 ,, 李莉, 郑先波, 王苗苗, 宋尚伟, 焦健, 宋春晖
,, 李莉, 郑先波, 王苗苗, 宋尚伟, 焦健, 宋春晖 ,河南农业大学园艺学院/河南省果树瓜类生物学重点实验室,郑州 450002
,河南农业大学园艺学院/河南省果树瓜类生物学重点实验室,郑州 450002Screening and Expression Analysis of Co Candidate Genes in Columnar Apple
BAI TuanHui ,, LI Li, ZHENG XianBo, WANG MiaoMiao, SONG ShangWei, JIAO Jian, SONG ChunHui
,, LI Li, ZHENG XianBo, WANG MiaoMiao, SONG ShangWei, JIAO Jian, SONG ChunHui ,College of Horticulture, Henan Agricultural University/Henan Key Laboratory of Fruit and Cucurbit Biology, Zhengzhou 450002
,College of Horticulture, Henan Agricultural University/Henan Key Laboratory of Fruit and Cucurbit Biology, Zhengzhou 450002通讯作者:
责任编辑: 赵伶俐
收稿日期:2019-07-17接受日期:2019-09-6网络出版日期:2019-12-01
| 基金资助: |
Received:2019-07-17Accepted:2019-09-6Online:2019-12-01

摘要
关键词:
Abstract
Keywords:
PDF (776KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
白团辉, 李莉, 郑先波, 王苗苗, 宋尚伟, 焦健, 宋春晖. 柱状苹果Co基因的筛选与候选基因分析[J]. 中国农业科学, 2019, 52(23): 4350-4363 doi:10.3864/j.issn.0578-1752.2019.23.015
BAI TuanHui, LI Li, ZHENG XianBo, WANG MiaoMiao, SONG ShangWei, JIAO Jian, SONG ChunHui.
0 引言
【研究意义】苹果(Malus×domestica Borkh.)是我国主要栽培的果树树种之一,其面积和产量均居世界首位,发展苹果产业,对增加农民收入、调整产业结构具有重要的意义。树型是苹果重要的农艺性状之一,对定植密度、机械采收、整形修剪以及栽培模式都有至关重要的作用[1]。柱状苹果是一种特殊的矮生突变类型,其节间非常短、侧生分枝少,且几乎不需修剪,被认为是苹果矮化密植栽培、集约化生产的理想树型[2,3,4]。开展柱状苹果Co的鉴定与分离,对实现定向遗传改良苹果树型和分子标记辅助选育柱状苹果具有重要意义。【前人研究进展】柱状苹果是1969年发现于McIntosh上的一个芽变,后来命名为McIntosh Wijcik[5]。最早研究报告显示柱状苹果是受显性单基因控制的质量性状[2]。随着生物技术的快速发展,国内外研究者相继开展了柱状苹果的分子标记和基因定位研究。CONNER等[6]用集群分类法(bulked segregant analysis,BSA)首次找到一个与Co连锁RAPD 标记 OA111000。CONNER等[7]用两个柱状杂交群体后代分离数据构建了3个亲本的遗传连锁图,包含283个标记,分布在17个连锁群上,Co被定位在第10个连锁群上。王彩虹等[8]利用AFLP结合BSA法在Spur Fuji×Telamon杂交组合中,筛选到了2个AFLP标记,分别与Co位点遗传距离为3.3和27.5 cM。TIAN等[9]报道了Co所在区域分子图谱,将Co界定在两个稳定的SSR标记(COL和CH02a10)之间,覆盖基因组长度为54.6 cM,标记间平均距离为4.55 cM。MORIYA等[10]将Co定位在两个SSR(CH03d11和Hi01a03)之间,进一步缩短了目的基因和标记的距离。随后,自2010年‘金冠’苹果基因组测序公布[11],研究者开始对Co基因开展了精细定位研究。笔者课题组前期研究将Co定位到第10条染色上物理距离75 kb以内[12]。BALDI等[13]分析了Co区域内的ORFs,发现Co区域内存在若干个潜在的Co候选基因,并对该区域内的36个基因进行了预测和注释。OTTO等[14]研究发现,在18.73—18.92 Mb区域内有一个柱型特异的8.2 kb的反转录转座子的插入,预测其可能与柱状树形有关。【本研究切入点】目前,柱状苹果Co基因的精细定位仍存在一些差异[15,16,17,18,19],还未能从分子水平上阐明柱状苹果形成机制。【拟解决的关键问题】本研究利用已发布的苹果参考基因组GDDH13(Golden Delicious doubled-haploid tree 13),通过NCBI中的序列阅读存档/表达序列标签数据库检索,对Co精细定位区间的基因进行注释分类、预测,选择目标基因,利用实时荧光定量PCR技术分析目标基因在不同组织器官中的表达特征并筛选出差异基因。1 材料与方法
试验于2018—2019年在河南农业大学果树生物学实验室进行。1.1 植物材料
本试验以3年生柱状苹果‘舞佳’(WJ)和‘润太一号’(RT),普通型苹果‘富士’(FJ)和‘华硕’(HS)植株为试材,于2018年3月萌芽后开始,分别采集萌芽期的顶芽和侧芽、生长期的主枝茎尖和侧枝茎尖、叶片、花、根和果实,其中植株的根部样品为组培苗的根,用锡箔纸包好放入液氮中速冻,迅速带回试验室于-80℃中保存。1.2 试验方法
1.2.1 总RNA的提取、检测及反转录 使用改良CTAB法[20]提取样品总RNA。使用分光光度计(NanoDrop 2000,Thermo Scientific)检测RNA溶液的浓度及纯度,并使用1%的琼脂糖凝胶电泳检测RNA的质量和完整性。使用RNA反转录试剂盒Prime ScriptTM 1st Strand cDNA Synthesis Kit(大连宝生物科技有限公司)进行反转录,严格按照说明书进行操作。1.2.2 预测基因的遴选和生物信息学分析 根据分子标记与目标基因的遗传距离和物理距离[12,13,14],在苹果基因组注释数据库(
1.2.3 引物设计与验证 根据苹果参考基因组中mRNA序列,按照荧光定量PCR引物设计原则使用Premier 5.0软件设计引物,引物信息见表1,由生工生物工程(上海)有限公司合成。以柱状和普通型苹果的cDNA为模板,对设计的引物进行PCR特异性检测。PCR体系为Taq Mix 10 μL,正、反向引物(10 mmol?L-1)各1 μL,100 ng·μL-1 cDNA 1 μL,ddH2O 7 μL,总体系20 μL。PCR反应程序为:94℃预变性2 min;94℃变性20 s,50—60℃退火20 s,72℃延伸30 s,28—32个循环,72℃延伸5 min,4℃保存。退火温度依据引物自身的Tm值进行相应的设置。使用1%的琼脂糖凝胶电泳检测扩增产物。
Table 1
表1
表114个预测基因相对表达实时荧光定量PCR引物序列
Table 1
| 编号 No | 基因名称 Gene name | 正向及反向引物序列 Forward and reverse primer sequences (5'-3') | 扩增产物大小 Amplified product size (bp) | |
|---|---|---|---|---|
| A | MD10G1184100 | F: | AGCAGGGAAGAAGAAGAAGACGC | 201 |
| R: | GGTCGTGACAGATGACGTGT | |||
| B | MD10G1185400 | F: | CTTGCTGAAGGGTTGGGACTGAG | 218 |
| R: | GAAATAGGCTCAACACCAATCCAA | |||
| C | MD10G1185600 | F: | TCAACCACCTCTGCCAAGTC | 244 |
| R: | GACTGATTTGTCAAGCCCGC | |||
| D | MD10G1186300 | F: | CATGAACTCCGGAGATGTGCTTGA | 237 |
| R: | CTTGTCACCACGGGCTTCTGTC | |||
| E | MD10G1186400 | F: | CACAACCAAGCATTCGGATCCT | 259 |
| R: | TTTGAATCTTTGCGTAGCACTTGGA | |||
| F | MD10G1187000 | F: | CGATCTCGGCTTTCTAACCTTGCT | 232 |
| R: | GTCGTCCGTGCCAACAAAACAT | |||
| G | MD10G1190500 | F: | TTCAGTCACCCACAATCGTAAACAA | 204 |
| R: | ACCTGCCACTACTCCAAACAAAAAC | |||
| G | MD10G1190700 | F: | TTGTTGGCGAGATACCGTTAGAAAT | 264 |
| R: | AGATTACAAAGGCATGAACAAGGGA | |||
| I | MD10G1191100 | F: | AGGGTGGGCAGGCTAAATGAC | 209 |
| R: | GAAGTTTAAAATGCTACCTGGCTGG | |||
| J | MD10G1192300 | F: | CACCCGACAGCCGCAGAAG | 232 |
| R: | GTCATGCCGAATACTCCAATACCAG | |||
| K | MD10G1192400 | F: | CAGAGGAACCTTCTAATGGTCCGATA | 247 |
| R: | TTCGCCTTCACTAAGTCAAAGCTGTAA | |||
| L | MD10G1192900 | F: | GAAACGCGCAGATGGATTGAG | 201 |
| R: | AAGCAAACACCAAGCCGTCCT | |||
| M | MD10G1193400 | F: | CGCAGCAGTAGCTCTCTCTC | 213 |
| R: | GGAGTTCTCCTCTGAGCTGC | |||
| N | MD10G1194200 | F: | CTCGAGCAGCCCACCATGATG | 222 |
| R: | GAGGTGGAAGCAGAAGCAGTCG | |||
| MdActin | F: | GGATTTGCTGGTGATGATGCT | 178 | |
| R: | AGTTGCTCACTATGCCGTGC | |||
新窗口打开|下载CSV
1.2.4 实时荧光定量PCR检测 荧光定量PCR检测在ABI7500荧光定量PCR仪上进行。使用SYBR? Green PCR Master Mix(美国应用生物系统公司)荧光定量试剂盒,反应体系为20 μL:10.0 μL SYBR? Green PCR Master Mix,2.0 μL 模板cDNA,上、下游引物各1.0 μL(引物浓度 10 μmol·L-1),6.0 μL ddH2O。荧光定量PCR的反应程序为:预变性过程,50℃ 20 s,95℃ 10 min,1次循环;95℃ 15 s,60℃ 1 min,并进行荧光信号采集;40次循环。每个样品设置3个生物学重复,所得数据使用2-ΔΔCT方法计算基因表达量[21,22]。
1.2.5 数据统计与分析 使用SPSS 17.0软件(IBM)对试验数据进行显著性分析(P<0.05)。用Excel 2010进行数据分析和作图。
2 结果
2.1 Co定位区间基因的注释和筛选
根据已经发表的柱状基因Co定位区间,提取‘金冠’参考基因组GDDH13 10号染色体27.66—29.05 Mb区域内基因组信息,结果表明,这一段序列包含67个基因(表2),其中12个为非编码RNA(ncRNA),其余为编码蛋白质的基因。根据柱状和普通型苹果SRA RNA-seq数据,差异倍数1倍以上的有25个基因,其中13个基因在柱状苹果中上调表达,12个基因在柱状苹果中下调表达。通过比较67个基因在柱状和普通型苹果中的表达并结合基因功能预测,筛选到14个预测基因(表1)。其中,MD10G1192100、MD10G1192900、MD10G1188000、MD10G1194100、MD10G1193400、MD10G1189000和MD10G1192200在叶片、顶梢、茎、腋芽、根、果实和花中均表达(图1)。而MD10G1192900和MD10G1192100的表达量在各个器官中均较高,MD10G1194100和MD10G1187000在根中表达量高,MD10G1192200和MD10G1188000在果实中表达量较高。Table 2
表2
表2苹果Co定位区间的基因初步筛选
Table 2
| 基因编号 Gene ID | 预测功能 Putative function | 基因表达量Gene expression | ||
|---|---|---|---|---|
| 普通型Normal | 柱状Columnar | |||
| MD10G1183600 | 肽酶S24/S26A/S26B/S26C家族蛋白 Peptidase family protein | 1040.4 | 1080.7 | |
| MD10G1183700 | 染色体(SMC)家族蛋白的结构维持 (SMC) family protein | 777.0 | 1174.6 | |
| MD10G1183800 | 非编码RNA ncRNA | - | - | |
| MD10G1183900 | 非编码RNA ncRNA | - | - | |
| MD10G1184000 | 前mRNA剪接Prp 18-相互作用因子 Pre-mRNA splicing Prp18-interacting factor | 949.5 | 1446.9 | |
| MD10G1184100 | MYB结构域蛋白15 MYB domain protein 15 | 11.4 | 230.1 | |
| MD10G1184200 | 非编码RNA ncRNA | - | - | |
| MD10G1184300 | 未知功能蛋白 Protein of unknown function | 331.7 | 335.6 | |
| MD10G1184400 | 未知功能蛋白 Protein of unknown function | 270.1 | 240.6 | |
| MD10G1184500 | 肌动蛋白解聚因子6 Actin depolymerizing factor 6 | 1209.1 | 1275.9 | |
| MD10G1184600 | 磷酸盐转运蛋白1 Phosphate transporter 1 | - | - | |
| MD10G1184700 | 磷酸盐转运蛋白1 Phosphate transporter 1 | 472.8 | 519.2 | |
| MD10G1184800 | 乙烯响应因子1 Ethylene response factor 1 | 2.8 | 0.0 | |
| MD10G1184900 | 非编码RNA ncRNA | - | - | |
| MD10G1185000 | 整合酶型DNA结合超家族蛋白 Integrase-type DNA-binding superfamily protein | 2.8 | 0.0 | |
| MD10G1185100 | 整合酶型DNA结合超家族蛋白 Integrase-type DNA-binding superfamily protein | - | - | |
| MD10G1185200 | 未知功能蛋白 Protein of unknown function | 518.3 | 640.6 | |
| MD10G1185300 | 环/U盒超家族蛋白 RING/U-box superfamily protein | - | - | |
| MD10G1185400 | 2-氧戊二酸(2OG)和铁(II)依赖性加氧酶超家族蛋白 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | 0.0 | 26.4 | |
| MD10G1185500 | 2-氧戊二酸(2OG)和铁(II)依赖性加氧酶超家族蛋白 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | - | - | |
| MD10G1185600 | 环/U盒超家族蛋白RING/U-box superfamily protein | 0.9 | 3.2 | |
| MD10G1185700 | 四三肽重复序列(TPR)样超家族蛋白 Tetratricopeptide repeat (TPR)-like superfamily protein | - | - | |
| MD10G1185800 | 四三肽重复序列(TPR)样超家族蛋白 Tetratricopeptide repeat (TPR)-like superfamily protein | - | - | |
| MD10G1185900 | 自噬9 Autophagy 9 (APG9) | 420.7 | 515.0 | |
| MD10G1186000 | 结节蛋白MTN21/EAMA样转运蛋白家族 Nodulin MTN21/EAMA-like transporter family protein | 84.3 | 108.7 | |
| MD10G1186100 | 无顶端分生组织结构域转录调节超家族蛋白 NAC (No Apical Meristem) domain transcriptional regulator superfamily protein | - | - | |
| MD10G1186200 | 非编码RNA ncRNA | - | - | |
| MD10G1186300 | 开放阅读框7上游保守肽 Conserved peptide upstream open reading frame 7 | 966.5 | 1175.6 | |
| MD10G1186400 | 2-氧戊二酸(2OG)和铁(II)依赖性加氧酶超家族蛋白 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | 41.7 | 11.6 | |
| MD10G1186500 | 糖基水解酶9B6 Glycosyl hydrolase 9B6 | - | - | |
| MD10G1186600 | 糖基水解酶9B1 Glycosyl hydrolase 9B1 | - | - | |
| MD10G1186700 | 2-氧戊二酸(2OG)和铁(II)依赖性加氧酶超家族蛋白 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | - | - | |
| MD10G1186800 | 碱性螺旋-环-螺旋(bHLH)DNA结合超家族蛋白 Basic helix-loop-helix (bHLH) DNA-binding superfamily protein | - | - | |
| MD10G1186900 | 碱性螺旋-环-螺旋(bHLH)DNA结合超家族蛋白 Basic helix-loop-helix (bHLH) DNA-binding superfamily protein | - | - | |
| MD10G1187000 | 2-氧戊二酸(2OG)和铁(II)依赖性加氧酶超家族蛋白 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | 0.9 | 11.6 | |
| MD10G1187100 | 非编码RNA ncRNA | - | - | |
| MD10G1187200 | α/β水解酶超家族蛋白Alpha/beta-Hydrolases superfamily protein | 2.8 | 0.0 | |
| MD10G1187300 | 侧生器官边界结构域29 Lateral organ boundaries-domain 29 | - | - | |
| MD10G1187400 | 锌指Zinc finger | - | - | |
| MD10G1187500 | 光系统I亚单位H2 Photosystem I subunit H2 | - | - | |
| MD10G1187600 | 侧生器官边界结构域29 Lateral organ boundaries-domain 29 | - | - | |
| MD10G1187700 | 侧生器官边界结构域16 Lateral organ boundaries-domain 16 | 1.9 | 0.0 | |
| MD10G1187800 | 多聚ADP核糖聚合酶2 Poly(ADP-ribose) polymerase 2 | 869.9 | 577.3 | |
| MD10G1187900 | ARM重复超家族蛋白 ARM repeat superfamily protein | 560.0 | 506.6 | |
| MD10G1188000 | BET1P/SFT1P样蛋白14A BET1P/SFT1P-like protein 14A | 915.4 | 391.5 | |
| MD10G1188100 | C2H2和C2HC锌指超家族蛋白C2H2 and C2HC zinc fingers superfamily protein | 5.7 | 12.7 | |
| MD10G1188200 | C2H2和C2HC锌指超家族蛋白C2H2 and C2HC zinc fingers superfamily protein | - | - | |
| MD10G1188300 | 核糖体S17家族蛋白 Ribosomal S17 family protein | 0.0 | 2.1 | |
| MD10G1188400 | 具有Dil结构域的肌球蛋白家族蛋白 Myosin family protein with Dil domain | 110.9 | 186.8 | |
| MD10G1188500 | 半乳糖氧化酶/kelch重复超家族蛋白 Galactose oxidase/kelch repeat superfamily protein | - | - | |
| MD10G1188600 | 未知Unknown | - | - | |
| MD10G1188700 | 2-氧戊二酸(2OG)和铁(II)依赖性加氧酶超家族蛋白 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | - | - | |
| MD10G1188800 | 2-氧戊二酸(2OG)和铁(II)依赖性加氧酶超家族蛋白 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | - | - | |
| MD10G1188900 | 未知Unknown | 34.1 | 51.7 | |
| MD10G1189000 | 未知Unknown | 292.8 | 93.9 | |
| MD10G1189100 | 未知Unknown | 154.5 | 228.0 | |
| MD10G1189200 | 光合电子转移C Photosynthetic electron transfer C | - | - | |
| MD10G1189300 | 未知Unknown | 145.0 | 162.5 | |
| MD10G1189400 | 2-氧戊二酸(2OG)和铁(II)依赖性加氧酶超家族蛋白 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | - | - | |
| MD10G1189500 | 未知Unknown | 37.9 | 19.0 | |
| MD10G1189600 | 2-氧戊二酸(2OG)和铁(II)依赖性加氧酶超家族蛋白 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | - | - | |
| MD10G1189700 | 半乳糖氧化酶/kelch重复超家族蛋白 Galactose oxidase/kelch repeat superfamily protein | - | - | |
| MD10G1189800 | 未知Unknown | - | - | |
| MD10G1189900 | 2-氧戊二酸(2OG)和铁(II)依赖性加氧酶超家族蛋白 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | - | - | |
| MD10G1190000 | 碱性螺旋-环-螺旋(bHLH)DNA结合超家族蛋白 Basic helix-loop-helix (bHLH) DNA-binding superfamily protein | 668.0 | 853.8 | |
| MD10G1190100 | 未知Unknown | 245.4 | 51.7 | |
| MD10G1190200 | 细胞色素P450 Cytochrome P450 | 531.6 | 331.4 | |
| MD10G1190300 | 未知Unknown | - | - | |
| MD10G1190400 | 细胞色素P450 Cytochrome P450 | 661.4 | 798.9 | |
| MD10G1190500 | 未知功能蛋白 Protein of unknown function | 69.2 | 180.5 | |
| MD10G1190600 | 非编码RNA ncRNA | 6.6 | 5.3 | |
| MD10G1190700 | IND1(NADH脱氢酶所需的铁硫蛋白)-类 IND1(iron-sulfur protein required for NADH dehydrogenase)-like | 225.5 | 520.3 | |
| MD10G1190800 | 类pfkb碳水化合物激酶家族蛋白 Pfkb-like carbohydrate kinase family protein | 193.3 | 224.8 | |
| MD10G1190900 | Mudr家族转座酶 Mudr family transposase | 389.5 | 707.1 | |
| MD10G1191000 | 非编码RNA ncRNA | 1.9 | 3.2 | |
| MD10G1191100 | 含核苷三磷酸水解酶超家族蛋白的P环 P-loop containing nucleoside triphosphate hydrolases superfamily protein | 2.8 | 9.5 | |
| MD10G1191200 | 翼螺旋DNA结合转录因子家族蛋白 Winged-helix DNA-binding transcription factor family protein | 10231.9 | 5165.8 | |
| MD10G1191300 | 乙烯响应元件结合因子3 Ethylene responsive element binding factor 3 | 1089.7 | 547.7 | |
| MD10G1191400 | WRKY DNA结合蛋白21 WRKY DNA-binding protein 21 | 839.5 | 546.7 | |
| MD10G1191500 | 信号识别粒子 Signal recognition particle | 508.8 | 376.8 | |
| MD10G1191600 | 跨膜氨基酸转运体家族蛋白 Transmembrane amino acid transporter family protein | — | — | |
| MD10G1191700 | PRA1家族蛋白 PRA1 (Prenylated rab acceptor) family protein | 321.2 | 246.9 | |
| MD10G1191800 | DREB2A相互作用蛋白2 DREB2A-interacting protein 2 | 338.3 | 338.8 | |
| MD10G1191900 | 镁转运蛋白9 Magnesium transporter 9 | 364.8 | 421.1 | |
| MD10G1192000 | 半乳糖醛糖基转移酶6 Galacturonosyltransferase 6 | 188.6 | 193.1 | |
| MD10G1192100 | 光系统II反应中心W Photosystem II reaction center W | 6877.5 | 1885.9 | |
| MD10G1192200 | 非编RNA ncRNA | 7.6 | 2.1 | |
| MD10G1192300 | 谷胱甘肽还原酶 Glutathione reductase | 665.2 | 1506.0 | |
| MD10G1192400 | Transferin/WD 40重复超家族蛋白 Transducin/WD40 repeat-like superfamily protein | 11.4 | 58.0 | |
| MD10G1192500 | α/β水解酶超家族蛋白 Alpha/beta-Hydrolases superfamily protein | 303.2 | 237.4 | |
| MD10G1192600 | 非编码RNA ncRNA | |||
| MD10G1192700 | 非编码RNA ncRNA | - | - | |
| MD10G1192800 | 类RNI超家族蛋白 RNI-like superfamily protein | 1226.2 | 1437.4 | |
| MD10G1192900 | 吲哚-3-乙酸7 Indole-3-acetic acid 7 | 3281.4 | 8731.8 | |
| MD10G1193000 | AUX/IAA转录调节家族蛋白 AUX/IAA transcriptional regulator family protein | - | - | |
| MD10G1193100 | 原体prib/单链DNA结合 Primosome prib/single-strand DNA-binding | 302.3 | 378.9 | |
| MD10G1193200 | 碱性螺旋-环-螺旋(bHLH)DNA结合超家族蛋白 Basic helix-loop-helix (bHLH) DNA-binding superfamily protein | 525.0 | 624.8 | |
| MD10G1193300 | 色氨酸合酶α链 Tryptophan synthase alpha chain | 329.8 | 215.3 | |
| MD10G1193400 | 碱性亮氨酸拉链25 Basic leucine zipper 25 | 145.9 | 575.2 | |
| MD10G1193500 | GNAT家族1组蛋白乙酰转移酶 Histone acetyltransferase of the GNAT family 1 | 547.7 | 491.8 | |
| MD10G1193600 | 非编码RNA ncRNA | - | - | |
| MD10G1193700 | 泛素结合酶11 Ubiquitin-conjugating enzyme 11 | - | - | |
| MD10G1193800 | SOUL血红素结合家族蛋白 SOUL heme-binding family protein | 1072.7 | 1758.2 | |
| MD10G1193900 | 泛素特异性蛋白酶15 Ubiquitin-specific protease 15 | 810.2 | 1333.9 | |
| MD10G1194000 | 阳离子氨基酸转运蛋白8 Cationic amino acid transporter 8 | 532.5 | 255.4 | |
| MD10G1194100 | 拟南芥赤霉素2-氧化酶1 Arabidopsis thaliana gibberellin 2-oxidase 1 | 1936.8 | 795.7 | |
新窗口打开|下载CSV
图1
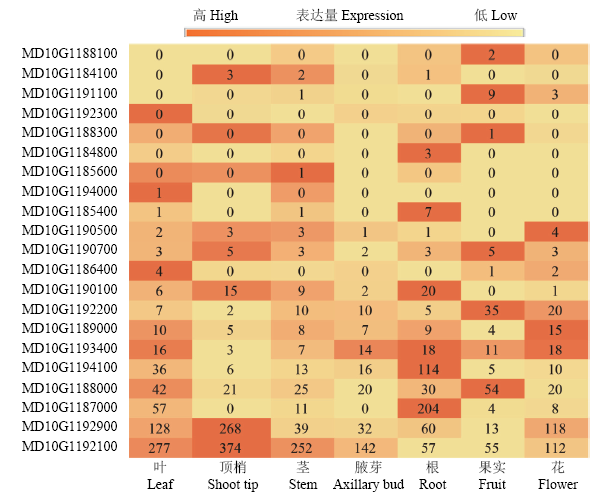 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1初步筛选基因组织表达
Fig. 1Tissue expression of preliminary screening gene
2.2 预测基因在苹果主枝茎尖中的表达特征
对扩增到cDNA序列的14个预测基因,利用克隆到的基因序列设计qRT-PCR特异引物(表1),在柱状苹果‘舞佳’和‘润太’,普通型苹果‘富士’和‘华硕’的主枝茎尖进行表达分析。结果表明,MD10G1184100和MD10G1185600在两个柱状苹果主枝茎尖中的相对表达量均显著高于两个普通型苹果(图2-A和C)。MD10G1186300在‘华硕’中的表达量显著高于‘舞佳’和‘润太’,而在‘润太’与‘富士’中的表达量则无显著差异(图2-D)。MD10G1187000在‘润太’中的表达量显著高于‘富士’和‘华硕’,但是在‘舞佳’中的表达量又显著低于‘富士’和‘华硕’(图2-F)。MD10G1190500在‘润太’中的表达量显著高于其他3个品种,但在这3个品种之间则无显著差异(图2-G)。MD10G1190700在‘润太’中的表达量最低,在‘舞佳’中的表达量显著高于‘华硕’,但其与‘富士’之间无显著差异(图2-H);MD10G1191100在两个柱状苹果以及‘富士’中的表达量显著低于‘华硕’,但在‘舞佳’与‘富士’之间,其表达量无显著差异(图2-I);MD10G1192900在‘润太’中的表达量显著高于其他3个品种,但在‘舞佳’中的表达量显著低于普通型(图2-L)。则是在‘华硕’中的表达量显著高于其他3个品种,但其表达量在这3个品种间无显著差异(图2-M)。MD10G1192300和MD10G1192400在柱状与普通型中的表达量无显著差异。MD10G1194200在两个柱状苹果中的表达量显著低于两个普通型苹果(图2-N)。图2
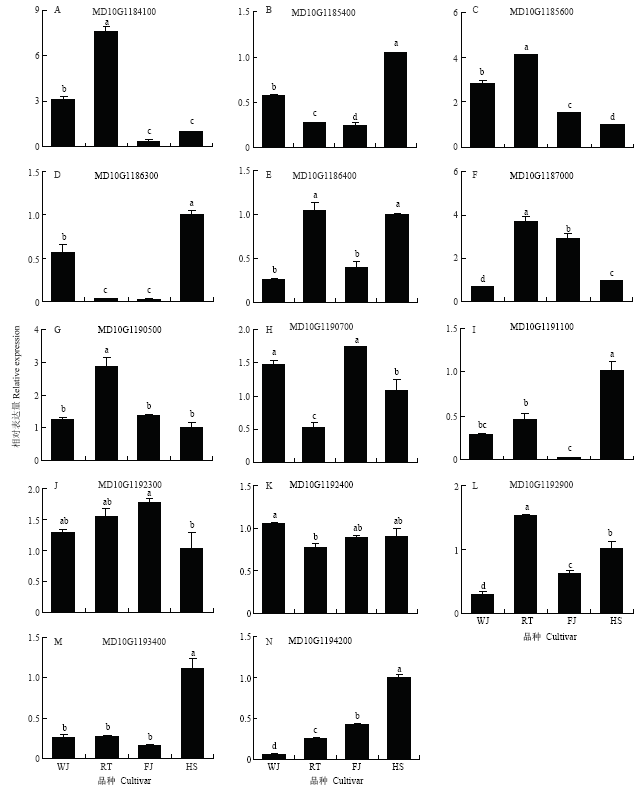 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2预测基因在柱状和普通型苹果主枝茎尖中的相对表达水平
WJ:舞佳 Wujia;RT:润太 Runtai;FJ:富士 Fuji;HS:华硕 Huashuo。不同小写字母表示差异显著(P<0.05)。下同
Fig. 2Expression of putative genes in stem tips of columnar and standard apple
Different lowercase letters indicate significant difference (P<0.05). The same as below
2.3 预测基因在苹果侧枝茎尖中表达特征
对14个预测基因在柱状苹果‘舞佳’和‘润太’,普通型苹果‘富士’和‘华硕’的侧枝茎尖进行了表达分析。结果表明,MD10G1185400和MD10G1190500在2个柱状苹果侧枝茎尖的表达量显著高于两个普通型苹果(图3-B和G),而MD10G1184100和MD10G1186400在2个柱状苹果中的表达量分别显著低于2个普通型苹果(图3-A和E)。MD10G1185600在‘舞佳’与‘富士’中的表达量显著高于另外2个苹果品种,但是这两者之间无显著差异,并且在‘润太’中的表达量显著高于‘华硕’(图3-C),MD10G1186300在‘华硕’中的表达量显著高于其他3个苹果,但除‘华硕’外的3个品种间无显著差异(图3-D)。其他基因则是在品种间的表达无差异或在柱状与普通型苹果中的表达量存在不一致的现象,综合考虑,选择在柱状苹果‘舞佳’和‘润太’,普通型苹果‘富士’和‘华硕’的主枝茎尖以及侧枝茎尖中差异表达的4个基因MD10G1184100、MD10G1185400、MD10G1185600和MD10G1190500作为候选基因。图3
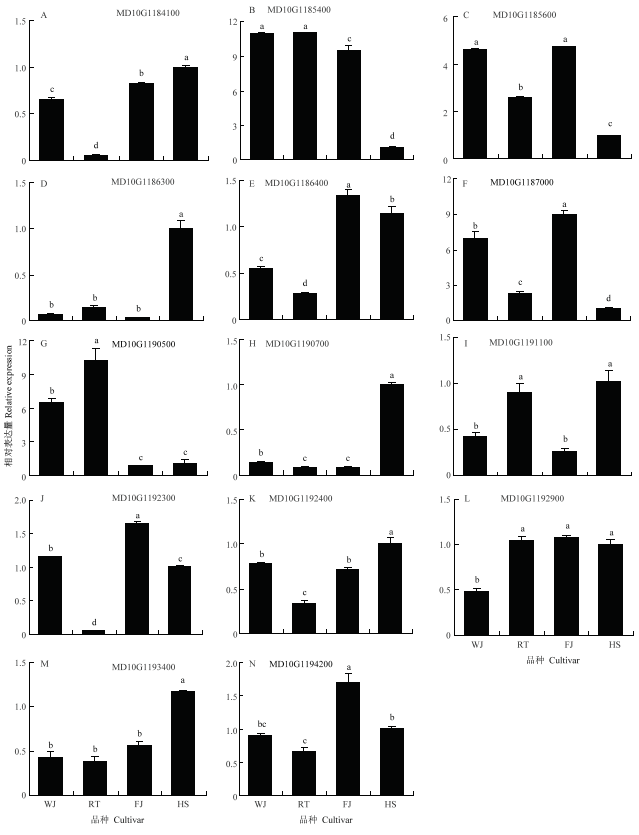 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3预测基因在柱状和普通型苹果侧枝茎尖中的相对表达水平
Fig. 3Expression of putative gene in lateral shoot tips of lateral buds of columnar and standard apple
2.4 候选基因在不同组织部位中的表达分析
通过实时荧光定量PCR检测4个Co候选基因(MD10G1184100、MD10G1185400、MD10G1185600和MD10G1190500)在柱状和普通型苹果顶芽、侧芽、主枝茎尖、侧枝茎尖、根、叶、花、果实中的相对表达水平。结果表明,4个候选基因在顶芽、侧芽、主枝茎尖、侧枝茎尖、根、叶、花和果实中均有表达,但表达特征有差异(图4)。MD10G1184100在柱状苹果根部的表达量最高,显著高于其他部位,为普通型苹果根部表达量的65倍。MD10G1184100在顶芽、主枝茎尖、根和果实中的表达量显著大于普通型苹果。MD10G1185400在柱状苹果侧枝茎尖中高表达,在柱状苹果的侧枝茎尖、根、果实和叶片中的表达量显著高于普通型;而在花、侧芽和侧枝茎尖的表达量显著小于普通型。MD10G1185600在柱状苹果的侧枝茎尖中高表达,在主枝茎尖、侧枝茎尖、果实和叶片中的表达量显著大于普通型,而在顶芽、侧芽、根和花等组织中的表达量显著小于普通型。MD10G1190500在柱状苹果顶芽中的表达量最高,在顶芽、侧芽、侧枝茎尖、花、果实和叶片中表达量显著高于普通型。图4
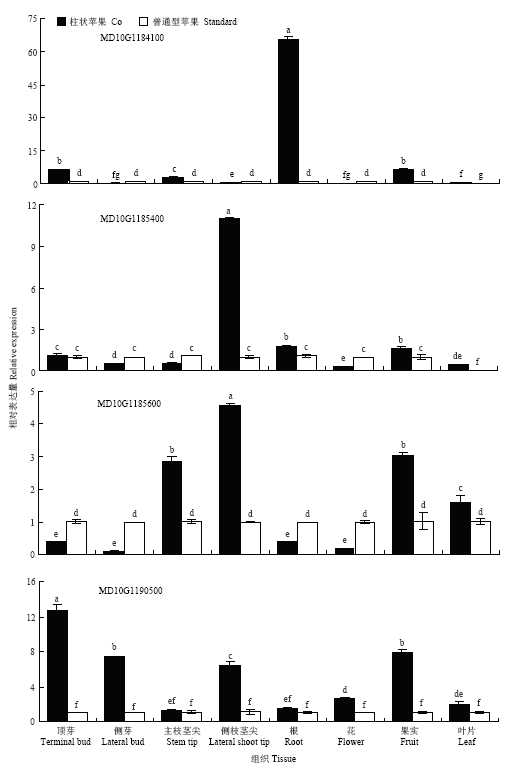 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4候选基因在柱状和普通型苹果不同组织中的相对表达水平
Fig. 4Expression of candidate genes in different tissues of columnar and standard apple
3 讨论
苹果是世界上最重要的经济作物之一。近30多年来,世界苹果栽培制度发生了深刻的变化,欧美等发达国家苹果园的矮砧栽培比例高达90%以上,而中国苹果矮化栽培的比例不足10%[23]。随着农业产业结构的调整和城市化进程的日益加快,农村劳动力向城市大量转移,苹果种植从业人员不足的现象日益严重,矮砧密植栽培是我国苹果产业发展的必然趋势[22,23,24]。柱状苹果因其节间短、侧生分枝少,且几乎不需修剪,被认为是现代苹果矮砧密植栽培的理想树型[25,26,27]。但到目前为止,柱状苹果形成的分子调控机制尚不清楚。从分子水平上揭示柱状苹果独特的生长特性是实现苹果定向遗传改良树型的基础。前期研究表明,苹果柱状树型是受位于10号染色体上的单显性基因Co控制,并且在Co区域内有多个预测基因[6,28-30]。笔者课题组前期研究开发了7个与Co紧密连锁的SSRs标记,并构建了Co遗传连锁和物理图谱,将Co定位到第10条染色体分子标记16470_23262和C5629_32009之间物理距离75 kb以内[12]。本研究以柱状苹果与普通型苹果为试材,对定位区间的基因进行注释分类,发现在10号染色体27.66—29.05 Mb区域内包含67个基因,进一步发现4个基因(MD10G1185400、MD10G1184100、MD10G1185600和MD10G1190500)在柱状和普通型苹果的主枝茎尖或侧枝茎尖基因表达存在显著差异,推测这4个基因可能是Co候选基因。在所预测的4个候选基因中,已有研究表明MD10G1185400可能参与调控苹果柱状树型[19,31]。经序列比对发现,MD10G1185400与WOLTERS等[31]报道的MdCo31 dmr6-like(downy mildew resistance 6-like)和OKADA等[19]报道的91071是同一个基因。WOLTERS等[31]发现,与普通型苹果相比,MdCo31在柱状苹果腋芽部位显著上调表达。OKADA等[19]发现基因91071也是仅在柱状苹果中表达,而在普通型苹果中不表达。本研究与其相似,该基因在柱状苹果‘舞佳’和‘润太’的侧枝茎尖中高表达,并且在柱状苹果和普通型苹果不同部位以及不同时期中的表达量均有显著差异。OKADA等[19]将91071转入烟草中,发现转基因烟草的株高降低和节间长度变短,在苹果中过表达该基因也引起节间变短,依此推断,91071可能是Co候选基因,但柱状苹果的生长表型不仅仅是节间短。进一步通过对MD10G1185400进行分析,表明MD10G1185400属于2OGDs的DOXC41亚家族,DOXC41亚家族成员基因功能多样,参与各种特殊代谢,如莨菪碱6β-羟化酶(H6H)和大麦铁缺乏,但苹果柱状性状与这几种物质代谢好像又关系不大,MD10G1185400的具体功能依然未知,其是否在激素通路或者其他通路中发挥作用,有待进一步研究。另外一个候选基因MD10G1184100是一个转录因子,为MYB domain protein 15,在柱状苹果中显著上调表达。拟南芥的同源基因AtMYB15在防御诱导的木质化和基础免疫中发挥着作用,沉默AtMYB15降低了防御诱导的木质素的含量,而过表达AtMYB15增加了木质素含量[32]。推测柱状苹果树主干上萌发出的直立侧枝,可能与木质素合成异常有关,因此,这个基因也有待验证。PETERSEN等[18]研究发现,MDP0000163720在状状苹果‘P28’的顶端分生组织中上调表达。通过BLAST比对发现,MDP0000163720与本研究中的MD10G1185600是同一个基因,且表达特征与其研究结果一致。MD10G1190500编码一个肌球结合蛋白基因(myosin-binding protein 1-like,Myob),可直接与肌球蛋白结合,并将其募集到囊泡样内膜膜室中,从而快速沿着F-肌动蛋白轨迹移动[33]。MD10G1190500在柱状苹果侧枝茎尖、花、果实和叶片中的表达量显著大于普通型苹果。肌球结合蛋白基因在植物中研究较少,MD10G1190500是否与苹果柱状树形有关,有待深入研究。具体哪一个基因调控柱状苹果树形,还需进一步验证候选基因的生物学功能,可能还与植物激素IAA、CK和GA的代谢和信号转导有关;同时,笔者在柱状的杂交群体中发现,杂交后代中柱状的植株比例通常小于50%的理论值。另外,柱状苹果的杂交后代中,有些植株携带着反转座子插入标记,但却表现出普通型的表型,表明其他基因可能对Co起修饰作用。
4 结论
通过对苹果Co定位区域进行注释分类和筛选,检测到67个基因,其中12个为非编码RNA,其余为编码蛋白质的基因。根据RNA-seq分析67个基因在柱状和普通型苹果表达特征并结合基因功能预测,筛选到14个预测基因,通过qRT-PCR分析发现4个基因(MD10G1184100、MD10G1185400、MD10G1185600和MD10G1190500)在柱状和普通型苹果的主枝茎尖或侧枝茎尖存在显著表达差异,可作为Co候选基因。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.3864/j.issn.0578-1752.2017.22.018URL [本文引用: 1]

Columnar apple is a dwarfed mutant with thick, upright main stems and shortened internodes, and generates short fruit spurs instead of long lateral branches. It is a good resource for high dense planting and high yield production in the modern apple industry. Therefore, to understand its unique growth habit was highly interested for all research groups. The current research achievements are summarized as follows: (1) The growth habits of columnar apple is closely related with its endogenous hormones. The free IAA to total IAA ratio was found higher in the axillary buds of columnar apple trees than in the standard type apple trees. The columnar apple producing high number of spurs is because the higher level of zeatin-like growth substances exists in both apical and lateral shoots. The dwarfed growth phenotype is probably correlated with the lower GA level in the columnar apple trees. (2) The columnar phenotype is controlled by a single dominant allele of the columnar gene, which is clustered with the genes controlling main stem growth, branching habit, leaf feature and fruit quality. The Co gene has been fine-mapped to chromosome 10 within the region of 18.51-19.09 Mb. (3) Five Co candidate genes has been reported till today. As the observation that expression of 91071 in apple and tobacco led to shortened internodes in transgenic plants, 91071 was taken as the most promising Co candidate gene, although more studies are needed to clarify whether the 91071 gene also causes the reduced lateral branches and increased spurs in columnar apple. Since co gene is closed related with both plant hormone metabolism and signal transduction, studies on its biological functions by RNAi and transgenic technologies can not only reveal the molecular mechanism of the unique growth characteristics of columnar apple tree, but also provide the theoretical basis for the molecular breeding of columnar apple with improved quality.
DOI:10.3864/j.issn.0578-1752.2017.22.018URL [本文引用: 1]

Columnar apple is a dwarfed mutant with thick, upright main stems and shortened internodes, and generates short fruit spurs instead of long lateral branches. It is a good resource for high dense planting and high yield production in the modern apple industry. Therefore, to understand its unique growth habit was highly interested for all research groups. The current research achievements are summarized as follows: (1) The growth habits of columnar apple is closely related with its endogenous hormones. The free IAA to total IAA ratio was found higher in the axillary buds of columnar apple trees than in the standard type apple trees. The columnar apple producing high number of spurs is because the higher level of zeatin-like growth substances exists in both apical and lateral shoots. The dwarfed growth phenotype is probably correlated with the lower GA level in the columnar apple trees. (2) The columnar phenotype is controlled by a single dominant allele of the columnar gene, which is clustered with the genes controlling main stem growth, branching habit, leaf feature and fruit quality. The Co gene has been fine-mapped to chromosome 10 within the region of 18.51-19.09 Mb. (3) Five Co candidate genes has been reported till today. As the observation that expression of 91071 in apple and tobacco led to shortened internodes in transgenic plants, 91071 was taken as the most promising Co candidate gene, although more studies are needed to clarify whether the 91071 gene also causes the reduced lateral branches and increased spurs in columnar apple. Since co gene is closed related with both plant hormone metabolism and signal transduction, studies on its biological functions by RNAi and transgenic technologies can not only reveal the molecular mechanism of the unique growth characteristics of columnar apple tree, but also provide the theoretical basis for the molecular breeding of columnar apple with improved quality.
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.4141/cjps69-130URL [本文引用: 1]
[J]
[本文引用: 2]
DOI:10.1007/s001220050835URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s10681-005-1163-9URL [本文引用: 1]
DOI:10.2503/jjshs1.78.279URL [本文引用: 1]
DOI:10.1038/ng.654URLPMID:20802477 [本文引用: 1]

We report a high-quality draft genome sequence of the domesticated apple (Malus × domestica). We show that a relatively recent (&gt;50 million years ago) genome-wide duplication (GWD) has resulted in the transition from nine ancestral chromosomes to 17 chromosomes in the Pyreae. Traces of older GWDs partly support the monophyly of the ancestral paleohexaploidy of eudicots. Phylogenetic reconstruction of Pyreae and the genus Malus, relative to major Rosaceae taxa, identified the progenitor of the cultivated apple as M. sieversii. Expansion of gene families reported to be involved in fruit development may explain formation of the pome, a Pyreae-specific false fruit that develops by proliferation of the basal part of the sepals, the receptacle. In apple, a subclade of MADS-box genes, normally involved in flower and fruit development, is expanded to include 15 members, as are other gene families involved in Rosaceae-specific metabolism, such as transport and assimilation of sorbitol.
DOI:10.1007/s00438-012-0689-5URL [本文引用: 3]

Tree architecture is an important, complex and dynamic trait affected by diverse genetic, ontogenetic and environmental factors. 'Wijcik McIntosh', a columnar (reduced branching) sport of 'McIntosh' and a valuable genetic resource, has been used intensively in apple-breeding programs for genetic improvement of tree architecture. The columnar growth habit is primarily controlled by the dominant allele of gene Co (columnar) on linkage group-10. But the Co locus is not well mapped and the Co gene remains unknown. To precisely map the Co locus and to identify candidate genes of Co, a sequence-based approach using both peach and apple genomes was used to develop new markers linked more tightly to Co. Five new simple sequence repeats markers were developed (C1753-3520, C18470-25831, C6536-31519, C7223-38004 and C7629-22009). The first four markers were obtained from apple genomic sequences on chromosome-10, whereas the last (C7629-22009) was from an unanchored apple contig that contains an apple expressed sequence tag CV082943, which was identified through synteny analysis between the peach and apple genomes. Genetic mapping of these five markers in four F-1 populations of 528 genotypes and 290 diverse columnar selections/cultivars (818 genotypes in total) delimited the Co locus in a genetic interval with 0.37 % recombination between markers C1753-3520 and C7629-22009. Marker C18470-25831 co-segregates with Co in the 818 genotypes studied. The Co region is estimated to be 193 kb and contains 26 predicted gene in the 'Golden Delicious' genome. Among the 26 genes, three are putative LATERAL ORGAN BOUNDARIES (LOB) DOMAIN (LBD) containing transcription factor genes known of essential roles in plant lateral organ development, and are therefore considered as strong candidates of Co, designated MdLBD1, MdLBD2, and MdLBD3. Although more comprehensive studies are required to confirm the function of MdLBD1-3, the present work represents an important step forward to better understand the genetic and molecular control of tree architecture in apple.
DOI:10.1007/s11032-012-9800-1URL [本文引用: 2]

A better understanding of the genetic control of tree architecture would potentially allow improved tailoring of newly bred apple cultivars in terms of field management aspects, such as planting density, pruning, pest control and disease protection. It would also have an indirect impact on yield and fruit quality. The Columnar (Co) locus strongly suppresses lateral branch elongation and is the most important genetic locus influencing tree architecture in apple. Co has previously been mapped on apple linkage group (LG) 10. In order to obtain fine mapping of Co, both genetically and physically, we have phenotypically analysed and screened three adult segregating experimental populations, with a total of 301 F-1 plants, and one substantial 3-year old population of 1,250 F-1 plants with newly developed simple sequence repeat (SSR) markers, based on the 'Golden delicious' apple genome sequence now available. Co was found to co-segregate with SSR marker Co04R12 and was confined in a region of 0.56 cM between SSR markers Co04R11 and Co04R13, corresponding to 393 kb on the 'Golden delicious' genome sequence. In this region, 36 genes were predicted, including at least seven sequences potentially belonging to genes that could be considered candidates for involvement in control of shoot development. Our results provide highly reliable, virtually co-segregating markers that will facilitate apple breeding aimed at modifications of the tree habit and lay the foundations for the cloning of Co.
DOI:10.1007/s11032-013-0001-3URL [本文引用: 2]

The columnar growth habit of apple trees (Malus x domestica Borkh.) is a unique plant architecture phenotype that arose as a bud sport mutation of a McIntosh tree in the 1960s. The mutation ("Co gene") led to trees (McIntosh Wijcik) with thick, upright main stems and short internodes that generate short fruit spurs instead of long lateral branches. Although Co has been localized to chromosome 10, in a region approximately between 18.5 and 19 Mb, its molecular nature is unknown. In a classical positional cloning approach in combination with the analysis of NGS data, we cloned and analyzed the Co region. Our results show that the insertion of a Ty3/Gypsy retrotransposon into a non-coding region at position 18.8 Mb is the only detectable genomic difference between McIntosh and McIntosh Wijcik and is found in all columnar cultivars. The genetic effect of the insertion is unclear; however, Illumina(A (R)) RNA-seq data of McIntosh and McIntosh Wijcik suggest that the columnar growth habit is associated with differential expression of the retrotransposon transcript, causing changes in the expression levels of many protein coding genes. The mechanism by which the Gypsy retrotransposon is involved in generating the columnar habit is not yet clear; our findings form the basis for tackling this question.
DOI:10.1111/pbr.2012.131.issue-5URL [本文引用: 1]
DOI:10.1387/ijdb.190239gsURLPMID:31840777 [本文引用: 1]

The social amoeba Dictyostelium discoideum is a tractable model organism to study cellular allorecognition, which is the ability of a cell to distinguish itself and its genetically similar relatives from more distantly related organisms. Cellular allorecognition is ubiquitous across the tree of life and affects many biological processes. Depending on the biological context, these versatile systems operate both within and between individual organisms, and both promote and constrain functional heterogeneity. Some of the most notable allorecognition systems mediate neural self-avoidance in flies and adaptive immunity in vertebrates. D. discoideum's allorecognition system shares several structures and functions with other allorecognition systems. Structurally, its key regulators reside at a single genomic locus that encodes two highly polymorphic proteins, a transmembrane ligand called TgrC1 and its receptor TgrB1. These proteins exhibit isoform-specific, heterophilic binding across cells. Functionally, this interaction determines the extent to which co-developing D. discoideum strains co-aggregate or segregate during the aggregation phase of multicellular development. The allorecognition system thus affects both development and social evolution, as available evidence suggests that the threat of developmental cheating represents a primary selective force acting on it. Other significant characteristics that may inform the study of allorecognition in general include that D. discoideum's allorecognition system is a continuous and inclusive trait, it is pleiotropic, and it is temporally regulated.
DOI:10.1007/s10725-018-0400-xURL [本文引用: 1]
DOI:10.1186/s12870-014-0356-6URLPMID:25648715 [本文引用: 2]

Primary roots (radicles) represent the first visible developmental stages of the plant and are crucial for nutrient supply and the integration of environmental signals. Few studies have analyzed primary roots at a molecular level, and were mostly limited to Arabidopsis. Here we study the primary root transcriptomes of standard type, heterozygous columnar and homozygous columnar apple (Malus x domestica) by RNA-Seq and quantitative real-time PCR. The columnar growth habit is characterized by a stunted main axis and the development of short fruit spurs instead of long lateral branches. This compact growth possesses economic potential because it allows high density planting and mechanical harvesting of the trees. Its molecular basis has been identified as a nested Gypsy-44 retrotransposon insertion; however the link between the insertion and the phenotype as well as the timing of the phenotype emergence are as yet unclear. We extend the transcriptomic studies of columnar tissues to the radicles, which are the earliest developmental stage and investigate whether homozygous columnar seedlings are viable.
DOI:10.1007/s10265-016-0863-7URLPMID:27650512 [本文引用: 5]

Determining the molecular mechanism of fruit tree architecture is important for tree management and fruit production. An apple mutant 'McIntosh Wijcik', which was discovered as a bud mutation from 'McIntosh', exhibits a columnar growth phenotype that is controlled by a single dominant gene, Co. In this study, the mutation and the Co gene were analyzed. Fine mapping narrowed the Co region to a 101?kb region. Sequence analysis of the Co region and the original wild-type co region identified an insertion mutation of an 8202?bp long terminal repeat (LTR) retroposon in the Co region. Segregation analysis using a DNA marker based on the insertion polymorphism showed that the LTR retroposon was closely associated with the columnar growth phenotype. RNA-seq and RT-PCR analysis identified a promising Co candidate gene (91071-gene) within the Co region that is specifically expressed in 'McIntosh Wijcik' but not in 'McIntosh'. The 91071-gene was located approximately 16?kb downstream of the insertion mutation and is predicted to encode a 2-oxoglutarate-dependent dioxygenase involved in an unknown reaction. Overexpression of the 91071-gene in transgenic tobaccos and apples resulted in phenotypes with short internodes, like columnar apples. These data suggested that the 8202?bp retroposon insertion in 'McIntosh Wijcik' is associated with the short internodes of the columnar growth phenotype via upregulated expression of the adjacent 91071-gene. Furthermore, the DNA marker based on the insertion polymorphism could be useful for the marker-assisted selection of columnar apples.
DOI:10.1007/BF02772687URL [本文引用: 1]

Recovering RNA of high quality and quantity is a prerequisite for ensuring representation of all expressed genes in a cDNA library. An efficient procedure for isolating RNA from bud, internodal shoot, flower, and fruit tissues of apple has been developed. This protocol does not involve the use of phenol, lyophilization, or ultracentrifugation. In addition, this protocol overcomes problems of both RNA degradation and low yield attributed to oxidation by polyphenolic compounds and coprecipitation with polysaccharides, both abundant components in apple fruit and flower tissues. Isolated RNA is of high quality and is undegraded as assessed by spectrophotometric readings and electrophoresis in denaturing agarose gels. RNA quality is further assessed following its use in reverse transcription and cDNA library construction, and it can be used for a number of downstream analyses, including Northern blot hybridization and reverse transcription-polymerase chain reaction (RT-PCR). With this modified protocol, 25–900 μg of total RNA is routinely obtained from 1 g of fresh material. This method is of low cost and easy to perform.
[本文引用: 1]
[本文引用: 1]
DOI:10.1006/meth.2001.1262URLPMID:11846609 [本文引用: 2]

The two most commonly used methods to analyze data from real-time, quantitative PCR experiments are absolute quantification and relative quantification. Absolute quantification determines the input copy number, usually by relating the PCR signal to a standard curve. Relative quantification relates the PCR signal of the target transcript in a treatment group to that of another sample such as an untreated control. The 2(-Delta Delta C(T)) method is a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments. The purpose of this report is to present the derivation, assumptions, and applications of the 2(-Delta Delta C(T)) method. In addition, we present the derivation and applications of two variations of the 2(-Delta Delta C(T)) method that may be useful in the analysis of real-time, quantitative PCR data.
DOI:10.3864/j.issn.0578-1752.2017.19.015URL [本文引用: 2]

【Objective】 Effect of different tree shapes on growth, yield and fruit quality in ‘Fuji’ apple trees was studied in-depth to provide the theoretical basis and mentoring programs for the selection of tree shapes on dwarfing interstock.【Method】 The ‘Fuji’ apple with SH6 dwarf interstock planted in spring 2010 was used to investigate the effect of three different tree shapes (Spindle 2 m×4.5 m, V-trellis 1.5 m×6 m, Hedgerow 3 m×3 m) on growth, yield and quality in ‘Fuji’ apple trees from 2011 to 2016.【Result】The tree growth, fruit yield and quality of different tree shapes were quite different. Since the first 3 years after planting, the total branches and coverage rate of different tree shapes showed significant differences, spindle maximum, followed by V-trellis, Hedgerow minimum. There was no significant difference on the proportion of different branches among different tree shapes. There was also no significant difference on leaf fresh weight, chlorophyll content and photosynthetic rate among them. All trees began to have production in the fourth year. The cumulative average yield per plant of Spindle was significantly higher than that of V-trellis and Hedgerow. Average fruit weight of Spindle was the biggest, with V-trellis smallest. Firmness of V-trellis was the highest, with Spindle lowest. There was no significant difference on the fruit figure index, soluble solids, titratable acidity and TSS/TA among different tree shapes. 【Conclusion】 According to the survey data of seven years, the ‘Fuji’ apple with SH6 dwarf interstock of Spindle shows large total branches amount, moderate growth, early bearing, high and stable yield, and good fruit quality compared with that of V-trellis and Hedgerow. Spindle is the suitable shape for apple dwarf stock cultivation model.
DOI:10.3864/j.issn.0578-1752.2017.19.015URL [本文引用: 2]

【Objective】 Effect of different tree shapes on growth, yield and fruit quality in ‘Fuji’ apple trees was studied in-depth to provide the theoretical basis and mentoring programs for the selection of tree shapes on dwarfing interstock.【Method】 The ‘Fuji’ apple with SH6 dwarf interstock planted in spring 2010 was used to investigate the effect of three different tree shapes (Spindle 2 m×4.5 m, V-trellis 1.5 m×6 m, Hedgerow 3 m×3 m) on growth, yield and quality in ‘Fuji’ apple trees from 2011 to 2016.【Result】The tree growth, fruit yield and quality of different tree shapes were quite different. Since the first 3 years after planting, the total branches and coverage rate of different tree shapes showed significant differences, spindle maximum, followed by V-trellis, Hedgerow minimum. There was no significant difference on the proportion of different branches among different tree shapes. There was also no significant difference on leaf fresh weight, chlorophyll content and photosynthetic rate among them. All trees began to have production in the fourth year. The cumulative average yield per plant of Spindle was significantly higher than that of V-trellis and Hedgerow. Average fruit weight of Spindle was the biggest, with V-trellis smallest. Firmness of V-trellis was the highest, with Spindle lowest. There was no significant difference on the fruit figure index, soluble solids, titratable acidity and TSS/TA among different tree shapes. 【Conclusion】 According to the survey data of seven years, the ‘Fuji’ apple with SH6 dwarf interstock of Spindle shows large total branches amount, moderate growth, early bearing, high and stable yield, and good fruit quality compared with that of V-trellis and Hedgerow. Spindle is the suitable shape for apple dwarf stock cultivation model.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.21273/JASHS.122.3.347URL [本文引用: 1]
DOI:10.1016/j.gene.2012.01.078URL [本文引用: 1]

Columnar apple trees (Malus x domestica) provide several economic advantages due to their specific growth habit. The columnar phenotype is the result of the dominant allele of the gene Co and is characterized by thick stems with short internodes and reduced lateral branching. Co is located on chromosome 10 and often appears in a heterozygous state (Co/co). The molecular explanation of columnar growth is not well established. Therefore, we studied the transcriptomes of columnar and standard type apple trees using 454 and Illumina next generation sequencing (NGS) technologies. We analyzed the transcriptomes of shoot apical meristems (SAMs) because we expect that these organs are involved in forming the columnar growth phenotype. The results of the comparative transcriptome analysis show significant differences in expression levels of hundreds of genes. Many of the differentially expressed genes are associated with membrane and cell wall growth or modification and can be brought in line with the columnar phenotype. Additionally, earlier findings on the hormonal state of shoots of columnar apples could be affirmed. Our study resulted in a large number of genes differentially expressed in columnar vs. standard type apple tree SAMs. Although we have not unraveled the nature of the Co gene, we could show that the modified expression of these genes, most likely due to the presence of Co, can determine the columnar phenotype. Furthermore, the usefulness of NGS for the analysis of the molecular basis of complex phenotypes is discussed. (C) 2012 Elsevier B.V.
DOI:10.1111/nph.12580URL [本文引用: 3]

Understanding the genetic mechanisms controlling columnar-type growth in the apple mutant Wijcik will provide insights on how tree architecture and growth are regulated in fruit trees.
In apple, columnar-type growth is controlled by a single major gene at the Columnar (Co) locus. By comparing the genomic sequence of the Co region of Wijcik with its wild-type McIntosh, a novel non-coding DNA element of 1956 bp specific to Pyreae was found to be inserted in an intergenic region of Wijcik.
Expression analysis of selected genes located in the vicinity of the insertion revealed the upregulation of the MdCo31 gene encoding a putative 2OG-Fe(II) oxygenase in axillary buds of Wijcik.
Constitutive expression of MdCo31 in Arabidopsis thaliana resulted in compact plants with shortened floral internodes, a phenotype reminiscent of the one observed in columnar apple trees. We conclude that MdCo31 is a strong candidate gene for the control of columnar growth in Wijcik.
DOI:10.1111/j.1744-7909.2006.00311.xURL [本文引用: 1]

The MYB transcription factor genes play important roles in many developmental processes and various defense responses of plants. The shikimate pathway is a major biosynthetic pathway for the production of three aromatic amino acids and other aromatic compounds that are involved in multiple responses of plants, including protection against UV and defense. Herein, we describe the characterization of the R2R3-MYB gene AtMYB15 as an activator of the shikimate pathway in Arabidopsis. The AtMYB15 protein is nuclear localized and a transcriptional activation domain is found in its C-terminal portion. Northern blots showed that AtMYB15 is an early wounding-inducible gene. Resutls of microarray analysis, confirmed using quantitative real-time polymerase chain reaction, showed that overexpression of AtMYB15 in transgenic plants resulted in elevated expression of almost all the genes involved in the shikimate pathway. Bioinformatics analysis showed that one or more AtMYB15-binding AC elements were detected in the promoters of these upregulated genes. Furthermore, these genes in the shikimate pathway were also found to be induced by wounding. These data suggest an important role of AtMYB15 as a possible direct regulator of the Arabidopsis shikimate pathway in response to wounding.(*Author for correspondence. Tel: +86 (0)10 6275 1847; Fax: +86 (0)10 6275 3339; E-mail: qulj@pku.edu.cn)
[本文引用: 1]
