 ,, 汤继华
,, 汤继华 ,河南农业大学农学院/省部共建小麦玉米作物学国家重点实验室,郑州 450002
,河南农业大学农学院/省部共建小麦玉米作物学国家重点实验室,郑州 450002Identification of miRNAs and tRFs in Response to Salt Stress in Rice Roots
MENG ShuJun, ZHANG XueHai, WANG QiYue, ZHANG Wen, HUANG Li, DING Dong ,, TANG JiHua
,, TANG JiHua ,College of Agronomy, Henan Agricultural University/National Key Laboratory of Wheat and Maize Crop Science, Zhengzhou 450002
,College of Agronomy, Henan Agricultural University/National Key Laboratory of Wheat and Maize Crop Science, Zhengzhou 450002通讯作者:
责任编辑: 李莉
收稿日期:2019-07-7接受日期:2019-08-6网络出版日期:2020-02-16
| 基金资助: |
Received:2019-07-7Accepted:2019-08-6Online:2020-02-16
作者简介 About authors
孟淑君,E-mail:18237116524@163.com。
张雪海,E-mail:xuehai85@126.com。

摘要
关键词:
Abstract
Keywords:
PDF (995KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
孟淑君, 张雪海, 王琪月, 张稳, 黄力, 丁冬, 汤继华. 水稻根系盐胁迫响应miRNA和tRF的鉴定[J]. 中国农业科学, 2020, 53(4): 669-682 doi:10.3864/j.issn.0578-1752.2020.04.001
MENG ShuJun, ZHANG XueHai, WANG QiYue, ZHANG Wen, HUANG Li, DING Dong, TANG JiHua.
0 引言
【研究意义】水稻(Oryza sativa L.)是中国最重要的粮食作物,随着全球气候变暖及生态环境恶化,水稻生长过程中遭受的干旱、高盐等非生物胁迫日益增加。盐胁迫作为主要的非生物胁迫之一,严重影响水稻的生长发育。植物盐胁迫一般表现为渗透胁迫和离子毒害2个阶段,引起植株体内活性氧破坏、膜系统损伤、光合和呼吸作用受抑制、酶活性降低等;表现为植株矮小,叶片黄化,最终导致粮食产量、品质的下降[1]。在高盐环境下,水稻会产生2种生理反应:快速的渗透反应,能抑制新梢/根的生长;较慢的离子反应,加速成熟叶片的衰老或加快细胞程序性死亡[2]。【前人研究进展】MicroRNAs(miRNAs)是一类普遍存在于生物体内,5'端带有磷酸基团,3'端带有羟基,长度为20—24 nt的内源性非编码小分子RNA[3],在转录后水平调控其靶基因的表达。miRNA通过剪切靶基因转录本或抑制靶基因翻译行使其生物学功能[4,5]。在植物中,miRNA通过对靶基因的调控,参与植物的生长发育、器官形态建成、生物及非生物胁迫响应等多种生物学进程[6,7]。GAO等[8]发现miR393的表达水平在盐碱胁迫下明显上调,而过表达miR393的转基因水稻对盐碱处理比野生型更敏感,表明miR393是水稻盐碱胁迫的负调控miRNA。YANG等[9]研究表明,过表达miR171c可降低水稻的盐胁迫耐受能力,说明miR171c是水稻耐盐胁迫的负调控因子。此外,miRNA156是调控水稻干旱和盐胁迫的重要因子[10],水稻miR393可以和miR390在不同胁迫条件下协同调控水稻侧根生长[11]。tRF是一种最近被发现的、在动植物中广泛存在的小分子量非编码RNA,其结构、分布及其生物学功能正日益受到研究者关注。tRF序列全长20—40 nt,由前体tRNA或成熟tRNA经核苷酸酶加工和修饰而成[12]。tRF最先在人类细胞中被鉴定获得[13]。根据来源不同,tRF可被分为3'端tRF[14]、不包含5'端和3'端结构的tRF[15]、含有CCA结构的tRF[16]、5'端tRF[17]、tRNA半分子tiR(tRNA-derived stress-induced RNAs)[18]等5种类型。功能研究表明,tRF具有类似于miRNA的调控功能,是一种潜在的转录后基因表达调控因子。tRF在调控mRNA翻译、细胞增殖、应激条件下细胞应答和人类疾病方面均具有重要作用[19]。除了在四膜虫中[20],tRF的表达与它们各自的前体tRNA的丰度无关[21,17]。迄今为止,tRF在植物中的功能研究相对较少。CHEN等[22]在水稻胚愈伤组织中鉴定到大量tRF,其中表达量最高的tRF来自于Ala-AGC tRNA的5'端,且未分化愈伤中的tRF含量显著高于已分化愈伤。【本研究切入点】根系是水稻直接接收盐胁迫信号的部位。除功能基因外,调控性核酸分子在植物逆境响应过程中也发挥着不可替代的作用。miRNA是植物胁迫响应的上游调控因子,通过调节靶基因的表达响应胁迫信号。而作为新近发现的小分子量调控RNA,tRF在植物胁迫响应方面的鉴定及功能研究鲜见报道。【拟解决的关键问题】本研究以水稻粳稻品种日本晴为试验材料,对盐胁迫处理的水稻根系材料进行了小分子量RNA测序,并利用qRT-PCR方法对测序结果进行了验证。鉴定和筛选到水稻盐胁迫响应miRNA、tRF及其靶基因;分析盐胁迫处理/非盐处理水稻根系miRNA、tRF差异表达情况,并建立了水稻非编码小分子量RNA参与的盐胁迫响应基因表达调控网络。1 材料与方法
1.1 试验材料
挑选饱满且均匀一致的水稻粳稻品种日本晴种子60粒,剥去种子的颖壳,在超净工作台内经75%酒精浸泡清洗5 min,用0.1%升汞处理20 min,灭菌去离子水清洗4—5次,并用滤纸吸取多余的水分。将灭菌处理过的种子分别铺于1/2MS和含150 mmol·L-1 NaCl的1/2MS培养基上,每个直径90 mm的培养皿中铺放30粒种子。用灭菌的镊子夹取种子,将水稻种子按照从左到右的顺序均匀排列。封口膜封口后的培养皿放置于28℃恒温培养箱中进行暗培养,待种子露白后于光照培养箱中培养(每日光照28℃ 14 h/黑暗25℃ 10 h)。处理21 d后,取盐胁迫处理/非盐处理水稻植株根部样品液氮中速冻后置于-80℃保存备用。每份材料取3个生物学重复,分别用于提取根系组织总RNA进行小分子量RNA测序,每个生物学重复由3个植株的根系样品等量混合组成。1.2 水稻根系RNA提取
使用冷冻的TriZol(Invitrogen,Carlsbad,CA,USA)根据RNeasy minikit(Qiagen, Hilden, Germany)说明书提取水稻根系总RNA,利用NanoDrop2000分光光度计检测RNA浓度,并利用Agilent2100检测RNA样品的质量及完整性。1.3 Small RNA建库测序
使用15%PAGE胶分离不同片段大小的RNA,取14—40 nt条带回收小分子量RNA;使用RNA连接酶将回收的小分子量RNA加上3'和5'接头,每次连接后进行切胶回收。将最终回收到的小分子量RNA反转录成cDNA,PCR扩增16个循环;用PAGE胶对PCR产物切胶回收,完成文库构建,质检后用于高通量测序。利用illumina Nextseq500仪器,75SE测序模式对样品进行小分子量RNA测序。1.4 Small RNA 数据分析方法
首先对测序得到的原始数据进行过滤,去除有5'接头污染的、长度小于18 nt、polyA和低质量(Q<=5)的reads,得到clean reads用于数据分析。1.4.1 miRNA数据分析 利用bowtie软件[23]将数据与miRBase数据库(水稻)(http://www.miRBase.org/, Version 21;包括172个成熟的miRNA)进行比对。当测序片段与数据库中已知的miRNA序列错配数小于2时认为该测序片段为已知的miRNA。为了判断样品之间miRNA的表达量差异,用featurecounts软件[24]统计样本reads数。用DESeq2进行差异基因分析[23]。将每个样品的所有miRNA进行归一化处理,处理后如果miRNA在2个材料内的表达量都小于1RPM,则不予考虑[25]。两材料之间表达量差异大于2倍(log2FC>1或<-1),则认为是差异表达的小分子量RNA。
1.4.2 tRF数据分析 从Ensembl数据库(http://grch37. ensembl.org/index.html)下载水稻参考基因组数据的注释信息,从中筛选出tRNA的注释信息,将tRNA序列从反密码子处分开,区分为tRNA的5'和3'端,构建tRF序列比对文件。根据构建的5'和3'的注释信息,使用featurecounts软件统计与tRNA5’及3’序列匹配的reads数目,并计算每种长度的tRF所占的比例。
1.5 miRNA和tRF及其靶基因表达量分析
分别以日本晴盐处理/非盐处理根系总RNA为模板,利用Mir-XTM miRNA First-Strand Synthesis试剂盒(TaKaRa)加A法进行反转录合成cDNA第一链。利用TB GreenTM Premix Ex TaqTM Ⅱ(TaKaRa)荧光定量试剂盒在CFX96 Real-Time PCR Detection System实时定量PCR仪上对转录组数据表达差异的3个miRNA基因(miR156、miR167和miR396)进行定量分析。miRNA的定量引物使用其成熟序列(U用T取代)作为qRT-PCR的正链引物(F)(表1),试剂盒中的mRQ3'作为负链引物(R)。内参为U6,qRT-PCR引物列于表1中,每个反应设有3个生物学重复和3次技术重复。采用2-ΔΔCt方法[26]计算基因的相对表达量,并利用T测验分析基因的表达差异。Table 1
表1
表1水稻盐胁迫相关基因qRT-PCR引物序列
Table 1
| 名称 Name | 引物序列 Primer sequence (5'-3') |
|---|---|
| osa-miR156a-F | TGACAGAAGAGAGTGAGCAC |
| osa-miR167f-F | TGAAGCTGCCAGCATGATCTG |
| osa-miR396e-5p-F | TCCACAGGCTTTCTTGAACTG |
| EPlORYSAT000373812-F | GCGGATGTAGCCAAGTGGATCA |
| EPlORYSAT000373840-F | GGATTGTAGTTCAATTGGTCAGAGC |
| T156-OsSPL11-F | ACCATGCAAACACCACTTCA |
| T156-OsSPL11-R | TTGGCAAGAGCTCATTTGTG |
| T156-OsSPL13-F | GTGCCAGGTGGAGAGGTG |
| T156-OsSPL13-R | GTCGAACTCCGTCAGCTCAT |
| T167-OsARF6-F | AGCCTGAGTACCTCCAGCAA |
| T167-OsARF6-R | GGTGTAGACTGAGGGGTGGA |
| T167-Os06g03830-F | GATGACCTTCGCCACAAACT |
| T167-Os06g03830-R | CGTCGGATCGTACGGTATCT |
| T396-OsGRF2-F | GTTGTCCAAGGAGCACTGC |
| T396-OsGRF2-R | GTGGGGATGGAGATGGAGAG |
| Os01g0810100-F | ATTCTGGGCACTGTTTGGAG |
| Os01g0810100-R | GCAACATCTTGCCATGTGAG |
| Os04g0531300-F | TGCCCACAAGAAAGGGATAG |
| Os04g0531300-R | GCTCCCATTCCACCACTAAG |
| Osβ-Actin-F | GGAAGTACAGTGTCTGGATTGGAG |
| Osβ-Actin-R | TCTTGGCTTAGCATTCTTGGGT |
新窗口打开|下载CSV
选取测序结果中差异表达的来源于tRNA EPlORYSAT000373840和EPlORYSAT000373812的2个tRF,截取特异序列作为正链引物(F)(表1),进行qRT-PCR分析,方法同miRNA定量。
靶基因定量分析利用TaKaRa PrimerScriptTM RT reagent Kit with gDNA Eraser试剂盒(TaKaRa)对总RNA进行反转录。利用Primer3(http://primer3.ut.ee/)在靶基因外显子区设计特异定量引物(表1),qRT-PCR方法同上,内参基因为Osβ-Actin。
1.6 保守性miRNA及tRF靶基因分析
利用psRNATarget网站(https://plantgrn.noble. org/psRNATarget/analysis)预测保守Small RNA靶基因及其功能,在Gramene网站(http://www.gramene. org/)使用BLAST功能分析tRF靶基因并进行功能注释。2 结果
2.1 水稻盐胁迫响应表型
在1/2MS培养基中对水稻进行盐胁迫处理21 d,150 mmol·L-1 NaCl处理与非盐处理相比,水稻日本晴幼苗明显受到盐胁迫的抑制(图1)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1NaCl处理21 d的水稻苗期表型
A:盐处理的日本晴;B:非盐处理的日本晴;比例尺:1 cm
Fig. 1Phenotypes of rice seedlings at 21 days under NaCl
A: Salt treatment of Nipponbare; B: Non salt treatment of Nipponbare; Bar: 1 cm
2.2 RNA-seq测序数据保守miRNA的鉴定
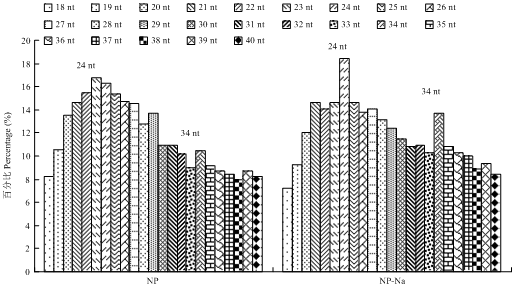
2.2.1 水稻小分子量RNA测序 对日本晴盐处理和非盐处理21 d根系的小分子量RNA文库进行高通量测序,以鉴定影响水稻盐胁迫响应的miRNA和tRF。为消除不同个体间的差异,每个样品设置3个生物学重复用于RNA提取及后续测序分析。盐处理和非盐处理21 d的水稻根系每个样品设置3个生物学重复,共6个样品,共得到150 501 099条测序数据。平均每个样品有25 083 517个读数,范围从15 848 571到54 767 011。测序所得的小分子量RNA几乎涵盖所有RNA类型,包括tRNA、rRNA、snRNA、snoRNA、siRNA、miRNA、外显子和内含子降解片段等。片段大小在18—75 bp,根据小分子量RNA的长度获得分类柱状图(图2)。在盐处理水稻根系中24 nt的小分子量RNA的量最多,占其测序数据的18.40%,非盐处理水稻根系中24 nt的小分子量RNA的量占其测序数据的16.27%。盐处理水稻根系中34—38 nt的小分子量RNA的量明显多于非盐处理水稻根系。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2小分子量RNA的长度分类柱状图
NP:日本晴非盐处理;NP-Na:日本晴盐处理 NP: Non salt treatment of Nipponbare; NP-Na: Salt treatment of Nipponbare
Fig. 2Histogram of the length of small molecular RNA
2.2.2 水稻盐胁迫响应miRNA的鉴定 将测序数据与水稻miRNA数据库miRBase(http://www.mirbase. org)中的成熟序列及前体序列进行比对,鉴定出保守的miRNA。以至少有一个样品RPM>1为筛选条件,共比对到451个miRNA。为了寻找表达量较高的miRNA,以至少有一个样品RPM>50为筛选条件,共比对到149个miRNA(电子附表1)。
2.2.3 保守miRNA差异表达分析 为了寻找水稻盐胁迫响应相关的差异miRNA,对检测到的149个保守miRNA进行均一化后的差异分析,以至少一组数据RPM>500为筛选条件,共得到54个miRNA,分属于25个miRNA家族。对在盐胁迫处理和非盐处理条件下水稻日本晴根系的miRNA的表达量进行分析,以log2FC>1或<-1为筛选条件,共得到8个表达量显著上调的miRNA和23个表达量显著下调的miRNA(表2),属于12个miRNA家族。
Table 2
表 2
表 2水稻盐胁迫响应差异表达miRNA
Table 2
| miRNA | 染色体 Chr. | 起始 Start | 终止 End | 长度 Length (bp) | 序列 Sequence (5'-3') | NP-RPM 平均值 Average of NP-RPM | NP-Na-RPM 平均值 Average of NP-Na-RPM | Log2 (NP-Na/NP) | 表达差异 Differential expression |
|---|---|---|---|---|---|---|---|---|---|
| osa-miR397a | 6 | 28489785 | 28489898 | 21 | UCAUUGAGUGCAGCGUUGAUG | 3385.00 | 273.09 | -3.631709 | 下调Down |
| osa-miR397b | 2 | 3280781 | 3280898 | 21 | UUAUUGAGUGCAGCGUUGAUG | 14209.57 | 1461.91 | -3.280937 | 下调Down |
| osa-miR1882e-3p | 10 | 10320904 | 10321044 | 24 | GAAAUGAUCUUGGACGUAAUCUAG | 3760.37 | 649.34 | -2.533830 | 下调Down |
| osa-miR396f-5p | 2 | 35636546 | 35636721 | 22 | UCUCCACAGGCUUUCUUGAACU | 1173.18 | 321.53 | -1.867414 | 下调Down |
| osa-miR396e-5p | 4 | 34436820 | 34437003 | 21 | UCCACAGGCUUUCUUGAACUG | 1173.18 | 321.53 | -1.867414 | 下调Down |
| osa-miR156a | 1 | 22524147 | 22524246 | 20 | UGACAGAAGAGAGUGAGCAC | 2155.81 | 688.99 | -1.645684 | 下调Down |
| osa-miR156b-5p | 1 | 4666341 | 4666516 | 20 | UGACAGAAGAGAGUGAGCAC | 2155.81 | 688.99 | -1.645684 | 下调Down |
| osa-miR156c-5p | 1 | 4665975 | 4666123 | 20 | UGACAGAAGAGAGUGAGCAC | 2155.81 | 688.99 | -1.645684 | 下调Down |
| osa-miR156d | 2 | 4512884 | 4513012 | 20 | UGACAGAAGAGAGUGAGCAC | 2155.81 | 688.99 | -1.645684 | 下调Down |
| osa-miR156e | 4 | 25026327 | 25026430 | 20 | UGACAGAAGAGAGUGAGCAC | 2155.81 | 688.99 | -1.645684 | 下调Down |
| osa-miR156f-5p | 8 | 21478230 | 21478415 | 20 | UGACAGAAGAGAGUGAGCAC | 2155.81 | 688.99 | -1.645684 | 下调Down |
| osa-miR156g-5p | 2 | 8412516 | 8412618 | 20 | CGACAGAAGAGAGUGAGCAC | 2155.81 | 688.99 | -1.645684 | 下调Down |
| osa-miR156h-5p | 8 | 21491232 | 21491417 | 20 | UGACAGAAGAGAGUGAGCAC | 2155.81 | 688.99 | -1.645684 | 下调Down |
| osa-miR156i | 2 | 24119995 | 24120084 | 20 | UGACAGAAGAGAGUGAGCAC | 2155.81 | 688.99 | -1.645684 | 下调Down |
| osa-miR156j-5p | 6 | 26554795 | 26554959 | 22 | GCUCGCUCCUCUUUCUGUCAGC | 2155.81 | 688.99 | -1.645684 | 下调Down |
| osa-miR167d-5p | 7 | 4166404 | 4166295 | 21 | UGAAGCUGCCAGCAUGAUCUG | 2924.08 | 1169.08 | -1.322607 | 下调Down |
| osa-miR167e-5p | 2 | 3742241 | 3742513 | 21 | UGAAGCUGCCAGCAUGAUCUG | 2924.08 | 1169.08 | -1.322607 | 下调Down |
| osa-miR167f | 10 | 14723044 | 14723156 | 21 | UGAAGCUGCCAGCAUGAUCUG | 2924.08 | 1169.08 | -1.322607 | 下调Down |
| osa-miR167g | 3 | 3347682 | 3347763 | 21 | UGAAGCUGCCAGCAUGAUCUG | 2924.08 | 1169.08 | -1.322607 | 下调Down |
| osa-miR167h-5p | 12 | 25480618 | 25480737 | 21 | UGAAGCUGCCAGCAUGAUCUG | 2924.08 | 1169.08 | -1.322607 | 下调Down |
| osa-miR167i-5p | 6 | 27674749 | 27674949 | 21 | UGAAGCUGCCAGCAUGAUCUG | 2924.08 | 1169.08 | -1.322607 | 下调Down |
| osa-miR167j | 1 | 32686068 | 32686227 | 21 | UGAAGCUGCCAGCAUGAUCUG | 2924.08 | 1169.08 | -1.322607 | 下调Down |
| osa-miR1432-5p | 7 | 23401702 | 23401810 | 21 | AUCAGGAGAGAUGACACCGAC | 485.58 | 219.53 | -1.145259 | 下调Down |
| osa-miR159f | 1 | 6693112 | 6693299 | 21 | CUUGGAUUGAAGGGAGCUCUA | 874.14 | 1817.05 | 1.055665 | 上调Up |
| osa-miR159a.1 | 1 | 17681923 | 17682194 | 21 | UUUGGAUUGAAGGGAGCUCUG | 15079.03 | 32657.51 | 1.114871 | 上调Up |
| osa-miR159b | 1 | 1215030 | 1215217 | 21 | UUUGGAUUGAAGGGAGCUCUG | 15079.03 | 32657.51 | 1.114871 | 上调Up |
| osa-miR168a-5p | 2 | 1553154 | 1553240 | 21 | UCGCUUGGUGCAGAUCGGGAC | 398.64 | 916.89 | 1.201650 | 上调Up |
| osa-miR1876 | 10 | 4833365 | 4833521 | 24 | AUAAGUGGGUUUGUGGGCUGGCCC | 1462.10 | 3609.01 | 1.303566 | 上调Up |
| osa-miR1423-5p | 4 | 19715117 | 19715252 | 24 | AGGCAACUACACGUUGGGCGCUCG | 265.74 | 1720.74 | 2.694957 | 上调Up |
| osa-miR164e | 3 | 10542157 | 10542288 | 21 | UGGAGAAGCAGGGCACGUGAG | 75.61 | 558.04 | 2.883783 | 上调Up |
| osa-miR5077 | 3 | 14094752 | 14094842 | 19 | GUUCGCGUCGGGUUCACCA | 93.53 | 856.96 | 3.195773 | 上调Up |
新窗口打开|下载CSV
水稻盐胁迫相关的miRNA的差异还反映在每个家族成员的数目上(图3)。测序结果检测到数目最多的miRNA家族是osa-miR156,由10个成员组成;随后是osa-miR167、osa-miR397、osa-miR396和osa-miR159,分别有7、2、2和3个成员。其他的miRNA家族,如osa-miR1882、osa-miR1432、osa-miR168、osa-miR1432、osa-miR1876、osa-miR164和osa-miR5077等,只检测到一个盐胁迫响应成员。说明不同miRNA家族成员对盐胁迫信号响应情况存在差异。
图3
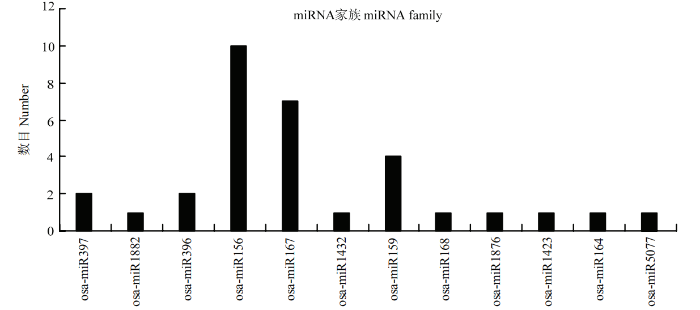 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3检测到的每个miRNA家族成员数
Fig. 3The number of detected family members per miRNA family
2.2.4 差异miRNA靶基因预测 大多数植物内保守的miRNA与其靶基因具有高度的匹配,一个miRNA可以靶定多个靶基因。利用miRNA预测软件psRNA Target program(http://plantgrn.noble.org/psRNATarget/)对在水稻盐胁迫响应的31个差异表达的miRNA(12个miRNA家族)的靶基因进行了预测,共得到162个靶基因(电子附表2)。
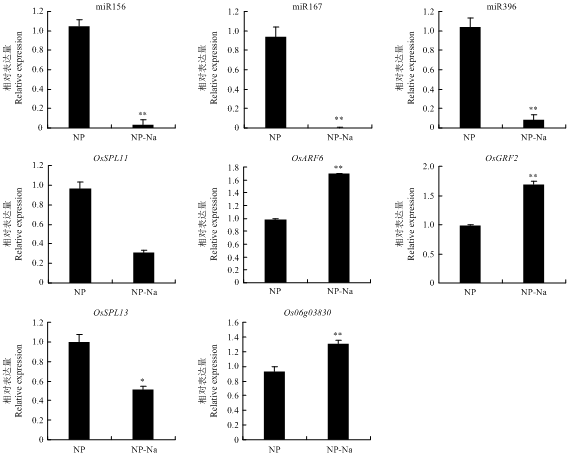
2.2.5 差异miRNA及其靶基因的qRT-PCR表达分析 选择在本研究中被检测到的家族成员较多,且在其他物种中已有其与盐胁迫相关报道的3个差异表达的miRNA,包括miR156、miR167和miR396,利用qRT-PCR对其成熟序列进行检测验证,结果表明,高通量测序与qRT-PCR虽然存在相对倍数的差异,但所有检测miRNA的相对表达差异与测序数据相一致(表2和图4)。这种表达量差异倍数差异也许是由于2种技术的操作原理与精度不同导致的。利用qRT-PCR对选择的miRNA对应的靶基因进行表达量分析。在盐胁迫条件下,miR156的靶基因OsSPL11(Os06g45310)和OsSPL13(Os07g32170)的相对表达量都较对照有显著降低;miR167的靶基因Os06g03830相对表达量显著上调,而OsARF6(Os02g0164900)的相对表达量较对照极显著下调;miR396的靶基因OsGRF2(Os06g10310)的相对表达量和对照相比极显著上调(图4)。
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4水稻盐胁迫相关miRNA及其靶基因的qRT-PCR表达分析
NP:非盐处理日本晴;NP-Na:盐处理日本晴;*表示0.05水平差异;**表示0.01水平差异。下同
Fig. 4qRT-PCR expression analysis of salt stress miRNAs and target genes in rice
NP: Non salt treatment Nipponbare; NP-Na: Salt treatment Nipponbare; * indicates difference at the 0.05 level; ** indicates difference at the 0.01 level. The same as below
2.2.6 水稻盐胁迫响应tRF的鉴定及靶基因预测 通过与tRNA数据库进行比较,以至少有一个样品RPM>1为筛选条件[25],与tRNA 5'端序列共比对到145个tRF。与非盐处理的日本晴相比,在盐胁迫条件下的日本晴有77个差异表达的tRF,其中有75个上调表达的tRF,2个下调表达的tRF(电子附表3),以至少一组数据RPM>50数据为筛选条件,共比对到6个tRF,其中有3个tRF的表达量是上升的,其余3个tRF无差异(表3)。以至少有一个样品RPM >1为筛选条件,3'端共比对到78个tRF,与非盐处理的日本晴根系相比,在盐胁迫条件下的日本晴根系有58个上调表达的tRF,其余的tRF无表达差异(电子附表3),以至少一组数据RPM>50数据为筛选条件,共比对到4个tRF,其中3个tRF的表达量上调(表3)。与非盐处理的日本晴根系相比,在盐胁迫条件下的日本晴根系在3'和5'端比对到tRF共有的来源tRNA有2个,分别为tRNA-Glu的EPlORYSAT000373797以及tRNA-Asp的EPlORYSAT000373840。由此可见,大多数tRF受盐胁迫诱导表达,推测这些差异tRF可能是水稻盐胁迫响应通路中的重要调节因子。
Table 3
表3
表3水稻盐胁迫相关tRF的来源tRNA
Table 3
| 来源tRNA基因编号 Gene ID of source tRNA | 基因注释 Gene expression | 位置 Loction | 长度 Length (bp) | 序列 Sequence (5'-3') | NP-RPM 平均值 NP-RPM average value | NP-Na-RPM 平均值 NP-Na-RPM average value | Log2 (NP-Na/NP) |
|---|---|---|---|---|---|---|---|
| ENSRNA049444301 | 反密码子为UUC的谷氨酸 tRNA tRNA-Glu for anticodon UUC | 12:25043392..25043429 | 38 | GAAAGCCAGATAT CCTAACCGGACTA GACGACAATGGA | 54.91457551 | 79.26862858 | 0.529560894 |
| ENSRNA049444701 | 反密码子为UUC的谷氨酸 tRNA tRNA-Glu for anticodon UUC | 12:2735598..2735634 | 37 | GCCATTGTCGTCTA GTCCGGTTAGGAT ACCTGGCTTT | 280.0756319 | 532.1395357 | 0.925988127 |
| EPlORYSAT000373797 | 反密码子为UCC的谷氨酸 tRNA tRNA tRNA-Glu (UCC) | Pt:15650..15686 | 37 | GCCCCTATCGTCTA GTGGTTCAGGACAT CTCTCTTTC | 51.33846231 | 85.59087607 | 0.737416929 |
| EPlORYSAT000373812 | 反密码子为GUG的组氨酸 tRNA tRNA-His (GUG) | Pt:81050..81087 | 38 | GGCGGATGTAGC CAAGTGGATCAAG GCAGTGGATTGTG | 27.04710805 | 62.75454797 | 1.214245673 |
| EPlORYSAT000373840 | 反密码子为GUC的天门 冬氨酸 tRNA tRNA-Asp (GUC) | Pt:16231..16267 | 37 | GGGATTGTAGTTC AATTGGTCAGAGC ACCGCCCTGTC | 45.81324826 | 183.8090975 | 2.004371412 |
| ENSRNA049445145 | 反密码子为CGC的丙氨酸 tRNA tRNA-Ala for anticodon CGC | 8:22194244..22194280 | 37 | GGGGACGTAGCTCATATGGTAGAGCGCTCGCTTCGCA | 3.948187725 | 64.83354259 | 4.037477913 |
| ENSRNA049444418 | 反密码子为UUC的谷氨酸 tRNA tRNA-Glu for anticodon UUC | 12:27018602..27018639 | 38 | TCACCCAGACGACCCGGGTTCAAATCCCGGCAATGGAA | 8.877923176 | 86.56822355 | 3.285543425 |
| ENSRNA049446755 | 反密码子为GCC的甘氨酸 tRNA tRNA-Gly for anticodon GCC | 3:25619176..25619211 | 36 | ACGGTACAGACCC GGGTTCGATTCCC GGCTGGTGCA | 5.511015306 | 72.72375321 | 3.722036619 |
| EPlORYSAT000373797 | 反密码子为UCC的谷氨酸 tRNA tRNA-Glu(UCC) | Pt:15686..15722 | 37 | CAAGGAGGCAGCG GGGATTCGACTTC CCCTGGGGGTA | 24.28546159 | 47.55822306 | 0.969601905 |
| EPlORYSAT000373840 | 反密码子为GUC的天门 冬氨酸 tRNA tRNA-Asp(GUC) | Pt:16267..16304 | 38 | CAAGGCGGAAGCT GCGGGTTCGAGCC CCGTCAGTCCCG | 14.74937311 | 52.34254363 | 1.827330398 |
新窗口打开|下载CSV
利用Gramene网站(http://gramene.org/)的BLAST功能对盐胁迫诱导的6个tRF进行靶基因预测,共得到29个靶基因(表4)。
Table 4
表4
表4水稻盐胁迫差异表达tRF预测靶基因
Table 4
| 来源tRNA基因编号 Gene ID of source tRNA | 靶基因编号 Target gene ID | 位置 Location | 基因注释 Gene description |
|---|---|---|---|
| EPlORYSAT000373812 | Os01g0810100 | 1:34412916..34416988 | 叶绿体核糖核酸酶III蛋白 Chloroplast ribonuclease III domain protein |
| Os10g0119300 | 10:1234067..1241670 | FH2结构域结合肌动蛋白 Actin-binding FH2 domain containing protein | |
| Os03g0301700 | 3:10621704..10625395 | 钙调蛋白结合蛋白磷酸酶 Calmodulin-binding protein phosphatase | |
| Os03g0435200 | 3:18355628..18365524 | 五肽重复蛋白 Pentatricopeptide repeat domain containing protein | |
| EPlORYSAT000373840 | Os04g0531300 | 4:26565544..26571685 | tRNA -二氢吡啶合酶蛋白 tRNA-dihydrouridine synthase domain containing protein |
| Os11g0593500 | 11:22604874..22606587 | 环状F-box蛋白 Cyclin-like F-box domain containing protein | |
| Os06g0128200 | 6:1485977..1490634 | LMBR1膜内在蛋白 LMBR1 integral membrane protein | |
| Os03g0562200 | 3:20188178..20194997 | 环状F-box蛋白 Cyclin-like F-box domain containing protein | |
| Os03g0860700 | 3:36320679..36333253 | 肌球蛋白Myosin | |
| Os01g0850100 | 1:36540164..36545196 | 磷脂酸类磷脂 Phosphatidic acid phosphatase-like protein | |
| ENSRNA049445145 | Os08g0389300 | 8:18398361..18407618 | UbiA-异戊二烯转移酶家族蛋白 UbiA prenyltransferase family domain containing protein |
| Os10g0464400 | 10:17133478..17137843 | 卤酸脱卤酶类水解酶蛋白 Haloacid dehalogenase-like hydrolase domain containing protein | |
| ENSRNA049444418 | Os08g0134900 | 8:1988346..1990818 | 保守假设蛋白Conserved hypothetical protein |
| Os01g0267800 | 1:9192164..9193439 | 丝氨酸/苏氨酸蛋白激酶蛋白 Serine/threonine protein kinase domain containing protein | |
| Os03g0320100 | 3:11549564..11551862 | α-N-阿拉伯呋喃糖苷酶 A Alpha-N-arabinofuranosidase A | |
| Os07g0484800 | 7:17787901..17801227 | 腺嘌呤磷酸核糖转移酶蛋白 Adenine phosphoribosyltransferase like protein | |
| Os09g0333600 | 9:10071433..10085739 | 多效性耐药蛋白4 Pleiotropic drug resistance protein 4 | |
| Os09g0529700 | 9:20744838..20748509 | 液泡蛋白分选;内吞体分选转运复合体 Vacuolar protein sorting; endocyte sorting and transport complex | |
| Os02g0250700 | 2:8518506..8535685 | 表达蛋白Expressed protein | |
| Os12g0256600 | 12:8809699..8811919 | 保守假设蛋白Conserved hypothetical protein | |
| Os08g0226100 | 8:7698342..7699299 | 保守假设蛋白Conserved hypothetical protein | |
| Os05g0400200 | 5:19470226..19475222 | 逆转录转座子蛋白Ty1亚类 Retrotransposon protein Ty1-copia subclass | |
| Os05g0126200 | 5:1497942..1499638 | 保守假设蛋白Conserved hypothetical protein | |
| ENSRNA049446755 | Os08g0387300 | 8:18299461..18299571 | 膜蛋白质Membrane protein |
| Os03g0320100 | 3:11549564..11551862 | α-N-阿拉伯呋喃糖苷酶A Alpha-N-arabinofuranosidase A | |
| Os05g0400200 | 5:19470226..19475222 | 逆转录转座子蛋白Ty1亚类 Retrotransposon protein Ty1-copia subclass | |
| EPlORYSAT000373840 | Os03g0165300 | 3:3513371..3517214 | 保守假设蛋白Conserved hypothetical protein |
| Os12g0274450 | 12:10072058..10075165 | 保守假设蛋白Conserved hypothetical protein | |
| Os04g0271700 | 4:11375483..11377317 | 玉米素葡萄糖基转移酶UDP-glycosyltransferase |
新窗口打开|下载CSV
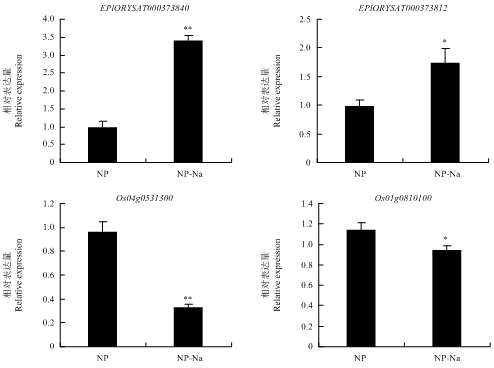
2.2.7 盐胁迫诱导差异表达tRF及其靶基因的qRT-PCR表达分析 对水稻盐胁迫诱导差异表达的2个tRF(EPlORYSAT000373812和EPlORYSAT000373840)进行qRT-PCR验证,结果表明,检测tRF的相对表达差异与测序数据相一致(图5),与非盐处理相比,盐处理水稻根系中2个tRF的相对表达量均显著升高。利用qRT-PCR对EPlORYSAT000373812和EPlORYSAT000373840靶基因Os01g0810100、Os04g0531300的表达量进行分析发现(图5),与非盐处理相比,盐处理水稻根系中靶基因的相对表达量均显著降低。
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5水稻盐胁迫相关tRF及其靶基因的qRT-PCR表达分析
Fig. 5qRT-PCR expression analysis of salt stress tRFs and target genes in rice
3 讨论
3.1 使用RNA-seq方法,可高通量获得水稻盐胁迫响应small RNA
已有研究表明,植物遭受非生物胁迫会诱导相应miRNA的表达变化,通过对其靶基因的转录后表达调控影响植株形态、生理和代谢以对逆境做出响应。RNA-Seq技术已经成为目前研究胁迫相关的miRNA功能及调控路径的主要方法[27,28,29]。水稻是盐敏感作物,高盐环境能够影响水稻正常的代谢活动,引起细胞壁的破坏、膜系统破坏、细胞质溶解,产生渗透胁迫甚至离子毒害,并影响基因组的稳定性,最终引起细胞凋亡或植株死亡,严重影响水稻的产量和品质。miRNA在水稻盐胁迫响应通路中发挥重要的调控作用。本研究通过RNA-Seq手段共检测到水稻根系盐胁迫差异表达的miRNA 54个,分属于25个miRNA家族,其中8个miRNA表达量上调,23个miRNA表达量下调。利用实时荧光定量PCR方法对测序结果进行了验证,miRNA定量结果与测序结果基本一致,说明RNA-Seq能够高通量、准确可靠地鉴定盐胁迫下水稻根系miRNA的表达情况。
由于miRNA的碱基序列较短,靶定的靶基因较多,且参与许多较复杂的生物学过程并起很重要的调控作用,因而预测miRNA的靶基因是研究miRNA作用机制的基础,研究miRNA的功能及作用机制就要研究靶基因与其miRNA的相互作用方式。本研究运用psRNATarget网站分析鉴定了RNA-Seq中筛选得到的盐胁迫相关差异表达的miRNA靶基因162个。这些miRNA通过对其靶基因的转录后表达调控影响水稻盐胁迫响应的生理生化过程。
3.2 miRNA在不同植物生长发育和胁迫响应中功能的通用性
在本研究检测到的差异表达基因中,有8个miRNA家族在其他物种中也被报道为盐胁迫响应miRNAs,如表达量下调的osa-miR397、osa-miR396、osa-miR156、osa-miR167、osa-miR1432和表达量上调的osa-miR159、osa-miR168、osa-miR164。研究表明,红花miR397a表达可增强植物对NaCl的敏感性[30];番茄的miR397过表达植株中,LeLACmiR397表达量降低,从而使番茄出现盐敏感表型[31]。烟草中Sp-miR396a-5p通过作用于NtGRF1和NtGRF3的调控网络,靶向NtGRF7调控渗透应激反应基因的表达和病原菌感染,在非生物胁迫中发挥重要作用[32]。紫花苜蓿在重度盐胁迫下,miR156下调了SPL转录因子家族基因,修饰了重要转录因子的表达,并下调下游盐胁迫应答基因,说明miR156在紫花苜蓿对盐胁迫的生理反应和转录过程中起着调节作用[33]。在番茄中盐、干旱和热处理导致了miR167表达量下调,表明不同胁迫激活了miR167a调控替代机制[34];拟南芥中miR167与其靶基因ARF6和ARF8之间存在反馈调节作用,ARF6激活miR167表达,ARF8抑制miR167表达,它们之间的彼此激活或抑制作用调控不定根的形成[35]。毛竹经强光、黑暗、高温、低温、NaCl等胁迫处理后,叶片中phe-miR1432的表达下调[36]。YIN等[37]利用棉花耐盐品种和盐敏感品种研究miRNA的表达差异,发现在盐胁迫条件下,盐敏感品种中的miR159表达下调;而在耐盐品种中miR156a/d/e、miR169显著下调,miR167a、miR397a/b上调。miR168介导的反馈调控环调节ARGONAUTE1(AGO1)的稳态对基因表达调控和植物发育至关重要,过表达miR168a的植株和AGO1功能缺失突变体均表现出ABA超敏性和耐旱性,而mir168a突变体表现出ABA低敏性和干旱超敏性[38];高盐条件下拟南芥中miR168的表达量在初期上升,但6 h后恢复正常水平[39]。李贺春等[40]利用miRNA基因芯片杂交技术对盐胁迫条件下耐盐棉花品系和盐敏感棉花品种的miRNA进行差异表达发现,在盐胁迫条件下miR156、miR164、miR167、miR397和miR399在耐盐棉花品系较盐敏感品系表达上调。以上分析表明,部分水稻中的盐胁迫响应miRNA在其他植物的盐胁迫信号通路中依然适用,说明了这些miRNA在调节不同植物生长发育和胁迫响应中功能的通用性及进化的整体性。在miRNA与下游靶基因的对应关系中,某一miRNA可能有不止一个靶基因,这些靶基因可能含有类似结构域,也可能隶属不同家族的蛋白,涉及植株不同的发育进程和调控路径。如本研究中的miR156参与了水稻盐胁迫响应过程,其靶基因有OsSPL11和OsSPL12,说明miRNA与其靶基因共同参与的植物逆境应答是一个极其复杂多变的动态调控网络。
3.3 tRF在水稻盐胁迫条件下差异表达
tRF作为新近发现的含量丰富的非编码小分子调控性RNA,日益受到研究者的关注。尽管tRF已被证明在生物细胞中普遍存在,其产生机制、结构、分布、生物学功能和作用机制等方面的研究仍处于起步阶段。已有研究表明,tRF可调控基因的转录和翻译[41,42,43]、应激条件下的细胞应答[44,45,46]、恶性肿瘤[47,48]等人类疾病,但目前植物中tRF的研究鲜有报道。本研究通过RNA-Seq技术手段,在全基因组层面挖掘了水稻盐胁迫响应tRF。盐胁迫处理后,水稻根系产生的34—38nt tRF显著多于未处理材料,说明tRF的产生并非是随机的,而是通过某种特定机制或特定信号诱发tRNA的加工[41,17]。与对照组相比,共检测到盐胁迫响应的6个表达量很高且由盐胁迫诱导的tRF,而未检测到表达量下调的tRF。推测这些差异tRF是潜在的水稻盐胁迫响应tRF。利用实时荧光定量PCR手段证实了它们的存在和验证了测序结果的可靠性。进一步证明tRF可能受到盐胁迫诱导,tRNA加工增强,导致tRF表达量上升。靶基因分析发现,这些tRF的靶基因为叶绿体核糖核酸酶Ⅲ域蛋白、F-box和DUF结构域蛋白质、LMBR1膜内蛋白、内吞体分选转运复合体及玉米素葡萄糖基转移酶等,表明其参与了水稻不同的调控路径,也说明了tRF在水稻盐胁迫调控网络中的功能重要性。而不同结构的tRF与其靶基因之间的对应关系,tRF表达量与其靶基因表达量之间的对应关系有待进一步的研究。
根据以上的研究结果,构建了依赖miRNA和tRF的水稻盐胁迫响应调控网络(图6)。
图6
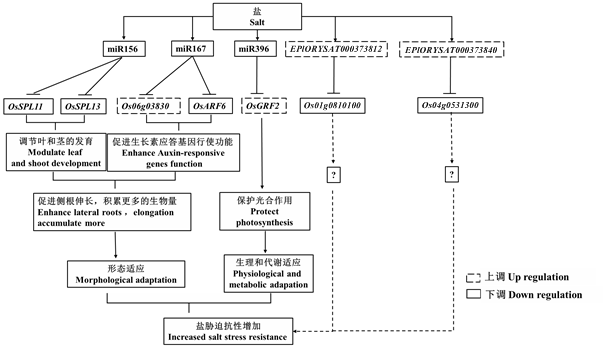 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6水稻根系中可能的盐胁迫响应miRNA及tRF调控网络
Fig. 6The potential regulating network of salt-responsive miRNAs and tRFs in rice roots
4 结论
水稻中大多数tRF受盐胁迫诱导表达;共检测到12种水稻根系中盐胁迫响应miRNA,其靶基因多为转录因子编码基因,推测其通过对其靶基因转录因子的转录后调控参与了盐胁迫响应的表达调控。其中8个水稻盐胁迫响应miRNA家族是不同物种间保守的通用盐胁迫响应miRNA。另外,从转录组水平挖掘出水稻盐胁迫响应tRF,并鉴定了6个盐胁迫诱导表达的tRF。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
