 ,, 丛佩华
,, 丛佩华 ,中国农业科学院果树研究所/农业部园艺作物种质资源利用重点实验室/国家苹果育种中心,辽宁兴城 125100
,中国农业科学院果树研究所/农业部园艺作物种质资源利用重点实验室/国家苹果育种中心,辽宁兴城 125100Bioinformatics and Expression Analysis of the LIM Gene Family in Apple
YUAN GaoPeng, HAN XiaoLei, BIAN ShuXun, ZHANG LiYi, TIAN Yi, ZHANG CaiXia ,, CONG PeiHua
,, CONG PeiHua ,Institute of Pomology, Chinese Academy of Agricultural Sciences/Key Laboratory of Fruit Germplasm Resources Utilization, Ministry of Agriculture/National Apple Breeding Center, Xingcheng 125100, Liaoning
,Institute of Pomology, Chinese Academy of Agricultural Sciences/Key Laboratory of Fruit Germplasm Resources Utilization, Ministry of Agriculture/National Apple Breeding Center, Xingcheng 125100, Liaoning通讯作者:
责任编辑: 赵伶俐
收稿日期:2019-03-19接受日期:2019-06-19网络出版日期:2019-12-01
| 基金资助: |
Received:2019-03-19Accepted:2019-06-19Online:2019-12-01
作者简介 About authors
袁高鹏,E-mail:yuangp329@163.com
韩晓蕾,E-mail:hanxiaolei@caas.cn

摘要
关键词:
Abstract
Keywords:
PDF (3552KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
袁高鹏, 韩晓蕾, 卞书迅, 张利义, 田义, 张彩霞, 丛佩华. 苹果LIM基因家族生物信息学及表达分析[J]. 中国农业科学, 2019, 52(23): 4322-4332 doi:10.3864/j.issn.0578-1752.2019.23.013
YUAN GaoPeng, HAN XiaoLei, BIAN ShuXun, ZHANG LiYi, TIAN Yi, ZHANG CaiXia, CONG PeiHua.
0 引言
【研究意义】LIM蛋白是一类半胱氨酸富集的蛋白(cysteine-rich proteins,CRPs),在动物[1,2,3]、酵母和植物[4,5,6]中广泛存在。LIM蛋白广泛参与细胞的发育及分化调节,并且能够调控多种基因的转录,在细胞核或细胞质中均有分布,也可穿梭于两者之间[5]。我国苹果栽培面积和产量均居世界第一,但生产上常面临诸多问题,其中苹果果锈作为一种非侵染性生理病害,在沿海和内陆多湿地区苹果主栽品种都易发生,正严重威胁我国苹果产业发展[7]。苹果果锈的发生与苯丙烷途径中相关基因的表达有关[8],而LIM蛋白可以调控苯丙烷生物合成通路中相关基因的表达[9,10]。开展苹果LIM家族的生物信息学及表达分析研究,可为苹果LIM蛋白功能的深入研究,以及今后培育无锈苹果新种质奠定理论基础。【前人研究进展】真核生物的LIM蛋白分子结构中往往包含1个或多个锌指结构,并且都含有高度保守的LIM结构域,通常由50余个氨基酸残基构成,其序列为:CX2CX16-23HX2CX2CX2CX16-21CX2-3(C/H/D)[11]。被子植物LIM蛋白可以划分为4个亚组(部分具有小亚组):αLIM1(FLIM1、WLIM1、PLIM1)、βLIM1、γLIM2(WLIM2)、δLIM2(PLIM2-I、PLIM2-II)[12]。关于植物LIM蛋白的早期研究主要集中于向日葵[13,14]、烟草[15]和拟南芥[16]等模式物种,随后在棉花[17,18,19]、杨树[20]、百合[21]、橡胶树[22]、玉米[23,24]、水稻[25]、高粱[10]等物种上也分离获得LIM蛋白。植物中的LIM蛋白一般具有两大功能:(1)作为转录因子调控靶基因,如作为转录因子调节苯丙烷生物合成途径相关基因的表达[3,9-10];(2)作为肌动蛋白的结合蛋白,调节肌动蛋白细胞骨架结构,如细胞骨架形成的调节功能[26,27]。首次得到的LIM蛋白是来自向日葵的HaPLIM1蛋白,该蛋白在花粉中特异表达,并结合肌动蛋白细胞骨架,另外,向日葵原生质中的HaWLIM1蛋白与皮层微管相关联,有助于细胞骨架稳定[13,14]。罗明等[17]将棉花GhLIM1在棉花植株中过表达,结果发现棉花木质素含量增加,表明其作为转录因子对木质素合成途径中相关基因(PAL、4CL和CAD)的表达起正调控作用。李彤[10]将高粱SbLIM1在拟南芥中过表达,发现转基因植株中苯丙烷代谢途径相关基因(At4CL、AtSGT、AtF5H、AtMYB46和AtMYB83)的表达均受到抑制,并且发现过表达株系中木质素的含量明显降低。【本研究切入点】苹果果锈的发生与苯丙烷代谢途径多个基因的表达有关[8],但目前对于苹果果锈的关注重点大多在MYB转录因子的调控作用[8,28],对于LIM蛋白如何调控苹果中苯丙烷相关基因表达的分子机制尚不清楚。【拟解决的关键问题】利用生物信息学的方法,鉴定并分析苹果基因组中全部LIM,从基因组水平上分析苹果LIM家族的启动子作用元件、染色体定位、基因结构和组织表达模式。1 材料与方法
试验于2017—2019年在中国农业科学院果树研究所/农业部园艺作物种质资源利用重点实验室/苹果育种中心实验室进行。1.1 材料
苹果材料来源于中国农业科学院果树研究所国家苹果种质资源圃(辽宁兴城)17年生‘金冠’苹果(Golden Delicious),在花后20 d对果实进行套袋和不套袋处理,不套袋果实有重度果锈,套袋果实无果锈;‘短枝旭’苹果(Macspur,无果锈);‘斯塔克金矮生’苹果(Starkspur Golden Delicious,重度果锈)。果锈程度的划分根据聂继云等[29]制定的标准。套袋‘金冠’苹果取其花、叶、果皮、茎用于不同组织的基因表达分析,其余品种在成熟时取其果皮用于不同程度带锈品种的基因表达分析,材料经液氮速冻后快速转移至-80℃冰箱保存备用。1.2 MdLIM家族成员鉴定及LIM结构域分析
苹果LIM家族序列信息来自GDR数据库(利用在线软件SMS2(
1.3 MdLIM启动子元件分析
从苹果GDR数据库(1.4 系统进化树构建
利用MEGA7.0软件比对分析苹果中11个LIM蛋白以及拟南芥(AtWLIM1,At1g10200,gi|837558;AtWLIM2a,At2g39900,gi|818577;AtWLIM2b,At3g55770,gi|818577;AtPLIM2,gi|819188,NC_003071.7;AtPLIM2b,At1g01780,gi|839267;AtPLIM2c,At3g61230,gi|825295)、烟草(NtWLIM1,AAD56948,gi|AAD56948;NtPLIM1a,AAF13231,gi|107810326;NtPLIM1b,AAF13232,gi|107802998;
NtLIM1a,DV999848.1;NtLIM1b,DW004416.1;NtWLIM2,CAA71891,gi|CAA71891;NtPLIM2,AAF75828,gi
|107796302;NtLIM1,BAA82827,gi|107824896)、陆地棉(GhWLIM2a,AAL38006,gi|107962044)、油菜(BnPLIM2a,CX191460.1)、甜橙(CsWLIM1,CK933805.1;CsLIM1,CV720040.1)、蓝桉(EgWLIM1,BAD91878.1)、野草莓(FvWLIM2,DY672997.1)、大豆(GmWLIM1a,BU084575.1;GmWLIM1b,BU089834.1;GmLIM1,CX709969.1;GmWLIM2,CX703844.1)、雷蒙德氏棉(GrPLIM2a,CO128503.1;GrPLIM2b,CO122392.1;GrPLIM2c,CO128788.1)、苜蓿(MtWLIM1,AJ498553.1;MtWLIM2,BG585852.1)、小立碗藓(PpLIM,BJ946868.1)、高粱(SbWLIM2,CN127879.1)、马铃薯(StWLIM1,BQ115637.1;StLIM1b,DN908198.1;StWLIM2,CV504017.1)、葡萄(VvLIM1,CF213597.1;VvWLIM2,CB974935.1)和玉米(ZmWLIM1,DR826954.1;ZmWLIM2,AY103686.2;ZmPLIM2a,EB167114.1;ZmPLIM2b,DV169657.1)的LIM蛋白氨基酸序列,进化树使用邻接法(Neighbor-Joining,NJ)构建,Bootstrap设置为1 000次[36]。
1.5 MdLIM结构分析及染色体定位
利用在线软件GSDS(1.6 MdLIM定量表达分析
用于荧光定量PCR分析的苹果材料如1.1所述。RNA提取试剂盒购自北京华越洋生物科技有限公司,cDNA第一条链合成所采用的反转录试剂盒购自宝生物工程(大连)有限公司(TaKaRa)。各MdLIM的荧光定量引物如表1所示,选用MdActin作为内参基因。反应体系为:SYBR Green 12.5 μL,上、下游引物(10 μmol·L-1)各0.75 μL,ddH2O 9 μL,cDNA 2 μL,总体积25 μL。反应条件为:95℃预变性3 min;95℃变性5 s,58℃退火30 s,72℃延伸30 s,40个循环。样品均为3次重复。试验所得数据采用2-ΔΔCT计算[37],利用SPSS18.0进行差异显著性分析。Table 1
表1
表1荧光定量PCR引物序列
Table 1
| 基因 Gene | 上游引物Forward primer (5′-3′) | 下游引物Reverse primer (5′-3′) |
|---|---|---|
| MdLIM1 | ATGCAGCGCCTGTGATAAGA | GCTCGAAATGAGGTTTGCAGT |
| MdLIM2 | GCTACTTGTGGTAAAACCGCT | CCAAAGCCGCGTAGTTTGAG |
| MdLIM3 | ATGCCAAGTCAGTCTCAAACT | TGGTATGGAGTCCCGTTG |
| MdLIM4 | GGGACCGTAGACAAATGTGC | AGTTGGGCAAAGTGGTGCTT |
| MdLIM5 | AAAACTGTTCGAGAAAGATCCG | GAAGAGTTGGCTATGGTGATGC |
| MdLIM6 | GACAGTCTATCCGCTGGAGA | ACGATGAATGTGTAAGGGCA |
| MdLIM7 | GATGGGATTTCCTACCACAA | CAGTCTCCTTGAACAGTTGCT |
| MdLIM8 | TCTCCTTTCTCAGGGACACC | GCACTTATCTCTGGTTCCACC |
| MdLIM9 | TGCCTACCATAAGACCTGCT | TCTCCTTGAAGAGTTGCTCG |
| MdLIM10 | ACTAGTTCCAGTACACAGACCAAC | CTCCATGAGTGCACCGGAAA |
| MdLIM11 | TCTACCACAAAGCCTGCTTCC | CAAGACTGCCGGTTCTTTTGA |
| MdActin | TGACCGAATGAGCAAGGAAATTACT | TACTCAGCTTTGGCAATCCACATC |
新窗口打开|下载CSV
2 结果
2.1 苹果LIM家族成员及LIM结构域
对苹果全基因组进行分析,共鉴定得到11个MdLIM成员(表2)。利用在线工具Smart(Table 2
表2
表2苹果中LIM家族信息
Table 2
| 基因 Gene | 基因登录号 Gene accession No. | 染色体定位 Chromosome location | 大小 Size (aa) | 分子量 Molecular weight (D) | 等电点 Isoelectric point | 亚细胞定位 Subcellular Localization |
|---|---|---|---|---|---|---|
| MdLIM1 | MD01G1054500 | Chr01:15895562..15897359 | 219 | 23939.82 | 6.14 | 细胞核Nucleus |
| MdLIM2 | MD01G1145700 | Chr01:25554814..25556454 | 199 | 21649.78 | 9.01 | 细胞核Nucleus |
| 高尔基体Golgi apparatus | ||||||
| MdLIM3 | MD05G1101500 | Chr05:20964629..20966107 | 197 | 22242.34 | 8.73 | 细胞核Nucleus |
| MdLIM4 | MD06G1078000 | Chr06:19291196..19292843 | 213 | 23595.59 | 7.51 | 细胞核Nucleus |
| MdLIM5 | MD06G1178800 | Chr06:31881986..31883872 | 188 | 21297.27 | 8.86 | 细胞核Nucleus |
| MdLIM6 | MD07G1140900 | Chr07:20424546..20426430 | 221 | 24195.19 | 6.31 | 细胞核Nucleus |
| MdLIM7 | MD07G1212800 | Chr07:29070702..29074084 | 200 | 21720.86 | 9.01 | 细胞核Nucleus |
| MdLIM8 | MD08G1081300 | Chr08:6745950..6747349 | 96 | 10755.12 | 7.03 | 细胞核Nucleus |
| MdLIM9 | MD14G1098800 | Chr14:15093296..15094757 | 222 | 24598.9 | 8.49 | 细胞核Nucleus |
| MdLIM10 | MD14G1185200 | Chr14:27756572..27758485 | 188 | 21254.15 | 8.77 | 细胞核Nucleus |
| MdLIM11 | MD15G1068200 | Chr15:4725341..4726746 | 195 | 21931.05 | 8.87 | 细胞核Nucleus |
| 高尔基体Golgi apparatus |
新窗口打开|下载CSV
图1
 新窗口打开|下载原图ZIP|生成PPT
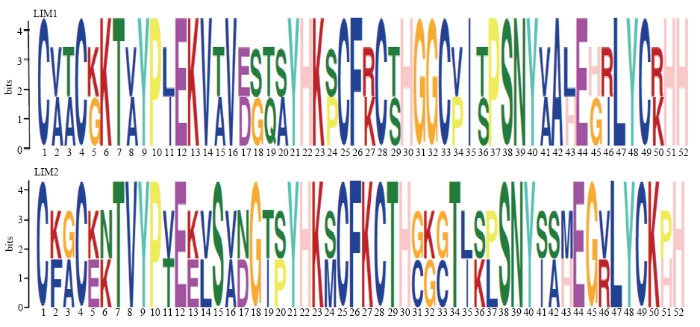
新窗口打开|下载原图ZIP|生成PPT图1LIM蛋白motifs
Fig. 1The motifs of LIM1 and LIM2
通过Conserved Domains和TBtools软件对苹果LIM蛋白保守结构域进行比对发现,除MdLIM8只含有1个LIM结构域外,其余10个MdLIM蛋白均含有两个LIM结构域(图2)
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2MdLIM蛋白保守结构域
Fig. 2Conserved Domains of MdLIM proteins
2.2 苹果LIM家族成员启动子作用元件分析
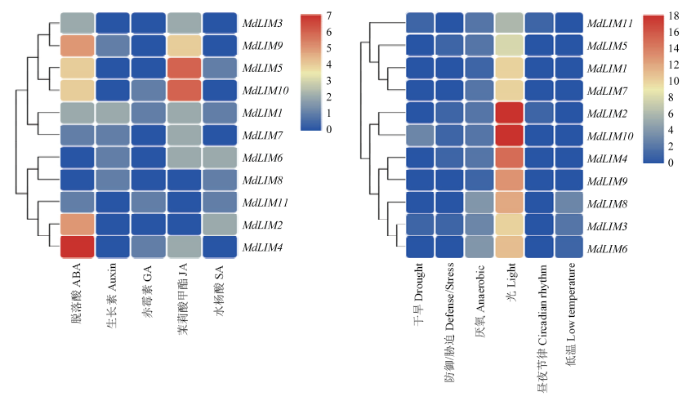
通过PlantCARE对LIM翻译起始位点上游2 000 bp的启动子顺式作用元件进行分析,然后对激素诱导表达元件、环境适应及逆境诱导表达元件在启动子上的数量进行聚类分析(图3)。对激素诱导表达的相关元件分析发现,脱落酸、生长素、赤霉素、茉莉酸甲酯和水杨酸等5种激素诱导表达元件在11个MdLIM启动子上均有2—5种分布;在环境适应和逆境诱导表达元件中,光诱导元件分布最多,其次为厌氧、防御和胁迫、低温、干旱、生理节律等诱导元件。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3基于苹果基因组GDDH13数据库的MdLIM家族基因启动子元件聚类分析
Fig. 3Clustering Analysis of Promoter Elements based on database of GDDH13 apple genome
2.3 苹果LIM家族成员系统进化及基因结构分析
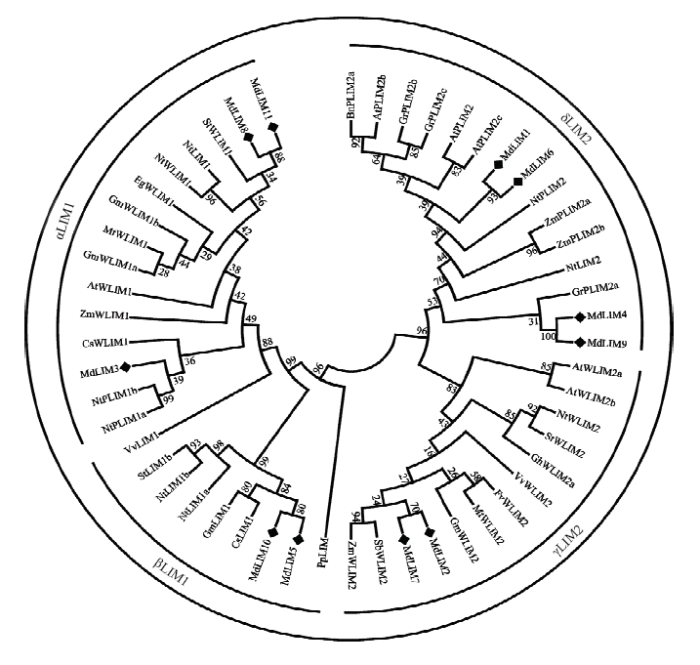
ARNAUD等[20]根据编码LIM蛋白的基因序列构建的系统树表现出的亲缘关系,将植物LIM蛋白重新分为4类:αLIM1(FLIM1、WLIM1、PLIM1)、βLIM1、γLIM2(WLIM2)和δLIM2(PLIM2-I、PLIM2-II)。将苹果中LIM蛋白的氨基酸序列与其他植物LIM蛋白的氨基酸序列构建进化树(图4)。结果显示,苹果中的LIM蛋白可分为4类,其中,MdLIM3、MdLIM8和MdLIM11属于αLIM1蛋白,MdLIM5和MdLIM10属于βLIM1蛋白,MdLIM2和MdLIM7属于γLIM2蛋白,MdLIM1、MdLIM4、MdLIM6、MdLIM9属于δLIM2蛋白。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4苹果LIM家族进化树
Fig. 4The phylogenetic tree of LIM family
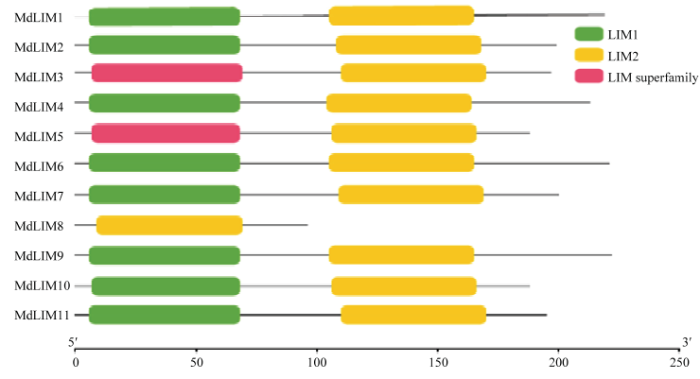
对MdLIM家族成员的基因结构进行分析(图5),结果表明,MdLIM8只含有1个外显子,MdLIM11含有4个外显子,MdLIM1、MdLIM4、MdLIM5、MdLIM6、MdLIM9、MdLIM10含有5个外显子,MdLIM2、MdLIM3、MdLIM7含有6个外显子。
图5
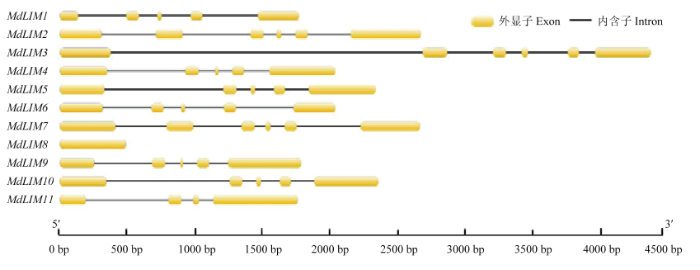 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5苹果LIM基因家族结构
Fig. 5The gene structures of LIM family
2.4 苹果LIM的染色体定位
为了解苹果LIM在染色体上的分布情况,对11个MdLIM进行了染色体定位(图6)。11个MdLIM在苹果7条染色体上呈不均匀分布,其中1号、6号、7号和14号染色体均含有2个MdLIM成员;5号、8号和15号染色体各自只含有1个MdLIM成员;其余10条染色体未有基因定位。图6
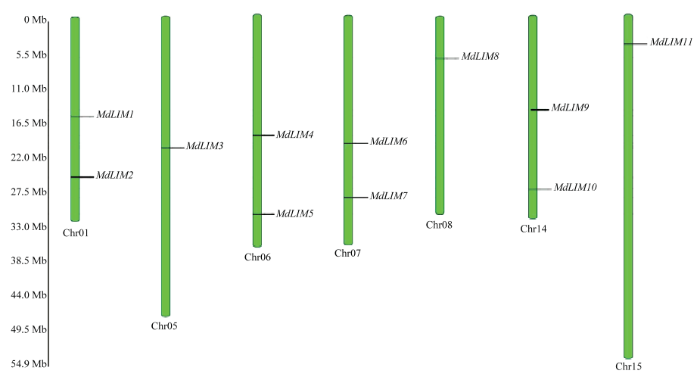 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6苹果LIM在染色体上的位置
Fig. 6Genetic map position of apple LIM genes
2.5 苹果LIM的组织表达
通过实时荧光定量RT-PCR分析11个MdLIM在苹果不同组织中的表达特性。结果表明,MdLIMs在苹果的花、叶、果皮(果皮来自套袋果实花后150 d的果皮)和茎中均有表达,但其表达量具有一定差异(图7)。MdLIM1、MdLIM3、MdLIM4、MdLIM7和MdLIM9在花中具有较高的表达量,与在其他组织中的表达量相比有显著差异(P<0.05),并且MdLIM1、MdLIM4、MdLIM7和MdLIM9在其他组织中几乎不表达。MdLIM2、MdLIM5、MdLIM6、MdLIM8、MdLIM10和MdLIM11在茎中的表达量和其他组织相比有显著差异(P<0.05)。图7
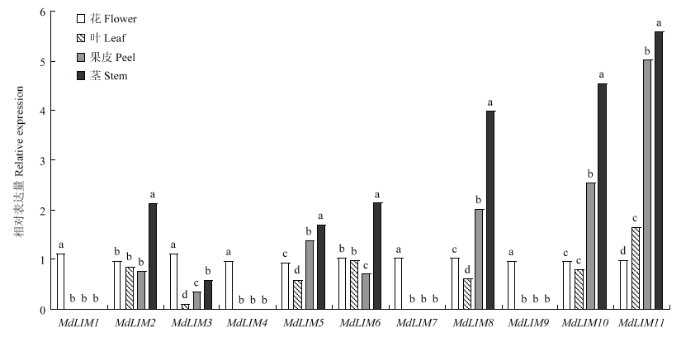 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7‘金冠’苹果不同组织中LIM的相对表达量
不同小写字母表示差异显著(P<0.05)。下同
Fig. 7Relative expression of MdLIMs in different tissues of Golden Delicious apple
Different lowercase letters indicate significant differences (P<0.05). The same as below
2.6 苹果LIM在不同品种果皮中的表达
通过实时荧光定量PCR分析MdLIM8和MdLIM11(在进化树分析中与NtLIM1聚为一类,而NtLIM1已被证明直接调控木质素合成途径相关基因的表达)在不同程度果锈品种中的表达量(图8)。结果表明,MdLIM8和MdLIM11在无锈果实(Macspur和GD T)上的表达量均高于带锈果实(Starspur GD和GD CK),且差异达到显著水平(P<0.05)。图8
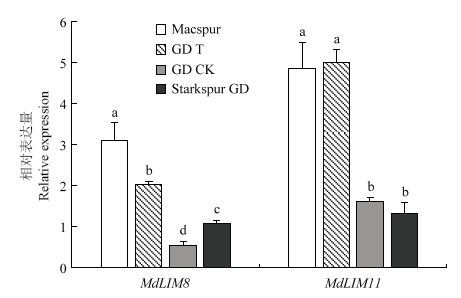 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8苹果LIM在不同品种果皮间的表达
Macspur为‘短枝旭’苹果,GD T为‘金冠’套袋果实,GD CK为‘金冠’不套袋果实,Starspur GD为‘斯塔克金矮生’苹果
Fig. 8The expression of LIM in peel of different varieties
The Macspur represent Macspur fruit, GD T represents bagging Golden Delicious fruit, GD CK represents non-bagged Golden Delicious fruit, Starspur GD represents Starspur Golden Delicious fruit
3 讨论
不同植物的LIM家族基因成员数目差异不大,杨树[20,24]中有12个LIM蛋白,水稻[20]中有6个,拟南芥中有9个[23],本研究通过对苹果全基因组进行分析,共获得11个LIM家族基因,比拟南芥中的LIM多2个,可能是因为苹果具有17条染色体,LIM在苹果基因组中得到了扩展。本研究所获得的11个苹果LIM蛋白中,10个含双LIM结构域,1个含单LIM结构域。单LIM结构域蛋白只保留了N端的LIM结构域,虽然C端产生了高度保守的结构域,并且依然富含半肌氨酸残基,但是其并没有形成典型的LIM结构域,这可能是LIM结构域发生了异化。在苹果LIM蛋白的结构域中,无论是单LIM结构域还是双LIM结构域,其Motif均含有52个氨基酸残基,具有完整的植物LIM蛋白保守结构域,具有完整的LIM蛋白功能[23]。本研究发现,苹果LIM启动子上的一些顺式作用元件数量相差不多(如光诱导和厌氧诱导元件),前人对玉米的LIM蛋白研究结果也表明,单、双LIM结构域的基因启动子上供分析的顺式元件相差不大[23],这在一定程度上表明单、双结构域LIM在基因水平上的亲缘关系较为密切。
系统进化树结果表明,聚类关系越近,具有类似功能的可能性越大[38]。在本研究所构建的进化树中,LIM蛋白可分为4类,每类中的MdLIM蛋白极有可能与该类中其他物种已知的LIM蛋白具有类似功能。如MdLIM8和MdLIM11与NtLIM1聚为一类,表明MdLIM8和MdLIM11可能与NtLIM1具有相似功能。研究表明,烟草的NtLIM1蛋白可以在体内激活含有PAL-box启动子所驱动的GUS表达,将NtLIM1在烟草中反义表达,导致苯丙烷代谢途径相关基因(PAL、C4H、4CL等)的表达量下降,并且木质素的含量降低20%以上[9]。果锈的发生与苯丙烷途径代谢产物木栓质和木质素密切相关[8,28-39],进一步对MdLIM8和MdLIM11的分析结果表明,不带锈果实MdLIM8和MdLIM11的表达量显著高于带锈果实。以上结果表明,苹果LIM蛋白可能参与苯丙烷途径相关基因的调控。
对苹果LIM结构分析发现,MdLIM的外显子数量具有差异(1—6个),可能是基因的结构在复制后产生了变化。ARNAUD等[20]对植物LIM结构的研究表明,大部分植物LIM含有4个内含子,本研究中大部分的MdLIM含有4—5个外显子,这一结果和前人研究结果类似[12,20],表明该类基因在进化上具有保守性。本研究获得的11个MdLIM在苹果7条染色体上呈不均衡分布,可能是由于苹果在进化过程中LIM发生了复制或重组,这些基因大多属于具有较高同源性的同一基因家族[23]。苹果LIM在复制的过程中也导致其结构发生了变化,这对基因的功能具有较强影响,表明片段重复和串联重复在MdLIM蛋白的多样性上具有重要的作用[40]。
基因的组织表达情况与其功能之间存在密切联系[41]。11个MdLIM在‘金冠’苹果的花、叶、果皮和茎中均有表达,但是在不同组织中的表达量存在一定差异,表明苹果LIM可能参与了不同器官的发育,其中MdLIM10和MdLIM11在茎中的表达量较高,表明其可能在木质素代谢过程中起重要作用。随着生物信息学和基因工程技术的快速发展,苹果LIM在苯丙烷代谢途径中的作用将可能成为今后研究的重要方向。
4 结论
鉴定得到11个苹果LIM,分为4类,定位到苹果7条染色体上。组织特异性表达分析表明,不同MdLIM在不同组织中的表达模式具有多样性和特异性,并且发现MdLIM8和MdLIM11可能与果锈的形成有关。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/0092-8674(88)90174-2URLPMID:2898300 [本文引用: 1]

The mec-3 gene is essential for proper differentiation of the set of six touch receptor neurons in C. elegans. In mutants lacking mec-3 activity, the touch receptors express none of their unique differentiated features and appear to be transformed into other types of neurons. We cloned the mec-3 gene by transposon tagging and showed that a mec-3 mutant can be rescued by germ line transformation using a 5.6 kb genomic DNA fragment. In a strain in which transforming mec-3 DNA is present in about 50 copies per haploid genome, additional cells express a mec-3-dependent phenotype. The putative coding sequence of mec-3 contains a homeobox, suggesting that the mec-3 protein specifies the expression of touch cell differentiation by binding to DNA and regulating transcription of genes that encode the differentiated features of these cells.
DOI:10.1038/344876a0URLPMID:1970421 [本文引用: 1]

The gene lin-11 is required for the asymmetric division of a vulval precursor cell type in the nematode Caenorhabditis elegans. Putative lin-11 complementary DNAs were sequenced and found to encode a protein that contains both a homeodomain and two tandem copies of a novel cysteine-rich motif: C-X2-C-X17-19-H-X2-C-X2-C-X2-C-X7-11-(C)-X8-C. Two tandem copies of this motif are also present amino-terminal to the homeodomains in the proteins encoded by the genes mec-3, which is required for C. elegans touch neuron differentiation, and isl-1, which encodes a rat insulin I gene enhancer-binding protein. The arrangement of cysteine residues in this motif, referred to as LIM (for lin-11 isl-1 mec-3), suggests that this region is a metal-binding domain. The presence in these three proteins of both a potential metal-binding domain and a homeodomain distinguishes them from previously characterized proteins.
DOI:10.1038/344879a0URLPMID:1691825 [本文引用: 2]

The activity of the rat insulin I gene enhancer is mainly dependent on two cis-acting protein-binding domains. Here we report the isolation of a complementary DNA encoding a protein, Isl-1, that binds to one of these domains. Isl-1 contains a homeodomain with greatest similarity to those of the Caenorhabditis elegans proteins encoded by mec-3 and lin-11. In addition, Isl-1, like the lin-11 and mec-3 gene products, contains a novel Cys-His domain which is reminiscent of known metal-binding regions. Together these proteins define a novel class of proteins containing both a homeo- and a Cys His-domain. Isl-1 is preferentially expressed in cells of pancreatic endocrine origin. If the structural homologies between Isl-1 and the C. elegans gene products reflect functional similarities, a role for Isl-1 in the development of pancreatic endocrine cells could be envisaged.
DOI:10.1093/nar/22.15.3151URLPMID:8065929 [本文引用: 1]

We here report the sequence of a yeast gene LRG1 whose deduced amino acid sequence contains sequence motifs similar to LIM domain proteins in the amino-terminal, and to rho/rac GTPase activating proteins (rho/racGAP's) in the carboxy-terminal part. LRG1 expression is differentially regulated showing a peak of expression in sporulating cells. Gene disruption experiments indicate that the gene is not essential and may play a role during mating.
DOI:10.1023/A:1006333611189URL [本文引用: 2]

LIM proteins are important eucaryotic developmental regulators characterized by the presence of one or several double zinc finger motifs, the LIM domains, which are protein-interacting domains. Using the cDNA of the previously described pollen LIM protein PLIM1 from sunflower as a hybridization probe we have isolated the coding sequence for a related protein from cDNA libraries from various sunflower organs. This protein, WLIM1, is 188 amino acids long and, like the pollen protein PLIM1, contains two LIM domains, separated by a 48 residue spacer region. The two sunflower proteins are structurally related to the animal LIM proteins CRP and MLP. A WLIM1 gene transcript was detected by RT-PCR in all vegetative and reproductive plant organs tested. Polyclonal antibodies raised against the bacterially expressed and affinity-purified protein recognize a polypeptide of ca. 50kDa in these organs. Immunocytochemical studies detect the protein in many cell types in each of these organs where it is localized either to the cytoplasm, the nucleus, or both. The protein is often associated with plastids and smaller cellular structures or organelles. In late anaphase and early telophase of dividing cells from ovaries, stems and roots it accumulates in the phragmoplast, and may therefore also play a role in cytokinesis.
DOI:10.1042/BST20190869URLPMID:31829417 [本文引用: 1]

Ubiquitin modifications of target proteins act to localise, direct and specify a diverse range of cellular processes, many of which are biomedically relevant. To allow this diversity, ubiquitin modifications exhibit remarkable complexity, determined by a combination of polyubiquitin chain length, linkage type, numbers of ubiquitin chains per target, and decoration of ubiquitin with other small modifiers. However, many questions remain about how different ubiquitin signals are specifically recognised and transduced by the decoding ubiquitin-binding domains (UBDs) within ubiquitin-binding proteins. This review briefly outlines our current knowledge surrounding the diversity of UBDs, identifies key challenges in their discovery and considers recent structural studies with implications for the increasing complexity of UBD function and identification. Given the comparatively low numbers of functionally characterised polyubiquitin-selective UBDs relative to the ever-expanding variety of polyubiquitin modifications, it is possible that many UBDs have been overlooked, in part due to limitations of current approaches used to predict their presence within the proteome. Potential experimental approaches for UBD discovery are considered; web-based informatic analyses, Next-Generation Phage Display, deubiquitinase-resistant diubiquitin, proximity-dependent biotinylation and Ubiquitin-Phototrap, including possible advantages and limitations. The concepts discussed here work towards identifying new UBDs which may represent the 'dark matter' of the ubiquitin system.
[本文引用: 1]
[本文引用: 1]
DOI:10.1105/tpc.16.00490URLPMID:27604696 [本文引用: 4]

Suberin, a polymer composed of both aliphatic and aromatic domains, is deposited as a rough matrix upon plant surface damage and during normal growth in the root endodermis, bark, specialized organs (e.g., potato [Solanum tuberosum] tubers), and seed coats. To identify genes associated with the developmental control of suberin deposition, we investigated the chemical composition and transcriptomes of suberized tomato (Solanum lycopersicum) and russet apple (Malus x domestica) fruit surfaces. Consequently, a gene expression signature for suberin polymer assembly was revealed that is highly conserved in angiosperms. Seed permeability assays of knockout mutants corresponding to signature genes revealed regulatory proteins (i.e., AtMYB9 and AtMYB107) required for suberin assembly in the Arabidopsis thaliana seed coat. Seeds of myb107 and myb9 Arabidopsis mutants displayed a significant reduction in suberin monomers and altered levels of other seed coat-associated metabolites. They also exhibited increased permeability, and lower germination capacities under osmotic and salt stress. AtMYB9 and AtMYB107 appear to synchronize the transcriptional induction of aliphatic and aromatic monomer biosynthesis and transport and suberin polymerization in the seed outer integument layer. Collectively, our findings establish a regulatory system controlling developmentally deposited suberin, which likely differs from the one of stress-induced polymer assembly recognized to date.
DOI:10.1016/s0031-9422(01)00054-1URLPMID:11430987 [本文引用: 3]

Lignin is a complex phenolic plant polymer that is essential for mechanical support, defense, and water transport in higher plants. The AC-rich motif, Pal-box is an important cis-acting element for gene expression in phenylpropanoid biosynthesis. We isolated a cDNA clone (Ntlim1) encoding a Pal-box binding protein by Southwestern screening. The deduced amino acid sequence of Ntlim1 is highly similar to members of the LIM protein family that contain a zinc finger motif. Moreover, Ntlim1 had a specific DNA-binding ability and transiently activated transcription of a beta-glucuronidase reporter gene driven by the Pal-box sequence. The results of transient expression assays with tobacco cultured cells showed that fusion proteins between GFP and Ntlim1 can enter nuclei. Transgenic tobacco plants with antisense Ntlim1 showed low levels of transcripts from some key phenylpropanoid pathway genes such as phenylalanine ammonia-lyase, hydroxycinnamate CoA ligase and cinnamyl alcohol dehydrogenase. Furthermore, a greater than 20% reduction in lignin content was observed in transgenic tobacco with antisense Ntlim1.
[D].
[本文引用: 4]
[D].
[本文引用: 4]
DOI:10.1083/jcb.119.6.1573URLPMID:1469049 [本文引用: 1]

Interaction with extracellular matrix can trigger a variety of responses by cells including changes in specific gene expression and cell differentiation. The mechanism by which cell surface events are coupled to the transcriptional machinery is not understood, however, proteins localized at sites of cell-substratum contact are likely to function as signal transducers. We have recently purified and characterized a low abundance adhesion plaque protein called zyxin (Crawford, A. W., and M. C. Beckerle. 1991. J. Biol. Chem. 266:5847-5853; Crawford, A. W., J. W. Michelsen, and M. C. Beckerle. 1992. J. Cell Biol. 116:1381-1393). We have now isolated and sequenced zyxin cDNA and we report here that zyxin exhibits an unusual proline-rich NH2-terminus followed by three tandemly arrayed LIM domains. LIM domains have previously been identified in proteins that play important roles in transcriptional regulation and cellular differentiation. LIM domains have been proposed to coordinate metal ions and we have demonstrated by atomic absorption spectroscopy that purified zyxin binds zinc, a result consistent with the idea that zyxin has zinc fingers. In addition, we have discovered that zyxin interacts in vitro with a 23-kD protein that also exhibits LIM domains. Microsequence analysis has revealed that the 23-kD protein (or cCRP) is the chicken homologue of the human cysteine-rich protein (hCRP). By double-label indirect immunofluorescence, we found that zyxin and cCRP are extensively colocalized in chicken embryo fibroblasts, consistent with the idea that they interact in vivo. We conclude that LIM domains are zinc-binding sequences that may be involved in protein-protein interactions. The demonstration that two cytoskeletal proteins, zyxin and cCRP, share a sequence motif with proteins important for transcriptional regulation raises the possibility that zyxin and cCRP are components of a signal transduction pathway that mediates adhesion-stimulated changes in gene expression.
[本文引用: 2]
[本文引用: 2]
URLPMID:1302629 [本文引用: 2]

We have isolated, via differential hybridization screening of a floral cDNA library from sunflower, a cDNA clone that hybridizes to a 1100 nucleotide-long mRNA found exclusively in mature pollen grains. The cDNA encodes a 219 amino acid-long polypeptide containing two potential zinc fingers alternating with two basic domains. A similar organization is found in the erythroid-specific transcription factor Eryf1 from chicken and its murine homolog GF-1. The C-terminus of the protein contains a sixfold repeat of the pentapeptide sequence (S,T,A)(E,D)TQN. These features suggest that the SF3 protein is a transcription factor required for the expression of late pollen genes. The SF3 gene is a member of a multicopy gene family. A genomic copy of the gene has been isolated and sequenced; it is split by four short, AT-rich introns.
DOI:10.1105/tpc.4.12.1465URLPMID:1467648 [本文引用: 2]
DOI:10.1105/tpc.106.040956URLPMID:16905656 [本文引用: 1]

We used confocal microscopy and in vitro analyses to show that Nicotiana tabacum WLIM1, a LIM domain protein related to animal Cys-rich proteins, is a novel actin binding protein in plants. Green fluorescent protein (GFP)-tagged WLIM1 protein accumulated in the nucleus and cytoplasm of tobacco BY2 cells. It associated predominantly with actin cytoskeleton, as demonstrated by colabeling and treatment with actin-depolymerizing latrunculin B. High-speed cosedimentation assays revealed the ability of WLIM1 to bind directly to actin filaments with high affinity. Fluorescence recovery after photobleaching and fluorescence loss in photobleaching showed a highly dynamic in vivo interaction of WLIM1-GFP with actin filaments. Expression of WLIM1-GFP in BY2 cells significantly delayed depolymerization of the actin cytoskeleton induced by latrunculin B treatment. WLIM1 also stabilized actin filaments in vitro. Importantly, expression of WLIM1-GFP in Nicotiana benthamiana leaves induces significant changes in actin cytoskeleton organization, specifically, fewer and thicker actin bundles than in control cells, suggesting that WLIM1 functions as an actin bundling protein. This hypothesis was confirmed by low-speed cosedimentation assays and direct observation of F-actin bundles that formed in vitro in the presence of WLIM1. Taken together, these data identify WLIM1 as a novel actin binding protein that increases actin cytoskeleton stability by promoting bundling of actin filaments.
DOI:10.1105/tpc.110.075960URLPMID:20817848 [本文引用: 1]

Recently, a number of two LIM-domain containing proteins (LIMs) have been reported to trigger the formation of actin bundles, a major higher-order cytoskeletal assembly. Here, we analyzed the six Arabidopsis thaliana LIM proteins. Promoter-β-glucuronidase reporter studies revealed that WLIM1, WLIM2a, and WLIM2b are widely expressed, whereas PLIM2a, PLIM2b, and PLIM2c are predominantly expressed in pollen. LIM-green fluorescent protein (GFP) fusions all decorated the actin cytoskeleton and increased actin bundle thickness in transgenic plants and in vitro, although with different affinities and efficiencies. Remarkably, the activities of WLIMs were calcium and pH independent, whereas those of PLIMs were inhibited by high pH and, in the case of PLIM2c, by high [Ca(2+)]. Domain analysis showed that the C-terminal domain is key for the responsiveness of PLIM2c to pH and calcium. Regulation of LIM by pH was further analyzed in vivo by tracking GFP-WLIM1 and GFP-PLIM2c during intracellular pH modifications. Cytoplasmic alkalinization specifically promoted release of GFP-PLIM2c but not GFP-WLIM1, from filamentous actin. Consistent with these data, GFP-PLIM2c decorated long actin bundles in the pollen tube shank, a region of relatively low pH. Together, our data support a prominent role of Arabidopsis LIM proteins in the regulation of actin cytoskeleton organization and dynamics in sporophytic tissues and pollen.
URLPMID:12776607 [本文引用: 2]

LIM-domain protein plays an important role in various cellular processes, including construction of cytoskeleton, transcription control and signal transduction. Based on cotton fiber EST database and contig analysis, the coding region of a cotton LIM-domain protein gene (GhLIM1) was obtained by RT-PCR from 4DPA (day post anthesis) ovule with fiber. The cloned fragment of 848 bp contains an open reading frame of 570 bp, coding for a polypeptide of 189 amino acids. It was demonstrated that the deduced GhLIM1 protein was highly homologous to the LIM-domain protein of sunflower (Helianthus annuus), tobacco (Nicotiana tabacum) and Arabidopsis thaliana. Two intact LIM-domains, with the conserved sequence of a double zinc-finger structure (C-X2-C-X17-19-H-X2-C-X2-C-X2-C-X16-24-C-X2-H), were found in the GhLIM1 protein. RT-PCR and Northern blot analysis showed that GhLIM1 gene expressed in root, shoot tip, hypocotyls, bud, leaf, anther, ovule and fiber (4DPA, 12DPA, 18DPA). However it was preferentially expressed in the shoot tip, fiber and ovule. It was proposed that the express of GhLIM1 gene is related to cotton fiber development.
URLPMID:12776607 [本文引用: 2]

LIM-domain protein plays an important role in various cellular processes, including construction of cytoskeleton, transcription control and signal transduction. Based on cotton fiber EST database and contig analysis, the coding region of a cotton LIM-domain protein gene (GhLIM1) was obtained by RT-PCR from 4DPA (day post anthesis) ovule with fiber. The cloned fragment of 848 bp contains an open reading frame of 570 bp, coding for a polypeptide of 189 amino acids. It was demonstrated that the deduced GhLIM1 protein was highly homologous to the LIM-domain protein of sunflower (Helianthus annuus), tobacco (Nicotiana tabacum) and Arabidopsis thaliana. Two intact LIM-domains, with the conserved sequence of a double zinc-finger structure (C-X2-C-X17-19-H-X2-C-X2-C-X2-C-X16-24-C-X2-H), were found in the GhLIM1 protein. RT-PCR and Northern blot analysis showed that GhLIM1 gene expressed in root, shoot tip, hypocotyls, bud, leaf, anther, ovule and fiber (4DPA, 12DPA, 18DPA). However it was preferentially expressed in the shoot tip, fiber and ovule. It was proposed that the express of GhLIM1 gene is related to cotton fiber development.
DOI:10.1105/tpc.113.116970URLPMID:24220634 [本文引用: 1]

LIN-11, Isl1 and MEC-3 (LIM)-domain proteins play pivotal roles in a variety of cellular processes in animals, but plant LIM functions remain largely unexplored. Here, we demonstrate dual roles of the WLIM1a gene in fiber development in upland cotton (Gossypium hirsutum). WLIM1a is preferentially expressed during the elongation and secondary wall synthesis stages in developing fibers. Overexpression of WLIM1a in cotton led to significant changes in fiber length and secondary wall structure. Compared with the wild type, fibers of WLIM1a-overexpressing plants grew longer and formed a thinner and more compact secondary cell wall, which contributed to improved fiber strength and fineness. Functional studies demonstrated that (1) WLIM1a acts as an actin bundler to facilitate elongation of fiber cells and (2) WLIM1a also functions as a transcription factor to activate expression of Phe ammonia lyase-box genes involved in phenylpropanoid biosynthesis to build up the secondary cell wall. WLIM1a localizes in the cytosol and nucleus and moves into the nucleus in response to hydrogen peroxide. Taken together, these results demonstrate that WLIM1a has dual roles in cotton fiber development, elongation, and secondary wall formation. Moreover, our study shows that lignin/lignin-like phenolics may substantially affect cotton fiber quality; this finding may guide cotton breeding for improved fiber traits.
DOI:10.1016/j.plaphy.2013.01.018URLPMID:23466745 [本文引用: 1]

LIM-domain proteins play important roles in cellular processes in eukaryotes. In this study, a LIM protein gene, GhWLIM5, was identified in cotton. Quantitative RT-PCR analysis showed that GhWLIM5 was expressed widely in different cotton tissues and had a peak in expression during fiber elongation. GFP fluorescence assay revealed that cotton cells expressing GhWLIM5:eGFP fusion gene displayed a network distribution of eGFP fluorescence, suggesting that GhWLIM5 protein is mainly localized to the cell cytoskeleton. When GhWLIM5:eGFP transformed cells were stained with rhodamine-phalloidin there was consistent overlap in eGFP and rhodamine-palloidin signals, demonstrating that GhWLIM5 protein is colocalized with the F-actin cytoskeleton. In addition, high-speed cosedimentation assay verified that GhWLIM5 directly bound actin filaments, while low cosedimentation assay and microscopic observation indicated that GhWLIM5 bundled F-actin in vitro. Increasing amounts of GhWLIM5 protein were able to protect F-actin from depolymerization in vitro in the presence of Lat B (an F-actin depolymerizer). Our results contribute to a better understanding of the biochemical role of GhWLIM5 in modulating the dynamic F-actin network in cotton.
DOI:10.1093/dnares/dsm013URLPMID:17573466 [本文引用: 6]

In Eukaryotes, LIM proteins act as developmental regulators in basic cellular processes such as regulating the transcription or organizing the cytoskeleton. The LIM domain protein family in plants has mainly been studied in sunflower and tobacco plants, where several of its members exhibit a specific pattern of expression in pollen. In this paper, we finely characterized in poplar six transcripts encoding these proteins. In Populus trichocarpa genome, the 12 LIM gene models identified all appear to be duplicated genes. In addition, we describe several new LIM domain proteins deduced from Arabidopsis and rice genomes, raising the number of LIM gene models to six for both species. Plant LIM genes have a core structure of four introns with highly conserved coding regions. We also identified new LIM domain proteins in several other species, and a phylogenetic analysis of plant LIM proteins reveals that they have undergone one or several duplication events during the evolution. We gathered several LIM protein members within new monophyletic groups. We propose to classify the plant LIM proteins into four groups: alphaLIM1, betaLIM1, gammaLIM2, and deltaLIM2, subdivided according to their specificity to a taxonomic class and/or to their tissue-specific expression. Our investigation of the structure of the LIM domain proteins revealed that they contain many conserved motifs potentially involved in their function.
DOI:10.1104/pp.108.118604URLPMID:18480376 [本文引用: 1]

Actin microfilaments are crucial for polar cell tip growth, and their configurations and dynamics are regulated by the actions of various actin-binding proteins (ABPs). We explored the function of a lily (Lilium longiflorum) pollen-enriched LIM domain-containing protein, LlLIM1, in regulating the actin dynamics in elongating pollen tube. Cytological and biochemical assays verified LlLIM1 functioning as an ABP, promoting filamentous actin (F-actin) bundle assembly and protecting F-actin against latrunculin B-mediated depolymerization. Overexpressed LlLIM1 significantly disturbed pollen tube growth and morphology, with multiple tubes protruding from one pollen grain and coaggregation of FM4-64-labeled vesicles and Golgi apparatuses at the subapex of the tube tip. Moderate expression of LlLIM1 induced an oscillatory formation of asterisk-shaped F-actin aggregates that oscillated with growth period but in different phases at the subapical region. These results suggest that the formation of LlLIM1-mediated overstabilized F-actin bundles interfered with endomembrane trafficking to result in growth retardation. Cosedimentation assays revealed that the binding affinity of LlLIM1 to F-actin was simultaneously regulated by both pH and Ca(2+): LlLIM1 showed a preference for F-actin binding under low pH and low Ca(2+) concentration. The potential functions of LlLIM1 as an ABP sensitive to pH and calcium in integrating endomembrane trafficking, oscillatory pH, and calcium circumstances to regulate tip-focused pollen tube growth are discussed.
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 5]
[D].
[本文引用: 5]
[本文引用: 2]
[本文引用: 2]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1074/jbc.M102820200URLPMID:11395501 [本文引用: 1]

The localization of proteins to particular intracellular compartments often regulates their functions. Zyxin is a LIM protein found prominently at sites of cell adhesion, faintly in leading lamellipodia, and transiently in cell nuclei. Here we have performed a domain analysis to identify regions in zyxin that are responsible for targeting it to different subcellular locations. The N-terminal proline-rich region of zyxin, which harbors binding sites for alpha-actinin and members of the Ena/VASP family, concentrates in lamellipodial extensions and weakly in focal adhesions. The LIM region of zyxin displays robust targeting to focal adhesions. When overexpressed in cells, the LIM region of zyxin causes displacement of endogenous zyxin from focal adhesions. Upon mislocalization of full-length zyxin, at least one member of the Ena/VASP family is also displaced, and the organization of the actin cytoskeleton is perturbed. Zyxin also has the capacity to shuttle between the nucleus and focal adhesion sites. When nuclear export is inhibited, zyxin accumulates in cell nuclei. The nuclear accumulation of zyxin occurs asynchronously with approximately half of the cells exhibiting nuclear localization of zyxin within 2.3 h of initiating leptomycin B treatment. Our results provide insight into the functions of different zyxin domains.
DOI:10.1083/jcb.200212057URLPMID:12566430 [本文引用: 1]

Epithelial protein lost in neoplasm (EPLIN) is a cytoskeleton-associated protein encoded by a gene that is down-regulated in transformed cells. EPLIN increases the number and size of actin stress fibers and inhibits membrane ruffling induced by Rac. EPLIN has at least two actin binding sites. Purified recombinant EPLIN inhibits actin filament depolymerization and cross-links filaments in bundles. EPLIN does not affect the kinetics of spontaneous actin polymerization or elongation at the barbed end, but inhibits branching nucleation of actin filaments by Arp2/3 complex. Side binding activity may stabilize filaments and account for the inhibition of nucleation mediated by Arp2/3 complex. We propose that EPLIN promotes the formation of stable actin filament structures such as stress fibers at the expense of more dynamic actin filament structures such as membrane ruffles. Reduced expression of EPLIN may contribute to the motility of invasive tumor cells.
DOI:10.1111/nph.14170URLPMID:27716944 [本文引用: 2]

A comparison of the transcriptomes of russeted vs nonrusseted apple skins previously highlighted a tight relationship between a gene encoding an MYB-type transcription factor, MdMYB93, and some key suberin biosynthetic genes. The present work assesses the role of this transcription factor in the suberization process. A phylogenetic analysis of MdMYB93 and Arabidopsis thaliana MYBs was performed and the function of MdMYB93 was further investigated using Agrobacterium-mediated transient overexpression in Nicotiana benthamiana leaves. An RNA-Seq analysis was performed to highlight the MdMYB93-regulated genes. Ultraperformance liquid chromatography-triple time-of-flight (UPLC-TripleTOF) and GC-MS were used to investigate alterations in phenylpropanoid, soluble-free lipid and lipid polyester contents. A massive accumulation of suberin and its biosynthetic precursors in MdMYB93 agroinfiltrated leaves was accompanied by a remobilization of phenylpropanoids and an increased amount of lignin precursors. Gene expression profiling displayed a concomitant alteration of lipid and phenylpropanoid metabolism, cell wall development, and extracellular transport, with a large number of induced transcripts predicted to be involved in suberin deposition. The present work supports a major role of MdMYB93 in the regulation of suberin deposition in russeted apple skins, from the synthesis of monomeric precursors, their transport, polymerization, and final deposition as suberin in primary cell wall.
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/ng.3886URLPMID:28581499 [本文引用: 1]

Using the latest sequencing and optical mapping technologies, we have produced a high-quality de novo assembly of the apple (Malus domestica Borkh.) genome. Repeat sequences, which represented over half of the assembly, provided an unprecedented opportunity to investigate the uncharacterized regions of a tree genome; we identified a new hyper-repetitive retrotransposon sequence that was over-represented in heterochromatic regions and estimated that a major burst of different transposable elements (TEs) occurred 21 million years ago. Notably, the timing of this TE burst coincided with the uplift of the Tian Shan mountains, which is thought to be the center of the location where the apple originated, suggesting that TEs and associated processes may have contributed to the diversification of the apple ancestor and possibly to its divergence from pear. Finally, genome-wide DNA methylation data suggest that epigenetic marks may contribute to agronomically relevant aspects, such as apple fruit development.
DOI:10.1093/nar/gku986URLPMID:25324309 [本文引用: 1]

Comparative sequence analysis has significantly altered our view on the complexity of genome organization and gene functions in different kingdoms. PLAZA 3.0 is designed to make comparative genomics data for plants available through a user-friendly web interface. Structural and functional annotation, gene families, protein domains, phylogenetic trees and detailed information about genome organization can easily be queried and visualized. Compared with the first version released in 2009, which featured nine organisms, the number of integrated genomes is more than four times higher, and now covers 37 plant species. The new species provide a wider phylogenetic range as well as a more in-depth sampling of specific clades, and genomes of additional crop species are present. The functional annotation has been expanded and now comprises data from Gene Ontology, MapMan, UniProtKB/Swiss-Prot, PlnTFDB and PlantTFDB. Furthermore, we improved the algorithms to transfer functional annotation from well-characterized plant genomes to other species. The additional data and new features make PLAZA 3.0 (http://bioinformatics.psb.ugent.be/plaza/) a versatile and comprehensible resource for users wanting to explore genome information to study different aspects of plant biology, both in model and non-model organisms.
DOI:10.3390/biology8030063URLPMID:31470601 [本文引用: 1]

Lycoris radiata belongs to the Amaryllidaceae family and is a bulbous plant native to South Korea, China, and Japan. Galantamine, a representative alkaloid of Amaryllidaceae plants, including L. radiata, exhibits selective and dominant acetylcholinesterase inhibition. In spite of the economic and officinal importance of L. radiata, the molecular biological and biochemical information on L. radiata is relatively deficient. Therefore, this study provides functional information of L. radiata, describe galantamine biosynthesis in the various organs, and provide transcriptomic and metabolic datasets to support elucidation of galantamine biosynthesis pathway in future studies. The results of studies conducted in duplicate revealed the presence of a total of 325,609 and 404,019 unigenes, acquired from 9,913,869,968 and 10,162,653,038 raw reads, respectively, after trimming the raw reads using CutAdapt, assembly using Trinity package, and clustering using CD-Hit-EST. All of the assembled unigenes were aligned to the public databases, including National Center for Biotechnology Information (NCBI) non-redundant protein (NR) and nucleotide (Nt) database, SWISS-PROT (UniProt) protein sequence data bank, The Arabidopsis Information Resource (TAIR), the Swiss-Prot protein database, Gene Ontology (GO), and Clusters of Orthologous Groups (COG) database to predict potential genes and provide their functional information. Based on our transcriptome data and published literatures, eight full-length cDNA clones encoding LrPAL2, LrPAL3, LrC4H2, LrC3H, LrTYDC2, LrNNR, LrN4OMT, and LrCYP96T genes, involved in galantamine biosynthesis, were identified in L. radiata. In order to investigate galantamine biosynthesis in different plant parts of L. radiata grown in a growth chamber, gene expression levels were measured through quantitative real-time polymerase chain reaction (qRT-PCR) analysis using these identified genes and galantamine levels were quantified by high-performance liquid chromatography (HPLC) analysis. The qRT-PCR data revealed high expression levels of LrNNR, LrN4OMT, and LrCYP96T in the bulbs, and, as expected, we observed higher amounts of galantamine in the bulbs than in the root and leaves. Additionally, a total of 40 hydrophilic metabolites were detected in the different organs using gas-chromatography coupled with time-of-flight mass spectrometry. In particular, a strong positive correlation between galantamine and sucrose, which provides energy for the secondary metabolite biosynthesis, was observed.
DOI:10.1002/prca.201700069URLPMID:28975713 [本文引用: 1]

PepSweetener is a web-based visualization tool designed to facilitate the manual annotation of intact glycopeptides from MS data regardless of the instrument that produced these data.
DOI:10.2144/00286ir01URLPMID:10868275 [本文引用: 1]
[本文引用: 2]
DOI:10.1093/molbev/msr121URL [本文引用: 1]

Comparative analysis of molecular sequence data is essential for reconstructing the evolutionary histories of species and inferring the nature and extent of selective forces shaping the evolution of genes and species. Here, we announce the release of Molecular Evolutionary Genetics Analysis version 5 (MEGA5), which is a user-friendly software for mining online databases, building sequence alignments and phylogenetic trees, and using methods of evolutionary bioinformatics in basic biology, biomedicine, and evolution. The newest addition in MEGA5 is a collection of maximum likelihood (ML) analyses for inferring evolutionary trees, selecting best-fit substitution models (nucleotide or amino acid), inferring ancestral states and sequences (along with probabilities), and estimating evolutionary rates site-by-site. In computer simulation analyses, ML tree inference algorithms in MEGA5 compared favorably with other software packages in terms of computational efficiency and the accuracy of the estimates of phylogenetic trees, substitution parameters, and rate variation among sites. The MEGA user interface has now been enhanced to be activity driven to make it easier for the use of both beginners and experienced scientists. This version of MEGA is intended for the Windows platform, and it has been configured for effective use on Mac OS X and Linux desktops. It is available free of charge from http://www.megasoftware.net.
DOI:10.1006/meth.2001.1262URLPMID:11846609 [本文引用: 1]

The two most commonly used methods to analyze data from real-time, quantitative PCR experiments are absolute quantification and relative quantification. Absolute quantification determines the input copy number, usually by relating the PCR signal to a standard curve. Relative quantification relates the PCR signal of the target transcript in a treatment group to that of another sample such as an untreated control. The 2(-Delta Delta C(T)) method is a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments. The purpose of this report is to present the derivation, assumptions, and applications of the 2(-Delta Delta C(T)) method. In addition, we present the derivation and applications of two variations of the 2(-Delta Delta C(T)) method that may be useful in the analysis of real-time, quantitative PCR data.
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00299-016-1998-7URLPMID:27255339 [本文引用: 1]

Differential genes of suberin, polyamine and transcription factors in transcriptome sequences and the contents of H 2 O 2 , spermidine, spermine, and putrescine changed significantly after treating with MGBG. Russeting is a commercially important process that restores the control of water loss through the skin via the formation of a waterproofing periderm just beneath the microcracked skin of pear primary fruit. A spontaneous russet skin mutant, the yellow-green 'Dangshansuli' pear, has been identified. To understand the role of polyamines in the formation of the russet skin of the mutant-type (MT) pear, it was treated with methylglyoxal-bis-(guanylhydrazone) (MGBG) for 4?weeks after full bloom. One week later, differentially expressed genes among the wild-type (WT), MT, and MGBG-treated MT pears were screened, hydrogen peroxide (H2O2) was localized using CeCl3, and the contents of H2O2 and polyamine were measured. A total of 57,086,772, 61,240,014, and 67,919,420 successful reads were generated from the transcriptomes of WT, MT, and MGBG-treated MT, with average unigene lengths of 701, 720, and 735?bp, respectively. Differentially expressed genes involved in polyamine metabolism and suberin synthesis were screened in 'Dangshansuli' and in the mutant libraries, and their relative expression was found to be significantly altered after treatment with MGBG, which was confirmed by real-time PCR. The expression patterns of differentially expressed transcription factors were identified and were found to be similar to those of the polyamine- and suberin-related genes. The results indicated that the H2O2 generated during polyamine metabolism might contribute to russet formation on the exocarp of the mutant pear. Furthermore, the contents of H2O2, spermidine, spermine, and putrescine and H2O2 localization provided a comprehensive transcriptomic view of russet formation in the mutant pear.
DOI:10.1186/1471-2164-14-652URLPMID:24063813 [本文引用: 1]

Divergence in gene structure following gene duplication is not well understood. Gene duplication can occur via whole-genome duplication (WGD) and single-gene duplications including tandem, proximal and transposed duplications. Different modes of gene duplication may be associated with different types, levels, and patterns of structural divergence.
[本文引用: 1]
[本文引用: 1]
