| 内源参考基因Actin Endogenous gene Actin | GmActin11F | ATCTTGACTGAGCGTGGTTATTCC | 126 |
| GmActin11R | GCTGGTCCTGGCTGTCTCC | ||
| 侧翼序列ZH10P1/GAT-2 Flanking sequence ZH10P1/GAT-2 | ZH10P1 | TAATAGTAGAATGGGACTGGTGGAT | 810 |
| GAT | GCGGACTTGCTTTGGTGTAAT | ||
| 侧翼序列G2EPSPS-2/ZH10P2 Flanking sequence G2EPSPS-2/ZH10P2 | G2 | CCCGAATCATCAGGCAAACA | 1626 |
| ZH10P2 | AACACATCATAGTATTCTAAAACGCTT | ||
| 外源基因G2-EPSPS Exogenous gene G2-EPSPS | G2-EPSPS F | CAAATCCATTACCAACCGTGC | 430 |
| G2-EPSPS R | ACCACCATCAATCTCGAAACG | ||
| 外源基因GAT Exogenous gene GAT | GAT F | CTCAGACCAAACCAGCCGATAG | 338 |
| GAT R | GTGTCGAATACCTCTCCCTGCTC |
新窗口打开|下载CSV
1.4 引物特异性和适用性检测
单重PCR反应体系为25 μL,包括DNA 2 μL(50 ng·μL-1)、Ex Taq酶0.2 μL、10×ExTaq Buffer 2.5 μL、dNTP 2 μL、引物(10 μmol·L-1)0.6 μL和ddH2O 17.7 μL。多重PCR反应体系为25 μL,包括DNA 2 μL(50 ng·μL-1)、Ex Taq酶 0.2 μL、10×ExTaq Buffer 2.5 μL、dNTP 2 μL、引物(10 μmol·L-1,GmActin11 F/R 0.6 μL、G2-EPSPS F/R 0.6 μL、GAT F/R 0.6 μL、ZH10P1/GAT 0.6 μL、G2/ZH10P2 0.6 μL)和ddH2O 15.3 μL。
单重和多重PCR反应程序为95℃ 5 min;95℃ 30 s,58℃ 30 s,72℃ 1 min20 s 35个循环;72℃ 8 min;4℃保存。
用1%琼脂糖凝胶电泳检测多重PCR产物,并利用凝胶成像系统观察。
1.5 多重PCR反应体系和程序的优化
以转基因大豆ZH10-6和受体中黄10的DNA为模板,将5对引物的用量设置为GmActin11 F/R 0.4 μL、G2-EPSPS F/R 0.6 μL、GAT F/R 0.4 μL、ZH10P1/GAT 0.6 μL和G2/ZH10P2 0.6 μL,DNA模板量设置为100、150、200和250 ng 4个梯度,退火温度设置为55℃、58℃和60℃,延伸温度设置为68℃和72℃ 2个梯度,dNTP含量为2、3、4和5 μL 4个梯度,分别进行多重PCR扩增,筛选出五重PCR体系的最优反应条件。1.6 多重PCR灵敏度检测
为了验证多重PCR体系灵敏性,将相同浓度的转基因大豆ZH10-6和受体中黄10的基因组DNA按质量比混合,制备成100%、50%、10%、5%、1%、0.5%、0.1%和0的DNA样品。使用1.5建立的多重PCR体系及程序进行多重PCR方法灵敏度检测。1.7 多重PCR体系的应用
用建立的多重PCR体系对转基因大豆ZH10-6衍生品系进行检测,包括东北地区和黄淮地区材料,进一步验证多重PCR体系的实用性。2 结果
2.1 样品DNA检测
使用NanoDrop 2000/2000c超微量分光光度计测定样品DNA浓度和质量,经测定,样品OD260/OD230比值均为2.0—2.2,OD260/OD280比值均为1.8—2.0,说明提取的样品DNA质量较好。用1%琼脂糖凝胶电泳对DNA样品进行检测,仅1号样品第三条带有降解,该样品不在后续试验中使用,其余样品几乎无降解,符合多重PCR试验要求(图1)。将样品DNA浓度稀释至50 ng·μL-1,备用。图1
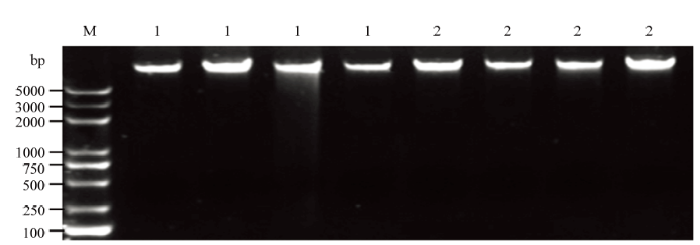 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图11%琼脂糖凝胶电泳DNA检测结果
M:2K Plus II;1:中黄10;2:ZH10-6
Fig. 1DNA detection results of 1% agarose gel electrophoresis
M: 2K Plus II; 1: ZH10; 2: ZH10-6
2.2 引物特异性和适用性检测
以转基因大豆ZH10-6和受体中黄10的DNA为模板进行5对引物特异性和适用性检测。单引物扩增时,5对引物均可在目的位置处观察到特异性条带,未观察到明显的非特异性条带。5对引物同时扩增时,转基因大豆ZH10-6可以扩增出多条特异性条带,但个别条带相对微弱(图2),表明这5对引物可以用于多重PCR区别ZH10-6和受体中黄10,但是需要进一步调整扩增条件。图2
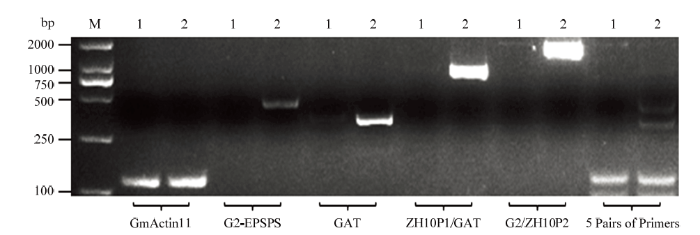 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2单基因特异性检测结果
M:2K Plus II;1:中黄10;2:ZH10-6;GmActin11、G2-EPSPS、GAT、ZH10P1/GAT、G2/ZH10P2为PCR扩增引物,5 Pairs of Primers是GmActin11、G2-EPSPS、GAT、ZH10P1/GAT、G2/ZH10P2等量混合
Fig. 2Single gene specific test results
M: 2K Plus II; 1: ZH10; 2: ZH10-6; GmActin11, G2-EPSPSP, GAT, ZH10P1/GAT and G2/ZH10P2 were PCR amplification primers, and 5 Pairs of Primers were equal combinations of GmActin11, G2-EPSPSP, GAT, ZH10P1/GAT and G2/ZH10P2
2.3 多重PCR检测体系的建立
多重PCR体系中,将5对引物用量均设置为0.6 μL时,部分条带不能被扩增(图2)。可以看出在多重PCR中,引物存在强弱之分,需要减少相对较强引物GmActin11 F/R和GAT F/R的用量。以条带亮度作为参考,最终将5对引物的用量设置为GmActin11 F/R 0.4 μL、G2-EPSPS F/R 0.6 μL、GAT F/R 0.4 μL、ZH10P1/GAT 0.6 μL和G2/ZH10P2 0.6 μL。将DNA模板量、退火温度、延伸温度和dNTP含量均设置为不同梯度,进行同时扩增(图3),筛选多重PCR最适温度和体系。图3
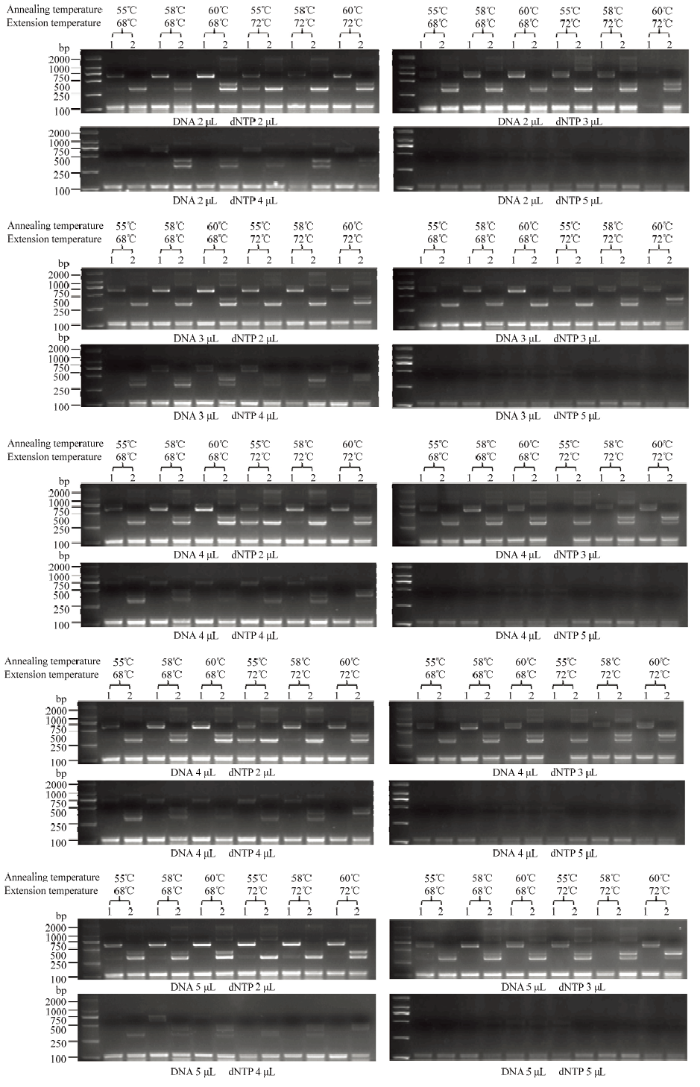 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3梯度多重PCR检测结果
M:2K Plus II;1:中黄10;2:ZH10-6
Fig. 3Results of gradient multiple PCR
M: 2K Plus II; 1: ZH10; 2: ZH10-6
通过不同体系检测(图4),多重PCR扩增最适体系为:DNA 2—4 μL、Ex Taq 0.2 μL、10×ExTaq Buffer 2.5 μL、dNTP 2 μL、引物(GmActin11 F/R 0.4 μL、G2-EPSPS F/R 0.6 μL、GAT F/R 0.4 μL、ZH10P1/GAT 0.6 μL和G2/ZH10P2 0.6 μL),ddH2O补足25 μL。多重PCR扩增最适程序为95℃ 5 min;95℃ 30 s,60℃ 30 s,68℃ 1 min20 s,35个循环;72℃ 12 min;4℃保存。最终扩增产物长度为内参基因126 bp、外源基因G2-EPSPS 430 bp、外源基因GAT 338 bp、侧翼序列ZH10P1/GAT-2 810 bp、侧翼序列G2EPSPS-2/ZH10P2 1 626 bp。其中,受体中黄10除了可以扩增出Actin内参序列以外,还可以扩增出ZH10-6 2对侧翼序列的上游引物ZH10P1和下游引物ZH10P2的632 bp片段,而在ZH10-6中,由于插入外源基因过大,此段序列无法被扩增出。因此,受体中黄10可以被扩增出2条带,而转基因材料ZH10-6可以被扩增出5条带,此体系可以用于鉴定ZH10-6和受体中黄10。
图4
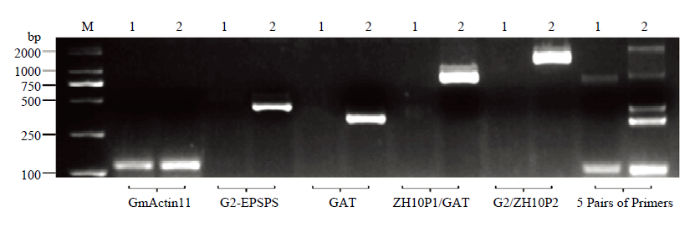 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4单重及多重PCR检测ZH10-6及中黄10
M:2K Plus II;1:中黄10;2:ZH10-6;GmActin11、G2-EPSPS、GAT、ZH10P1/GAT、G2/ZH10P2为PCR扩增引物,5 Pairs of Primers是GmActin11、G2-EPSPS、GAT、ZH10P1/GAT、G2/ZH10P2等量混合
Fig. 4Single and multiple PCR detection of ZH10-6 and ZH10
M: 2K Plus II; 1: ZH10; 2: ZH10-6; GmActin11, G2-EPSPSP, GAT, ZH10P1/GAT and G2/ZH10P2 were PCR amplification primers, and 5 Pairs of Primers were equal combinations of GmActin11, G2-EPSPSP, GAT, ZH10P1/GAT and G2/ZH10P2
2.4 多重PCR体系灵敏度检测
将模板DNA稀释至设定浓度,通过多重PCR灵敏度试验(图5),随着受体中黄10质量分数的上升,特异性片段632 bp的条带逐渐变亮,而ZH10-6的5条带则出现不同程度的减弱,特别是外源基因G2-EPSPS的430 bp条带和侧翼序列G2EPSPS-2/ ZH10P2为1 626 bp条带。因为在转基因大豆ZH10-6和非转基因大豆中黄10混合后,相对于原有的多重PCR体系中增添了一重PCR反应。灵敏度是非转基因样品中转基因成分的最低检测限,虽然图5转外源基因G2-EPSPS的430 bp条带和侧翼序列G2EPSPS-2/ ZH10P2为1 626 bp条带相比于其他条带较弱,但是仍可以看出条带所在位置。因此,只要样品中含有0.5%以上的ZH10-6转基因成分,利用此体系都能检测出目标条带。说明建立的多重PCR体系灵敏度可达0.5%,符合欧盟有关转基因产品标识的要求。图5
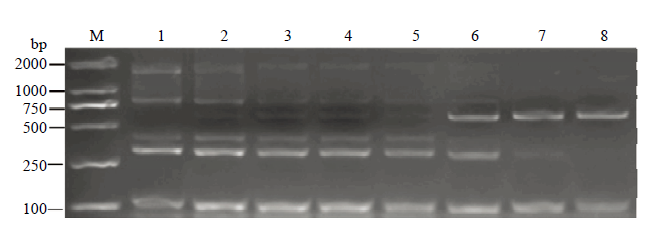 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5多重PCR灵敏度检测
M:2K Plus II;1—7:ZH10-6的质量分数分别为100%、50%、10%、5%、1%、0.5%和0.1%;8:中黄10
Fig. 5Results of multiple PCR sensitivity detection
M: 2K Plus II; 1-7: The mass fraction of ZH10-6: 100%, 50%, 10%, 5%, 1%, 0.5%, 0.1%; 8: ZH10
2.5 多重PCR体系的应用
以转基因大豆ZH10-6衍生品系的DNA为模板,将DNA浓度稀释成50 ng·μL-1。用建立的多重PCR体系待测大豆进行多重扩增,根据不同的条带类型,判断ZH10-6衍生品系中外源基因和侧翼序列情况(图6)。主要检测到5种带型(表2),类型1出现与受体中黄10相同带型,认为是未发生杂交的野生型;类型2出现与ZH10-6相同带型;类型3和类型4分别以NZC400和NZK001为例,认为是其他转化事件;第5类带型包含ZH10-6和受体ZH10的全部6条带型,认为是转基因杂合材料。在11份ZH10-6衍生品系中,NZC100和NZC400为黄淮材料,剩余为东北材料。用单重PCR对该扩增结果进行验证,与多重PCR鉴定结果完全一致,证明建立的多重PCR体系可以鉴定转基因大豆ZH10-6和受体中黄10,也可以鉴定不同地区的ZH10-6衍生品系,应用性较为广泛。图6
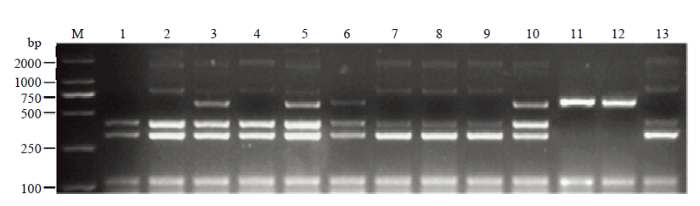 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6ZH10-6衍生品系检测
M:2K Plus II;1:NZK001;2:NZK100;3:NZK150;4: NZK180;5:NZC100;6:NZC400;7:NZJ77;8:NZJ110;9:SUN100;10:NZM150;11:NZG700;12:ZH10;13:ZH10-6
Fig. 6Test results of ZH10-6 derivative lines
Table 2
表2
表2ZH10-6衍生品系类型
Table 2
| 衍生品系类型 Derived strain type | 引物名称 The name of the primers | 衍生品系名称 The name of the derivative lines | |||||
|---|---|---|---|---|---|---|---|
| GmActin11 | G2-EPSPS | GAT | ZH10P1/GAT | G2/ZH10P2 | ZH10P1/ZH0P2 | ||
| 类型1 Type 1 | √ | √ | NZG700、ZH10 | ||||
| 类型2 Type 2 | √ | √ | √ | √ | √ | NZK100、NZK180、NZJ77、NZJ110、SUN100、ZH10-6 | |
| 类型3 Type 3 | √ | √ | √ | √ | NZC400 | ||
| 类型4 Type 4 | √ | √ | √ | NZK001 | |||
| 类型5 Type 5 | √ | √ | √ | √ | √ | √ | NZC100、NZK150、NZM150 |
新窗口打开|下载CSV
3 讨论
PCR检测技术是转基因作物及产品中外源基因检测的重要手段[24,25],相比于普通PCR,多重PCR只需一个反应和一次电泳就可同时检出多个目标片段,可极大地节约试剂和时间[26,27],目前,已被广泛应用到医学及生物等领域[28]。在研究过程中发现,不同引物序列的多重PCR在实际应用中会受各项参数直接影响。3.1 多重PCR体系中引物选择的直接依据
本研究中选择特异性强、引物二聚体少的5对引物进行扩增,其中包括起质量控制作用的内源基因,以避免转基因检测的假阴性结果出现。多重PCR扩增时,各个引物的扩增并非均一的,本研究通过5对引物等量加入时的条带亮度对引物进行强弱判断,选择各基因扩增效率相对一致的引物浓度配比条件建立了多重PCR体系。引物设计是多重PCR反应成败的关键,需要遵循PCR引物设计通用准则,即单条引物长度一般为18—25个碱基,且4种碱基的比例要适当,G+C含量一般为40%—60%。同时,选择的引物必须高度特异,彼此之间无同源性,且连续互补的碱基不超过4个,避免出现交叉错配等现象[29]。引物之间TM值尽可能相差较小,可以实现在同一退火温度下的扩增。3.2 不同反应体系和条件对多重PCR体系建立的影响
在建立多重PCR反应体系时,反应体系和条件是关键因素[30,31]。反应体系主要包括DNA模板量、dNTP含量等。本研究将DNA模板量从100 ng增加至250 ng时,扩增效果没有明显变化,说明DNA模板量对多重PCR体系扩增影响较小。同时发现在多重PCR扩增中,dNTP含量可以同单重PCR扩增设置一致,dNTP含量过高反而会抑制反应。反应条件是另一个关键因素,主要包括退火温度和延伸温度。一般情况下,可以依据引物的TM值减少5℃直接选择退火温度,但有时结果与预期并不一致。较为简单的方法就是设置54℃为初始退火温度,如果多重PCR扩增产生非特异性条带,需适当提高引物退火温度,反之则降低引物退火温度[32]。退火温度应在Tm值允许范围内选择较高的温度,以确保PCR反应的特异性。但在这个过程中会出现目标片段扩增变弱的现象,可以通过延长延伸时间来消除。同时适当降低延伸温度有利于目标条带的产生。本研究中设置了55℃、58℃和60℃ 3个退火温度,68℃和72℃ 2个延伸温度,最终在退火温度为60℃,延伸温度为68℃时,多重PCR体系扩增效果最好。
3.3 目标片段的选择对多重PCR体系灵敏度的影响
在多重PCR目标片段的选择时,目标片段必须具有高度特异性,避免目标片段之间的竞争性扩增,才能实现高效灵敏的扩增反应,建议PCR产物片段大小在200 bp以内时,片段间隔大于30 bp;产物在500 bp以上时,片段间隔要大于70 bp[33]。此外,各个目的片段长度应差异明显,以便于鉴别,但最大和最小片段最好相差不超过400 bp,否则会导致多重PCR体系灵敏度降低,且体系建立的难度较大。目前转基因大豆多重PCR技术中,董立明等[34]选择商品化早、种植面积大的5种转基因大豆品系305423、MON89788、CV127、GTS40-3-2、356043及大豆内源基因Lectin为研究对象,选用相应的国家标准和文献中的引物,建立了能同时检测5种转基因大豆品系和大豆内源基因的六重PCR检测体系,检测最大片段和最小片段相差400 bp,该体系能够从0.1%及以上含量的阳性对照样品中检测出全部6种靶标成分。本研究检测的目标片段是外源基因和宿主DNA之间的连接区域,具有更高的特异性,扩增最大目标片段与最小片段之间高达1 500 bp,但灵敏度也能达到0.5%,完全符合欧盟有关转基因标识的要求。同时,该多重PCR体系在ZH10-6和受体中黄10混合体系中,实现了六重PCR扩增。4 结论
通过设计内参基因、外源基因和侧翼序列引物并对反应体系和条件进行优化,建立的转EPSPS/GAT大豆多重PCR检测体系具有高通量、特异性强、操作简便和应用广泛的优点,并且能够快速、准确地检测转基因大豆ZH10-6及其衍生品系。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/0924-2244(94)90197-XURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1111/j.1365-294X.2008.03663.xURLPMID:18302682 [本文引用: 1]

Whether the potential costs associated with broad-scale use of genetically modified organisms (GMOs) outweigh possible benefits is highly contentious, including within the scientific community. Even among those generally in favour of commercialization of GM crops, there is nonetheless broad recognition that transgene escape into the wild should be minimized. But is it possible to achieve containment of engineered genetic elements in the context of large scale agricultural production? In a previous study, Warwick et al. (2003) documented transgene escape via gene flow from herbicide resistant (HR) canola (Brassica napus) into neighbouring weedy B. rapa populations (Fig. 1) in two agricultural fields in Quebec, Canada. In a follow-up study in this issue of Molecular Ecology, Warwick et al. (2008) show that the transgene has persisted and spread within the weedy population in the absence of selection for herbicide resistance. Certainly a trait like herbicide resistance is expected to spread when selected through the use of the herbicide, despite potentially negative epistatic effects on fitness. However, Warwick et al.'s findings suggest that direct selection favouring the transgene is not required for its persistence. So is there any hope of preventing transgene escape into the wild?
DOI:10.1007/BF02857926URL [本文引用: 1]
DOI:10.1038/s41598-017-09377-wURLPMID:28819142 [本文引用: 1]

Quantification of genetically modified organisms (GMOs) in food and feed products is often required for their labelling or for tolerance thresholds. Standard-curve-based simplex quantitative polymerase chain reaction (qPCR) is the prevailing technology, which is often combined with screening analysis. With the rapidly growing number of GMOs on the world market, qPCR analysis becomes laborious and expensive. Innovative cost-effective approaches are therefore urgently needed. Here, we report the development and inter-laboratory assessment of multiplex assays to quantify GMO soybean using droplet digital PCR (ddPCR). The assays were developed to facilitate testing of foods and feed for compliance with current GMO regulations in the European Union (EU). Within the EU, the threshold for labelling is 0.9% for authorised GMOs per ingredient. Furthermore, the EU has set a technical zero tolerance limit of 0.1% for certain unauthorised GMOs. The novel multiplex ddPCR assays developed target 11 GMO soybean lines that are currently authorised, and four that are tolerated, pending authorisation in the EU. Potential significant improvements in cost efficiency are demonstrated. Performance was assessed for the critical parameters, including limits of detection and quantification, and trueness, repeatability, and robustness. Inter-laboratory performance was also determined on a number of proficiency programme and real-life samples.
DOI:10.1016/j.biotechadv.2009.05.025URL [本文引用: 1]

Abstract
This paper presents an overview of GMO testing methodologies and how these have evolved and may evolve in the next decade. Challenges and limitations for the application of the test methods as well as to the interpretation of results produced with the methods are highlighted and discussed, bearing in mind the various interests and competences of the involved stakeholders. To better understand the suitability and limitations of detection methodologies the evolution of transformation processes for creation of GMOs is briefly reviewed.URL [本文引用: 1]

转基因玉米(Zea mays) MON88017是美国孟山都公司通过DNA重组技术开发的具有抗玉米根叶甲虫(Diabrotica virgifera)和抗草甘膦类除草剂特性的玉米品系, 本研究的目的是建立该品系的转化事件特异性定性检测方法的标准。根据转基因玉米MON88017的旁侧序列信息,在左边界的结合位点处设计一系列引物组合,在筛选出最佳引物组合后,优化了该引物组合的反应体系,建立了转基因玉米MON88017转化事件特异性定性PCR检测方法。全国7家实验室的验证结果表明,该方法能够特异性检测样品中MON88017转化事件,检测灵敏度均达到0.10%以上,且检测结果具有良好的可重复性、可重现性。本方法符合标准检测特异性强、灵敏度高、重复性好的要求,为我国抗虫耐除草剂玉米MON88017检测和转基因生物安全管理制度的实施提供了相应的技术支撑。
URL [本文引用: 1]

转基因玉米(Zea mays) MON88017是美国孟山都公司通过DNA重组技术开发的具有抗玉米根叶甲虫(Diabrotica virgifera)和抗草甘膦类除草剂特性的玉米品系, 本研究的目的是建立该品系的转化事件特异性定性检测方法的标准。根据转基因玉米MON88017的旁侧序列信息,在左边界的结合位点处设计一系列引物组合,在筛选出最佳引物组合后,优化了该引物组合的反应体系,建立了转基因玉米MON88017转化事件特异性定性PCR检测方法。全国7家实验室的验证结果表明,该方法能够特异性检测样品中MON88017转化事件,检测灵敏度均达到0.10%以上,且检测结果具有良好的可重复性、可重现性。本方法符合标准检测特异性强、灵敏度高、重复性好的要求,为我国抗虫耐除草剂玉米MON88017检测和转基因生物安全管理制度的实施提供了相应的技术支撑。
DOI:10.1021/jf035052xURLPMID:15161182 [本文引用: 2]

The detection of genetically modified organisms (GMOs) in food and feed is an important issue for all the subjects involved in raw material control, food industry, and distribution. Because the number of GMOs authorized in the EU increased during the past few years, there is a need for methods that allow a rapid screening of products. In this paper, we propose a method for the simultaneous detection of four transgenic maize (MON810, Bt11, Bt 176, and GA21) and one transgenic soybean (Roundup Ready), which allows routine control analyses to be sped up. DNA was extracted either from maize and soybean seeds and leaves or reference materials, and the recombinant DNA target sequences were detected with 7 primer pairs, accurately designed to be highly specific for each investigated transgene. Cross and negative controls were performed to ensure the specificity of each primer pair. The method was validated on an interlaboratory ring test and good analytical parameters were obtained (LOD = 0.25%, Repeatability, (r) = 1; Reproducibility, (R) = 0.9). The method was then applied to a model biscuit made of transgenic materials baked for the purpose and to real samples such as feed and foodstuffs. On account of the high recognition specificity and the good detection limits, this multiplex PCR represents a fast and reliable screening method directly applicable in all the laboratories involved in raw material and food control.
URLPMID:11835531 [本文引用: 1]
URL [本文引用: 1]

应用PCR检测方法 ,以大豆凝集素 (lectin)基因为内置标准 ,检测了抗草甘膦转基因大豆中的外源CaMV35S启动子、NOS终止子和CP4 EPSPS成分 ,并通过Southern杂交进一步证实了PCR检测结果的可靠性 ,建立了基于PCR的抗草甘膦转基因大豆检测方法 ,最低检测限度为 0 .0 5 %。用此方法检测转基因情况未知 ,分别来自欧盟、美国、阿根廷、未知国别的进口大豆及未知国别的进口豆粕 ,除从欧盟进口的大豆无CaMV35S启动子、NOS终止子、CP4 EPSPS基因成分外 ,其余样品均含有上述成分 ;而从国内非转基因大豆中未检出上述成分。用此方法检测抗草甘膦大豆京引D1×豫豆 12的F2 和F2∶5群体植株 ,PCR检测结果与大田施药 (草甘膦有效成分 1.0kg·ha-1)检测结果一致率分别达 10 0 %和 93.0 %。
URL [本文引用: 1]

应用PCR检测方法 ,以大豆凝集素 (lectin)基因为内置标准 ,检测了抗草甘膦转基因大豆中的外源CaMV35S启动子、NOS终止子和CP4 EPSPS成分 ,并通过Southern杂交进一步证实了PCR检测结果的可靠性 ,建立了基于PCR的抗草甘膦转基因大豆检测方法 ,最低检测限度为 0 .0 5 %。用此方法检测转基因情况未知 ,分别来自欧盟、美国、阿根廷、未知国别的进口大豆及未知国别的进口豆粕 ,除从欧盟进口的大豆无CaMV35S启动子、NOS终止子、CP4 EPSPS基因成分外 ,其余样品均含有上述成分 ;而从国内非转基因大豆中未检出上述成分。用此方法检测抗草甘膦大豆京引D1×豫豆 12的F2 和F2∶5群体植株 ,PCR检测结果与大田施药 (草甘膦有效成分 1.0kg·ha-1)检测结果一致率分别达 10 0 %和 93.0 %。
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1002/jsfa.6625URL

BACKGROUND: There are about 80 biotech crop events that have been approved by safety assessment in Korea. They have been controlled by genetically modified organism (GMO) and livingmodified organism (LMO) labeling systems. The DNA-based detection method has been used as an efficient scientific management tool. Recently, the multiplex polymerase chain reaction (PCR) and DNA chip have been developed as simultaneous detection methods for several biotech crops' events.
RESULTS: The event-specific multiplex PCR method was developed to detect five biotech maize events: MIR604, Event 3272, LY 038, MON 88017 and DAS-59122-7. The specificity was confirmed and the sensitivity was 0.5%. The screening DNA chip was developed from four endogenous genes of soybean, maize, cotton and canola respectively along with two regulatory elements and seven genes: P35S, tNOS, pat, bar, epsps1, epsps2, pmi, cry1Ac and cry3B. The specificity was confirmed and the sensitivity was 0.5% for four crops' 12 events: one soybean, six maize, three cotton and two canola events.
CONCLUSION: The multiplex PCR and DNA chip can be available for screening, gene-specific and event-specific analysis of biotech crops as efficient detection methods by saving on workload and time. (C) 2014 Society of Chemical Industry
DOI:10.1016/j.foodchem.2011.11.096URL

Currently, the detection methods with lower cost and higher throughput are the major trend in screening genetically modified (GM) food or feed before specific identification. In this study, we developed a quadruplex degenerate PCR screening approach for more than 90 approved GMO events. This assay is consisted of four PCR systems targeting on nine DNA sequences from eight trait genes widely introduced into GMOs, such as CP4-EPSPS derived from Acetobacterium tumefaciens sp. strain CP4, phosphinothricin acetyltransferase gene derived from Streptomyces hygroscopicus (bar) and Streptomyces viridochromogenes (pat), and Cry1Ab, Cry1Ac, Cry1A(b/c), mCry3A, and Cry3Bb1 derived from Bacillus thuringiensis. The quadruplex degenerate PCR assay offers high specificity and sensitivity with the absolute limit of detection (LOD) of approximate 80 target copies. Furthermore, the applicability of the quadruplex PCR assay was confirmed by screening either several artificially prepared samples or samples of Grain Inspection, Packers and Stockyards Administration (GIPSA) proficiency program. (C) 2011 Elsevier Ltd.
DOI:10.1021/jf0341159URLPMID:13129280 [本文引用: 1]

Multiplex PCR procedures were developed for simultaneously detecting multiple target sequences in genetically modified (GM) soybean (Roundup Ready), maize (event 176, Bt11, Mon810, T14/25), and canola (GT73, HCN92/28, MS8/RF3, Oxy 235). Internal control targets (invertase gene in corn, lectin and beta-actin genes in soybean, and cruciferin gene in canola) were included as appropriate to assess the efficiency of all reactions, thereby eliminating any false negatives. Primer combinations that allowed the identification of specific lines were used. In one system of identification, simultaneous amplification profiling (SAP), rather than target specific detection, was used for the identification of four GM maize lines. SAP is simple and has the potential to identify both approved and nonapproved GM lines. The template concentration was identified as a critical factor affecting efficient multiplex PCRs. In canola, 75 ng of DNA template was more effective than 50 ng of DNA for the simultaneous amplification of all targets in a reaction volume of 25 microL. Reliable identification of GM canola was achieved at a DNA concentration of 3 ng/microL, and at 0.1% for GM soybean, indicating high levels of sensitivity. Nonspecific amplification was utilized in this study as a tool for specific and reliable identification of one line of GM maize. The primer cry1A 4-3' (antisense primer) recognizes two sites on the DNA template extracted from GM transgenic maize containing event 176 (European corn borer resistant), resulting in the amplification of products of 152 bp (expected) and 485 bp (unexpected). The latter fragment was sequenced and confirmed to be Cry1A specific. The systems described herein represent simple, accurate, and sensitive GMO detection methods in which only one reaction is necessary to detect multiple GM target sequences that can be reliably used for the identification of specific lines of GMOs.
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.foodcont.2011.03.018URL [本文引用: 1]

The use of genetically modified organisms (GMOs) as food products becomes more and more widespread. The European Union has implemented a set of very strict procedures for the approval to grow, import and/or utilize GMOs as food or food ingredients. Thus, analytical methods for detection of the GMOs are necessary in order to verify compliance with labeling requirements. There are few effective screening methods for highly processed GM (genetically modified) products. Four genes (CP4-EPSPS, Cry1A(b), BAR, and, PAT) are common exogenous genes used in commercialized transgenic soybean, maize, and rice. In the present study, a multiplex nested polymerase chain reaction (PCR) method was developed to simultaneously detect the four exogenous genes and one endogenous gene in two runs. We tested eleven representative highly processed products samples (soya lecithin, soya protein powder, chocolate beverage, infant rice cereal, soybean refine oil, soybean salad oil, maize oil, maize protein powder, maize starch, maize jam) using the developed method, and amplicons of endogenous gene and transgenic fragments were obtained from all the processed products except for soybean refined oil, soybean salad oil and maize oil, and the sensitivity was 0.005%. These results indicate that multiplex nested PCR is appropriate for qualitative detection of transgenic soybean, maize and rice in highly processed products except for refined oil. (C) 2011 Elsevier Ltd.
DOI:10.1016/j.foodcont.2012.10.015URL [本文引用: 1]

Genetically modified (GM) soybean and derived products make up a large part of the biotech-derived food and feed market. As more GM soybean varieties have been approved for commercialization, labeling requirement by South Korea and other countries needs the technical testing methods. This paper reports the development of a multiplex PCR method for identifying six commercialized GM soybean events using the event-specific fragment. Event specific primers targeting Roundup Ready Soybean (RRS, GTS40-3-2), A2704-12, DP356043-5, MON89788, A5547-127, and DP305423-1 were designed, and a multiplex PCR assay consisting of six event-specific fragments and one endogenous lectin fragment was developed. The specificity of the event-specific PCR method was confirmed using 20 GM events of maize, soybean, cotton, and canola. The limit of detection (LOD) for each event in the multiplex PCR is approximately 0.05%. Intra-lab validation by two different operators,confirmed the specificity and LOD of this multiplex PCR method. The method was used to test 30 soybean-derived foods from South Korean and US markets, and results revealed three varieties of GM soybean (RRS, A2704-12, and MON89788) in 19 of the 30 food samples tested. This work provides an efficient and cost-effective approach for event-specific analysis of six commercialized GM soybean varieties and related processed foods in Korea. (c) 2012 Elsevier Ltd.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.biotechadv.2009.05.025URL [本文引用: 1]

Abstract
This paper presents an overview of GMO testing methodologies and how these have evolved and may evolve in the next decade. Challenges and limitations for the application of the test methods as well as to the interpretation of results produced with the methods are highlighted and discussed, bearing in mind the various interests and competences of the involved stakeholders. To better understand the suitability and limitations of detection methodologies the evolution of transformation processes for creation of GMOs is briefly reviewed.URLPMID:17199308 [本文引用: 1]
DOI:10.1002/jsfa.7913URLPMID:27436567 [本文引用: 1]

BACKGROUND: As event-specific sequence information for most unauthorised genetically modified organisms (GMOs) is currently still unavailable, detecting unauthorised GMOs remains challenging. Here, we used insect-resistant rice TT51-1 as an example to develop a novel approach via detecting GMOs by RNA-seq (sequencing) and PCR. RNA-seq of TT51-1 generated 4.8 million (M) 21-nt cDNA tags. Alignment to the Oryza sativa subsp. japonica reference genome revealed 24 098 unmapped tags. Foreign tags from the nopaline synthetic enzyme gene (NOS) terminator and insect-resistant genes were then identified by searching against the NCBI VecScreen and NT databases. RESULTS: To further isolate foreign DNA sequences, putative NOS terminator and insect-resistant gene tags were combined and used directly as primer pairs for long-range PCR, producing a 5016-bp fragment. The inserted DNA sequence of TT51-1 has been submitted to a database, and thus, similarity analysis using the database could identify a test sample. CONCLUSION: The novel approach has a great potential for application to the detection and identification of unauthorised GMOs in food and feed products. (c) 2016 Society of Chemical Industry.
DOI:10.1021/jf100466nURLPMID:20687600 [本文引用: 1]

The genetically modified (GM) Bt crops expressing delta-endotoxins from Bacillus thuringiensis provide protection against a wide range of lepidopteron insect pests throughout the growing season of the plant. Bt cotton is the only commercialized crop in India that is planted on an area of 7.6 million hectares. With the increase in development and commercialization of transgenic crops, it is necessary to develop appropriate qualitative and quantitative methods for detection of different transgenic events. The present study reports on the development of a decaplex polymerase chain reaction (PCR) assay for simultaneous detection of transgene sequences, specific transgene constructs, and endogenous stearoyl acyl desaturase (Sad1) gene in two events of Bt cotton, i.e., MON531 and MON15985. The decaplex PCR assay is an efficient tool to identify and discriminate the two major commercialized events of Bt cotton, i.e., MON531 and MON15985, in India. Real-time PCR assays were also developed for quantification of cry1Ac and cry2Ab genes being employed in these two events. The quantitative method was developed using seven serial dilutions containing different levels of Bt cotton DNA mixed with a non-Bt counterpart ranging from 0.01 to 100%. The results revealed that the biases from the true value and the relative standard deviations were all within the range of +/-20%. The limit of quantification (LOQ) of the developed real-time PCR method has also been established up to 0.01%.
URLPMID:15951030 [本文引用: 1]
URLPMID:25697653 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
