 ,1
,1Identification and Characterization of the Expansin Gene Family in Upland Cotton (Gossypium hirsutum)
ZHANG QiYan1, LEI ZhongPing2, SONG Yin1, HAI JiangBo1, HE DaoHua ,1
,1通讯作者:
收稿日期:2019-04-29接受日期:2019-06-18网络出版日期:2019-11-08
| 基金资助: |
Received:2019-04-29Accepted:2019-06-18Online:2019-11-08
作者简介 About authors
张奇艳,E-mail:
雷忠萍,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (3863KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张奇艳, 雷忠萍, 宋银, 海江波, 贺道华. 陆地棉扩展蛋白基因的鉴定与特征分析[J]. 中国农业科学, 2019, 52(21): 3713-3732 doi:10.3864/j.issn.0578-1752.2019.21.001
ZHANG QiYan, LEI ZhongPing, SONG Yin, HAI JiangBo, HE DaoHua.
0 引言
【研究意义】棉花是一种重要的纤维作物。棉纤维是由胚珠外表皮的单个细胞发育而成,是高度伸长、增厚、没有分支的单细胞表皮毛,与拟南芥表皮毛的发生机制相似。细胞壁对棉纤维的品质起决定作用,是改良棉花品质的重要途径之一。棉纤维的伸长与细胞壁松弛联系密切,扩展蛋白能够打断纤维素微丝间的氢键,从而使细胞壁延展疏松[1]。研究表明,扩展蛋白在植物的生长发育与逆境胁迫应答等方面具有重要作用,对棉花的遗传改良具有潜在价值。【前人研究进展】1992年,MC QUEEN-MASON等[2]从黄瓜下胚轴细胞壁中分离纯化得到2个分子量为29和30 kD能诱导细胞壁恢复酸生长的蛋白。这种蛋白粗提液可在体外诱导热失活细胞壁的重新伸展,但伸展过程受到pH和金属离子等因素的影响。研究发现,这种现象是由细胞壁上的一类细胞壁疏松蛋白介导的,因而将其命名为扩展蛋白(Expansin)[3]。扩展蛋白能够打断细胞壁多糖之间的非共价键,从而使细胞壁聚合物发生膨压诱导的蠕动,对细胞壁起到疏松作用,使细胞壁的柔韧性增加而松弛[4,5]。随后,人们相继从水稻[6]、玉米[7]、拟南芥[6]、大豆[8]、番茄[9]、葡萄[10]等100多个物种的细胞壁中也鉴定出扩展蛋白,推测扩展蛋白广泛存在于双子叶植物和单子叶植物中。研究表明,扩展蛋白能影响植物的生长发育,诸如促进组织细胞的生长[11]、种子发育[12]、根毛起始和根系生长[13,14]、叶和茎的发育[15,16,17,18,19]、花粉管伸长[20,21]、促进叶柄脱落[16]、果实成熟[22,23]和降低番茄的裂果率[24]等。另外,在植物抗逆性方面,扩展蛋白也发挥着重要作用,如抗旱性[25,26]、耐盐性[27,28]、抗高温[29]和抗病性[30]等。棉花为人类提供了一种优良的天然纤维,其发育过程包含5个彼此重叠的时期,依次为起始期(initiation)、伸长期(elongation)、初生壁向次生壁发育的转换期(transition)、次生壁合成期(secondary wall biosynthesis)和脱水成熟期(maturation)[31]。研究表明,扩展蛋白在转录水平上与棉纤维的伸长联系紧密[32,33],许多扩展蛋白基因在纤维细胞的伸长中高度表达[34,35]。XU等[36]发现在棉花中过表达GhEXPA1可以显著增加棉铃数量,单株棉花纤维产量可增加40%,对纤维品质和棉株生长没有不利的影响。LI等[37]发现GbEXPATR(来自At亚基因组,是Dt亚基因组中GbEXPA2的部分同源基因)在次生壁合成和代谢中发挥重要作用。BAJWA等[38]发现,转GhEXPA8棉株的纤维长度和马克隆值均有显著提高。以上研究结果均表明扩展蛋白在棉花中具有重要的作用。【本研究切入点】2012年,WANG等[39]和PATERSON等[40]分别完成了棉花D基因组雷蒙德氏棉(G. raimondii)的全基因组测序工作;2014年,LI等[41]完成了棉花A基因组亚洲棉(G. arboretum)石系亚1号的全基因组测序及组装工作,绘制出的亚洲棉基因组约为1 694 Mb。在上述成果的基础上,2015年,LI等[42]和ZHANG等[43]分别独立完成了异源四倍体陆地棉(G. hirsutum)TM-1的全基因组测序及组装。随后,CHEN(https://www.cottongen.org/species/Gossypium_ hirsutum/jgi-AD1_genome_v1.1)、WANG等[44]和HU等[45]分别对棉属的基因组序列图谱进行了更新和完善,为在陆地棉全基因组范围内系统进行扩展蛋白基因家族的鉴定、进化和功能分析提供了条件。【拟解决的关键问题】本研究基于陆地棉及相关二倍体棉种的基因组测序的序列信息,对棉花扩展蛋白基因家族进行筛选鉴定,并从DNA水平(基因外显子-内含子结构、密码子偏好性、同线性)、RNA水平(基于RNA-seq和qRT-PCR的基因表达模式、部分同源基因间表达丰度差异)和氨基酸水平(信号肽、模体结构、理化性质及亚细胞定位)对其进化及功能进行分析,为进一步研究扩展蛋白在棉花(纤维)生长发育等生物学过程中的功能奠定基础。1 材料与方法
1.1 棉花中扩展蛋白家族成员的鉴定
陆地棉基因组数据下载于1.2 棉属扩展蛋白成员系统发育树的构建及基因命名
利用软件ClustalW对所鉴定的棉花、拟南芥和水稻的扩展蛋白氨基酸序列进行多序列比对分析;利用软件MEGA v5.0,采用Neighbor-Joining法构建扩展蛋白氨基酸序列的系统发育树。参照拟南芥及水稻的扩展蛋白的亚家族分类,将棉花的扩展蛋白家族成员归类到EXPA、EXPB、EXLA和EXLB 4个亚家族中,根据陆地棉、亚洲棉各扩展蛋白基因与雷蒙德氏棉Expansin基因之间的同线性关系、系统发育树、及其在染色体上的位置顺序等信息对各基因进行命名。1.3 同线性及基因家族成员间进化关系分析
通过BLAST比对,将所获得的各扩展蛋白基因定位到相应的染色体上。利用MCScanX软件分析各扩展蛋白基因之间的同线性关系。利用Circos软件图形化展示扩展蛋白基因在染色体上的物理位置及基因间的同线性关系。1.4 理化性质、信号肽、模体结构及亚细胞定位分析
利用在线软件Prot Param(1.5 基因结构和密码子偏好性分析
通过对各扩展蛋白基因的CDS序列与DNA序列进行BLAST比对,利用1.6 扩展蛋白基因的表达特征分析
1.6.1 转录组(RNA-seq)数据分析 为了获得扩展蛋白基因在不同时空条件的表达特征,参考HE等[47]的方法,从NCBI SRA数据库下载2组深度测序(RNA-seq)数据:SRP059947和SRP017168。利用Tophat将RNA-seq的短读序列mapping到陆地棉基因组上,利用cufflinks函数估算扩展蛋白基因在不同时空上的表达水平(fragments per kilobase of exon model per million reads mapped,FPKM值),通过cuffdiff函数筛出在不同时空之间存在表达差异的基因。根据RNA-seq数据的分析结果,挑选13个扩展蛋白基因进行实时荧光定量PCR分析,以便进一步验证其表达特征。1.6.2 实时荧光定量PCR(quantitative real-time PCR,qRT-PCR) 于陆地棉材料N874系的花铃期,取-3、-1、0、3、5、10和15 DPA(days post-anthesis)的棉铃分离出胚珠,取5、10、15和20 DPA的棉铃分离出纤维。利用总RNA提取试剂盒(天根,北京)从棉花胚珠和纤维中提取RNA,用NanoDrop 2000和1%的琼脂糖凝胶电泳检测RNA的含量和质量。以RNA为模板反转录合成cDNA。qRT-PCR引物下载于西南大学(
Table 1
表1
表1实时荧光定量PCR引物
Table 1
| 基因名称Gene name | 正向引物Forward primers (5′-3′) | 反向引物Reverse primers (5′-3′) |
|---|---|---|
| GhEXLA1A | TCACTGACATTGCAAAAGAAGG | GAACACCATTGATTGTGTAGCA |
| GhEXLA3A | GTGATTACAGCCATTGGTGATG | CCAAAACAAAAGCTCTCAAAGACTC |
| GhEXLA2D | TAACATGCAAAGAAAGTGAGGC | AATCCATGGTTTTCCGGAAATG |
| GhEXLA3D | CTTTCGGGTCACTCAAAAATGT | AATGCTTCTCAACATCCCAAAC |
| GhEXPA06A | CTTTCGGGTCACTCAAAAATGT | AATGCTTCTCAACATCCCAAAC |
| GhEXPA16A | GAATCGTCCCCATTTCATTCAG | CACCACCAACATTTGTAGTGAG |
| GhEXPA25A | CACCATCATTGTAACTGCTACG | ATTTTGAACTTAACACCACCGG |
| GhEXPA06D | GCACTGCTTTGTTTAACAATGG | ATTAGGGAGAGCATTGTTTGGA |
| GhEXPA17D | TTGTTGGGTTTTCTAGCAATGG | CTGCAGTATTTGTTCCATAGCC |
| GhEXPA19D | GGAAACCTGTACAGTCAAGGTA | GTTATGGTTCGACTAATGCACC |
| GhEXPA20D | CCTAACTTAGCATTGTCCAACG | TTTCTTCATGCAGGGTACTCTT |
| GhEXPA23D | CGGCCACTCATACTTTAACTTG | CTTCGTATGTTTGTCCGAACTG |
| GhEXPA24D | GAACGAAGAAAACACCCATCTT | CTCTTCAGCCAAAGCATCTTAC |
| UBQ7 | GAAGGCATTCCACCTGACCAAC | CTTGACCTTCTTCTTCTTGTGCTTG |
新窗口打开|下载CSV
2 结果
2.1 扩展蛋白家族成员的鉴定
根据拟南芥和水稻扩展蛋白的基因序列(作为query),通过BLAST比对搜索过程,初步鉴定出棉属准扩展蛋白家族成员。然后通过HMMER检测pfam01357和pfam03330 2个结构域存在与否,最终确认为扩展蛋白。在陆地棉中,共鉴定出72个扩展蛋白家族成员(表2);同时,在二倍体物种亚洲棉中,鉴定出39个扩展蛋白基因,与在雷蒙德氏棉中鉴定出的扩展蛋白数量一致[46]。其中,同为双子叶植物的拟南芥、杨树和大豆中,拟南芥和杨树的扩展蛋白基因数量与二倍体棉相近,大豆的扩展蛋白数量则与四倍体棉相近(表3)。基于本研究所鉴定的棉花、拟南芥和水稻扩展蛋白的系统发育树分析,将陆地棉和亚洲棉扩展蛋白划分为4个亚家族(EXPA、EXPB、EXLA和EXLB)。陆地棉的72个扩展蛋白成员中,46个属于EXPA亚家族,8个属于EXPB亚家族,6个属于EXLA亚家族,12个属于EXLB亚家族。亚洲棉39个扩展蛋白成员的亚家族分布与雷蒙德氏棉类似,EXPA、EXPB、EXLA和EXLB 4个亚家族的成员数分别为26、4、3和6个。数据显示,四倍体陆地棉中扩展蛋白成员的数量几乎是二倍体棉种(亚洲棉与雷蒙德氏棉)的2倍(四倍体陆地棉中EXPA亚家族的成员数量(46)比二倍体的数量(26)的2倍少),说明二倍体经过远缘杂交和基因组加倍形成四倍体后,扩展蛋白家族内的成员得到继承,仅亚家族EXPA中的个别成员发生了丢失。在各个亚家族内,根据各基因在染色体上的位置顺序,对陆地棉扩展蛋白基因进行了命名(表2)。Table 2
表2
表2陆地棉扩展蛋白基因家族成员的鉴定
Table 2
| 基因 Gene | 序列号 Phytozome ID | 染色体位置 Location | 基因长度Length of gene (bp) | 外显子个数Number of exon | 氨基酸个数 Number of amino acids | 信号 肽长度Signal peptide (aa) | 分子量Molecular weight (kD) | 理论等电点Theoretical pI | 蛋白不稳定指数Instability index | 脂溶指数Aliphatic index | 总平均 疏水性GRAVY |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GhEXPA03A | Gh_A01G0477 | GhA01: 7603263..7604881(+) | 1619 | 3 | 256 | 27 | 28.40 | 9.90 | 43.09 | 67.85 | -0.118 |
| GhEXPA06A | Gh_A03G0885 | GhA03: 56794871..56796095(-) | 1225 | 3 | 248 | 20 | 26.59 | 9.30 | 32.13 | 62.94 | -0.140 |
| GhEXPA16A | Gh_A05G2385 | GhA05: 29485491..29486507(-) | 1017 | 3 | 253 | 25 | 26.94 | 7.53 | 30.72 | 59.8 | -0.166 |
| GhEXPA22-1A | Gh_A05G3210 | GhA05: 83889906..83890791(+) | 886 | 2 | 262 | 22 | 29.18 | 9.25 | 20.09 | 68.47 | -0.167 |
| GhEXPA22-2A | Gh_A05G3211 | GhA05: 83905450..83906383(+) | 934 | 2 | 278 | 23 | 30.96 | 9.24 | 18.98 | 74.64 | 0.008 |
| GhEXPA21A | Gh_A05G3493 | GhA05: 90545166..90546162(+) | 997 | 3 | 253 | 25 | 27.07 | 6.78 | 31.44 | 65.57 | -0.066 |
| GhEXPA14A | Gh_A06G0018 | GhA06: 82222..83190(+) | 969 | 3 | 255 | 23 | 27.38 | 9.15 | 26.88 | 67.76 | -0.015 |
| GhEXPA01A | Gh_A07G1208 | GhA07: 26997118..26998121(-) | 1004 | 3 | 258 | NO | 27.92 | 9.70 | 38.52 | 65.81 | -0.140 |
| GhEXPA02A | Gh_A07G1881 | GhA07: 74202971..74204340(-) | 1370 | 3 | 253 | 21 | 27.52 | 9.40 | 38.41 | 77.79 | -0.025 |
| GhEXPA05A | Gh_A08G1595 | GhA08: 94460350..94461323(+) | 974 | 3 | 267 | NO | 29.21 | 8.38 | 30.56 | 65.43 | 0.045 |
| GhEXPA07A | Gh_A09G0135 | GhA09: 3242057..3243766(+) | 1710 | 3 | 256 | 21 | 28.16 | 9.22 | 41.30 | 68.52 | -0.160 |
| GhEXPA08A | Gh_A09G1727 | GhA09: 70679591..70680590(-) | 1000 | 3 | 265 | 28 | 28.73 | 8.59 | 32.02 | 60.79 | -0.005 |
| GhEXPA20A | Gh_A10G1606 | GhA10: 86603436..86604481(-) | 1046 | 3 | 253 | 25 | 27.00 | 7.55 | 33.15 | 62.89 | -0.118 |
| GhEXPA19A | Gh_A10G2323 | scaffold2672_A10: 149124..150059(-) | 936 | 3 | 258 | 30 | 27.94 | 8.36 | 29.01 | 62.83 | -0.090 |
| GhEXPA09A | Gh_A11G0187 | GhA11: 1736060..1737333(+) | 1274 | 4 | 265 | NO | 29.26 | 9.42 | 29.16 | 66.26 | -0.040 |
| GhEXPA10A | Gh_A11G0964 | GhA11: 10482352..10483322(-) | 971 | 3 | 258 | NO | 28.42 | 9.43 | 33.85 | 66.98 | -0.229 |
| GhEXPA11A | Gh_A11G2155 | GhA11: 71713430..71714457(-) | 1028 | 3 | 247 | 20 | 26.39 | 9.32 | 38.00 | 69.55 | -0.027 |
| GhEXPA13A | Gh_A12G1619 | GhA12: 77299134..77300408(-) | 1275 | 3 | 248 | 20 | 26.62 | 9.19 | 31.75 | 69.6 | -0.030 |
| GhEXPA23A | Gh_A13G0050 | GhA13: 540164..541646(+) | 1483 | 3 | 254 | 26 | 27.28 | 8.41 | 33.34 | 63.39 | -0.134 |
| GhEXPA24A | Gh_A13G0672 | GhA13: 20543406..20544365(-) | 960 | 3 | 248 | 17 | 26.26 | 9.28 | 32.55 | 67.3 | -0.111 |
| GhEXPA25A | Gh_A13G0719 | GhA13: 23714615..23715449(+) | 835 | 2 | 254 | 27 | 28.23 | 8.33 | 27.99 | 75.24 | -0.061 |
| GhEXPA26A | Gh_A13G2059 | GhA13: 79891203..79892227(+) | 1025 | 3 | 249 | 23 | 26.93 | 8.36 | 29.07 | 68.59 | -0.064 |
| GhEXPA03D | Gh_D01G0492 | GhD01: 5972132..5973758(+) | 1627 | 3 | 252 | NO | 27.98 | 9.90 | 39.24 | 70.48 | -0.078 |
| GhEXPA06D | Gh_D02G1269 | GhD02: 41719506..41720746(-) | 1241 | 3 | 267 | NO | 28.95 | 9.78 | 35.24 | 68.31 | -0.104 |
| 基因 Gene | 序列号 Phytozome ID | 染色体位置 Location | 基因长度Length of gene (bp) | 外显子个数Number of exon | 氨基酸个数 Number of amino acids | 信号 肽长度Signal peptide (aa) | 分子量Molecular weight (kD) | 理论等电点Theoretical pI | 蛋白不稳定指数Instability index | 脂溶指数Aliphatic index | 总平均 疏水性GRAVY |
| GhEXPA22D | Gh_D04G0396 | GhD04: 6280383..6281268(-) | 886 | 2 | 262 | 23 | 29.21 | 9.18 | 20.03 | 68.47 | -0.143 |
| GhEXPA21D | Gh_D04G1924 | scaffold3943_D04: 15458..16477(-) | 1020 | 3 | 253 | 25 | 27.08 | 8.02 | 32.54 | 65.57 | -0.065 |
| GhEXPA15D | Gh_D05G2433 | GhD05: 24370173..24371133(+) | 961 | 3 | 255 | 23 | 27.45 | 8.88 | 29.40 | 67.02 | -0.077 |
| GhEXPA16D | Gh_D05G2650 | GhD05: 27568523..27569536(-) | 1014 | 3 | 252 | 24 | 26.92 | 7.53 | 30.80 | 60.79 | -0.130 |
| GhEXPA17D | Gh_D05G2934 | GhD05: 34837918..34838746(-) | 829 | 2 | 253 | 29 | 27.21 | 9.04 | 31.30 | 64.78 | -0.122 |
| GhEXPA01D | Gh_D07G1309 | GhD07: 20886446..20887308(-) | 863 | 3 | 207 | NO | 22.54 | 9.90 | 35.72 | 67.83 | -0.247 |
| GhEXPA02D | Gh_D07G2095 | GhD07: 51074809..51076181(-) | 1373 | 3 | 253 | 21 | 27.49 | 9.30 | 38.37 | 76.25 | -0.044 |
| GhEXPA04D | Gh_D08G1362 | GhD08: 44540800..44541775(-) | 976 | 254 | 23 | 28.01 | 9.31 | 35.94 | 67.68 | -0.187 | |
| GhEXPA05D | Gh_D08G1905 | GhD08: 56979061..56980034(+) | 974 | 3 | 267 | NO | 29.32 | 8.08 | 29.33 | 66.14 | 0.037 |
| GhEXPA07D | Gh_D09G0128 | GhD09: 3266295..3268335(+) | 2041 | 4 | 303 | NO | 33.80 | 9.57 | 43.94 | 72.38 | -0.164 |
| GhEXPA08D | Gh_D09G1840 | GhD09: 45961349..45962346(-) | 998 | 3 | 265 | 28 | 28.74 | 8.38 | 34.27 | 59.32 | -0.006 |
| GhEXPA19D | Gh_D10G1145 | GhD10: 18608318..18609246(+) | 929 | 3 | 258 | 30 | 27.95 | 8.05 | 29.69 | 62.44 | -0.106 |
| GhEXPA20D | Gh_D10G1861 | GhD10: 52037144..52038189(-) | 1046 | 3 | 253 | 25 | 27.07 | 7.55 | 33.33 | 65.97 | -0.085 |
| GhEXPA09D | Gh_D11G0198 | GhD11: 1728429..1729700(+) | 1272 | 4 | 265 | NO | 29.17 | 9.41 | 28.62 | 64.83 | -0.051 |
| GhEXPA10D | Gh_D11G1115 | GhD11: 10243558..10244528(-) | 971 | 3 | 258 | NO | 28.42 | 9.43 | 33.85 | 66.98 | -0.229 |
| GhEXPA11D | Gh_D11G2459 | GhD11: 49988580..49989605(-) | 1026 | 3 | 247 | 20 | 26.35 | 9.30 | 36.98 | 69.15 | -0.039 |
| GhEXPA13D | Gh_D12G1759 | GhD12: 49966434..49967705(-) | 1272 | 3 | 248 | 20 | 26.77 | 9.19 | 34.40 | 69.19 | -0.016 |
| GhEXPA23D | Gh_D13G0060 | GhD13: 622156..623599(+) | 1444 | 3 | 254 | 26 | 27.36 | 8.61 | 29.33 | 58.78 | -0.198 |
| GhEXPA24D | Gh_D13G0786 | GhD13: 13599569..13600499(-) | 931 | 3 | 248 | 17 | 26.33 | 9.30 | 31.82 | 65.69 | -0.111 |
| GhEXPA25D | Gh_D13G0843 | GhD13: 15386343..15387204(+) | 862 | 2 | 254 | 27 | 28.19 | 8.53 | 28.55 | 73.7 | -0.042 |
| GhEXPA26D | Gh_D13G2460 | GhD13: 60483670..60484694(+) | 1025 | 3 | 249 | 23 | 26.86 | 8.36 | 29.68 | 68.59 | -0.067 |
| GhEXPA17A | Gh_Sca005793G01 | scaffold5793: 5707..6535(+) | 829 | 2 | 253 | 29 | 27.17 | 8.88 | 29.73 | 68.62 | -0.076 |
| GhEXPB2A | Gh_A03G2139 | scaffold729_A03: 85394..86864(+) | 1471 | 4 | 267 | 26 | 29.31 | 8.63 | 35.29 | 75.96 | -0.247 |
| GhEXPB1A | Gh_A08G1231 | GhA08: 82800735..82801892(-) | 1158 | 4 | 273 | NO | 28.60 | 5.19 | 37.99 | 70.07 | 0.000 |
| GhEXPB3A | Gh_A12G0861 | GhA12: 57318819..57321212(-) | 2394 | 5 | 256 | NO | 26.80 | 4.88 | 35.29 | 74.69 | -0.083 |
| 基因 Gene | 序列号 Phytozome ID | 染色体位置 Location | 基因长度Length of gene (bp) | 外显子个数Number of exon | 氨基酸个数 Number of amino acids | 信号 肽长度Signal peptide (aa) | 分子量Molecular weight (kD) | 理论等电点Theoretical pI | 蛋白不稳定指数Instability index | 脂溶指数Aliphatic index | 总平均 疏水性GRAVY |
| GhEXPB4A | Gh_A12G1270 | GhA12: 67653347..67655408(-) | 2062 | 4 | 267 | 27 | 29.12 | 8.71 | 31.28 | 77.75 | -0.137 |
| GhEXPB2D | Gh_D02G1839 | GhD02: 61451593..61453105(+) | 1513 | 4 | 275 | 34 | 30.25 | 8.98 | 34.08 | 76.22 | -0.242 |
| GhEXPB1D | Gh_D08G1516 | GhD08: 48665574..48666719(-) | 1146 | 4 | 273 | NO | 28.61 | 5.03 | 33.99 | 69.71 | -0.015 |
| GhEXPB3D | Gh_D12G0941 | GhD12: 34578510..34580023(-) | 1514 | 3 | 180 | NO | 18.92 | 5.03 | 30.62 | 83.44 | 0.032 |
| GhEXPB4D | Gh_D12G1393 | GhD12: 43009111..43011170(-) | 2060 | 4 | 267 | 27 | 29.09 | 8.71 | 31.74 | 76.29 | -0.155 |
| GhEXLA1A | Gh_A01G1439 | GhA01: 89104034..89105296(+) | 1263 | 5 | 259 | 17 | 28.02 | 8.26 | 34.36 | 83.28 | 0.000 |
| GhEXLA2A | Gh_A03G2186 | scaffold739_A03: 13564..15228(-) | 1665 | 4 | 260 | 20 | 28.26 | 8.24 | 35.24 | 82.92 | -0.029 |
| GhEXLA3A | Gh_A04G1065 | GhA04: 60713119..60715274(+) | 2156 | 5 | 259 | 18 | 28.10 | 8.59 | 29.34 | 81.39 | -0.019 |
| GhEXLA1D | Gh_D01G1679 | GhD01: 52783404..52784664(+) | 1261 | 5 | 259 | 17 | 28.01 | 8.26 | 33.65 | 81.78 | 0.001 |
| GhEXLA2D | Gh_D02G2280 | GhD02: 66566105..66567707(+) | 1603 | 4 | 260 | 20 | 28.21 | 8.24 | 33.56 | 81.42 | -0.050 |
| GhEXLA3D | Gh_D04G1670 | GhD04: 48878933..48881117(+) | 2185 | 5 | 259 | 18 | 28.13 | 8.59 | 29.82 | 80.27 | -0.027 |
| GhEXLB1A | Gh_A03G0046 | GhA03: 643878..645995(+) | 2118 | 4 | 250 | 24 | 27.75 | 6.09 | 28.76 | 74.12 | -0.153 |
| GhEXLB2A | Gh_A08G1444 | GhA08: 90327706..90329016(-) | 1311 | 5 | 256 | 24 | 27.85 | 4.81 | 31.94 | 82.62 | -0.099 |
| GhEXLB3A | Gh_A08G1477 | GhA08: 90994259..90995358(+) | 1100 | 4 | 249 | 25 | 27.51 | 6.40 | 29.96 | 77.87 | -0.139 |
| GhEXLB4A | Gh_A08G1478 | GhA08: 91000911..91001970(+) | 1060 | 4 | 248 | NO | 27.67 | 9.00 | 23.00 | 80.52 | -0.158 |
| GhEXLB6A | Gh_A12G1923 | GhA12: 81945320..81946466(-) | 1147 | 5 | 249 | 21 | 27.08 | 4.65 | 35.69 | 84.98 | -0.109 |
| GhEXLB1D | Gh_D03G1609 | GhD03: 46020990..46023128(-) | 2139 | 4 | 250 | 24 | 27.69 | 5.64 | 26.36 | 71.76 | -0.174 |
| GhEXLB2D | Gh_D08G1739 | GhD08: 53651905..53653218(-) | 1314 | 5 | 256 | 24 | 27.90 | 4.66 | 32.35 | 82.62 | -0.099 |
| GhEXLB3D | Gh_D08G1774 | GhD08: 54163295..54164384(+) | 1090 | 4 | 249 | 25 | 27.36 | 6.06 | 31.21 | 79.08 | -0.113 |
| GhEXLB4-1D | Gh_D08G1775 | GhD08: 54170354..54171413(+) | 1060 | 4 | 248 | NO | 27.67 | 9.00 | 23.00 | 80.52 | -0.158 |
| GhEXLB4-2D | Gh_D08G1776 | GhD08: 54175086..54176147(+) | 1062 | 4 | 248 | 25 | 27.55 | 8.96 | 25.58 | 80.52 | -0.147 |
| GhEXLB5D | Gh_D08G1777 | GhD08: 54180358..54181440(+) | 1083 | 4 | 250 | 24 | 27.57 | 6.12 | 28.08 | 77.96 | -0.104 |
| GhEXLB6D | Gh_D12G2103 | GhD12: 54022894..54024050(-) | 1157 | 5 | 249 | 21 | 27.12 | 4.65 | 36.35 | 85.38 | -0.098 |
新窗口打开|下载CSV
Table 3
表3
表3不同植物扩展蛋白及其各亚家族内基因数目
Table 3
| 物种Species | EXPA | EXPB | EXLA | EXLB | 总计Total | 参考文献Reference |
|---|---|---|---|---|---|---|
| 陆地棉G. hirsutum | 46 | 8 | 6 | 12 | 72 | 本研究This study |
| 亚洲棉G. arboreum | 26 | 4 | 3 | 6 | 39 | 本研究This study |
| 雷蒙德氏棉G. raimondii | 26 | 4 | 3 | 6 | 39 | [46] |
| 黄瓜Cucumis sativus | 21 | 3 | 9 | 2 | 35 | [48] |
| 拟南芥Arabidopsis thaliana | 26 | 6 | 3 | 1 | 36 | [6] |
| 水稻Oryza sativa | 34 | 19 | 4 | 1 | 58 | [6] |
| 玉米Zea mays | 36 | 48 | 4 | 0 | 88 | [7] |
| 大白菜B. rapa (Pekinensis) | 39 | 9 | 2 | 3 | 53 | [50] |
| 杨树Populus trichocarpa | 27 | 3 | 2 | 4 | 36 | [51] |
| 葡萄Vitis vinifera | 20 | 4 | 1 | 4 | 29 | [10] |
| 大豆Glycine max | 49 | 9 | 2 | 5 | 75 | [8] |
| 苹果Malus×Domestica (Borkh) | 34 | 1 | 2 | 4 | 41 | [52] |
| 菜豆Phaseolus vulgaris | 25 | 6 | 0 | 5 | 36 | [53] |
| 苜蓿Medicago truncatula | 16 | 1 | 0 | 1 | 18 | [53] |
| 小粒碗藓Physcomitrella patens | 27 | 7 | 0 | 0 | 34 | [54] |
| 共同祖先Last common ancestor | 10—12 | 2 | 1 | 2 | 15—17 | [6] |
新窗口打开|下载CSV
鉴定结果显示,扩展蛋白在陆地棉染色体上呈现不均匀分布,在大多染色体上有2—4个扩展蛋白基因。具有部分同源关系(homeologous)的GhA08和GhD08 2个染色体具有较多的扩展蛋白基因,分别为5和8个。染色体GhA02和GhD06上没有扩展蛋白基因。有的基因家族成员在染色体上呈现成簇(cluster)存在的现象,尤其是同一亚家族的成员,如GhD08染色体上的GhEXLB3D/GhEXLB4-1D/ GhEXLB4-2D/GhEXLB5D形成串联重复(tandem duplication)。GhEXPA22-1A/GhEXPA22-2A、GhEXLB3A/ GhEXLB4A分别形成了2个小的串联重复。
2.2 系统发育树的构建与同线性分析
陆地棉、亚洲棉和雷蒙德氏棉扩展蛋白的系统发育树显示,亚家族EXLA与EXLB的遗传距离较近,EXPA与其余3个亚家族的相似性较小,表明了各亚家族间的亲缘(起源)关系及进化关系(图1)。亚家族内的各成员聚集在同一分支中(聚集成群),拥有各自的进化枝。大部分的末端分支由来源于3个物种的4个(亚)基因组的4个基因组成,如:EXPA亚家族的Cotton_A_28454/Gh_A03G0885/ Gh_D02G1269/Gorai.005G142200,EXLB亚家族的Cotton_A_13148/Gh_A08G1444/Gh_D08G1379/Gorai.004G188800等。4个基因分别来自A基因组、At亚基因组、Dt亚基因组和D基因组,表明扩展蛋白的氨基酸序列在物种间/(亚)基因组间具有平行的相似性,四倍体陆地棉的2个亚基因组(At、Dt)与对应的二倍体基因组(A、D)有较近的亲缘关系。图1
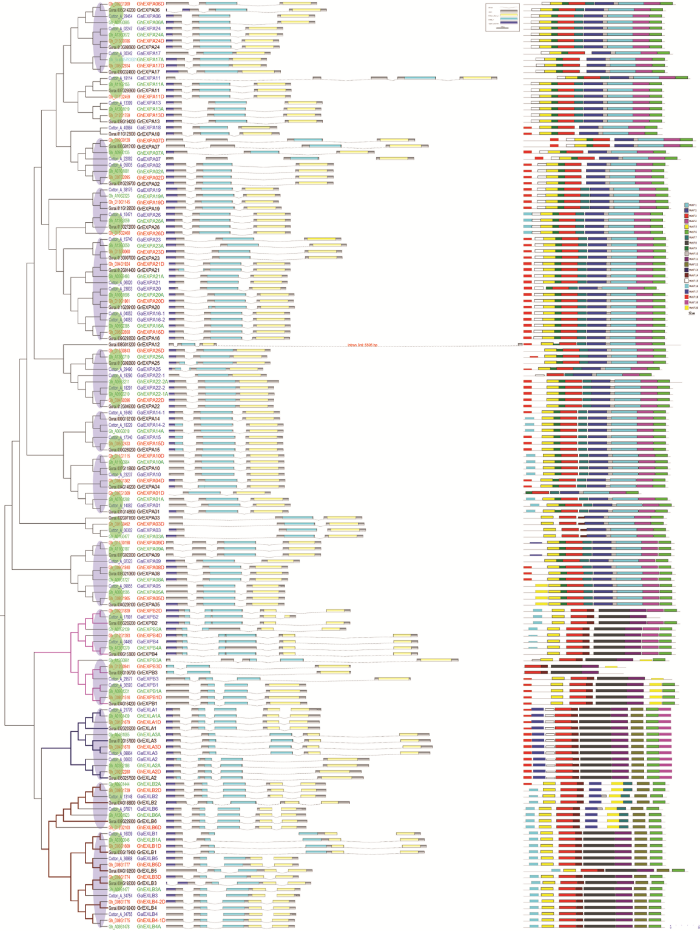 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1棉属扩展蛋白的系统发育树、外显子-内含子结构和氨基酸模体结构
Fig. 1Phylogenetic tree (left), exon-intron gene structures of the Expansin genes (middle), and schematic diagram of conserved motif (right) for the Expansin protein in Gossypium
利用软件MCScanX对棉属扩展蛋白在(亚)基因组间的同线性进行分析,结果显示,系统发育树中来源于A、D、At、Dt基因组/亚基因组的4个末端分支基因多具有同线性关系(图1系统发育树中绿色椭圆表示具有同线性关系),相互之间属于垂直同源基因(orthologous),陆地棉的2个亚基因组At与Dt在二倍体物种中也分别存在同线性同源区段。深入分析进一步表明,各基因组内部扩展蛋白之间存在一定的平行同源关系(paralogous),如图1系统发育树中紫色椭圆所示,显示了各基因组内部扩展蛋白基因的扩张(数量增加)历程,即大规模的染色体片段重复(segmental duplication)导致了基因家族的扩张,由共同祖先基因组中的15—17个[5]扩张到陆地棉中的72个。
四倍体陆地棉的2个亚基因组At与Dt间的同线性分析(图2)显示,除个别基因(GhEXPA19D)外,大多数扩展蛋白基因均位于同线性染色体区段上,说明AA与DD杂交加倍进化形成AtAtDtDt的陆地棉后,2个亚基因组At与Dt仍保持较高的相似性,并且遗传了其二倍体祖先基因组的染色体倍增的特点。值得注意的是,这种继承并不是完全的,扩展蛋白基因在染色体结构上发生了微小变化。
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2陆地棉扩展蛋白基因的染色体位置及亚基因组间的同线性关系
Fig. 2Chromosome location and collinearity between subgenome of G. hirsutum Expansin genes
2.3 理化性质、信号肽、模体结构及亚细胞定位分析
陆地棉72个扩展蛋白的氨基酸序列长度大多在247—278(除180(GhEXPB3D)、207(GhEXPA01D)和303(GhEXPA07D)外),分子量大多在26.39—30.96 kD(除18.92(GhEXPB3D)、22.54(GhEXPA01D)和33.80(GhEXPA07D)外),平均值为27.76 kD。等电点在4.65—9.90,平均值为8.18,75%的扩展蛋白的等电点大于8.0,即大多数陆地棉扩展蛋白偏碱性。除个别成员外,扩展蛋白不稳定指数在18.98—43.09,多数蛋白稳定性较好。脂溶指数在58.78—85.38,平均值达到71.39,其中有14个扩展蛋白的脂溶指数超过80,属于嗜热型蛋白。脂溶指数较高使得扩展蛋白可以较好地适应各种环境。总平均疏水性(GRAVY)在-0.247—0.045,均属于亲疏性相当的两性蛋白(大于0表示疏水性,小于0表示亲水性,介于±0.5之间为两性蛋白)[49](表3)。利用软件SignalP分析扩展蛋白信号肽,结果显示72个扩展蛋白中55个含有长度不等(17—34 aa)的信号肽。亚家族EXPB的信号肽长度均值为28.5 aa;亚家族EXLA的信号肽长度均值为18.3 aa,显著地(P< 0.01)低于其他3个亚家族。利用软件MEME对棉属3个物种的扩展蛋白氨基酸序列的保守基序(motif)进行分析(图3),获得20个(A—T)可靠的基序(e<1.20E-118)。其中EXPA亚家族的羧基端保守基序组合均为BJADF,而氨基端基序组合的种类则多达15种,该亚家族的氨基酸基序P、Q、R和T是不相容(兼容)的,其基序组合与系统发育树的呈现结果一致,均能说明基因的进化关系。EXPB亚家族的基序组合大多为PECNHKDF和RECNHKTF(除Gh_A12G0861例外)。EXLA亚家族的基序组合保守程度非常高,组合形式都为RQOCNHKLFS。EXLB亚家族的基序组合为PECNQTGLF和PECNHKLF(除Cotton_A_ 13148和Cotton_A_07871例外)。值得注意的是,EXPA亚家族含有不同于其他亚家族的模体I,EXLA亚家族含有不同于其他亚家族的模体S,该模体构成表明,各亚家族扩展蛋白在某些代谢途径中可能具有特殊功能。
图3
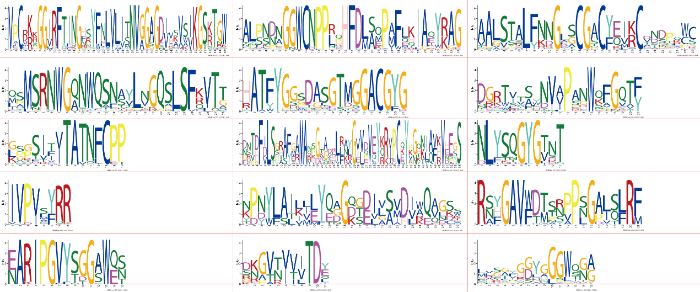 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3棉属扩展蛋白家族氨基酸序列模体(基序)
Fig. 3Sequence logo of conserved motifs in the family of Expansin proteins from Gossypium.
通过软件Euk-mPLoc 2.0进行亚细胞定位,结果表明,陆地棉72个扩展蛋白也全部定位于细胞外。
2.4 基因结构组成和密码子偏好性分析
为了在DNA水平进一步了解陆地棉扩展蛋白,对基因的“外显子-内含子”结构进行分析。陆地棉扩展蛋白基因的DNA长度在829—2 185 bp(Gh_A12G0861除外),其基因结构图(图1)表明,同一亚家族的扩展蛋白基因在“外显子-内含子”结构上非常相似。除个别基因(Gr08G013200和Cotton_A_ 18114)的内含子较长外,系统发育树中聚类在一起的基因的外显子数目、长度基本一致,内含子长度也很接近。表明扩展蛋白基因结构是相对较保守的,在进化上具有一定的保守性,且与氨基酸序列的多样性保持一致。密码子的偏好性分析既可以为基因表达选择适合的表达系统,又能为密码子的改造而改变基因表达量提供依据。生物的遗传密码中,除色氨酸和甲硫氨酸只有一个密码子外,其余氨基酸都有一个以上的简并密码子。简并密码子中,某一密码子相对同义密码子使用频率单值超过60%或者超过该组同义密码子平均占有频率的1.5倍的密码子即为高频密码子。对陆地棉72个扩展蛋白基因家族成员的18 455个密码子进行统计分析(表4),数据显示,UUG、CCU、GCU、AGA、AGG为高频密码子;UCG、CCG、ACG、GCG、CGG等第2、3位碱基为C和G的密码子为稀有密码子。4个亚家族之间的比较发现,AGG是4个亚家族共有的高频密码子,CGG是共有的低频密码子;EXPA和EXLA 2个亚家族具有较多相似的偏好性密码子。
Table 4
表4
表4陆地棉扩展蛋白基因密码子的相对使用频率
Table 4
| aa | Cod | Freq | aa | Cod | Freq | aa | Cod | Freq | aa | Cod | Freq | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | UUU | 0.95 | S | UCU | 1.35 | Y | UAU | 1.06 | C | UGU | 0.99 | |||
| UUC | 1.05 | UCC | 0.94 | UAC | 0.94 | UGC | 1.01 | |||||||
| L | UUA | 0.77 | UCA | 1.06 | * | UAA | 1.13 | * | UGA | 0.75 | ||||
| UUG | 1.54 | UCG | 0.49 | UAG | 1.13 | W | UGG | 1 | ||||||
| CUU | 1.22 | P | CCU | 1.69 | H | CAU | 1.26 | R | CGU | 0.54 | ||||
| CUC | 1.05 | CCC | 0.73 | CAC | 0.74 | CGC | 0.5 | |||||||
| CUA | 0.64 | CCA | 1.24 | Q | CAA | 1.2 | CGA | 0.63 | ||||||
| CUG | 0.78 | CCG | 0.34 | CAG | 0.8 | CGG | 0.29 | |||||||
| I | AUU | 1.3 | T | ACU | 1.28 | N | AAU | 1.02 | S | AGU | 0.86 | |||
| AUC | 1.05 | ACC | 1.14 | AAC | 0.98 | AGC | 1.31 | |||||||
| AUA | 0.64 | ACA | 1.33 | K | AAA | 1.11 | R | AGA | 1.8 | |||||
| M | AUG | 1 | ACG | 0.26 | AAG | 0.89 | AGG | 2.24 | ||||||
| V | GUU | 1.42 | A | GCU | 1.6 | D | GAU | 1.18 | G | GGU | 1.23 | |||
| GUC | 0.8 | GCC | 0.91 | GAC | 0.82 | GGC | 0.84 | |||||||
| GUA | 0.62 | GCA | 1.19 | E | GAA | 1.18 | GGA | 1.2 | ||||||
| GUG | 1.16 | GCG | 0.3 | GAG | 0.82 | GGG | 0.73 |
新窗口打开|下载CSV
2.5 基于转录组(RNA-seq)数据的Expansin基因表达模式
基因的表达与植物的生长发育密切相关,基于转录组(RNA-seq)数据的时空表达模式可以为探究扩展蛋白基因的功能提供线索。RNA-seq数据SRP059947中样品包括子叶、根、茎、叶(新叶、老叶)、苞叶、花萼、花冠、雌蕊雄蕊、胚珠(0、10、20、30和40 DPA)、纤维(10和20 DPA)共16个组织器官或发育阶段(图4)。RNA-seq分析表明,在不同组织、不同发育时期扩展蛋白基因的表达量存在较大差异,如:GhEXPA19A和GhEXPA19D在10和20 DPA纤维中的表达量很高,GhEXPA17D和GhEXPA17A在30和40 DPA胚珠中高量表达,GhEXPA19A和GhEXLA2A分别在10和20 DPA的胚珠中活跃表达,GhEXLB3A在20 DPA胚珠表达量特别高。GhEXPA19D在10和20 DPA的胚珠及10和20 DPA的纤维中均高量表达,表明GhEXPA19D对纤维的伸长(elongation (i.e., primary cell wall synthesis, 3-17 DPA))和纤维次生壁的合成阶段(secondary cell walls synthesis (17-23 DPA))有重要贡献。在根中,GhEXPA14A、GhEXLA3A、GhEXLA3D和GhEXLB5D表达量很高。在茎中,GhEXPA21D、GhEXLA1D和GhEXLB1A特高量表达;另外,GhEXPA14A和GhEXPA16D在茎中也有较高表达水平。在新叶中,GhEXPA06A、GhEXPA21D、GhEXPA06D及GhEXPA24D的表达量很高。在老叶中,GhEXPA24D、GhEXLA1D和GhEXLB2D高量表达。在子叶(cotyledon)中,GhEXPA21D、GhEXPA24D、GhEXLA1A及GhEXLA1D的表达量很高。值得注意的是,GhEXPA24D同时在子叶、新叶、老叶、苞叶(bracts)中均有很高的表达量,推测该基因可能对叶片组织的发育起到重要作用。在苞叶和花萼(calyxs)中,GhEXPA16A、GhEXPA16D有很高的表达量。在花冠(corollas)和雌雄蕊(gynoecium and androecium)中,GhEXPA16A、GhEXPA16D、GhEXPA11D、GhEXLA1A、GhEXLA2A、GhEXLA1D和GhEXLA2D具有较高的表达量。这些数据显示,不同的Expansin基因,在不同的时空条件下发挥功能,Expansin基因家族的成员在功能上既各司其职又相互补充。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4不同组织及不同时期扩展蛋白基因表达分析
Fig. 4Expression analysis of Expansin genes in diverse tissues and different stages of G. hirsutum
2.6 基于qRT-PCR的Expansin基因表达谱分析
基于转录组数据(RNA-seq)的表达模式分析初步提供了各基因在不同时空条件下的表达特征。对于至少在纤维和胚珠的发育的某个阶段表达丰度很高的基因,为了进一步明确其特异表达情况,本研究挑选13个扩展蛋白基因,通过qRT-PCR进行表达模式的验证(图5)。结果表明,qRT-PCR和RNA-seq均显示Expansin基因在各种组织/阶段中差异表达。GhEXLA3A和GhEXLA3D在纤维发育的伸长阶段表现出高表达,其中,在15 DPA表达量最高,GhEXLA2D有着同样的表达趋势。同时,GhEXLA1A的表达在整个纤维细胞起始和伸长过程中逐渐增加。GhEXPA16A和GhEXPA19D在纤维发育的10 DPA表达量较高。GhEXPA19D和GhEXLA2D在3 DPA的胚珠发育期间高度表达,GhEXLA3A在胚珠发育的10和15 DPA超高量表达,GhEXPA17D在胚珠发育的15 DPA高量表达,而GhEXPA19D在胚珠发育的3—10 DPA表达量都很高。而GhEXLA3D和GhEXPA25A主要在10—15 DPA胚珠和15—20 DPA纤维中表达。总的来说,除了少数基因外,RNA-seq数据和qRT-PCR结果基本一致,进一步显示Expansin基因在胚珠/纤维生长和发育中的潜在功能。图5
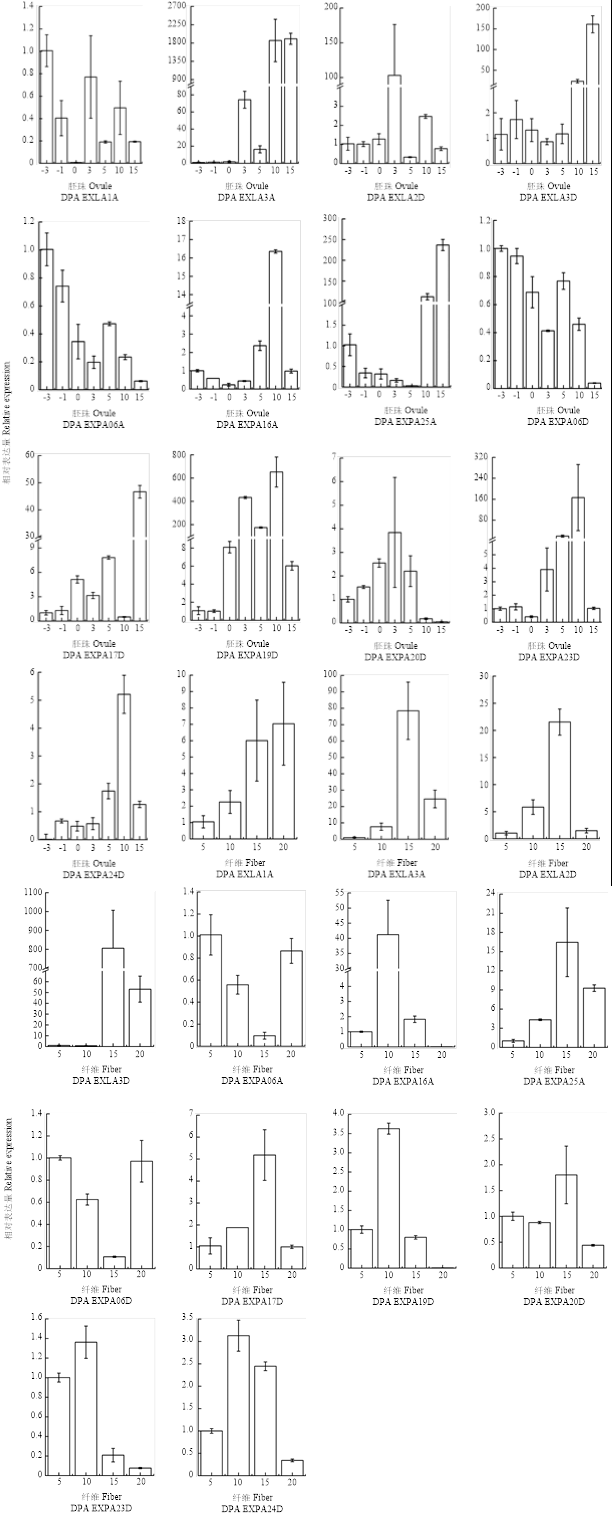 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5基于qRT-PCR的Expansin基因表达模式的验证
Fig. 5Validation of the expression patterns of thirteen selected GhExpansin gene by qRT-PCR
2.7 部分同源(homoeologous)基因间的表达丰度比较
异源四倍体的陆地棉,其基因组中每个基因都具有一个部分同源(homoeologous)基因。具有部分同源关系的基因对(pair)从同一祖先进化而来,经过了漫长的物种进化过程,基因的功能是否发生变化、表达模式是否相同,通过对RNA-seq数据(SRP017168和SRP059947)的分析,对陆地棉中具有同线性的直系同源基因的表达量进行比较,二项分布(binomial distribution)统计分析显示,具有部分同源关系的一对基因在相同的时空条件下,其表达模式具有差异。GhEXPA06D和GhEXPA06A、GhEXPA24A和GhEXPA24D、GhEXPA23A和GhEXPA23D、GhEXPA16A和GhEXPA16D、GhEXLA1A和GhEXLA1D、GhEXLB2A和GhEXLB2D这6个部分同源基因对(pair),各基因对之间在不同的时空条件下存在表达差异,At和Dt 2个亚基因组的表达丰度不一致。如:幼嫩叶片中,GhEXPA06D的表达量(位于Dt亚基因)>GhEXPA06A的表达量(位于At亚基因);在花萼中则相反,GhEXPA06D的表达量<GhEXPA06A的表达量。同时也存在一些部分同源基因对,如基因GhEXPA07A和GhEXPA07D,在部分时空条件下表达丰度均很低,但在茎、新叶、花冠中,基因GhEXPA07D的表达量显著低于GhEXPA07A的表达量。这些分析结果显示,异源四倍体陆地棉的部分同源基因对所发挥的作用可能存在差异,或是在进化过程中发生了功能异化(divergence)。3 讨论
3.1 扩展蛋白基因家族的成员鉴定及进化分析
在植物界,扩展蛋白基因家族包含4个亚家族:EXPA、EXPB、EXLA和EXLB。本研究在陆地棉全基因组水平上对Expansin基因家族进行鉴定与分析,以氨基酸序列中同时含有Pollen_allerg_1和DPPB_1这两个保守性特征结构域为标准,在陆地棉基因组中共鉴定出72个扩展蛋白基因。将陆地棉与拟南芥、水稻的扩展蛋白基因一起进行系统发育树分析,结果显示,陆地棉中4个亚家族EXPA、EXPB、EXLA、EXLB的基因分别有46、8、6和12个。陆地棉是由二倍体的祖先物种经过远缘杂交和染色体加倍进化而来,而二倍体的祖先物种的现代种是亚洲棉和雷蒙德氏棉。因此,为了研究陆地棉中Expansin基因家族的进化特征,本研究参阅了雷蒙德氏棉中Expansin基因鉴定的结果[46],同时在亚洲棉中进行了Expansin基因的鉴定(图1的系统发育树)。与雷蒙德氏棉和亚洲棉相比,陆地棉扩展蛋白各亚家族的基因数量几乎是二倍体棉种的2倍(EXPA亚家族46﹕26;EXPB亚家族8﹕4;EXLA亚家族6﹕3;EXLB亚家族12﹕6),但陆地棉扩展蛋白基因的数量(72个)少于二倍体雷蒙德氏棉和亚洲棉的扩展蛋白数量之和(39+39=78),可能原因是在四倍体形成后的进化过程中,染色体片段的复制(segmental duplication)和重排(chromosomal rearrangement)导致这些(6个)基因因异位(ectopy)而被淘汰或丢失(gene loss)。其他物种中的研究表明,双子叶植物中EXPA亚家族比EXPB亚家族的扩展蛋白数量多[48],表2显示陆地棉中EXPA与EXPB亚家族分别占总扩展蛋白数量的63.9%和11.1%,亚洲棉和雷蒙德氏棉EXPA与EXPB类扩展蛋白分别占总扩展蛋白的66.7%和10.3%,与前人研究结果一致。图1还显示,各亚家族内的亲缘关系相近的基因在基因的“外显子-内含子”结构及氨基酸序列模体(motif)组成等方面具有一定的相似性,表明在Expansin的进化方面,DNA水平的多样性与蛋白水平的多样性是基本吻合的。系统发育树的拓扑结构(topology)也和拟南芥、水稻等植物中的结果一致,这些是亚家族划分的有利依据。利用软件Euk-mPLoc 2.0对Expansin家族基因进行了亚细胞定位,结果显示所有的Expansin蛋白均定位于细胞外,推测它们均位于细胞壁中,可能与Expansin蛋白在棉属中所起的作用有关。
植物的共同祖先(last common ancestor)中,4个亚家族EXPA、EXPB、EXLA、EXLB基因的数量分别是10—12、2、1、2[5]。本研究发现,陆地棉扩展蛋白家族获得了极大的扩张。大规模的基因组重复(large-scale genomic duplication)、平行同源(paralogous)基因及部分同源(homoeologous)基因间功能多样化(diversification)是生物进化的动力之一[47,55],也是陆地棉扩展蛋白家族极大地扩张(contributed to the expansion of the Expansinfamily)的原因。利用软件MCScanX进行的同线性分析显示,陆地棉中除了3个串联重复(tandem duplications)区段(Gh_A05G3210/Gh_A05G3211;Gh_A08G1477/Gh_ A08G1478;Gh_D08G1774/Gh_D08G1775/Gh_D08G1776/ Gh_D08G1777)外,其余均为片段重复(dispersed segmental duplications)。陆地棉进化过程中的多次基因组/染色体片段重复(duplicated (syntenic) blocks)及微量的丢失(gene loss)使得Expansin基因家族成员扩张到72个。
3.2 Expansin家族基因的表达与功能
棉花纤维、胚珠等各种组织的生长发育与基因的表达丰度密切相关,Expansin基因在这些不同的生命活动阶段可能发挥着重要作用。ORFORD等[34]克隆了扩展蛋白基因GhExp1,该基因在纤维发育的伸长阶段特异表达;RUAN等[56]研究表明在纤维伸长阶段可观察到扩展蛋白基因的高量表达;LI等[37]发现GbEXPA2在纤维细胞伸长期间显著上调表达,表明GbEXPA2在纤维发育中具有重要的功能。BAJWA等[38]通过农杆菌介导的GhEXPA8转化,在棉花中过量表达GhEXPA8可以改善纤维品质。同样,INDRAIS等[57]通过RT-PCR和Northern印迹发现特异的扩展蛋白对纤维伸长具有贡献作用。本研究中,RNA-seq数据(SRP017168、SRP059947)分析结果同样表明(图4),各扩展蛋白基因在纤维和胚珠的不同发育时期特异表达。如GhEXPA19A和GhEXPA19D相比其他基因在纤维中的表达量很高;GhEXPA17D和GhEXPA17A在胚珠30和40 DPA高量表达;GhEXLB3A在胚珠20 DPA表达量特别高。基于RNA-seq数据的分析结果,对于那些在纤维和胚珠的某个时期高量表达的基因,选择了13个基因进行qRT-PCR定量验证,旨在分析Expansin基因对纤维发育过程中的影响和作用,大多数基因的表达模式与RNA-seq数据分析的结果一致。这些特定时空条件下活跃表达的基因,也许是棉花种质创新的重要候选基因。当然,少数基因的基于qRT-PCR的表达模式与RNA-seq数据分析结果存在差异,可能是因为RNA-seq测序样本与本研究中qRT-PCR棉花材料的不同所致。4 结论
陆地棉基因组中含有72个扩展蛋白基因,包括EXPA、EXPB、EXLA和EXLB 4个亚家族的46、8、6和12个基因。在DNA水平和氨基酸水平上具有进化的多样性与保守性。多数基因具有表达时空特异性。(责任编辑 李莉)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1134/S1021443708010123Magsci [本文引用: 1]

Effects of salinity of nutrient solution on expression of <i>ZmEXPA1</i> expansin gene and leaf growth were studied on maize plants (<i>Zea mays</i> L.). Rapid activation of the gene transcription was shown to precede the resumption of extension growth in leaf cells under water deficit induced by NaCl salinity. Auxins were found to accumulate in leaves during salinity treatment, the accumulation being faster than activation of <i>ZmEXPA1</i> transcription. In addition, exogenous IAA was shown to enhance the gene expression. Our results indicate that the hormone is involved in regulation of cell extension growth at high salinity through the expression of expansin gene.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
DOI:10.1093/mp/sss112Magsci [本文引用: 1]

Plant growth requires cell wall extension. The cotton AtRD22-Like 1 gene GhRDL1, predominately expressed in elongating fiber cells, encodes a BURP domain-containing protein. Here, we show that GhRDL1 is localized in cell wall and interacts with GhEXPA1, an -expansin functioning in wall loosening. Transgenic cotton overexpressing GhRDL1 showed an increase in fiber length and seed mass, and an enlargement of endopleura cells of ovules. Expression of either GhRDL1 or GhEXPA1 alone in Arabidopsis led to a substantial increase in seed size; interestingly, their co-expression resulted in the increased number of siliques, the nearly doubled seed mass, and the enhanced biomass production. Cotton plants overexpressing GhRDL1 and GhEXPA1 proteins produced strikingly more fruits (bolls), leading to up to 40% higher fiber yield per plant without adverse effects on fiber quality and vegetative growth. We demonstrate that engineering cell wall protein partners has a great potential in promoting plant growth and crop yield.
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
[本文引用: 3]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
