 ,, 刘澳星, 米思远, 李想, 罗汉鹏, 鄢新义, 王雅春
,, 刘澳星, 米思远, 李想, 罗汉鹏, 鄢新义, 王雅春 ,中国农业大学动物科学技术学院,北京 100193
,中国农业大学动物科学技术学院,北京 100193A Review on Longevity Trait in Dairy Cattle Breeding
ZHANG HaiLiang ,, LIU AoXing, MI SiYuan, LI Xiang, LUO HanPeng, YAN XinYi, WANG YaChun
,, LIU AoXing, MI SiYuan, LI Xiang, LUO HanPeng, YAN XinYi, WANG YaChun ,College of Animal Science and Technology, China Agricultural University, Beijing 100193
,College of Animal Science and Technology, China Agricultural University, Beijing 100193通讯作者:
责任编辑: 林鉴非
收稿日期:2019-11-8接受日期:2020-01-13网络出版日期:2020-10-01
| 基金资助: |
Received:2019-11-8Accepted:2020-01-13Online:2020-10-01
作者简介 About authors
张海亮,Tel:18813071851;E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (569KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张海亮, 刘澳星, 米思远, 李想, 罗汉鹏, 鄢新义, 王雅春. 奶牛育种中的长寿性状[J]. 中国农业科学, 2020, 53(19): 4070-4082 doi:10.3864/j.issn.0578-1752.2020.19.019
ZHANG HaiLiang, LIU AoXing, MI SiYuan, LI Xiang, LUO HanPeng, YAN XinYi, WANG YaChun.
长寿是奶牛生产盈利的基础要素,除了产量性状之外,长寿性状具有最大的经济价值,是奶牛育种中最重要的功能性状之一。每头奶牛只有弥补了其在后备牛阶段的培育成本之后,才有可能为牧场带来收益。一般认为奶牛在第二个泌乳期能够保持收支平衡,只有达到第三个泌乳期及以上,才能够获得较高的利润率[1]。提高奶牛长寿性,可以延长奶牛盈利期的长度,获得更高的收益;可以减少后备牛的饲养头数,降低牛群更新的成本;在牛群规模不变的情况下,可以允许牧场进行更高比例的主动淘汰,增加选择强度,进而加快牛群的遗传进展;牛群更加长寿意味着群体的健康水平更高,牧场的兽药和兽医投入也会更少[2,3,4]。
影响奶牛长寿性的因素有很多,所有能够影响奶牛淘汰的因素均会对奶牛长寿性产生影响。奶牛淘汰时所记录的淘汰原因多种多样,按照牧场管理者淘汰时的意愿,可以分为主动淘汰和被动淘汰[5]。主动淘汰的原因主要有年老、低产或育种计划等,被动淘汰则包括繁殖障碍、产后疾病、乳房疾病、肢蹄疾病、消化代谢疾病、外伤等,这些疾病和外界因素将导致奶牛生产能力下降或消失,甚至个体死亡,从而影响奶牛的长寿性。由于长寿性影响因素复杂、性状定义多样、数据分布特殊、性状表现晚、遗传力低(大多在0.02—0.15[6,7,8])等特点,长寿性状是奶牛育种中最复杂、选育最困难的性状之一。本文将从长寿性状的定义、遗传评估、与其他性状的关系、遗传标记、选育方法等方面对长寿性状进行概述,以期能帮助我国相关研究人员系统地了解奶牛长寿性状,对我国奶牛长寿性状的研究和选育提供参考和借鉴。
1 长寿性状的定义及遗传评估
1.1 长寿性状定义
根据观察值的数据类型,可将各种长寿性状的定义分为两类。第一类定义以奶牛在牧场中整个生存周期或某一(几)个生存阶段中的生存时间(单位为天、月、胎次等)作为观察值[9,10],包括在群寿命(herd life)、生产寿命(productive life)、产奶寿命(milking life)和产犊次数(胎次数)等。在群寿命代表了一头奶牛从出生到离群的整个过程中,在牧场中度过的时间;生产寿命代表了一头奶牛从头胎产犊到离群,在牧场中度过的时间;在生产寿命的基础上,去除奶牛各胎次的干奶期长度,即为产奶寿命。第二类定义以个体在某一(几)个生存阶段内生存或死亡的状态(survival)作为观察值,作为二分类阈性状(0或1)[11,12,13],可将此类性状称为某阶段(观测时间点)存活率。为了能与以时间长度定义的第一类长寿性状保持一致,通常将存活定义为1,淘汰或死亡定义为0。在计算个体的表型时,不同定义的性状所需要的信息不同;在选育中,不同定义的长寿性状对奶牛的侧重点也不尽相同。此外,对长寿性状进行遗传评估时,根据是否校正个体产量水平,可将长寿性状分为功能长寿(functional longevity,校正产量)和真实长寿(true longevity,不校正产量)[11]。功能长寿代表了奶牛抗被动淘汰的能力,而真实长寿则同时代表了奶牛抗被动淘汰和抗主动淘汰的能力。根据长寿性状的信息组成,可将长寿性状分为直接长寿和间接长寿。直接长寿利用奶牛的生存信息定义性状,直接描述了一头奶牛的存活状态或存活时间;而间接长寿则利用各种能够早期测定且与直接长寿遗传相关较高的其他性状,间接预测了一头奶牛的生存能力[13]。
1.2 长寿性状遗传评估
针对不同定义的各种长寿性状,不同国家和研究单位使用不同的方法对长寿性状进行遗传评估,常用的模型有线性模型、阈模型[14]、随机回归模型[15,16]和生存分析模型等[17,18]。线性模型可以对以时间长度定义的寿命类或以存活状态定义的生存率类长寿性状进行遗传评估,阈模型可以对存活率类性状进行遗传评估。这两种模型的数据准备过程简单,可选用的评估软件较多,各种软件的计算速度通常较快,其评估结果一般能够满足公牛排队中对育种值估计可靠性和参数估计稳定性的要求。对存活率类长寿性状进行遗传评估时,线性模型和阈模型评估结果之间的秩相关高达0.90以上[14],这两种评估模型对公牛排队的影响较小;使用相同软件时,线性模型的计算速度通常更快。生存分析是研究生存数据及其统计规律的一种分析方法,生存数据描述了从事件起始到事件结束之间的时间长度,长寿性状的观测值是一类典型的生存数据。根据模型特点,可将生存分析模型划分为参数模型、半参数模型和非参数模型,常用的参数模型有指数分布模型、对数逻辑斯谛分布模型、威布尔分布模型、伽玛分布模型、对数正态分布模型等。根据比例风险模型中基础风险函数的分布,可划分为基础风险函数为常数的指数比例风险模型、基础风险函数服从Weibull 分布的Weibull比例风险模型、基础风险函数任意的Cox比例风险模型等。生存分析能更好地拟合长寿性状的数据分布,且能利用存活个体的数据(删失数据),其评估结果的可靠性通常优于其他方法[7];但该方法也存在计算速度慢、表型数据完整度要求较高、数据准备复杂等限制因素。此外,受软件计算能力限制,使用参数模型的生存分析目前无法使用动物模型进行评估。评估生产寿命性状时,线性模型与生存分析模型评估结果的差异主要来自于所用数据的差异;数据相同时(例如,生存分析中不考虑删失数据),两种方法评估结果的秩相关可达0.90以上[19,20]。
2 长寿性状与其他性状的关系
出于对长寿性状进行间接选择的考虑,长寿性状与其他性状之间的关系一直是长寿性状的研究重点。通过不同的方法,长寿性状与体型、产量、繁殖、健康、管理等五大类性状之间的关系被大量报道。例如,采用多性状遗传分析的方法,探究长寿性状与其他性状之间的遗传相关;采用生存分析的方法,研究各性状的变异对奶牛淘汰风险的影响,确定各性状能够解释长寿性状变异的相对大小;利用单性状模型分别评估获得的育种值,计算长寿性状与其他性状之间的近似遗传相关;利用回归的方法确定长寿性状与其他性状的关系等。2.1 长寿性状与其他性状的遗传关系
在荷斯坦牛[21,22]、西门塔尔牛[23]、更赛牛[24]和瑞士褐牛[25]等常见乳用品种中,长寿性状与部分线性体型性状之间均存在低到中等的遗传相关。本文收集分析了各国奶牛群体中长寿性状与主要线性体型性状之间的遗传相关,长寿性状与各体型性状的遗传相关范围如图1所示。在不同群体中,泌乳系统相关的线性体型性状与长寿性状存在较高的遗传相关,如泌乳系统得分、乳房深度、前乳头位置、后乳房高度、后乳房宽度等[26,27,28];与运动相关的体型性状与长寿性之间的遗传相关较低,甚至接近于零,如肢蹄、骨质地、蹄角度、后肢后视等[29];与体躯大小相关的线性体型性状与长寿性状之间多呈低到中等的负遗传相关,如体躯容量、体高、体深等。同一群体中,随着牛只出生年份的变化,荷斯坦牛线性体型性状与长寿性状之间的遗传相关也在不断变化[21, 28]。图1
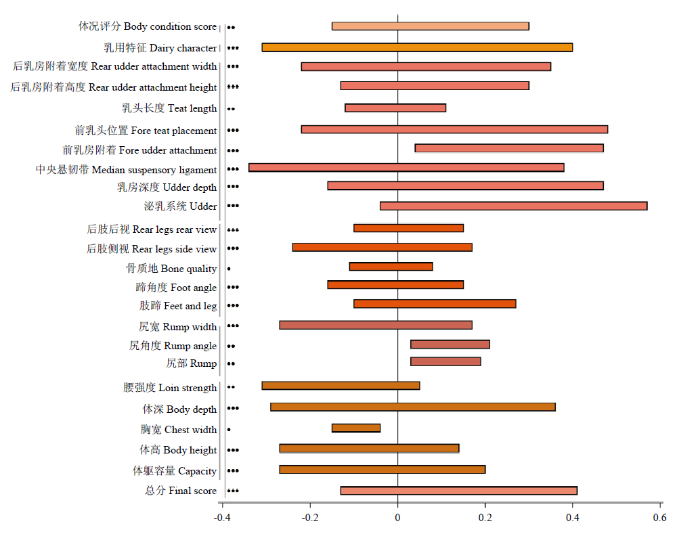 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1长寿性状与体型性状的遗传相关范围
各体型性状均为奶牛线性体型鉴定性状;以存活状态定义的长寿性状统一转换为存活率(将存活定义为1),其估计育种值的有利方向与以时间长度定义的长寿性状一致;图中“


Fig. 1Genetic correlations between longevity and type traits
All confirmation traits are linear type traits of dairy cattle. The longevity traits defined as the survival status are uniformly converted into the survival rate (the survival is defined as 1), and the favorable direction of its EBV is consistent with the longevity traits defined as the life length. In figure 1, “

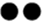

本文收集分析了各国奶牛群体中常见的产量、繁殖、健康和管理性状与长寿性状之间的遗传相关,长寿性状与各性状的遗传相关范围如图2所示。在各国群体中,产量性状与长寿性状间的遗传相关范围较大。在一些群体中,基于头胎产量水平进行的大量主动淘汰造成长寿性状和产量性状呈现较高的正遗传相关[26, 30];当使用校正头胎产量的功能长寿时,长寿性状与产量性状之间的遗传相关明显降低。随着对牛群产量的高强度选育,高产给予奶牛对抗主动淘汰的优势不再明显。例如,在1979—1993年间出生的美国荷斯坦牛中,产量性状与生产寿命之间的遗传相关逐渐减小[28]。与低产奶牛相比,高产奶牛抗被动淘汰的能力更差;在部分群体中,产量性状与功能长寿之间存在负遗传相关[25]。在各国群体中,繁殖性状与长寿性状存在低到中等的遗传相关,繁殖性能表现更好的母牛,其长寿性表现更好。疾病是引起奶牛淘汰的重要因素,奶牛乳房炎和部分繁殖疾病(繁殖障碍、卵巢囊肿)与长寿性状存在低到中等的负遗传相关[30,31,32,33],奶牛疾病抗性越强则长寿性表现越好。此外,与体细胞评分相比,临床乳房炎与长寿性之间的遗传相关更高[32,33,34]。
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2长寿性状与生产、繁殖、健康和管理性状的遗传相关范围
以存活状态定义的长寿性状统一转换为存活率(将存活定义为1),其估计育种值的有利方向与以时间长度定义的长寿性状一致;不返情率、临床乳房炎和繁殖疾病分别以不返情、临床乳房炎发病、繁殖疾病发病定义为1;图中“

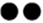

Fig. 2Genetic correlations between longevity and production, fertility, health and workability traits
The longevity traits defined as the survival status are uniformly converted into the survival rate (the survival is defined as 1), and the favorable direction of its EBV is consistent with the longevity traits defined as the life length. Among the non-return rate, clinical mastitis, and fertility diseases, cows with no return, clinical mastitis, or reproductive diseases respectively were defined as 1. In figure 2, “


2.2 长寿性状与其他性状之间的表型关系
采用生存分析的方法可以分析各性状的变异对长寿性的影响,尤其适用于各种分类性状,如线性体型性状、疾病性状和部分繁殖性状等。通过比较模型中纳入各性状时模型似然率的变化,可以估计各性状能够解释长寿性状变异的相对大小;通过计算各性状不同水平的相对淘汰风险(OR),可以反应各性状的变异对长寿性的具体影响。在各线性体型性状中,泌乳系统相关的性状对长寿性有最大的影响,如乳房深度、前乳房附着、后乳房高度、中央悬韧带等性状[35,36,37,38];除乳头长度外,泌乳系统相关性状表现更好的奶牛,其相对淘汰风险越低[35]。对于体躯容量性状,荷斯坦牛中,拥有中等体型的奶牛,其相对淘汰风险更低[35, 38];而在娟姗牛和爱尔夏等体型较小的品种中,体型更大的奶牛,其淘汰风险更低[39]。体躯容量相关的性状受牛群遗传改良方向的影响较大,在不同群体间,同一种性状对长寿性的影响差异较大。此外,乳用特征、泌乳速度等体型性状对长寿性也有较大的影响。
在荷斯坦牛、娟姗牛和爱尔夏牛中,产犊难易、犊牛大小、授精次数、产后首配日龄、首配至妊娠间隔及空怀天数等繁殖或产犊性状对功能生产寿命有显著影响,繁殖性状表现越好的奶牛,其相对淘汰风险越低[40]。例如,产犊间隔、空怀天数和首配至妊娠间隔越短,奶牛淘汰风险越低。体细胞评分对功能生产寿命有显著的影响,母牛的淘汰风险随体细胞评分的升高而增加,高体细胞评分奶牛的淘汰风险可达到牛群平均体细胞评分个体的4—6倍[41];与高体细胞数牛场相比,个体体细胞数相同幅度的增加,低体细胞数牛场的个体淘汰风险的增加幅度更大[42];与健康牛相比,泌乳期内任何时间患乳房炎均会造成奶牛淘汰风险的增加,泌乳高峰期时淘汰风险的增加幅度最大[34]。此外,通过生存分析的方法,发现泌乳性情[43]、泌乳速度[43]、出生时母亲年龄[44]、牛奶尿素氮含量[45]、乳糖率[45]、近交率[46]等性状的变异对荷斯坦牛、爱尔夏牛和娟姗牛的长寿性存在显著影响。
3 长寿性状的相关基因及分子标记
3.1 长寿性状候选基因关联分析
筛查前人通过QTL定位的结果或根据基因已知的生理功能初步筛选长寿性状潜在的候选基因,通过候选基因关联分析的方法,可以评估目标基因与长寿性状的关系,进而发现长寿性状的候选基因。在长寿性状的候选基因关联分析中,通常使用生产寿命育种值作为关联表型,使用线性混合模型或生存分析模型进行关联分析,各研究中选择进行关联分析的基因大多为已报道的影响奶牛产奶性能、健康状况和繁殖性能的基因。在不同群体中,α酪蛋白s1(casein alpha s1,CSN1S1)、催乳素受体(prolactin receptor,PRLR)、奶脂肪球-EGF因子蛋白8(milk fat globule-EGF factor 8 protein,MFGE8)、非受体酪氨酸激酶,SRC原癌基因(SRC proto-oncogene, non-receptor tyrosine kinase,SRC)、二酰基甘油O-酰基转移酶1(diacylglycerol O-acyltransferase 1,DGAT1)、β酪蛋白(casein beta,CSN2)[47]、瘦素(leptin,LEP)[48]、硬脂酰辅酶A去饱和酶(stearoyl-CoA desaturase,SCD1)[49]、丝氨酸蛋白酶抑制因子,分支A(α-1抗蛋白酶,抗胰蛋白酶),成员14 (serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin),member 14,SERPINA14,别称UTMP)[50]、钙抑素(calpastatin,CAST)[51]和serpin家族A成员1(serpin family A member 1,PI)[52]等基因存在与长寿性状显著关联的SNP或单倍型,这些基因可作为长寿性状的候选基因。在部分发达国家的奶牛群体中,因低产而进行的主动淘汰是影响奶牛长寿性的重要原因;由于对产量性状的影响,可能造成了FRLP[53]和DGAT1[54,55]等基因在特定群体中间接影响了长寿性状;在波兰群体中,当校正个体产量水平的影响后,DGAT1则对长寿性状没有显著的影响[48]。酪蛋白家族基因在奶牛的泌乳和健康中扮演着主要的角色[53,54,55,56],CSN1S1和CSN2与长寿性状间的关系进一步证明了这一点。在不同群体中,LEP与奶牛的适应性、代谢和健康之间的具体关系存在争议,其中一些研究报道了LEP与疾病风险[57]、死产[58]和犊牛存活率[58]等性状之间存在显著的关系,而这些性状导致的奶牛淘汰均是长寿性状的重要组成部分。
3.2 长寿性状全基因组关联分析
全基因组关联分析(genome-wide association analysis,GWAS)是一种鉴定目标性状相关分子标记的有效方法,各国研究人员采用不同的GWAS方法鉴定了大量与长寿性状显著相关的分子标记(表1),这些信息为长寿性状的标记辅助选择和基因组选择提供了有用信息,同时也为阐释长寿性状的分子遗传基础奠定了基础。在长寿性状的GWAS中,以存活时间长度为单位的生产寿命和在群寿命是最常用的性状定义方法,单标记回归是最常用的关联分析方法,逆回归育种值则是最常用的关联分析伪表型。Table 1
表1
表1长寿性状全基因组关联分析研究
Table 1
| 作者 Author | 品种 Breed | 性状定义 Trait definition | 关联分析表型 Phenotype | 分析方法 Analysis method | 显著SNP所在染色体 Chromosome of significant SNP | 候选基因 Candidate gene |
|---|---|---|---|---|---|---|
| STERI [61] | 荷斯坦牛 Holstein | 生产寿命 Productive life | 表型离差 Trait deviations | 简单回归 Simple regression | 16、30 | UBIAD1、MTOR、ANGPTL7、EXOSC10、SRM、MASP2、TARDBP、CASZ1、GPC3、PHF6 |
| 在群寿命 Herd life | — | |||||
| 产犊次数 The number of calving | 16、30 | |||||
| NAYERI [60] | 荷斯坦牛 Holstein | 在群寿命 Herd life | 逆回归育种值 Deregressed proof | 单标记回归 Single SNP regression | 5、6、7、14、18、 20、21 | CTU1、NPFFR2 |
| 间接长寿性 Indirect longevity | 5、6、18 | NPFFR2 | ||||
| 存活率 Survival | 6、7、14、18、20、 21 | — | ||||
| HAY [62] | 合成品种 Composite breed | 生产寿命 Productive life | 校正表型 Corrected phenotypes | 单标记回归 Single SNP regression | 1、2、3、8、9、 11、19、25 | GBE1、GTDC1、ZEB2、CTNNA2、REG3A、REG3G、BTBD17、CD300A、CD300LB、CDR2L、FADS6、FDXR、GPRC5C、GRIN2C、HID1 |
| 贝叶斯 Bayesian | 1、2、3、9、11、15、19、20、25、29 | |||||
| SAOWAPHAK[63] | 杂交荷斯坦牛 Cross Holstein | 生产寿命 Productive life | 性状表型值 Phenotype | 一步法 Single step GWAS | 5、X | SYT1、DOCK11、KLHL13、IL13RA1 |
| ZHANG[64] | 荷斯坦牛 Holstein | 多胎次生产寿命 Partial productive life | 逆回归育种值 Deregressed proof | 线性混合模型 Linear mixed model | 5、6、9、10、18、 23 | NPFFR2、vitamin D-buinding protein、PPP1R14C、GCH1、ZNF613 ZNF617 |
| 北欧红牛 Nordic Red dairy cattle | 6、21、23、25 | CCDC149、LGI2、KCNK16 | ||||
| 娟姗牛 Jersey | — | — | ||||
| MéSZáROS [65] | 西门塔尔牛 Simmental | 生产寿命 Productive life | 逆回归育种值 Deregressed proof | 单标记回归 Single SNP regression | 6、13、14、19、21 | DERL1、SNTG1、LOC614437、ALB、AFP、LOC100140503、ADAMTS3、E2F1、RALY、SYT10、NTRK2 |
| COLE[59] | 荷斯坦牛 Holstein | 生产寿命 Productive life | 估计传递力 Predicted transmitting ability | 单标记回归 Single SNP regression | 7、X | INSR、LOC520057 |
新窗口打开|下载CSV
在各关联分析中,在6、14、16、18和X染色体上发现的显著SNP最多,这些SNP大多定位在已知的与奶牛临床乳房炎、体细胞数(评分)、产犊性状(产犊难易、犊牛大小、死产)、繁殖性状、体型性状相关的QTL区间内。如,COLE等[59]利用1 654头荷斯坦母牛进行长寿性GWAS时,分别在7号染色体15.8 Mb处和X染色体106 Mb处发现了两个与体细胞评分、女儿妊娠率共同相关的遗传区域;NAYERI等[60]利用6 116头公牛进行长寿性GWAS时,在13号染色体59 Mb和18号染色体42 — 65 Mb处定位到的遗传区域均与前人发现的产犊性状QTL区域重合。在各国群体中,健康和繁殖问题均是引起奶牛淘汰的主要原因,各种长寿性状实际上度量了奶牛受产量、健康和繁殖等因素影响的存活时间(能力)。因此,对奶牛长寿性状进行的GWAS,大多发现了与上述性状共同相关的遗传标记。
4 长寿性状的选育
4.1 各国奶牛育种体系中的长寿性状
上世纪50年代以来,对长寿性状的大量研究已经证明长寿性是可遗传的,对牛群的长寿性进行选育提高是可能的。上世纪90年代之后,各国奶牛育种协会陆续将长寿性状纳入遗传评估计划,随着对长寿性状的持续研究,长寿性状的定义和遗传评估方法也在不断发展[66]。目前,世界各主要奶业发达国家对长寿性状的选育情况如表2所示。大多数国家的奶牛综合选育指数中都包含长寿性状,其权重多在5%—10%,德国尤其重视奶牛的长寿性选育,其总性能指数(RZG,2018)中长寿性状的权重高达20%。各国对长寿性状的定义和评估方法不尽相同,将长寿性状定义为奶牛在某几个生命阶段的存活状态是最常见的定义方法;大多数国家采用线性模型对各种长寿性状进行遗传评估,以法国为代表的欧洲联合遗传评估组织使用生存分析的方法进行遗传评估,荷兰则使用随机回归的方法对长寿性状进行遗传评估。4.2 长寿性状的间接选择
利用线性模型评估不同定义的直接长寿,需等到公牛的部分后裔淘汰或死亡后,才能获得表型信息;能够利用删失数据的生存分析方法,其估计育种值的可靠性也仅取决于公牛后裔中非删失的女儿数[67],这使得长寿性状的选育十分困难,长寿性状成为了奶牛育种中世代间隔最长的性状。因此,在利用直接长寿进行选择的同时,许多国家也考虑了一些与长寿性状遗传相关较高、并能早期测定的性状进行间接选择,如美国、加拿大、法国和新西兰等(表2)。Table 2
表2
表2各国长寿性状遗传评估及选育情况
Table 2
| 国家/地区 Country/region | 性状定义 Trait definition | 遗传评估方法 Methods of genetic evaluation | 指数中的权重 Weight in index |
|---|---|---|---|
| 加拿大 Canada | 直接长寿:头胎产犊 - 120 d、120- 240 d、240 d-2胎产犊、2胎产犊 - 3胎产犊、3胎产犊- 4胎产犊期间的存活状态,共5个性状 Direct longevity: a total 5 traits, survival status from the 1st calving to day 120, day 120 to day 240, day 240 to 2nd calving, 2nd calving to 3rd calving, and 3rd calving to 4th calving | 五性状 - BLUP - 动物模型 5 traits - BLUP - Animal model | 8%(LPI,2019) |
| 间接长寿:包括体细胞评分、泌乳速度、不返情率、产犊至首配间隔和部分体型性状 Indirect longevity: including somatic cell score, milking speed, non-return rate, calving to first service interval and some type traits | |||
| 美国 US | 直接长寿:生产寿命 Direct longevity: productive life | 单性状 - BLUP - 动物模型 Single trait - BLUP - Animal model | 4%(TPI,2017) |
| 间接长寿:包括体细胞评分、产奶量、乳脂量、乳蛋白量和部分体型性状 Indirect longevity: including somatic cell score, milk yield, milk fat yield, milk protein yield and some type traits | 3%(TPI,2017) | ||
| 荷兰 Netherlands | 头胎产犊后1 - 72月龄期间的存活状态 Survival status from the first calving to the age of 72 months | 单性状 - 随机回归 - BLUP - 动物模型 Single trait - Random regression - BLUP - Animal model | 8%(NVI,2019) |
| 德国 Germany | 前3胎次内泌乳0-49 d、50- 249 d、250 d -下次产犊期间的存活状态,共9个性状 A total of 9 traits, survival status from day 0 to 49, day 50 to 249 and day 250 to the next calving after the current calving among the first 3 lactations | 多性状 - BLUP - 动物模型 Muti-traits - BLUP - Animal model | 20%(RZG,2018) |
| 法国 France | 直接长寿:生产寿命 Direct longevity: productive life | 单性状-生存分析-公畜外祖父模型 Single trait - Survival analysis - Sire-maternal grand sire model | 5%(ISU,2019) |
| 间接长寿:包括体细胞数、临床乳房炎、泌乳速度、部分繁殖和体型性状 Indirect longevity: including somatic cell count, clinical mastitis, some reproduction and type traits | — | ||
| 北欧 Nodic | 头胎产犊至2、3、4、5、6胎产犊时生产寿命,共5个性状 A total of 5 traits, the partial productive life from the first calving to the 2nd, 3rd, 4th, 5th, 6th calving | 多性状 - BLUP - 动物模型 Muti-traits - BLUP - Animal model | 6%(NTM,2018) |
| 新西兰 New Zealand | 直接长寿:头胎产犊至2、3、4胎产犊的存活状态,共3个性状 Direct longevity: a total of 3 traits, survival status from the first calving to the 2nd, 3rd, 4th calving | 多性状 - BLUP - 动物模型 Muti-traits - BLUP - Animal model | 6%(BW,2016) |
| 间接长寿:包括头胎乳蛋白量、头胎体况评分、头胎泌乳速度、头胎体细胞数和部分体型性状等 Indirect longevity: including milk protein yield, body condition score, milking speed and somatic cell count in 1st lactation, and some type traits | — | ||
| 澳大利亚 Australia | 前7胎各胎次内母牛的存活状态,共7个性状 A total of 7 traits, survival status from the first calving to the 2nd, 3rd, 4th, 5th, 6th, 7th, 8th calving | 多性状 - BLUP - 动物模型 Muti-traits - BLUP - Animal model | 8%(BPI,2014) |
新窗口打开|下载CSV
在性状的选择上,早期测定、遗传力较高的线性体型性状成为了长寿性状间接选择的首选性状[68],其中体型总分、泌乳系统、腰强度、尻角度、肢蹄等最为常用;在各国群体中,体细胞评分(数)和临床乳房炎也是间接选择长寿性的常用性状;在美国群体中,因产量水平进行的主动淘汰比例较高,产奶量、乳脂量和乳蛋白量也是长寿性状间接选择的重要性状;此外,泌乳速度和部分繁殖性状也比较常用。
4.3 长寿性状的基因组选择
由于能够缩短世代间隔、提高选择的准确性,加快遗传进展,基因组选择方法已经在越来越多国家的奶牛育种中得以应用[69]。对于表现时间晚、遗传力低的长寿性状,基因组选择的应用潜力巨大。通过建立长寿性状基因组选择的参考群,使长寿性的选育不再依赖被选择个体的后裔淘汰信息,能够大幅缩短长寿性状的世代间隔。目前,许多国家都已经开展了长寿性状基因组选择的研究和应用,奶业发达国家的成功应用证明对长寿性状进行基因组选择是可行的。据报道,德国荷斯坦公牛长寿性状的基因组育种值可靠性可以达到51%,比系谱指数提高了17%[70];美国荷斯坦公牛长寿性状的基因组育种值可靠性可以达到45%,比系谱指数提高了18%[71]。5 展望
据调查,我国规模化奶牛场中荷斯坦牛的平均利用胎次不足三胎[72],远没有达到奶牛生产力最高的阶段,严重影响着奶牛养殖的效益。目前,在奶牛育种中,兼顾高产和健康长寿的平衡育种理念已经成为各国奶牛育种界的共识。在我国,长寿性状还没有纳入中国奶牛选育指数,对长寿性状的研究处于起步阶段,仅有使用局部地区的牛群数据进行的少量研究。例如,分别针对北京地区[10]和上海地区[73]牛群进行的遗传参数估计、使用微卫星标记定位方法[74]和候选基因关联分析方法[75,76,77,78]挖掘长寿性状的遗传标记。目前,我国奶牛群体的长寿性表现较差,不仅严重影响了生产效率,而且也挤压了进行高强度主动淘汰选育其他性状的空间,我国牛群的长寿性状选育刻不容缓。在常规遗传评估中,各国研究人员从模型和性状定义等角度提出了许多对长寿性状进行早期选择的方法;基因组选择技术在奶牛育种中成功应用之后,有效解决了该性状选择世代间隔长的问题;因此,在基因组评估中,长寿性状评估时所使用的定义和模型还需进一步探讨。有研究指出,在基因组选择中,将与目标性状相关的分子标记信息加入到基因组数据中可以提高基因组选择的准确性[79,80];在我国奶牛群体中,挖掘与长寿性状相关的遗传标记可以为该性状的基因组选择和标记辅助选择提供有用的信息。此外,长寿性状的遗传力较低,其受遗传与环境互作效应及气候条件、牧场管理等非遗传因素的影响较大;我国奶牛养殖在全国各地均有分布,不同地区之间生产环境条件和管理模式的差异较大,研究长寿性状中遗传与环境之间的互作具有较大意义。综上,针对我国奶牛群体,有必要统筹全国牛群数据,从长寿性状的遗传参数估计、遗传评估方法论证、遗传标记挖掘、基因组选择方法和选育策略等方面开展系统研究十分必要。随着我国奶牛养殖对长寿性状的不断认识,以及育种数据的不断积累,未来一段时间,长寿性状将会成为我国奶牛遗传育种领域的研究热点,将长寿性状纳入奶牛选育指数是我国奶牛育种的必然趋势。最终,通过遗传选育的手段,改善牛群的长寿性,进而提高奶牛的终生效益,不断提高我国奶牛养殖的竞争力。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
DOI:10.3168/jds.S0022-0302(94)76955-1URLPMID:8120199 [本文引用: 1]

Data on 82,835 Holstein cows, daughters of 703 sires and with first calving from September 1979 to December 1984 from 2384 herds enrolled in the Quebec Dairy Herd Analysis Service, were used to estimate genetic and phenotypic parameters of partial and total lifetime performance traits with REML. The model included herd-year-season of first calving, age at first calving, and proven sires as fixed effects and young sires and residuals as random effects. Individual lactation records were precorrected for year-month of calving before lifetime totals were calculated. Only cows with at least 5 yr of opportunity for production were analyzed. The ranges of heritability estimates were .11 to .13 for lifetime production and profit, .07 to .09 for measures of longevity, and .28 to .32 for yield per day of productive life. Correlations among total lifetime yield and profit traits and among measures of longevity were > or = .93. Genetic and phenotypic correlations, respectively, of early partial (two parities total) with total life-time yield and profit and longevity traits ranged from .81 to .94 and .66 to .78. Selection on early performance seems to be desirable and, given the high positive genetic correlations, should increase both lifetime yield and longevity.
DOI:10.1016/S0301-6226(98)00160-2URL [本文引用: 1]
DOI:10.1017/S135772980001626XURL [本文引用: 1]
URLPMID:12741574 [本文引用: 1]
DOI:10.3168/jds.S0022-0302(05)72696-5URLPMID:15591401 [本文引用: 1]

The objectives of this study were to identify the most important factors that influence functional survival and to estimate the genetic parameters of functional survival for Canadian dairy cattle. Data were obtained from lactation records extracted for the May 2002 genetic evaluation of Holstein, Jersey, and Ayrshire breeds that calved between July 1, 1985 and April 5, 2002. Analysis was performed using a Weibull proportional hazard model, and the baseline hazard function was defined on a lactation basis instead of the traditional analysis of the whole length of life. The statistical model included the effects of stage of lactation; season of production; the annual change in herd size; type of milk recording supervision; age at first calving; effects of milk, fat, and protein yields calculated within herd-year-parity deviations; and the random effects of herd-year-season of calving and sire. All effects fitted in the model had a significant effect on functional survival of cows in all breeds. Milk yield was by far the most important factor influencing survival, and the hazard increased as the milk production of the cows decreased. The hazard also increased as the fat content increased compared with the average group. Heifers that were older at calving were at higher risk of being culled, and expanding herds were at lower risk of being culled compared with stable herds. More culling was found in unsupervised herds than in supervised herds. The heritability values obtained were 0.14, 0.10, and 0.09 for Holstein, Jersey, and Ayrshire, respectively. Rank correlation between estimated breeding values (EBV) obtained from the current national genetic evaluation of direct herd life and the survival kit used in this study ranged from 0.65 to 0.87, depending on the number of daughters per sire. Estimated genetic trend obtained using the survival kit was overestimated.
DOI:10.3168/jds.S0022-0302(04)73298-1URLPMID:15290996 [本文引用: 2]

Predicted transmitting abilities (PTA) of US Jersey sires for daughter longevity were calculated using a Weibull proportional hazards sire model and compared with predictions from a conventional linear animal model. Culling data from 268,008 Jersey cows with first calving from 1981 to 2000 were used. The proportional hazards model included time-dependent effects of herd-year-season contemporary group and parity by stage of lactation interaction, as well as time-independent effects of sire and age at first calving. Sire variances and parameters of the Weibull distribution were estimated, providing heritability estimates of 4.7% on the log scale and 18.0% on the original scale. The PTA of each sire was expressed as the expected risk of culling relative to daughters of an average sire. Risk ratios (RR) ranged from 0.7 to 1.3, indicating that the risk of culling for daughters of the best sires was 30% lower than for daughters of average sires and nearly 50% lower than than for daughters of the poorest sires. Sire PTA from the proportional hazards model were compared with PTA from a linear model similar to that used for routine national genetic evaluation of length of productive life (PL) using cross-validation in independent samples of herds. Models were compared using logistic regression of daughters' stayability to second, third, fourth, or fifth lactation on their sires' PTA values, with alternative approaches for weighting the contribution of each sire. Models were also compared using logistic regression of daughters' stayability to 36, 48, 60, 72, and 84 mo of life. The proportional hazards model generally yielded more accurate predictions according to these criteria, but differences in predictive ability between methods were smaller when using a Kullback-Leibler distance than with other approaches. Results of this study suggest that survival analysis methodology may provide more accurate predictions of genetic merit for longevity than conventional linear models.
DOI:10.3168/jds.S0022-0302(88)79907-5URL [本文引用: 1]
URLPMID:15738249 [本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.3168/jds.S0022-0302(92)77958-2URLPMID:1500592 [本文引用: 2]

Genetic parameters were estimated simultaneously for 5 herd-life traits, 15 conformation (type) traits, and milk yield measured in first lactation for 128,601 Holstein cows. Heritabilities of all traits were higher in registered than in grade cows. Genetic correlations of linear type traits with first lactation yield ranged from -.48 for udder depth to .54 for dairy form. Genetic correlations among milk yield and herd-life traits were all positive except for milk-corrected herd life in grade cows. Udder traits had largest absolute genetic correlations with herd-life traits, followed by body traits and feet and leg traits. Some traits associated with body size and foot angle differed between registered and grade cows. Estimates of genetic trends from obtained parameters revealed greatest progress for milk yield from single-trait selection but also the largest changes for some type traits and milk-corrected herd life in an undesirable direction. Relative milk to type ratios of between 2:1 and 3:1 yielded 90% of the gain in milk yield with no change or slight improvement in type traits and functional herd life. Selection for type traits associated with herd life appears to be warranted to improve days of functional herd life or to decrease involuntary culling of dairy cows.
URLPMID:7738256 [本文引用: 1]
DOI:10.3168/jds.S0022-0302(98)75607-3URLPMID:9532510 [本文引用: 2]

Methods were developed for the national genetic evaluation of herd life for Canadian Holstein sires. The genetic evaluations incorporate information from survival (direct herd life) and information from conformation traits that are related to herd life (indirect herd life) after adjustment for production in first lactation to remove the effect of culling for production. Direct genetic evaluations for herd life were based on survival in each of the first three lactations, which was analyzed using a multiple-trait animal model. Sire evaluations thus obtained for survival in each of the first three lactations were combined based on their economic weights into an overall sire evaluation for direct herd life. Sire evaluations for indirect herd life were based on an index of sire evaluations for mammary system, feet and legs, rump, and capacity. A multiple-trait sire model based on multiple-trait across country evaluation methodology was used to combine direct and indirect genetic evaluations for herd life into an overall genetic evaluation for herd life. Sire evaluations for herd life were expressed in estimated transmitting ability as the number of lactations and represent expected differences among daughters in functional herd life (number of lactations); the average functional herd life was set equal to three lactations. Estimated transmitting abilities were normally distributed and ranged from 2.31 to 3.43 lactations.
DOI:10.3168/jds.S0022-0302(99)75324-5URLPMID:10342243 [本文引用: 2]

Several approaches for analysis of survival in the first three lactations were compared using data from approximately 700,000 Canadian Holsteins. Two approaches (linear model and threshold model) were used to analyze a binary measure of survival. Other approaches were survival analyses to evaluate two measures of the number of days that cows were in milk during their first three lactations. One measure restricted days per lactation to < or = 305; the other was based on the actual number of days in milk without an upper limit on days per lactation. Variance components and breeding values (EBV) were estimated. Sire models were used almost exclusively, but one set of EBV was obtained using a linear animal model. Effects in the models were herd-year of calving, age at first calving, interaction of several factors related to herd, and production. Thus, all EBV were for functional herd life. Heritabilities were approximately 0.04, 0.07, and 0.10 from linear, threshold, and survival analyses, respectively. Correlations among sire EBV from all analyses using sire models were high, particularly for linear and threshold models (0.98). In contrast, correlations of EBV from sire models with EBV from the linear animal model were less than 0.90, regardless of the approach taken. In Canada, the current linear animal model remains in use for sire evaluation of herd life, but research with survival analyses will continue.
DOI:10.1017/S1357729800055491URL [本文引用: 1]
DOI:10.3168/jds.2014-9090URLPMID:25892695 [本文引用: 1]

Longevity, productive life, or lifespan of dairy cattle is an important trait for dairy farmers, and it is defined as the time from first calving to the last test date for milk production. Methods for genetic evaluations need to account for censored data; that is, records from cows that are still alive. The aim of this study was to investigate whether these methods also need to take account of survival being genetically a different trait across the entire lifespan of a cow. The data set comprised 112,000 cows with a total of 3,964,449 observations for survival per month from first calving until 72 mo in productive life. A random regression model with second-order Legendre polynomials was fitted for the additive genetic effect. Alternative parameterizations were (1) different trait definitions for the length of time interval for survival after first calving (1, 3, 6, and 12 mo); (2) linear or threshold model; and (3) differing the order of the Legendre polynomial. The partial derivatives of a profit function were used to transform variance components on the survival scale to those for lifespan. Survival rates were higher in early life than later in life (99 vs. 95%). When survival was defined over 12-mo intervals survival curves were smooth compared with curves when 1-, 3-, or 6-mo intervals were used. Heritabilities in each interval were very low and ranged from 0.002 to 0.031, but the heritability for lifespan over the entire period of 72 mo after first calving ranged from 0.115 to 0.149. Genetic correlations between time intervals ranged from 0.25 to 1.00. Genetic parameters and breeding values for the genetic effect were more sensitive to the trait definition than to whether a linear or threshold model was used or to the order of Legendre polynomial used. Cumulative survival up to the first 6 mo predicted lifespan with an accuracy of only 0.79 to 0.85; that is, reliability of breeding value with many daughters in the first 6 mo can be, at most, 0.62 to 0.72, and changes of breeding values are still expected when daughters are getting older. Therefore, an improved model for genetic evaluation should treat survival as different traits during the lifespan by splitting lifespan in time intervals of 6 mo or less to avoid overestimated reliabilities and changes in breeding values when daughters are getting older.
DOI:10.4314/sajas.v45i2.1URL [本文引用: 1]
URLPMID:23607602 [本文引用: 1]
DOI:10.3168/jds.S0022-0302(98)75897-7URLPMID:9891278 [本文引用: 1]

A comparison was made among breeding values of sires for longevity that were obtained by different methods: phenotypic averages of daughters using only uncensored records, BLUP using only uncensored records, survival analysis using only uncensored records, and survival analysis using both censored and uncensored records. Two data files were used: one contained data from small herds, and the other contained data from large herds. The results from both data files were similar. Different methods of predicting breeding values resulted in different rankings of sires. The results obtained using phenotypic averages were weakly correlated (< or = 0.46) with those results obtained using the other methods of prediction. The REML BLUP had strong correlations (< or = -0.91) with the survival analysis predictor if the same data were used, and correlations weakened (< or = -0.60) when censored records were included in the survival analysis. The correlations are negative because the linear method analyzed longevity, and survival analysis measured the risk of being culled, which has an antagonistic relationship with longevity. The results from REML BLUP and survival analysis methods differed mainly because of the different data that were used (uncensored only versus both censored and uncensored).
DOI:10.1017/S1357729800054667URL [本文引用: 1]
URLPMID:9406094 [本文引用: 2]
DOI:10.17221/CJASURL [本文引用: 1]
DOI:10.17221/CJASURL [本文引用: 1]
DOI:10.3168/jds.S0022-0302(02)74195-7URLPMID:12086068 [本文引用: 1]

Production and type data were used to investigate the relationships of these traits with herd life data in US Guernsey cows that calved from 1985 through 1990. Two definitions of herd life were used: actual days from birth to disposal (true herd life) and herd life adjusted for milk production (functional herd life). Genetic parameters were calculated with data from cows that had an opportunity to reach 84 mo of age (n = 18,725). Linear type traits were preadjusted for stage of lactation and age at classification. True herd life was preadjusted for age at first calving and for functional herd life, within herd-year quartile ranking for milk yield. The (co)variance components for true and functional herd life, milk, fat, protein, and 15 linear type traits were estimated with multiple-trait REML in an animal model. Heritability estimates for true and functional herd life were 0.12 for both traits. Estimated genetic correlations of herd life with body size traits were from -0.14 to -0.29, with feet and leg traits were from -0.10 to 0.06, and with udder traits were from -0.09 to 0.24. These correlation parameters were used for indirect prediction of herd life from available production and type information in Guernseys.
DOI:10.1016/0301-6226(94)00044-8URL [本文引用: 2]
DOI:10.3168/jds.S0022-0302(98)75778-9URLPMID:9710774 [本文引用: 2]

Genetic evaluations for the length of productive life based on actual DHIA culling data have been available in the US since January 1994. Although substantial genetic variation in productive life exists, the reliability of selection is often low, particularly for recently progeny-tested bulls having daughters that have not yet had an opportunity to be culled. Correlated production and conformation traits, which have higher heritability than productive life and are available earlier in life, may be used to enhance evaluations of productive life for young bulls that have little or no direct culling information available. Genetic correlations between productive life and milk, fat, dairy form, and udder traits ranged from +0.22 to +0.46. The maximum reliability of the indirect prediction of productive life from 16 correlated type and production traits was 0.56, and the maximum reliability from a subset of 10 traits was 0.51. Indirect information about productive life that was derived from type and production traits was combined with actual culling information to increase the total amount of available information for many recently progeny-tested bulls. The procedures described herein for enhancing direct evaluations for the productive life of dairy sires with indirect information about production and type were implemented by the USDA Animal Improvement Programs Laboratory and the Holstein Association USA in July 1994.
DOI:10.1023/B:TROP.0000045965.99974.9cURLPMID:15643816 [本文引用: 1]

Relationships between longevity and linear type traits were estimated using data on 34 201 cows with lifetime information and linear type scores. The longevity trait considered was the number of lactations initiated and the linear type traits were rump height, body depth, angularity, rear udder height, fore udder attachment, udder depth, fore teat placement and fore teat length. Fixed effects included in the models were herd year, season of calving and herd–date of classification–classifier and days in milk. Age at first calving and age at classification were included as linear and quadratic covariates. Heritability estimates were low for longevity and moderate for most type traits except rump height and fore teat length. All the phenotypic correlations between longevity and the linear type traits were slightly positive (0.01 to 0.09) except the relationships with rump height and fore teat length which were –0.01 and –0.02, respectively. Genetic correlations between longevity and udder traits as well as angularity were moderate to high and positive (0.22 to 0.48). The only notable negative genetic correlations were longevity with body depth and fore teat length (–0.15 and –0.07, respectively). The genetic correlations suggest that selection for udder traits and angularity should improve longevity in the Holstein cattle population.
DOI:10.3168/jds.S0022-0302(04)73297-XURLPMID:15290995 [本文引用: 3]

Genetic correlations among milk, fat, and protein yields; body size composite (BSC); udder composite (UDC); and productive life (PL) in Holsteins were investigated over time. The data set contained 25,280 records of cows born in Wisconsin between 1979 and 1993. The multiple trait random regression (MT-RR) animal model included registration status, herd-year, age group, and stage of lactation as fixed effects; additive genetic effects with random regressions (RR) on year of birth using the first-order Legendre polynomial; and residual effects. Heterogeneous residual variances were considered in the model. Estimates of variance components and genetic correlations among traits from MT-RR were compared with those estimated with a multiple trait interval (MT-I) model, which assumed that every 3-yr interval was a separate trait and included the same effects as in the MT-RR model except for the RR. Genetic correlations estimated with MT-RR and MT-I models over time among all traits were compared with correlations among breeding values predicted with the single trait (ST) model without RR. Correlations among breeding values predicted with MT-RR, ST, and MT models were also calculated. Additive genetic and residual variances for all traits except PL increased over time; those for PL were constant. As a result, heritability estimates had no significant changes during the 15 yr. Genetic correlations of PL with milk, fat, protein, and BSC declined to zero or negative; those with UDC remained positive. Correlations among breeding values predicted with ST, MT, and MT-RR models were relatively high for all traits except PL. Genetic correlations between PL and other traits varied over time, with some correlations changing sign. For accurate indirect prediction of PL from other traits, the genetic correlations among the traits need to be re-estimated periodically.
DOI:10.3168/jds.S0022-0302(06)72246-9URLPMID:16606749 [本文引用: 1]

The phenotypic and genetic relationships of 3 locomotion traits with profit, production, longevity, and fertility traits were studied to determine the importance of locomotion traits for dairy producers. Two data sets including official milk records and type classification scores of 62,293 cows, and reproductive records of 24,561 cows from the Basque and Navarra Autonomous Regions were analyzed. Higher scores for feet and legs (FL), foot angle (FA), and rear legs set (RLS) were positively related to production and functional traits, whereas fertility was not significantly affected. The cows that scored the highest for FL were $213/yr more profitable, produced 575 kg more milk per year, and remained in the herd for 307 more functional days than the cows scoring the lowest. Feet and legs was the trait most genetically correlated to profit, although a low value (0.10) was obtained, whereas RLS was the trait most correlated to milk production (0.12). Genetic correlations among FL, FA, RLS, and longevity traits (from -0.10 to 0.05) were low. Quadratic curves were the best fit for both profit and functional herd life for EBV of each of the 3 locomotion traits. Further studies dealing with profitability and lameness, instead of using conformation traits, could be performed directly if a larger data pool of lameness was routinely recorded.
DOI:10.1016/S0301-6226(02)00188-4URL [本文引用: 2]
DOI:10.3168/jds.2007-0816URLPMID:18832226 [本文引用: 1]

First-lactation records on 836,452 daughters of 3,064 Norwegian Red sires were used to examine associations between culling in first lactation and 305-d protein yield, susceptibility to clinical mastitis, lactation mean somatic cell score (SCS), nonreturn rate within 56 d in heifers and primiparous cows, and interval from calving to first insemination. A Bayesian multivariate threshold-linear model was used for analysis. Posterior mean of heritability of liability to culling of primiparous cows was 0.04. The posterior means of the genetic correlations between culling and the other traits were -0.41 to 305-d protein yield, 0.20 to lactation mean SCS, 0.36 to clinical mastitis, 0.15 to interval from calving to first insemination, -0.11 to 56-d nonreturn as heifer, and -0.04 to 56-d nonreturn as primiparous cow. As much as 66% of the genetic variation in culling was explained by genetic variation in protein yield, clinical mastitis, interval of calving to first insemination, and 56-d nonreturn in heifers, whereas contribution from the SCS and 56-d nonreturn as primiparous cow was negligible, after taking the other traits into account. This implies that for breeds selected for a broad breeding goal, including functional traits such as health and fertility, most of the genetic variation in culling will probably be covered by other traits in the breeding goal. However, in populations where data on health and fertility is scarce or not available at all, selection against early culling may be useful in indirect selection for improved health and fertility. Regression of average sire posterior mean on birth-year of the sire indicate a genetic change equivalent to an annual decrease of the probability of culling in first-lactation Norwegian Red cattle by 0.2 percentage units. This genetic improvement is most likely a result of simultaneous selection for improved milk yield, health, and fertility over the last decades.
DOI:10.3168/jds.2015-9632URLPMID:26277309 [本文引用: 2]

The aim of this study was to conduct a multitrait 2-step approach applied to yield deviations and deregressed breeding values to get genetic parameters of functional longevity, clinical mastitis, early fertility disorders, cystic ovaries, and milk fever of Austrian Fleckvieh cattle. An approximate multitrait approach allows the combination of information from pseudo-phenotypes derived from different statistical models in routine genetic evaluation, which cannot be estimated easily in a full multitrait model. A total of 66,890 Fleckvieh cows were included in this study. For estimating genetic parameters, a simple linear animal model with year of birth as a fixed effect and animal as a random genetic effect was fitted. The joint analysis of yield deviations and deregressed breeding values was feasible. As expected, heritabilities were low, ranging from 0.03 (early fertility disorders) to 0.15 (functional longevity). Genetic correlations between functional longevity and clinical mastitis, early fertility disorders, cystic ovaries, and milk fever were 0.63, 0.29, 0.20, and 0.20, respectively. Within direct health traits genetic correlations were between 0.14 and 0.45. Results suggest that selecting for more robust disease-resistant cows would imply an improvement of functional longevity.
DOI:10.3168/jds.s0022-0302(98)75708-xURLPMID:9621248 [本文引用: 2]

Sire genetic evaluations for protein yield, somatic cell score (SCS), productive life, and udder type traits from the US were correlated with sire evaluations for udder health from Denmark and Sweden and then the correlations were adjusted for accuracies to approximate genetic correlations. Traits from Denmark and Sweden included somatic cell count (SCC) and clinical mastitis from single-trait analyses. In addition, evaluations for clinical mastitis from Denmark and Sweden were regressed on US traits to test for quadratic relationships. Information from 85 bulls with US and Danish evaluations (77 with US type) and from 80 bulls with US and Swedish evaluations (79 with US type) was used to calculate correlations. Genetic correlations of US protein yield with Danish and Swedish SCC and clinical mastitis were all unfavorable (-0.09 to -0.32). Genetic correlations of US productive life with Danish and Swedish SCC and clinical mastitis were all favorable (0.06 to 0.59). Genetic correlations between US SCS and Danish SCC and between US SCS and Swedish SCC were -0.87 and -0.99, respectively (favorable). Genetic correlations between US SCS and Danish clinical mastitis and between US SCS and Swedish clinical mastitis were -0.66 and -0.49, respectively (favorable). The US type traits that had the largest correlations with clinical mastitis from Denmark and Sweden, respectively, were udder composite (0.26, 0.47), udder depth (0.45, 0.52), and fore udder attachment (0.31, 0.34). In general, quadratic regressions indicated little nonlinearity between clinical mastitis and the US traits. Specifically, the US bulls with the lowest predicted transmitting abilities for SCS had the most favorable rates of daughter clinical mastitis in Denmark and Sweden. Selection for increased productive life, lower SCS, and more shallow udders should improve mastitis resistance.
DOI:10.3168/jds.S0022-0302(00)74970-8URLPMID:10821581 [本文引用: 2]

The relationship between mastitis and functional longevity was assessed with survival analysis on data of Danish Black and White dairy cows. Different methods of including the effect of mastitis treatment on the culling decision by a farmer in the model were compared. The model in which mastitis treatment was assumed to have an effect on functional longevity until the end of the lactation had the highest likelihood, and the model in which mastitis treatment had an effect for only a short period had the lowest likelihood. A cow with mastitis had 1.69 times greater risk of being culled than did a healthy herdmate with all other effects being the same. A model without mastitis treatment was used to predict transmitting abilities of bulls for risk of being culled, based on longevity records of their daughters, and was expressed in terms of risk of being culled. The correlation between the risk of being culled and the national evaluations of the bulls for mastitis resistance was approximately -0.4, indicating that resistance against mastitis was genetically correlated with a lower risk of being culled and, thus, a longer functional length of productive life.
DOI:10.3168/jds.S0022-0302(01)70187-7URLPMID:11417714 [本文引用: 3]

In survival analysis, type traits can be included as covariates to evaluate their use as predictors for survival. One problem in such an analysis is the availability of suitable data. Whereas data on the length of productive life (LPL) of individual cows can be retrieved from milk recording data, for type traits, all cows in the population must be scored for type at least once. In the present analysis, a dataset from the Osnabruck region in northwestern Germany, which fulfilled this requirement in recent years, was used. Data consisted of 169,733 cows with information on LPL for calving years 1980 to 1996 (dataset I) and of 39,233 cows with information on LPL and type for calving years 1990 to 1996 (dataset II). A further dataset (III) contained 43,116 cows from calving years 1987 to 1996 and included information on the housing system for each herd. The basic model included stage of lactation, relative production within herd, change of herd size, and year-season as time dependent effects; age at calving as a time-independent effect; and herd-year-season and sire as random effects. Other effects (information on type, housing system) were included additionally. For data-set II, the scores for 15 linear type traits were also included as corrected phenotypic values, estimated breeding values, and residuals from a previous BLUP analysis. The package Survival Kit 3.0 was used for all analyses. The results indicate a moderate heritability of 0.17 and 0.18 for true and functional LPL (dataset I). Almost all type traits analyzed (dataset II) exceeded a 0.001 level of significance in their effect on survival. The strongest relationships between survival and type were found for udder depth, fore udder attachment, and front teat placement. The main result from the comparison of housing systems (dataset III) was that bedding has a positive effect on survival.
DOI:10.1186/1297-9686-33-1-39URL [本文引用: 1]
DOI:10.3168/jds.S0022-0302(04)73533-XURLPMID:15483178 [本文引用: 1]

The aim of this study was to explore the impact of type traits on the functional survival of Canadian Holstein cows using a Weibull proportional hazards model. The data set consisted of 1,130,616 registered cows from 13,606 herds calving from 1985 to 2003. Functional survival was defined as the number of days from first calving to culling, death, or censoring. Type information consisted of phenotypic type scores for 8 composite traits (with 18 classes of each) and 23 linear descriptive traits (with 9 classes of each). The statistical model included the effects of stage of lactation, season of production, the annual change in herd size, type of milk recording supervision, age at first calving, effects of milk, fat and protein yields calculated within herd-year-parity deviations, herd-year-season of calving, each type trait, and the sire. Analysis was done one at a time for each of 31 type traits. The relative culling risk was calculated for animals in each class after accounting for the previously mentioned effects. Among the composite type traits with the greatest contribution to the likelihood function were final score, mammary system, and feet and legs, all having a strong relationship with functional survival. Cows with low scores for these traits had higher risk of culling compared with higher scores. For instance, cows classified as poor plus 1 vs. excellent plus 1 have a relative risk of culling 3.66 and 0.28, respectively. The corresponding figures for mammary system are 4.19 and 0.46 and for feet and legs are 2.34 and 0.50. Linear type traits with the greatest contribution to the likelihood function were fore udder attachment, udder texture, udder depth, rear udder attachment height, and rear udder attachment width. Stature and size had no strong relationship with functional survival.
DOI:10.1016/j.livsci.2008.01.024URL [本文引用: 2]
DOI:10.3168/jds.S0022-0302(05)72824-1URLPMID:15778325 [本文引用: 1]

The aim of this study was to use a Weibull proportional hazards model to explore the impact of type traits on the functional survival of Canadian Jersey and Ayrshire cows. The data set consisted of 49,791 registered Jersey cows from 900 herds calving from 1985 to 2003. The corresponding figures for Ayrshire were 77,109 cows and 921 herds. Functional survival was defined as the number of days from first calving to culling, death, or censoring. Type information consisted of phenotypic type scores for 8 composite traits and 19 linear descriptive traits. The statistical model included the effects of stage of lactation; season of production; annual change in herd size; type of milk recording supervision; age at first calving; effects of milk, fat, and protein yields calculated as within herd-year-parity deviations; herd-year-season of calving; each type trait; and the animal's sire. Analysis was done one trait at a time for each of 27 type traits in each breed. The relative culling risk was calculated for animals in each class after accounting for the previously mentioned effects. Among the composite type traits with the greatest contribution to the likelihood function was final score followed by mammary system for Jersey breed, while in Ayrshire breed feet and legs was the second most important trait next to final score. Cows classified as Poor for final score in both breeds were >5 times more likely to be culled compared with the cows classified as Good Plus. In both breeds, cows classified as Poor for feet and legs were 5 times more likely to be culled than were cows classified as Excellent, and cows classified as Excellent for mammary system were >9 times more likely to survive than were cows classified as Poor.
DOI:10.3168/jds.2007-0178URLPMID:18349259 [本文引用: 1]

The aim of this study was to use survival analysis to assess the relationship between reproduction traits and functional longevity of Canadian dairy cattle. Data consisted of 1,702,857; 67,470; and 33,190 Holstein, Ayrshire, and Jersey cows, respectively. Functional longevity was defined as the number of days from first calving to culling, death, or censoring; adjusted for the effect of milk yield. The reproduction traits included calving traits (calving ease, calf size, and calf survival) and female fertility traits (number of services, days from calving to first service, days from first service to conception, and days open). The statistical model was a Weibull proportional hazards model and included the fixed effects of stage of lactation, season of production, the annual change in herd size, and type of milk recording supervision, age at first calving, effects of milk, fat, and protein yields calculated as within herd-year-parity deviations for each reproduction trait. Herd-year-season of calving and sire were included as random effects. Analysis was performed separately for each reproductive trait. Significant associations between reproduction traits and longevity were observed in all breeds. Increased risk of culling was observed for cows that required hard pull, calved small calves, or dead calves. Moreover, cows that require more services per conception, a longer interval between first service to conception, an interval between calving to first service greater than 90 d, and increased days open were at greater risk of being culled.
DOI:10.3168/jds.S0022-0302(06)72400-6URLPMID:16899696 [本文引用: 1]

The aim of this study was to assess the level of somatic cell count (SCC) and to explore the impact of somatic cell score (SCS) on the functional longevity of Canadian dairy cattle by using a Weibull proportional hazards model. Data consisted of 1,911,428 cows from 15,970 herds sired by 7,826 sires for Holsteins, 80,977 cows in 2,036 herds from 1,153 sires for Ayrshires, and 53,114 cows in 1,372 herds from 1,758 sires for Jerseys. Functional longevity was defined as the number of days from the first calving to culling, death, or censoring. The test-day SCC was transformed to a linear score, and the resulting SCS were averaged within each lactation. The average SCS were grouped into 10 classes. The statistical model included the effects of stage of lactation; season of production; annual change in herd size; type of milk recording supervision; age at first calving; effects of milk, fat, and protein yields, calculated as within-herd-year-parity deviations; herd-year-season of calving; SCS class; and sire. The relative culling rate was calculated for animals in each SCS class after accounting for the aforementioned effects. The overall average SCC for Holsteins was 167,000 cells/mL, for Ayrshires was 155,000 cells/mL, and for the Jerseys was 212,000 cells/mL. In all breeds there were no appreciable differences in the relative risk of culling among classes of SCS breed averages (i.e., up to a SCS of 5). However, as the SCS increased beyond the breed average, the relative risk of cows being culled increased considerably. For instance, Holstein, Ayrshire, and Jersey cows with the highest classes of SCS had, respectively, a 4.95, 6.73, and 6.62 times greater risk of being culled than cows with average SCS.
DOI:10.3168/jds.S0022-0302(05)72745-4URLPMID:15653548 [本文引用: 1]

Survival analysis in a Weibull proportional hazards model was used to evaluate the impact of somatic cell count (SCC) on the involuntary culling rate of US Holstein and Jersey cows with first calvings from 1990 to 2000. The full data set, consisting of records from 978,043 Holstein and 250,835 Jersey cows, was divided into subsets (5 for Holsteins and 3 for Jerseys) based on herd average lactation SCC values. Functional longevity (also known as herd life or length of productive life) was defined as days from first calving until culling or censoring, after correcting for milk production. Our model included the time-dependent effects of herd-year-season, parity by stage of lactation interaction, within-herd-year quintile ranking for mature equivalent production, and lactation average SCC (rounded to the nearest 50,000 cells/mL), as well as the time-independent effect of age at first calving. Parameters of the Weibull distribution, as well as variance components for herd-year-season effects, were estimated within each group of herds. Mean failure and censoring times decreased as herd average SCC increased, and a nonlinear relationship was observed between SCC and longevity in all groups. The risk of culling for Holstein cows with lactation average SCC > 700,000 cells/mL was 3.4, 2.7, or 2.3 times greater, respectively, than that of Holstein cows with SCC of 200,000 to 250,000 cells/mL in herds with low, medium, or high average SCC. Likewise, the risk of culling for Jersey cows with lactation average SCC > 700,000 cells/mL was 4.0, 2.9, or 2.2 times greater, respectively, than that of Jersey cows with SCC of 200,000 to 250,000 cells/mL in low, medium, or high SCC herds. These trends may reflect more stringent culling of high SCC cows in herds with few mastitis problems. In addition, cows with lactation average SCC <100,000 cells/mL had a slightly higher risk of culling than cows with SCC of 100,000 to 200,000 cells/mL in both breeds, particularly in herds with high average SCC, where exposure to mastitis pathogens was likely.
DOI:10.3168/jds.2009-2969URLPMID:20723709 [本文引用: 2]

The aim of this study was to assess the effect of workability traits like milking speed and temperament on functional longevity of Canadian dairy cattle using a Weibull proportional hazards model. First-lactation data consisted of the following: 1,728,289 and 2,426,123 Holstein cows for milking temperament and milking speed, respectively, from 18,401 herds and sired by 8,248 sires; 39,618 and 60,121 Jersey cows for milking temperament and milking speed, respectively, from 1,845 herds and sired by 2,413 sires; and 54,391 and 94,847 Ayrshire cows for milking temperament and milking speed, respectively, from 1,316 herds and sired by 2,779 sires. Functional longevity was defined as the number of days from the first calving to culling, death, or censoring adjusted for production. Milking temperament and milking speed were recorded on a 1- to 5-point scale from very nervous to very calm and from very slow to very fast, respectively. The statistical model included the effects of stage of lactation; season of production; the annual change in herd size; type of milk recording supervision; age at first calving; effects of milk, fat, and protein yields calculated as within herd-year-parity deviations; herd-year-season of calving; sire; and milking temperament or milking speed class. The relative culling rate was calculated for animals in each milking temperament or milking speed class after accounting for the above-mentioned effects. The study showed that there was a statistically significant association between workability traits and functional longevity. Very nervous cows were 26, 23, and 46% more likely to be culled than very calm cows in Holstein, Ayrshire, and Jersey breeds, respectively. Similarly, very slow milkers were 36, 33, and 28% more likely to be culled than average milkers in Holstein, Ayrshire, and Jersey breeds, respectively. Additionally, very fast milkers were 11, 13, and 15% more likely to be culled than average milkers in Holstein, Ayrshire, and Jersey breeds, respectively. Producers might want to avoid consequences associated with the fast milkers such as udder health problems.
DOI:10.3168/jds.S0022-0302(04)70050-8URLPMID:15328244 [本文引用: 1]

Longevity is the economically most important functional trait in cattle populations. However, with an increased productive lifespan, the number of offspring born by older dams increases. A higher maternal age might have negative effects on the performance of offspring. The objective of this study was to investigate the effect of maternal age on production (energy-corrected milk yield [ECM]) and functional traits (fertility; somatic cell score, and functional longevity) in Austrian dual-purpose Simmental cows. Age of dam had a significant effect on ECM yield and longevity. The ECM yield of daughters decreased with age of dam. Although the risk of culling slightly increased with age of dam, it was lowest for daughters of oldest dams. Results for fertility were non-significant, and results for somatic cell scores were inconsistent across parities.
DOI:10.3168/jds.S0022-0302(06)72537-1URLPMID:17106119 [本文引用: 2]

The aim of this study was to assess the phenotypic level of lactose and milk urea nitrogen concentration (MUN) and the association of these traits with functional survival of Canadian dairy cattle using a Weibull proportional hazards model. A total of 1,568,952 test-day records from 283,958 multiparous Holstein cows from 4,758 herds, and 79,036 test-day records from 26,784 multiparous Ayrshire cows from 384 herds, calving from 2001 to 2004, were used for the phenotypic analysis. The overall average lactose percentage and MUN for Ayrshires were 4.49% and 12.20 mg/dL, respectively. The corresponding figures for Holsteins were 4.58% and 11.11 mg/dL. Concentration of MUN increased with parity number, whereas lactose percentage decreased in later parities. Data for survival analysis consisted of 39,536 first-lactation cows from 1,619 herds from 2,755 sires for Holsteins and 2,093 cows in 228 herds from 157 sires for Ayrshires. Test-day lactose percentage and MUN were averaged within first lactation. Average lactose percentage and MUN were grouped into 5 classes (low, medium-low, medium, medium-high, and high) based on mean and standard deviation values. The statistical model included the effects of stage of lactation, season of production, the annual change in herd size, type of milk-recording supervision, age at first calving, effects of milk, fat, and protein yields calculated as within herd-year-parity deviations, herd-year-season of calving, lactose percentage and MUN classes, and sire. The relative culling rate was calculated for animals in each class after accounting for the remaining effects included in the model. Results showed that there was a statistically significant association between lactose percentage and MUN in first lactation with functional survival in both breeds. Ayrshire cows with high and low concentration of MUN tended to be culled at a higher than average rate. Instead, Holstein cows had a linear association, with decreasing relative risk of culling with increasing levels of MUN concentration. The relationship between lactose percentage and survival was similar across breeds, with higher risk of culling at low level of lactose, and lower risk of culling at high level of lactose percentage.
DOI:10.3168/jds.S0022-0302(06)72291-3URLPMID:16702287 [本文引用: 1]

The aim of this study was to assess the level of inbreeding and its relationship to the functional survival of Canadian dairy breeds by using a Weibull proportional hazard model. Data consisted of records from 72,385 cows in 1,505 herds from 2,499 sires for Jerseys, 112,723 cows in 1,482 herds from 2,926 sires for Ayrshires, and 1,977,311 cows in 17,182 herds from 8,261 sires for Holsteins. Longevity was defined as the number of days from first calving to culling, death, or censoring. Inbreeding coefficients (F) were grouped into 7 classes (F = 0, 0 < F < 3.125, 3.125 < or = F < 6.25, 6.25 < or = F <12.5, 12.5 < or = F < 18.25, 18.25 < or = F < 25.0, and F > or = 25.0%). The statistical model included the effects of stage of lactation, season of production, the annual change in herd size, type of milk recording supervision, age at first calving, effects of milk, fat, and protein yields calculated as within herd-year-parity deviations, herd-year-season of calving, inbreeding, and sire. The relative culling rate was calculated for animals in each class after accounting for the above-mentioned effects. A trend toward increased risk of culling among more inbred animals was observed for all breeds. Little difference in survival was observed for cows with 0 < F <12.5%. The relative risk ratios (relative to F = 0) for cows with inbreeding coefficients up to 12.5% were 1.19, 1.16, and 1.14 for Jersey, Ayrshire, and Holstein cows, respectively. Greater effects of inbreeding were seen, however, when F increased beyond 12.5%.
DOI:10.1111/age.12164URLPMID:24796806 [本文引用: 1]

We genotyped 58 single nucleotide polymorphisms (SNPs) in 25 candidate genes in about 800 Italian Holstein sires. Fifty-six (minor allele frequency >0.02) were used to evaluate their association with single traits: milk yield (MY), milk fat yield (FY), milk protein yield (PY), milk fat percentage (FP), milk protein percentage (PP), milk somatic cell count (MSCC); and complex indexes: longevity, fertility and productivity-functionality type (PFT), using deregressed proofs, after adjustment for familial relatedness. Thirty-two SNPs were significantly associated (proportion of false positives <0.05) with different traits: 16 with MSCC, 15 with PY, 14 with MY, 12 with PFT, eight with longevity, eight with FY, eight with PP, five with FP and two with fertility. In particular, a SNP in the promoter region of the PRLR gene was associated with eight of nine traits. DGAT1 polymorphisms were highly associated with FP and FY. Casein gene markers were associated with several traits, confirming the role of the casein gene cluster in affecting milk yield, milk quality and health traits. Other SNPs in genes located on chromosome 6 were associated with PY, PP, PFT, MY (PPARGC1A) and MSCC (KIT). This latter association may suggest a biological link between the degree of piebaldism in Holstein and immunological functions affecting somatic cell count and mastitis resistance. Other significant SNPs were in the ACACA, CRH, CXCR1, FASN, GH1, LEP, LGB (also known as PAEP), MFGE8, SRC, TG, THRSP and TPH1 genes. These results provide information that can complement QTL mapping and genome-wide association studies in Holstein.
DOI:10.1186/1471-2156-12-30URLPMID:21392379 [本文引用: 2]

BACKGROUND: Longevity expressed as the number of days between birth and death is a trait of great importance for both human and animal populations. In our analysis we use dairy cattle to demonstrate how the association of Single Nucleotide Polymorphisms (SNPs) located within selected genes with longevity can be modeled. Such an approach can be extended to any genotyped population with time to endpoint information available. Our study is focused on selected genes in order to answer the question whether genes, known to be involved into the physiological determination of milk production, also influence individual's survival. RESULTS: Generally, the highest risk differences among animals with different genotypes are observed for polymorphisms located within the leptin gene. The polymorphism with a highest effect on functional longevity is LEP-R25C, for which the relative risk of culling for cows with genotype CC is 3.14 times higher than for the heterozygous animals. Apart from LEP-R25C, also FF homozygotes at the LEP-Y7F substitution attribute 3.64 times higher risk of culling than the YY homozygotes and VV homozygotes at LEP-A80V have 1.83 times higher risk of culling than AA homozygotes. Differences in risks between genotypes of polymorphisms within the other genes (the butyrophilin subfamily 1 member A1 gene, BTN1A1; the acyl-CoA:diacylglycerol acyltransferase 1 gene, DGAT1; the leptin receptor gene, LEPR; the ATP-binding cassette sub-family G member 2, ABCG2) are much smaller. CONCLUSIONS: Our results indicate association between LEP and longevity and are very well supported by results of other studies related to dairy cattle. In view of the growing importance of functional traits in dairy cattle, LEP polymorphisms should be considered as markers supporting selection decisions. Furthermore, since the relationship between both LEP polymorphism and its protein product with longevity in humans is well documented, with our result we were able to demonstrate that livestock with its detailed records of family structure, genetic, and environmental factors as well as extensive trait recording can be a good model organism for research aspects related to humans.
DOI:10.1007/BF03195663URLPMID:19433909 [本文引用: 1]

This study investigated the impact of 6 polymorphisms located in theABCG2, PPARGC1A, OLR1 andSCD1 genes on estimated breeding values for milk production, longevity, somatic cell count and reproductive traits. The analysis was conducted on 453 Polish Holstein-Friesian bulls. Genotypes were identified using PCR-RFLP, and haplotype inferences were performed for 3 linked mutations ofPPARGC1A. The most significant associations were found between the A/C polymorphism located in exon 14 ofABCG2 and milk fat production traits as well as calving-to-first insemination interval, and between the T/C substitution in intron 9 of thePPARGC1A and non-return rate in heifers.
DOI:10.3168/jds.2006-722URLPMID:17430947 [本文引用: 1]

Abstract
The uterine milk proteins (UTMP) are the major proteins secreted by the endometrium, primarily under the control of progesterone. Specific functions of UTMP are poorly understood, but may include protease inhibition, nutrition of the conceptus, growth control, and suppression of the maternal immune system. The uterine milk protein gene (UTMP) was chosen for this study because of its possible roles in health traits and results of previous studies on the association of the UTMP region with milk production and productive life in dairy cattle. Expression of UTMP was examined in 198 bovine tissues obtained from 10 fetuses and 17 cows. Transcripts of UTMP were found in all cotyledon tissues examined and to a lesser extent in ovary, pituitary, and spleen tissues obtained from fetuses. The UTMP gene was predominantly expressed in endometrium (17/17), ovary (15/16), and caruncle (12/12) tissues obtained from cows. The predominant expression of UTMP in reproductive tissues is consistent with an important role of this gene in reproductive success. To investigate the association between UTMP and production traits in cattle, we identified 2 synonymous single nucleotide polymorphisms (SNP) at positions 1179 (A/G) and 1296 (A/G) using the pooled DNA sequencing approach. The DNA was extracted from 28 Holstein sires and their 1,362 sons obtained from the Cooperative Dairy DNA Repository and from 913 cows from the University of Wisconsin resource population. Single nucleotide polymorphism 1296 was associated with a significant increase in productive life in both populations. This finding is similar to results previously obtained for the protease inhibitor gene, which is near UTMP and was also found to be associated with productive life in dairy cattle. Differential allelic expression of UTMP was observed in reproductive tissues obtained from 9 heterozygous individuals. The differential allelic expression observed in this study is consistent with other studies showing a correlation between allelic variation in gene expression and phenotypic variability.DOI:10.1111/j.1365-2052.2006.01443.xURLPMID:16734705 [本文引用: 1]
DOI:10.3168/jds.S0022-0302(05)72787-9URLPMID:15738254 [本文引用: 1]

Positional, comparative candidate gene analysis and previous quantitative trait loci linkage mapping results were used to search for candidate genes affecting milk production and reproduction traits in dairy cattle. The protease inhibitor (PI) gene was chosen for examination, and 5 single nucleotide polymorphisms were detected in coding regions of the gene by direct sequencing of reverse transcription-polymerase chain reaction products from a wide range of cattle tissues. A total of 6 different intragenic haplotypes were identified in North American Holstein population, and these were examined for associations with milk production traits in 24 half-sib families comprising 1007 sons utilizing a granddaughter design. One common haplotype was associated with increased milk and fat yields, increased productive life, and decreased somatic cell score. Another common haplotype was associated with decreased productive life and increased somatic cell score. One rare haplotype was associated with decreased milk, fat, and protein yields and increased milk protein percentage; another rare haplotype was associated with decreased milk yield, increased protein percentage, and decreased productive life. The observation that the PI gene is associated with analogous traits in humans demonstrates the effectiveness of the positional comparative candidate gene analysis that utilizes information about genes present in chromosomal regions with conserved synteny in other species.
DOI:10.1111/j.1365-2052.2011.02208.xURLPMID:22221027 [本文引用: 2]

Although genomic selection offers the prospect of improving the rate of genetic gain in meat, wool and dairy sheep breeding programs, the key constraint is likely to be the cost of genotyping. Potentially, this constraint can be overcome by genotyping selection candidates for a low density (low cost) panel of SNPs with sparse genotype coverage, imputing a much higher density of SNP genotypes using a densely genotyped reference population. These imputed genotypes would then be used with a prediction equation to produce genomic estimated breeding values. In the future, it may also be desirable to impute very dense marker genotypes or even whole genome re-sequence data from moderate density SNP panels. Such a strategy could lead to an accurate prediction of genomic estimated breeding values across breeds, for example. We used genotypes from 48 640 (50K) SNPs genotyped in four sheep breeds to investigate both the accuracy of imputation of the 50K SNPs from low density SNP panels, as well as prospects for imputing very dense or whole genome re-sequence data from the 50K SNPs (by leaving out a small number of the 50K SNPs at random). Accuracy of imputation was low if the sparse panel had less than 5000 (5K) markers. Across breeds, it was clear that the accuracy of imputing from sparse marker panels to 50K was higher if the genetic diversity within a breed was lower, such that relationships among animals in that breed were higher. The accuracy of imputation from sparse genotypes to 50K genotypes was higher when the imputation was performed within breed rather than when pooling all the data, despite the fact that the pooled reference set was much larger. For Border Leicesters, Poll Dorsets and White Suffolks, 5K sparse genotypes were sufficient to impute 50K with 80% accuracy. For Merinos, the accuracy of imputing 50K from 5K was lower at 71%, despite a large number of animals with full genotypes (2215) being used as a reference. For all breeds, the relationship of individuals to the reference explained up to 64% of the variation in accuracy of imputation, demonstrating that accuracy of imputation can be increased if sires and other ancestors of the individuals to be imputed are included in the reference population. The accuracy of imputation could also be increased if pedigree information was available and was used in tracking inheritance of large chromosome segments within families. In our study, we only considered methods of imputation based on population-wide linkage disequilibrium (largely because the pedigree for some of the populations was incomplete). Finally, in the scenarios designed to mimic imputation of high density or whole genome re-sequence data from the 50K panel, the accuracy of imputation was much higher (8696%). This is promising, suggesting that in silico genome re-sequencing is possible in sheep if a suitable pool of key ancestors is sequenced for each breed.
DOI:10.4238/gmr.15017198URLPMID:27051021 [本文引用: 2]

Sequence-related amplified polymorphism (SRAP) markers were used to evaluate the intra- and interspecific variation among 40 Lathyrus genotypes (four species) (Fabaceae). Ten SRAP primer combinations resulted in a total of 94 bands, and they exhibited high interspecific variability. The genetic differentiation among Lathyrus, estimated using AMOVA, was highly significant. The results indicated that 58% of the total genetic variation existed among species, and 42% of the differentiation was within species. This was explained by the high level of genome conservation of these species as well as the recent and slow evolution of this genus. These results were confirmed by the topology of the neighbor-joining cladogram and the results of the principal coordinate analysis. Our data support previous results based on seed protein diversity. These results make SRAP markers choice markers for the study of functional polymorphism that is directly related to the transcriptomic data. The SRAP markers used in this study provide an accurate picture of the population structure within Lathyrus germplasm, which is critically important information for the design of genetic diversity and structure analyses. Moreover, further extensive studies are necessary to fully examine other Lathyrus species and tests that adopt the SRAP technique to enrich the Lathyrus library for next-generation sequencing, thus providing a potent protocol for the study of polymorphism.
DOI:10.3168/jds.2006-707URLPMID:17517739 [本文引用: 2]

A quantitative trait locus (QTL) underlying different milk production traits has been identified with a high significance threshold value in the genomic region containing the acylCoA:diacylglycerol acyltransferase (DGAT1) gene, in the 3 main French dairy cattle breeds: French Holstein, Normande, and Montbeliarde. Previous studies have confirmed that the K232A polymorphism in DGAT1 is responsible for a major QTL underlying several milk production traits in Holstein dairy cattle and several other bovine breeds. In this study, we estimate the frequency of the 2 alternative alleles, K and A, of the K232A polymorphism in French Holstein, Normande, and Montbeliarde breeds. Although the K allele segregates in French Holstein and Normande breeds with a similar effect on production traits, the existence of additional mutations contributing to the observed QTL effect is strongly suggested in both breeds by the existence of sires heterozygous at the QTL but homozygous at the K232A polymorphism. One allele at a variable number of tandem repeats (VNTR) locus in the 5' noncoding region of DGAT1 has been recently proposed as a putative causative variant. In our study, this marker was found to present a high mutation rate of 0.8% per gamete and per generation, making the allele diversity observed compatible with that expected under neutrality. Moreover, among the sires homozygous at the K232A polymorphism, no allele at the VNTR can fully explain their QTL status. Finally, no allele at the VNTR was found to be significantly associated with the fat percentage variation in the 3 breeds simultaneously after correction for the effect of the K232A polymorphism. Therefore, our results suggest the existence of at least one other causative polymorphism not yet described. Because the A allele is nearly fixed in the Montbeliarde breed, this breed represents an interesting model to identify and confirm other mutations that have a strong effect on milk production traits.
DOI:10.3168/jds.2011-4757URLPMID:22192223 [本文引用: 1]

The objective of this study was to identify DNA markers in the 4 casein genes (CSN1S1, CSN1S2, CSN2, and CSN3) and the 2 major whey protein genes (LALBA and LGB) that show associations with milk protein profile measured by reverse-phase HPLC. Fifty-three single nucleotide polymorphisms (SNP) were genotyped for cows in a unique resource population consisting of purebred Holstein and (Holstein x Jersey) x Holstein crossbred animals. Seven traits were analyzed, including concentrations of alpha(S)-casein (CN), beta-CN, kappa-CN, alpha-lactalbumin, beta-lactoglobulin, and 2 additional secondary traits, the total concentration of the above 5 milk proteins and the alpha(S)-CN to beta-CN ratio. A substantial fraction of phenotypic variation could be explained by the additive genetic component for the 7 milk protein composition traits studied. Moreover, several SNP were significantly associated with all examined traits at an experiment-wise error rate of 0.05, except for alpha-lactalbumin. Importantly, the significant SNP explained a large proportion of the phenotypic variation of milk protein composition. Our findings could be used for selecting animals that produce milk with desired composition or desired processing and manufacturing properties.
DOI:10.3168/jds.2007-0891URLPMID:18565947 [本文引用: 1]

The objectives were to evaluate the associations among single nucleotide polymorphisms (SNP) in the R4C locus in exon 2 of the leptin gene and the lactational performance and health of Holstein cows. Eight hundred and fourteen lactating dairy cows had their DNA sequenced in exon 2 of the leptin gene to determine the presence of SNP in the R4C locus. Cows were milked 3 times daily, and yields of milk and milk components were recorded monthly individually during the first 305 d of lactation. Cows were examined daily by herd personnel for diagnosis of health events such as retained fetal membranes, displacement of abomasum, lameness, and mastitis. Resulting genotypes were CC (34.6%), CT (48.2%), and TT (17.2%). Cows bearing the CT genotype had lower body condition (2.98 +/- 0.02) during the first 62 d in milk (DIM) than cows homozygous for the C (3.02 +/- 0.02) and T (3.04 +/- 0.03) alleles. Leptin genotype was associated with yields of milk and milk components, and cows homozygous for the C allele were less productive than those carrying the CT and TT genotypes. The 305-d yields of 3.5% fat-corrected milk, milk fat, and milk true protein were less in CC compared with CT cows by 258, 12, and 10.7 kg, respectively. Cows carrying the TT genotype had increased incidence of displacement of abomasum (4.3%), but genotype did not affect the incidence of retained fetal membranes, clinical and subclinical mastitis, or lameness. Risk of developing at least one clinical health disorder was influenced by leptin genotype, and cows carrying the CT genotype had the lowest risk for developing any disease (19.6%). Mating decisions to increase the frequency of cows heterozygous in the R4C locus may improve productivity and health.
DOI:10.3168/jds.2009-2457URLPMID:20059932 [本文引用: 2]

Leptin is an important regulator of fetal and placental growth. This study evaluated the association of single nucleotide polymorphisms (SNP) in the leptin gene with perinatal mortality (stillbirths and mortality within 24h of parturition) in 385 Holstein-Friesian heifers on 18 dairy farms in the United Kingdom. The 3 SNP evaluated were exon 2FB, UASMS1, and UASMS2. The mean age at first calving was 27.0+/-0.2 mo. Associations between each SNP and perinatal mortality (calf alive or dead) were tested using a generalized linear model that included herd-year-season, calf sex, age at first calving, and age and pedigree of the dam. The overall level of perinatal mortality in the population was 16.9%, with significant allelic substitution effects for exon 2FB and UASMS1. These 2 SNP were in close linkage disequilibrium with each other (r(2)=0.98) but not with UASMS2 (r(2)=0.10). For exon 2FB, perinatal mortality was similar between heifers carrying the CT and TT alleles (20%), but was higher than in heifers carrying the CC allele (11%). For UASMS1, mortality was 21% with the CC and CT alleles but only 10% with the TT allele. No associations of perinatal mortality with SNP were found in the UASMS2 data set, possibly influenced by the low frequency (2%) of the TT genotype. No significant effects of herd-year-season, age at first calving, or calf sex were found. In conclusion, polymorphisms in the leptin gene were associated with 2-fold differences in perinatal mortality in dairy heifers.
DOI:10.1186/1471-2164-12-408URL [本文引用: 2]
DOI:10.3168/jds.2016-11770URLPMID:27889128 [本文引用: 2]

Female fertility in Holstein cattle can decline when intense genetic selection is placed on milk production. One approach to improving fertility is to identify the genomic regions and variants affecting fertility traits and then incorporate this knowledge into selection decisions. The objectives of this study were to identify or refine the positions of the genomic regions associated with lactation persistency, female fertility traits (age at first service, cow first service to conception, heifer and cow nonreturn rates), longevity traits (herd life, indirect herd life, and direct herd life), and lifetime profit index in the North American Holstein dairy cattle population. A genome-wide association study was performed for each trait, using a single SNP (single nucleotide polymorphism) regression mixed linear model and imputed high-density panel (777k) genotypes. No associations were identified for fertility traits. Several peak regions were detected for lifetime profit index, lactation persistency, and longevity. The results overlap with previous findings and identify some novel regions for lactation persistency. Previously proposed causative and candidate genes supported by this work include DGAT1, GRINA, and CPSF1, whereas new candidate genes are SLC2A4RG and THRB. Thus, the chromosomal regions identified in this study not only confirm several previous findings but also highlight new regions that may contribute to genetic variation in lactation persistency and longevity-associated traits in dairy cattle.
DOI:10.1017/S1751731118003191URLPMID:30501681 [本文引用: 1]

Longevity is one of the most important traits determining dairy cow profitability. In the last decades dairy cows suffered a lowering in the age at culling. With the aim to identify the genes involved in longevity, dates of birth, yields, dates of calving during lifespan and culling dates were collected for 946 culled cows which had been genotyped with the Bovine High Density panel. Using the GenABEL package in R, genome-wide association analysis was performed on three potential traits of longevity: (1) 'days in production,' (2) 'days in herd,' (3) number of calvings over lifespan.' Five genome-wide significant single nucleotide polymorphisms (SNPs) associated with all three longevity traits were detected. Several consecutive SNPs identified on chromosomes 16 and 30 indicated the presence of two suggestive quantitative trait loci (QTL). The genes comprised in the QTL regions had biological functions related to fertility, reproductive disorders, heat stress and welfare of cows. These findings might contribute to improving breeding strategies to improve longevity.
URLPMID:28464084 [本文引用: 1]
DOI:10.4238/gmr16029696URLPMID:28653742 [本文引用: 1]

The goals of this research were to evaluate the phenotypic and genotypic correlations between agronomic traits, to perform path analysis, having as main character grain yield, and to identify indirect selection criteria for grain yield. The experiment was carried out in an experimental area located at Capim Branco farm, which belongs to Federal University of Uberlandia, during the growing season of 2015/2016.Twenty-four soybean genotypes were evaluated under randomized complete block design with three repetitions, of which agronomic traits and grain yield were measured. There was genetic variability for all traits at 5% probability level through the F-test. Thirty significant phenotypic correlations were also observed with values oscillating from 0.42 to 0.87, which indicated a high level of association between some evaluated traits. Additionally, we verified that phenotypic and genotypic correlations were essential of the same direction, being the genotypic ones of superior magnitudes. Plants with superior vegetative cycle had longer life cycles; this fact could be explained by the significant phenotypic correlations between the number of days to the blooming and number of days to maturity (0.76). Significantly positive phenotypic and genotypic correlations for the total number of pods per plant and grain yield per plant (0.84) were observed. Through the path analysis, the trait that contributed the most over grain yield was the number of pods with three seeds as it showed the highest direct effect on grain yield per plant, as well as a strong indirect effect on the total number of pods. Therefore, the phenotypic and genotypic correlations suggested high correlations between grain yield and number of branched nodes, the number of pods with two and three seeds, and the total number of pods. Also, the path analysis determined the number of pods with three seeds as having the highest favorable effect on grain yield, and thus, being useful for indirect selection toward productive soybean genotypes.
DOI:10.3168/jds.2015-10697URLPMID:27289149 [本文引用: 1]

Longevity is an important economic trait in dairy production. Improvements in longevity could increase the average number of lactations per cow, thereby affecting the profitability of the dairy cattle industry. Improved longevity for cows reduces the replacement cost of stock and enables animals to achieve the highest production period. Moreover, longevity is an indirect indicator of animal welfare. Using whole-genome sequencing variants in 3 dairy cattle breeds, we carried out an association study and identified 7 genomic regions in Holstein and 5 regions in Red Dairy Cattle that were associated with longevity. Meta-analyses of 3 breeds revealed 2 significant genomic regions, located on chromosomes 6 (META-CHR6-88MB) and 18 (META-CHR18-58MB). META-CHR6-88MB overlaps with 2 known genes: neuropeptide G-protein coupled receptor (NPFFR2; 89,052,210-89,059,348 bp) and vitamin D-binding protein precursor (GC; 88,695,940-88,739,180 bp). The NPFFR2 gene was previously identified as a candidate gene for mastitis resistance. META-CHR18-58MB overlaps with zinc finger protein 717 (ZNF717; 58,130,465-58,141,877 bp) and zinc finger protein 613 (ZNF613; 58,115,782-58,117,110 bp), which have been associated with calving difficulties. Information on longevity-associated genomic regions could be used to find causal genes/variants influencing longevity and exploited to improve the reliability of genomic prediction.
DOI:10.4236/ojgen.2014.41007URL [本文引用: 1]
DOI:10.3168/jds.2017-12968URLPMID:29153164 [本文引用: 1]

Over the past 100 yr, the range of traits considered for genetic selection in dairy cattle populations has progressed to meet the demands of both industry and society. At the turn of the 20th century, dairy farmers were interested in increasing milk production; however, a systematic strategy for selection was not available. Organized milk performance recording took shape, followed quickly by conformation scoring. Methodological advances in both genetic theory and statistics around the middle of the century, together with technological innovations in computing, paved the way for powerful multitrait analyses. As more sophisticated analytical techniques for traits were developed and incorporated into selection programs, production began to increase rapidly, and the wheels of genetic progress began to turn. By the end of the century, the focus of selection had moved away from being purely production oriented toward a more balanced breeding goal. This shift occurred partly due to increasing health and fertility issues and partly due to societal pressure and welfare concerns. Traits encompassing longevity, fertility, calving, health, and workability have now been integrated into selection indices. Current research focuses on fitness, health, welfare, milk quality, and environmental sustainability, underlying the concentrated emphasis on a more comprehensive breeding goal. In the future, on-farm sensors, data loggers, precision measurement techniques, and other technological aids will provide even more data for use in selection, and the difficulty will lie not in measuring phenotypes but rather in choosing which traits to select for.
DOI:10.3168/jds.S0022-0302(02)74225-2URLPMID:12146488 [本文引用: 1]

Genetic evaluation for herd life based on survival analysis utilizes information available on all animals, dead (uncensored) and alive (censored), but the reliability of bulls' breeding values depends only on the number of uncensored daughters. Therefore, information on correlated conformation traits scored on daughters during their first lactation may be essential for the evaluation of young bulls with mostly censored daughters. Currently available programs for genetic evaluation based on survival analysis cannot combine indirect information on conformation traits with direct information on herd life, nor can they estimate genetic covariances between herd life and conformation traits. In this study, an alternative approach has been developed and tested using data on Swiss Simmental and Red & White cattle. Genetic covariances were approximated using breeding values for herd life from a survival analysis and BLUP breeding values for 26 linear conformation traits from a separate multivariate analysis. An index combining direct breeding values for herd life and indirect breeding values obtained from conformation traits was constructed. The relative weighting of both information sources varied depending on the amount of available information. The maximum reliability based only on conformation traits was 0.64. Except for old bulls with >100 uncensored daughters, the combined reliability was always higher than the direct reliability from survival analysis.
DOI:10.3168/jds.2006-719URLPMID:17369244 [本文引用: 1]

The national genetic evaluation of herd life for Canadian dairy breeds was modified from a 3-trait to a 5-trait animal model. The genetic evaluation incorporates information from daughter survival (direct herd life) and information from conformation, fertility, and udder health traits that are related to longevity (indirect herd life). Genetic evaluations for direct herd life were based on cows' survival from first calving to 120 days in milk (DIM), from 120 to 240 DIM, from 240 DIM to second calving, survival to third calving, and survival to fourth calving, which were analyzed using a multiple-trait animal model. Sire evaluations obtained for each of the 5 survival traits were combined into an overall sire evaluation for direct herd life. Sire evaluations for indirect herd life were based on an index of sire evaluations for dairy strength, feet and legs, overall mammary, rump angle, somatic cell score, milking speed, nonreturn rate in cows, and interval from calving to first service. A multiple-trait sire model based on multiple-trait across-country evaluation methodology was used to combine direct and indirect genetic evaluations for herd life into an overall genetic evaluation for herd life. Sire evaluations for herd life were expressed as an estimated transmitting ability for the number of lactations. The transmitting ability represents expected differences among daughters for herd life; and the average herd life was set to 3 lactations.
DOI:10.3168/jds.2007-0980URLPMID:18946147 [本文引用: 1]

Efficient methods for processing genomic data were developed to increase reliability of estimated breeding values and to estimate thousands of marker effects simultaneously. Algorithms were derived and computer programs tested with simulated data for 2,967 bulls and 50,000 markers distributed randomly across 30 chromosomes. Estimation of genomic inbreeding coefficients required accurate estimates of allele frequencies in the base population. Linear model predictions of breeding values were computed by 3 equivalent methods: 1) iteration for individual allele effects followed by summation across loci to obtain estimated breeding values, 2) selection index including a genomic relationship matrix, and 3) mixed model equations including the inverse of genomic relationships. A blend of first- and second-order Jacobi iteration using 2 separate relaxation factors converged well for allele frequencies and effects. Reliability of predicted net merit for young bulls was 63% compared with 32% using the traditional relationship matrix. Nonlinear predictions were also computed using iteration on data and nonlinear regression on marker deviations; an additional (about 3%) gain in reliability for young bulls increased average reliability to 66%. Computing times increased linearly with number of genotypes. Estimation of allele frequencies required 2 processor days, and genomic predictions required <1 d per trait, and traits were processed in parallel. Information from genotyping was equivalent to about 20 daughters with phenotypic records. Actual gains may differ because the simulation did not account for linkage disequilibrium in the base population or selection in subsequent generations.
DOI:10.1186/1297-9686-43-19URL [本文引用: 1]
DOI:10.3168/jds.2008-1514URLPMID:19109259 [本文引用: 1]

Genetic progress will increase when breeders examine genotypes in addition to pedigrees and phenotypes. Genotypes for 38,416 markers and August 2003 genetic evaluations for 3,576 Holstein bulls born before 1999 were used to predict January 2008 daughter deviations for 1,759 bulls born from 1999 through 2002. Genotypes were generated using the Illumina BovineSNP50 BeadChip and DNA from semen contributed by US and Canadian artificial-insemination organizations to the Cooperative Dairy DNA Repository. Genomic predictions for 5 yield traits, 5 fitness traits, 16 conformation traits, and net merit were computed using a linear model with an assumed normal distribution for marker effects and also using a nonlinear model with a heavier tailed prior distribution to account for major genes. The official parent average from 2003 and a 2003 parent average computed from only the subset of genotyped ancestors were combined with genomic predictions using a selection index. Combined predictions were more accurate than official parent averages for all 27 traits. The coefficients of determination (R(2)) were 0.05 to 0.38 greater with nonlinear genomic predictions included compared with those from parent average alone. Linear genomic predictions had R(2) values similar to those from nonlinear predictions but averaged just 0.01 lower. The greatest benefits of genomic prediction were for fat percentage because of a known gene with a large effect. The R(2) values were converted to realized reliabilities by dividing by mean reliability of 2008 daughter deviations and then adding the difference between published and observed reliabilities of 2003 parent averages. When averaged across all traits, combined genomic predictions had realized reliabilities that were 23% greater than reliabilities of parent averages (50 vs. 27%), and gains in information were equivalent to 11 additional daughter records. Reliability increased more by doubling the number of bulls genotyped than the number of markers genotyped. Genomic prediction improves reliability by tracing the inheritance of genes even with small effects.
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1111/j.1601-5223.1999.00337.xURLPMID:10509144 [本文引用: 1]
DOI:10.1111/j.1601-5223.1999.00337.xURLPMID:10509144 [本文引用: 1]
DOI:10.3864/j.issn.0578-1752.2017.12.016URL [本文引用: 1]

【Objective】 The objective of this study was to investigate the association between the SNPs in the coding sequence (CDS) of CXCR1 gene and the clinical mastitis and lifetime for Chinese Holstein.【Method】SNPs in the CDS of CXCR1 gene were screened by using PCR and direct sequencing for 20 cows with low SCS and 20 cows with high SCS. Finally, the selected SNPs of CXCR1 gene for 866 Chinese Holstein cows were detected using flight mass spectrometry. The clinical mastitis and productive life of tested cows were collected from the management system of dairy farm. The association between the SNPs and the clinical mastitis and productive life of tested cows was analyzed using multi factor variance analysis, Logistic regression, Cox regression.【Result】A total of 13 SNPs were found in the CDS region of CXCR1 gene (291 C>T, 333 C>G, 337 A>G, 365 C>T, 570 A>G, 642 A>G, 735 C>G, 816 A>C, 819 A>G, 980 A>G、, 995 A>G, 1008 C>T and 1068 A>G), and these SNPs were divided into 4 linkage groups, and four SNPs were selected from each linkage group for further analysis(642 A>G, 816 A>C, 980A>G and 1068 A>G). A total of 9 haplotypes were observed for 4 SNPs of CXCR1 gene, the GAGG haplotype frequency was the highest (0.3141), and the frequency of haplotype ACAA was the lowest (0.0017). CXCR1-642 A>G showed a significant association with the number of dairy cows suffering from clinical mastitis in second lactation (P<0.05). The number of dairy cows suffering from clinical mastitis for individual with AG genotype was significantly higher than that of AA genotype (P<0.05). The number of dairy cows suffering from clinical mastitis for individual with CXCR1-816 AA genotype was significantly higher than that of AC and CC genotypes (P<0.05). The other SNPs showed no significant association with the number of clinical mastitis. CXCR1-816 A>C mutation showed an extremely significant association with culling age (P<0.01) and a significant association with productive month and culling lactation (P<0.05). The productive month, culling age and culling lactation for the individual of CXCR1-816 AA were significantly higher than that of CC genotype (P<0.05). Cox regression showed that only CXCR1-816 showed significant association with the productive life(P<0.05), the survival probability of individual with CC genotype was significantly lower than that of AA and AC genotype in each time period. 【Conclusion】The mutation of CXCR1-816 A>C showed significant association with the clinical mastitis and lifetime for Chinese Holstein, and this SNP could be used for molecular marker-assisted selection of productive life for Chinese Holstein when the molecular function of this mutation were proved.
DOI:10.3864/j.issn.0578-1752.2017.12.016URL [本文引用: 1]

【Objective】 The objective of this study was to investigate the association between the SNPs in the coding sequence (CDS) of CXCR1 gene and the clinical mastitis and lifetime for Chinese Holstein.【Method】SNPs in the CDS of CXCR1 gene were screened by using PCR and direct sequencing for 20 cows with low SCS and 20 cows with high SCS. Finally, the selected SNPs of CXCR1 gene for 866 Chinese Holstein cows were detected using flight mass spectrometry. The clinical mastitis and productive life of tested cows were collected from the management system of dairy farm. The association between the SNPs and the clinical mastitis and productive life of tested cows was analyzed using multi factor variance analysis, Logistic regression, Cox regression.【Result】A total of 13 SNPs were found in the CDS region of CXCR1 gene (291 C>T, 333 C>G, 337 A>G, 365 C>T, 570 A>G, 642 A>G, 735 C>G, 816 A>C, 819 A>G, 980 A>G、, 995 A>G, 1008 C>T and 1068 A>G), and these SNPs were divided into 4 linkage groups, and four SNPs were selected from each linkage group for further analysis(642 A>G, 816 A>C, 980A>G and 1068 A>G). A total of 9 haplotypes were observed for 4 SNPs of CXCR1 gene, the GAGG haplotype frequency was the highest (0.3141), and the frequency of haplotype ACAA was the lowest (0.0017). CXCR1-642 A>G showed a significant association with the number of dairy cows suffering from clinical mastitis in second lactation (P<0.05). The number of dairy cows suffering from clinical mastitis for individual with AG genotype was significantly higher than that of AA genotype (P<0.05). The number of dairy cows suffering from clinical mastitis for individual with CXCR1-816 AA genotype was significantly higher than that of AC and CC genotypes (P<0.05). The other SNPs showed no significant association with the number of clinical mastitis. CXCR1-816 A>C mutation showed an extremely significant association with culling age (P<0.01) and a significant association with productive month and culling lactation (P<0.05). The productive month, culling age and culling lactation for the individual of CXCR1-816 AA were significantly higher than that of CC genotype (P<0.05). Cox regression showed that only CXCR1-816 showed significant association with the productive life(P<0.05), the survival probability of individual with CC genotype was significantly lower than that of AA and AC genotype in each time period. 【Conclusion】The mutation of CXCR1-816 A>C showed significant association with the clinical mastitis and lifetime for Chinese Holstein, and this SNP could be used for molecular marker-assisted selection of productive life for Chinese Holstein when the molecular function of this mutation were proved.
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1038/s41437-019-0246-7URLPMID:31278370 [本文引用: 1]

The availability of whole genome sequencing (WGS) data enables the discovery of causative single nucleotide polymorphisms (SNPs) or SNPs in high linkage disequilibrium with causative SNPs. This study investigated effects of integrating SNPs selected from imputed WGS data into the data of 54K chip on genomic prediction in Danish Jersey. The WGS SNPs, mainly including peaks of quantitative trait loci, structure variants, regulatory regions of genes, and SNPs within genes with strong effects predicted with variant effect predictor, were selected in previous analyses for dairy breeds in Denmark-Finland-Sweden (DFS) and France (FRA). Animals genotyped with 54K chip, standard LD chip, and customized LD chip which covered selected WGS SNPs and SNPs in the standard LD chip, were imputed to 54K together with DFS and FRA SNPs. Genomic best linear unbiased prediction (GBLUP) and Bayesian four-distribution mixture models considering 54K and selected WGS SNPs as one (a one-component model) or two separate genetic components (a two-component model) were used to predict breeding values. For milk production traits and mastitis, both DFS (0.025) and FRA (0.029) sets of additional WGS SNPs improved reliabilities, and inclusions of all selected WGS SNPs generally achieved highest improvements of reliabilities (0.034). A Bayesian four-distribution model yielded higher reliabilities than a GBLUP model for milk and protein, but extra gains in reliabilities from using selected WGS SNPs were smaller for a Bayesian four-distribution model than a GBLUP model. Generally, no significant difference was observed between one-component and two-component models, except for using GBLUP models for milk.
DOI:10.3168/jds.2014-9005URLPMID:25892697 [本文引用: 1]

This study investigated the effect on the reliability of genomic prediction when a small number of significant variants from single marker analysis based on whole genome sequence data were added to the regular 54k single nucleotide polymorphism (SNP) array data. The extra markers were selected with the aim of augmenting the custom low-density Illumina BovineLD SNP chip (San Diego, CA) used in the Nordic countries. The single-marker analysis was done breed-wise on all 16 index traits included in the breeding goals for Nordic Holstein, Danish Jersey, and Nordic Red cattle plus the total merit index itself. Depending on the trait's economic weight, 15, 10, or 5 quantitative trait loci (QTL) were selected per trait per breed and 3 to 5 markers were selected to tag each QTL. After removing duplicate markers (same marker selected for more than one trait or breed) and filtering for high pairwise linkage disequilibrium and assaying performance on the array, a total of 1,623 QTL markers were selected for inclusion on the custom chip. Genomic prediction analyses were performed for Nordic and French Holstein and Nordic Red animals using either a genomic BLUP or a Bayesian variable selection model. When using the genomic BLUP model including the QTL markers in the analysis, reliability was increased by up to 4 percentage points for production traits in Nordic Holstein animals, up to 3 percentage points for Nordic Reds, and up to 5 percentage points for French Holstein. Smaller gains of up to 1 percentage point was observed for mastitis, but only a 0.5 percentage point increase was seen for fertility. When using a Bayesian model accuracies were generally higher with only 54k data compared with the genomic BLUP approach, but increases in reliability were relatively smaller when QTL markers were included. Results from this study indicate that the reliability of genomic prediction can be increased by including markers significant in genome-wide association studies on whole genome sequence data alongside the 54k SNP set.
