 ,西南大学农学与生物科技学院,重庆400715
,西南大学农学与生物科技学院,重庆400715QTL Identification for Fatty Acid Content in Brassica napus Using the High Density SNP Genetic Map
YE Sang, CUI Cui, GAO HuanHuan, LEI Wei, WANG LiuYan, WANG RuiLi, CHEN LiuYi, QU CunMin, TANG ZhangLin, LI JiaNa, ZHOU QingYuan ,College of Agronomy and Biotechnology, Southwest University, Chongqing 400715
,College of Agronomy and Biotechnology, Southwest University, Chongqing 400715通讯作者:
收稿日期:2019-05-28接受日期:2019-07-11网络出版日期:2019-11-08
| 基金资助: |
Received:2019-05-28Accepted:2019-07-11Online:2019-11-08
作者简介 About authors
叶桑,Tel:13068300612;E-mail:
崔翠,Tel:13883787860;E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (5693KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
叶桑, 崔翠, 郜欢欢, 雷维, 王刘艳, 王瑞莉, 陈柳依, 曲存民, 唐章林, 李加纳, 周清元. 基于SNP遗传图谱对甘蓝型油菜部分脂肪酸 组成性状的QTL定位[J]. 中国农业科学, 2019, 52(21): 3733-3747 doi:10.3864/j.issn.0578-1752.2019.21.002
YE Sang, CUI Cui, GAO HuanHuan, LEI Wei, WANG LiuYan, WANG RuiLi, CHEN LiuYi, QU CunMin, TANG ZhangLin, LI JiaNa, ZHOU QingYuan.
0 引言
【研究意义】油菜是中国主要的油料作物,其种植面积和总产量居世界首位[1],目前,已经成为继水稻、小麦、玉米之后的第四大作物,是中国的优势油料作物[2]。菜籽油是中国主要的食用油之一,其主要脂肪酸组成包括硬脂酸(C18﹕0)、油酸(C18﹕1)、亚油酸(C18﹕2)、亚麻酸(C18﹕3)、廿碳烯酸(C20﹕1)和芥酸(C22﹕1)等。食用油中油酸具有极高的营养和应用价值,高油酸菜籽油不仅有利于人体心血管健康[3],还因其热稳定性高[4],利于纯化储存而被用于煎炸行业[5]以及生产、生活用品的原料[6]。亚油酸是一种必需脂肪酸,可防止动脉粥样硬化,但存在氧化稳定性差、储存时间短的缺点。亚麻酸属多烯类不饱和脂肪酸,因存在3个不饱和键而易氧化变质,故亚麻酸含量较高的菜籽油不耐储存[7]。芥酸因其碳链较长,在人体内不易被消化吸收,不适用于食品加工[8],然而芥酸是重要的工业原料,被广泛应用于众多工业领域[7]。因此,根据生产需要改善油菜脂肪酸的组成成分是油菜育种的重要目标,定位油菜脂肪酸组分的QTL和筛选影响脂肪酸组分的候选基因在油菜育种中都具有重要的指导作用。【前人研究进展】前人研究表明,在油菜主要脂肪酸的形成过程中,首先由棕榈酸在碳链延长酶的作用下合成硬脂酸[9],硬脂酸在质体基质中再被硬脂酰-ACP脱氢酶催化形成油酸[10],而油酸包含2条代谢途径:其一,油酸在碳链延长酶的作用下合成廿碳烯酸,廿碳烯酸进一步延长碳链生成芥酸[11];其二,油酸在△12减饱和酶作用下生成亚油酸[12],亚油酸在△15减饱和酶作用下进一步减饱和生成亚麻酸[13]。了解主要脂肪酸的代谢途径是改善油菜脂肪酸组成的前提,通过分子标记定位相关性状QTL并筛选出候选基因有利于更准确了解脂肪酸各组分受到影响的遗传因素。迄今为止,利用分子标记技术的QTL定位分析在油菜相关性状包括抗性[14,15,16]、育性[17,18]和品质[19,20,21]等方面广泛发展,为油菜分子的育种改良奠定了基础。油菜脂肪酸含量具有较高的遗传力,对其相关分子标记的确定是改善油菜脂肪酸组成的关键。BURNS等[22]利用RFLP分子标记进行油菜脂肪酸组成的QTL定位,获得4个与油酸含量相关的QTL,其中效应较大的QTL位于N3、N8、N18染色体,效应较小的位于N11染色体;获得5个与亚油酸含量相关的QTL,效应相对较大QTL位于N8和N14染色体;获得5个与亚麻酸含量相关的QTL,分别位于N6、N7、N18和N11染色体。张洁夫等[23]采用RAPD、SSR和SRAP 3种分子标记对油菜脂肪酸组成进行QTL定位,获得3个与硬脂酸含量相关的QTL,分别位于N1、N8和N16染色体;获得2个与油酸含量相关的主效QTL,分别位于N8和N13染色体;获得3个与亚油酸含量相关的QTL,其中2个位于N8染色体,1个位于N13染色体;获得3个与亚麻酸含量相关的微效QTL;获得4个与廿碳烯酸含量相关的QTL,分别位于N8、N13和N15染色体;在N8和N13染色体获得2个与芥酸含量相关的主效QTL。ZHAO等[24]利用SSR标记构建的遗传图谱找到了7个控制油酸含量的QTL,其中,主效QTL位于N18染色体;找到7个控制亚油酸含量的QTL,其中主效QTL位于N9染色体;并在N14染色体找到1个控制亚麻酸含量的QTL。与上述分子标记技术相比,SNP标记具有数量多、分布广和规模化等优点,已成为大家认可的第三代遗传标记,近几年被广泛应用于油菜产量、品质等性状的分子标记辅助育种及遗传定位等诸多领域。例如,LIU等[25]利用油菜SNP芯片技术构建高密度遗传图谱,对油菜籽木质素、纤维素和半纤维素等性状进行QTL定位。SHEN等[26]采用SNP分子标记技术定位了17个油菜分枝角度QTL,并在QTL区间获得27个候选基因。QU等[27]利用520份油菜资源群体对油菜籽脂肪酸组成进行全基因组关联分析,确定了62个与7种脂肪酸组成显著相关的基因组区域,并鉴定了24个参与脂肪酸生物合成的功能候选基因的同源基因。【本研究切入点】前人对油菜脂肪酸含量的定位研究主要是利用SSR、AFLP、RFLP和SRAP等分子标记,存在标记的多位点现象,给不同连锁图谱之间QTL位点的比较分析带来困难。另外,前人在QTL作图方法上大多采用区间作图法和复合区间作图法,致使QTL效应可能会被侧连标记区间之外的标记变量所吸收[28]。【拟解决的关键问题】本研究利用油菜6K SNP芯片构建高密度遗传连锁图谱,通过完备区间作图法对油菜主要脂肪酸组成进行QTL定位。结合2年的表型鉴定,发掘在不同环境条件下稳定表达的主效QTL,并利用甘蓝型油菜基因组序列,与拟南芥脂肪酸相关代谢基因进行同源性分析,筛选可能的候选基因,旨在探讨油菜主要脂肪酸之间的内在联系,为改善油菜脂肪酸组成进行分子标记辅助选择、克隆相关候选基因奠定基础。1 材料与方法
1.1 材料
重组自交系群体的母本为人工合成甘蓝型油菜新品系10D130,其芥酸含量50%左右,父本为高产优质的天然甘蓝型油菜常规品种中双11号(简称ZS11),其芥酸含量<1%。两亲本杂交F2通过单粒法连续自交7代,构建重组自交系群体,以其中的186个材料进行SNP标记分析,构建高密度SNP遗传连锁图谱。所有试验材料均由西南大学重庆市油菜工程技术研究中心提供。1.2 田间试验
2016-2017年和2017-2018年2个年度,在重庆市北碚区歇马镇油菜实验基地种植重组自交系群体及其亲本,2个环境分别记为17Cq和18Cq。田间播种采用随机区组试验设计,每个小区5行,每行10株。田间管理遵循常规生产方式,在油菜初花期逐株套袋自交,种子完全成熟后每家系收取5株正常植株的种子,自然风干后保存。1.3 脂肪酸组成的测定
根据文献[29]的方法,利用气相色谱仪GC-2010(Shimazu,JAP)测定2个环境下重组自交系群体种子中硬脂酸、油酸、亚油酸、亚麻酸、廿碳烯酸及芥酸的含量,每份材料重复测定3株,取其平均值。1.4 遗传连锁图谱构建及QTL分析
参考刘列钊等[30]方法,分别从每个株系的5个幼嫩植株叶片取混合样0.15 g,利用DNA提取试剂盒DP321-03(天根,中国北京)提取DNA并稀释至50 ng·μL-1用于SNP标记分析。严格按照Infinium HD Assay Ultra操作说明书(Illumina Inc 公司)进行DNA样品的预处理(等位扩增、片段化及富集)、与芯片杂交、洗涤、安装流动室、单碱基延伸、染色及包埋。芯片准备好后运用Illumina HiScan扫描仪的iScan Control Software软件扫描,然后利用GenomeStudio genotyping software v2011软件分析扫描结果,获取各个样本的SNP基因型数据,并为获得的SNP标记命名,命名方法以S-95505568为例,S表示SNP,95505568代表GenomeStudio genotyping software生成的相应SNP位点索引号。连锁图谱的构建采用JoinMap4.0软件[31],选用Kosambi[32]函数将重组值转换为图距单位(centiMorgan,cM),取最小阈值LOD2.0对所有标记进行分组,每个染色体上标记顺序通过两两标记之间最小重组频率计算,构建用于QTL定位的遗传连锁图谱。
采用软件QTL IciMapping V4.1[33]完备区间作图(inclusive composite interval mapping,ICIM)法,进行2个环境下硬脂酸、油酸、亚油酸、亚麻酸、廿碳烯酸及芥酸含量的QTL定位和检测。进行ICIM分析时,选用1 cM的步长(walking speed),作1 000次排列检验和显著水平0.01确定LOD临界值。软件运行结果可同时给出性状QTL的加性效应和表型贡献率。QTL命名时以q加上性状的英文缩写再加上染色体编号及QTL序号表示,字体斜体,如qC03LI-1代表亚油酸含量位于C03染色体上的第一个QTL。同一性状在染色体相同的位置检测到重复的QTL,且加性效应方向一致,认为是同一QTL。
1.5 候选基因筛选
为筛选出与脂肪酸组成相关的候选基因,首先将检测到的QTL置信区间在甘蓝型油菜基因组上对应的序列查询到,然后与BOTELLA等[34]、QU等[27]搜索出的拟南芥脂肪酸代谢相关基因进行BLASTN比对,将E值设定为E-20,最后筛选出每个QTL置信区间内匹配E值小于阈值的候选基因。2 结果
2.1 重组自交系群体脂肪酸含量的分析
两亲本和RIL群体的表型分析见表1。RIL群体的表型在2个环境中表现一致:6个性状的均值都介于两亲本之间,最大值和最小值都超过亲本值,存在明显的超亲分离现象。另外,两亲本各性状间差异及RIL群体各性状在株系间差异都达到显著或极显著水平,结合变异系数值反映出这几个性状在该群体中具有较广泛的遗传变异。由性状频率分布图(图1)可知,硬脂酸、亚油酸和亚麻酸呈现偏态分布的特征,油酸、廿碳烯酸和芥酸含量则表现为明显的多峰分布,结果表明,6种脂肪酸含量在2个环境中均表现为连续分布,适合进行QTL检测。Table 1
表1
表12个环境下油菜主要脂肪酸含量的变化
Table 1
| 环境 Environment | 性状 Trait | 亲本 Parents | RIL群体 RIL populations | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 10D130 | ZS11 | 最大值 Max | 最小值 Min | 均值 Average | 标准差 SE | 变异系数 CV (%) | |||
| 17Cq | C18:0 | 1.21a | 2.84b | 6.17 | 1.00 | 1.77** | 0.61 | 34.47 | |
| C18:1 | 18.83A | 64.27B | 83.15 | 13.17 | 38.25** | 19.11 | 49.95 | ||
| C18:2 | 10.86A | 23.27B | 31.18 | 6.15 | 14.49** | 4.50 | 31.04 | ||
| C18:3 | 7.68a | 9.55b | 12.79 | 4.04 | 8.65* | 1.59 | 18.35 | ||
| C20:1 | 8.08A | 0.07B | 19.33 | 0.00 | 10.26** | 5.86 | 57.12 | ||
| C22:1 | 53.34A | 0.00B | 57.96 | 0.00 | 26.56** | 17.70 | 66.62 | ||
| 18Cq | C18:0 | 1.52a | 2.68b | 4.73 | 0.47 | 1.98** | 0.56 | 28.13 | |
| C18:1 | 17.91A | 63.03B | 78.79 | 12.69 | 39.14** | 18.03 | 46.07 | ||
| C18:2 | 12.08A | 22.94B | 26.41 | 9.27 | 15.47** | 3.94 | 25.46 | ||
| C18:3 | 8.92a | 10.44b | 12.20 | 6.14 | 8.89* | 1.36 | 15.27 | ||
| C20:1 | 8.86A | 0.91B | 19.99 | 0.70 | 11.20** | 6.43 | 57.44 | ||
| C22:1 | 50.71A | 0.00B | 51.62 | 0.00 | 23.02** | 15.79 | 68.59 | ||
a, b and A, B indicate significance at P<0.05 and P<0.01, * and ** indicate significance at P<0.05 and P<0.01. C18:0, C18:1, C18:2, C18:3, C22:1 and C22:2 represent stearic acid, oleic acid, linoleic acid, linolenic acid, eicosenoic acid and erucic acid, respectively. The same as below
新窗口打开|下载CSV
由表2可知,在2个环境下油酸含量与硬脂酸及亚油酸含量均表现为极显著正相关(P<0.01),与廿碳烯酸及芥酸含量均表现为极显著负相关(P<0.01),与亚麻酸含量相关性不强。芥酸含量与廿碳烯酸含量在2个环境中均表现极显著正相关(P<0.01),相关系数分别为0.56和0.64;与亚油酸含量在2个环境中均表现极显著负相关(P<0.01),相关系数分别为-0.52和-0.54。结果表明,油酸转化为亚油酸的途径受到芥酸途径的掣肘,即油酸和亚油酸的含量受芥酸含量的变化影响较大。而亚麻酸受油酸和芥酸变化的影响较小,在遗传上具有相对独立的稳定性。
图1
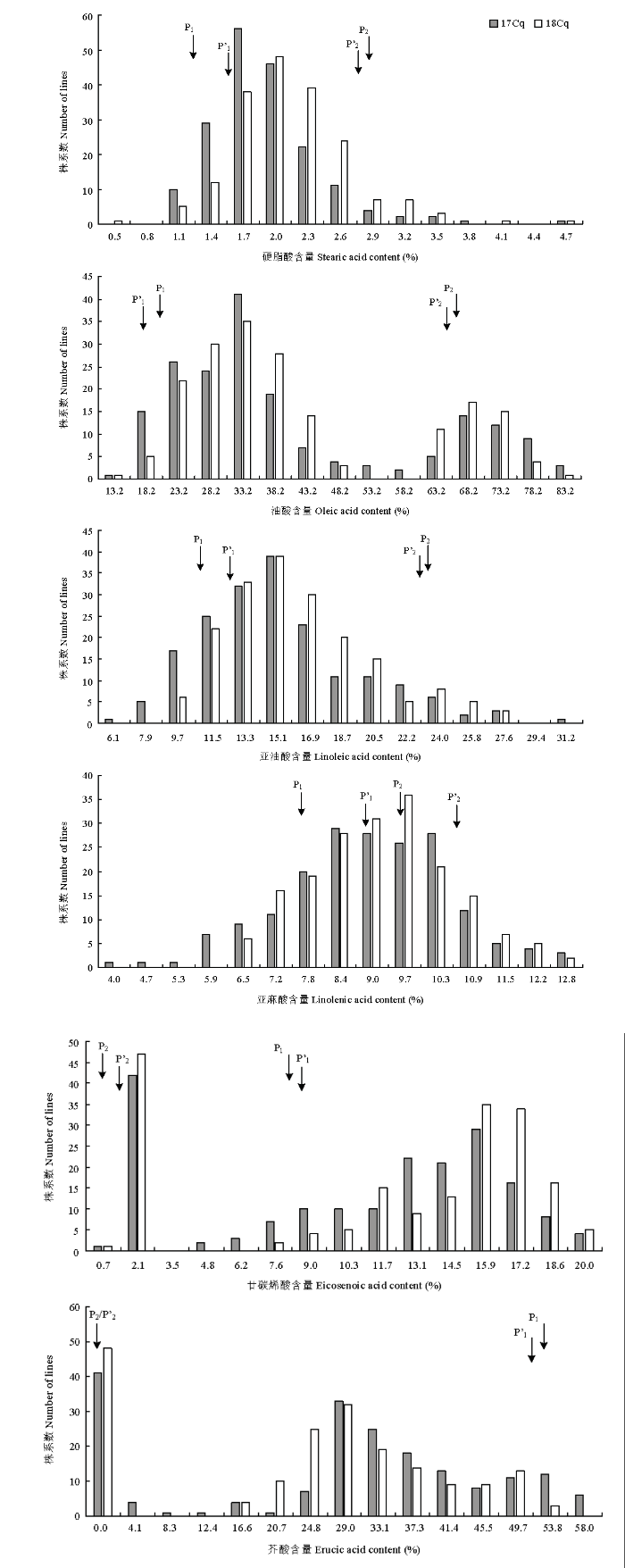 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1RIL群体硬脂酸、油酸、亚油酸、亚麻酸、廿碳烯酸及芥酸含量的频率分布
P1、P2和P’1、P’2分别标出了17Cq和18Cq环境下2个亲本的脂肪酸含量
Fig. 1Frequency distribution of stearic acid, oleic acid, linoleic acid, linolenic acid, eicosenoic acid and erucic acid content from the RIL population in two different environments
P1, P2 and P'1, P'2 indicated the fatty acid content of two parents under 17Cq and 18Cq environment respectively
Table 2
表2
表22个环境下主要脂肪酸含量相关分析
Table 2
| 测定指标 Index | C18:0 | C18:1 | C18:2 | C18:3 | C20:1 | C22:1 |
|---|---|---|---|---|---|---|
| C18:0 | 0.58** | 0.18* | -0.32** | -0.28** | -0.59** | |
| C18:1 | 0.65** | 0.36** | -0.13 | -0.68** | -0.95** | |
| C18:2 | 0.03 | 0.43** | 0.45** | -0.50** | -0.52** | |
| C18:3 | -0.29** | 0.11 | 0.49** | -0.12 | -0.01 | |
| C20:1 | -0.31** | -0.77** | -0.57** | -0.28** | 0.56** | |
| C22:1 | -0.64** | -0.96** | -0.54** | -0.20** | 0.64** |
新窗口打开|下载CSV
右上部代表17Cq环境,左下部代表18Cq环境The upper right represents the 17Cq environment and the lower left represents the 18Cq environment
2.2 遗传连锁图谱
利用包含5 058个标记的6K油菜芯片对186个RIL材料进行基因型鉴定,从中筛选出1 897个高质量多态性的SNP标记,约占37.5%,基于这些标记构建遗传连锁图谱。获得用于QTL定位的图谱覆盖甘蓝型油菜基因组3 214.19 cM,平均图距为1.69 cM。每条染色体长度在86.51-298.72 cM,平均长度为169.17 cM。染色体标记数目在24-153,平均数目为99.84个。但是从标记分布来看,各染色体分布不均,其中染色体A03、C03和C04上标记分布较多,分别为139、147和153个;而染色体C05、C08和C09上的标记数目较少,分别只有48、26和24个。此外各染色体标记密度也有较大差异,C04密度最大,平均间距仅1.10 cM,而密度最小的C08平均间距达6.13 cM。2.3 主要脂肪酸含量的QTL分析
2.3.1 硬脂酸含量的QTL 在2个环境中,硬脂酸含量共定位到6个QTL,解释表型变异5.08%-25.49%,其中主效QTL位点位于A08和C03上。C03上的主效位点在2个环境中能被重复检测到,分别解释表型变异的11.59%和9.12%;在A08上的2个QTL位点分别解释硬脂酸含量表型变异的14.00%和25.49%。且A08和C03上的QTL位点在2个环境中的加性效应均小于0(表3和图2),即加性效应均来源于亲本ZS11。Table 3
表3
表3油菜主要脂肪酸含量在2个环境中的QTL位点
Table 3
| 性状 Trait | 环境 Envi. | QTL | 染色体 Chr. | 标记区间 Position | 置信区间1) Confidence interval | LOD | 加性效应 Additive effect | 贡献率 R2(%) |
|---|---|---|---|---|---|---|---|---|
| C18:0 | 17Cq | qA08ST-1 | A08 | S-95505568-S-95506507 | 43.27-48.50 | 6.13 | -0.22 | 14.00 |
| qC03ST | C03 | S-177827612-S-95636886 | 216.25-219.73 | 5.11 | -0.20 | 11.59 | ||
| 18Cq | qA01ST | A01 | S-95637833-S-95664701 | 116.00-118.62 | 4.94 | 0.13 | 5.08 | |
| qA05ST | A05 | S-177830309-S-86232724 | 107.59-110.16 | 6.07 | 0.15 | 6.27 | ||
| qA08ST-2 | A08 | S-95506569-S-95506120 | 29.90-31.07 | 20.45 | -0.29 | 25.49 | ||
| qA09ST | A09 | S-182142581-S-182167880 | 224.50-229.51 | 8.76 | -0.18 | 9.45 | ||
| qC03ST | C03 | S-95665809-S-95636886 | 215.70-219.73 | 8.31 | -0.17 | 9.12 | ||
| C18:1 | 17Cq | qA05OL | A05 | S-182087654-S-86232724 | 106.33-110.16 | 4.59 | 3.81 | 3.80 |
| qA08OL | A08 | S-95507415-S-95663297 | 26.79-30.00 | 31.31 | -11.42 | 34.39 | ||
| qC03OL | C03 | S-105305847-S-182158964 | 218.48-222.84 | 29.65 | -10.95 | 31.80 | ||
| 18Cq | qA05OL | A05 | S-182087654-S-86232724 | 106.33-110.16 | 8.40 | 3.91 | 5.70 | |
| qA08OL | A08 | S-95507415-S-95663297 | 26.79-30.00 | 31.83 | -8.66 | 28.28 | ||
| qC03OL | C03 | S-105338742-S-182138971 | 219.51-224.50 | 42.44 | -10.75 | 43.68 | ||
| C18:2 | 17Cq | qA05LI | A05 | S-182087654-S-86232724 | 106.33-110.16 | 18.43 | -2.37 | 22.67 |
| qA08LI | A08 | S-177633794-S-95506569 | 25.61-29.50 | 8.45 | -1.48 | 8.91 | ||
| qC03LI-1 | C03 | S-105306222-S-95637726 | 235.80-239.65 | 14.65 | -2.06 | 17.00 | ||
| qC03LI-2 | C03 | S-105309588-S-105307276 | 250.03-253.95 | 6.28 | 1.28 | 6.69 | ||
| 18Cq | qA05LI | A05 | S-182087654-S-86232724 | 106.33-110.16 | 19.71 | -1.84 | 25.25 | |
| qA08LI | A08 | S-95507415-S-95663297 | 26.79-30.00 | 9.44 | -1.18 | 10.49 | ||
| qC03LI-1 | C03 | S-95637910-S-105307365 | 237.50-239.83 | 11.09 | -1.30 | 12.56 | ||
| C18:3 | 17Cq | qA02LN | A02 | S-95666343-S-95638378 | 55.50-64.85 | 4.96 | -0.44 | 8.49 |
| qC07LN | C07 | S-105339086-S-179307020 | 6.50-11.97 | 5.84 | 0.51 | 11.66 | ||
| 18Cq | qA05LN | A05 | S-182087654-S-86232724 | 106.33-110.16 | 33.12 | -1.12 | 23.66 | |
| qA09LN | A09 | S-182142581-S-182167880 | 224.50-229.51 | 8.28 | 0.47 | 4.24 | ||
| qA10LN | A10 | S-177910390-S-95636447 | 59.94-63.16 | 4.90 | 0.35 | 2.37 | ||
| C20:1 | 17Cq | qC03EI | C03 | S-105338742-S-182138971 | 219.51-224.50 | 7.45 | 2.27 | 17.23 |
| 18Cq | qA08EI | A08 | S-95507415-S-95663297 | 26.79-30.00 | 6.07 | 2.14 | 11.18 | |
| qC03EI | C03 | S-105338742-S-182138971 | 219.51-224.50 | 11.44 | 3.05 | 22.69 | ||
| C22:1 | 17Cq | qA08ER-1 | A08 | S-95506362-S-95664454 | 10.05-12.58 | 11.30 | 5.00 | 9.39 |
| qA08ER-2 | A08 | S-95507415-S-95663297 | 26.79-30.00 | 27.90 | 9.13 | 32.21 | ||
| qC03ER | C03 | S-105305847-S-182158964 | 218.48-222.84 | 32.17 | 9.62 | 36.01 | ||
| 18Cq | qA08ER-2 | A08 | S-95507415-S-95663297 | 26.79-30.00 | 96.68 | 17.53 | 67.26 | |
| qC03ER | C03 | S-105305847-S-182158964 | 218.48-222.84 | 50.41 | 8.64 | 16.45 |
新窗口打开|下载CSV
图2
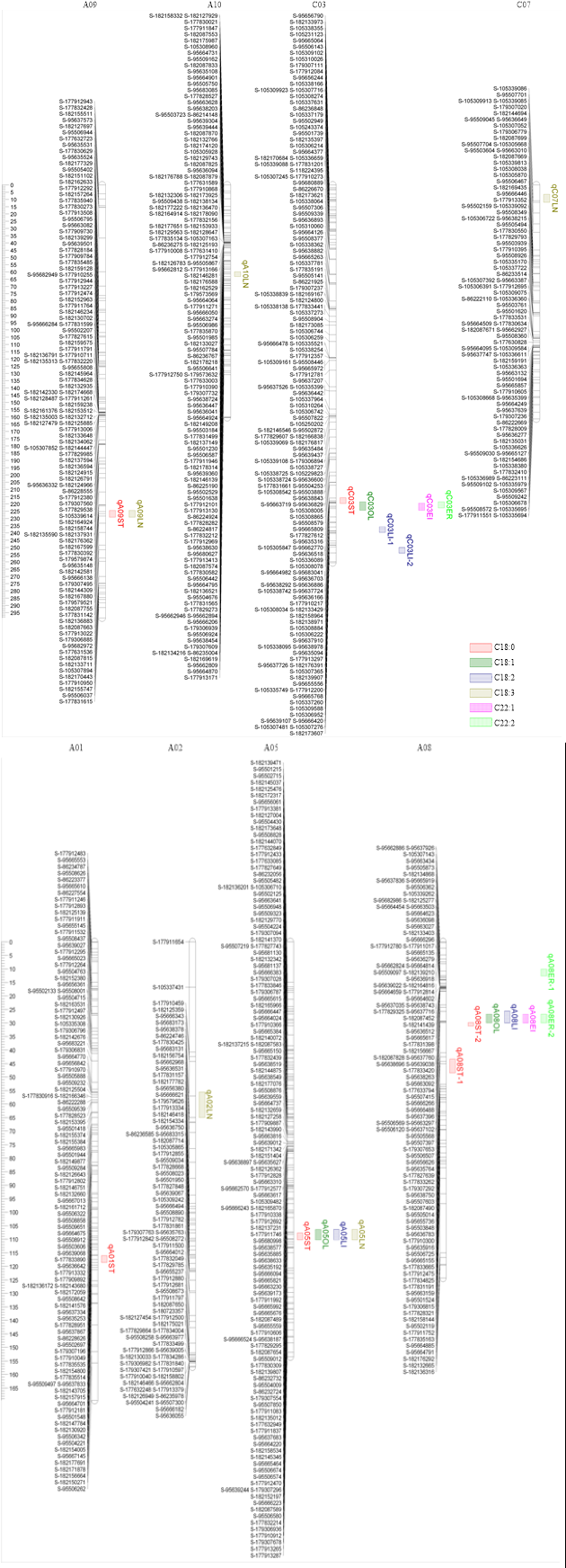 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2甘蓝型油菜主要脂肪酸含量QTL在SNP连锁群上的分布
Fig. 2Putative QTL locations of fatty acid content on the SNP genetic map
2.3.2 油酸含量的QTL 在2个环境中共检测到3个与油酸含量相关的 QTL,且3个位点在2个环境中均能被重复检测到,其中位于A08和C03染色体上的是主效位点,可解释油酸含量表型变异的28.28%-43.68%。在2个环境中,位于A05上的QTL位点加性效应分别为3.81和3.91,而A08和C03染色体上主效位点的效应值在-11.42--8.66(表3和图2),即前者的加性效应来源于亲本10D130,后两者的加性效应则来自亲本ZS11。
2.3.3 亚油酸含量的QTL 在2个环境中,亚油酸含量共检测到4个QTL,除了C03染色体上的qC03ST-2以外,其余3个QTL均能在2个环境中被重复检测到。其中主效QTL位于A05和C03上,可解释亚油酸含量表型变异的12.56%-25.25%;A08染色体上重复检测到的QTL位点在2个环境中的遗传贡献率分别为8.91%和10.49%。同时,在2个环境中重复检测到的3个QTL位点的效应值在-2.37--1.18(表3和图2),即加性效应均来自亲本ZS11。
2.3.4 芥酸含量的QTL 在2个环境中共定位到3个与芥酸含量相关的QTL,其中位于A08和C03上的主效QTL位点能够在2个环境中被重复检测到,可解释表型变异16.45%-67.26%。芥酸含量的3个QTL位点的加性效应均为正值,加性效应均来源于亲本10D130,其中,qA08ER的效应值最大,为17.53(表3和图2),属主基因座位。其他主要脂肪酸的QTL定位结果见表3和图2。
2.4 主要脂肪酸代谢候选基因筛选
将23个QTL置信区间序列与拟南芥脂肪酸代谢基因进行比对,分别在A01、A02、A05、A08、A09、A10和C03染色体上的17个QTL区间内检测到22个候选基因,匹配E值介于0-4E-17(表4)。位于A01染色体上的基因BnaA01g27100D和BnaA01g27120D分别与拟南芥基因AT3G18030和AT1G48605同源,参与辅酶A的生物合成。A02染色体上的基因BnaA02g13270D、BnaA02g13310D和BnaA02g13630D分别与拟南芥基因AT1G77590、AT1G67730和AT2G25710同源,分别编码长链酰基辅酶A合成酶、β-酮酰辅酶A还原酶和全羧化酶合酶。A05染色体上的基因BnaA05g26900D、BnaA05g25110D、BnaA05g25210D、BnaA05g25220D和BnaA05g26460D分别与拟南芥基因AT3G12120、AT3G14360、AT1G53990、AT3G14225和AT1G06800同源,第一个编码脂肪酸脱氢酶,后4个参与脂质代谢过程。在A09染色体上的基因BnaA09g39290D与拟南芥基因AT3G61580同源,编码脂肪酸去饱和酶。A10染色体上的基因BnaA10g10590D和BnaA10g10730D分别与拟南芥基因AT5G56350和AT5G56480同源,前者是丙酮酸激酶家族蛋白成员,后者是脂质转运蛋白家族成员。A08和C03染色体上存在多个同源基因,其中BnaA08g11130D和BnaC03g65980D与拟南芥基因AT4G34520同源,BnaA08g11140D和BnaC03g66040D与拟南芥基因AT4G34510同源,两者同属3-酮酰辅酶A合成酶家族;BnaA08g11440D和BnaC03g66380D与拟南芥基因AT4G33790同源,编码一种脂肪酰辅酶A还原酶。Table 4
表4
表4在甘蓝型油菜脂肪酸含量QTL置信区间比对拟南芥相关基因获得的候选基因
Table 4
| 染色体 Chr. | 预测基因 Gene prediction | 物理位置 Physical position (Mb) | 拟南芥相关基因 Related genes in A. thaliana | |||
|---|---|---|---|---|---|---|
| 基因名称 Gene name | 基因登录号 Gene accession | E值 E-value | 描述 Description | |||
| A01 | BnaA01g27100D | 18.94 | HAL3A | AT3G18030 | E-60 | 辅酶A生物合成过程 Coenzyme A biosynthetic process |
| BnaA01g27120D | 18.94 | HAL3B | AT1G48605 | 5E-23 | 辅酶A生物合成过程 Coenzyme A biosynthetic process | |
| A02 | BnaA02g13270D | 7.29 | LACS9 | AT1G77590 | 2E-52 | 长链酰基辅酶A合成酶9 Long chain acyl-CoA synthetase 9 |
| BnaA02g13310D | 7.31 | KCR1 | AT1G67730 | 2E-74 | β-酮酰辅酶A还原酶1 Beta-ketoacyl-CoA reductase 1 | |
| BnaA02g13630D | 7.49 | HCS1 | AT2G25710 | 2E-58 | 全羧化酶合成酶1 Holocarboxylase synthase 1 | |
| A05 | BnaA05g26430D | 19.37 | ALIS1 | AT3G12740 | 2E-93 | 参与磷脂转运 Involved in phospholipid transport |
| BnaA05g26900D | 19.54 | FAD2 | AT3G12120 | 0 | 不饱和脂肪酸生物合成过程 Unsaturated fatty acid biosynthetic process | |
| BnaA05g25110D | 18.65 | ATOBL1 | AT3G14360 | E-156 | 脂质代谢过程 Lipid metabolic process | |
| BnaA05g25210D | 18.70 | GLIP3 | AT1G53990 | 4E-17 | 脂质分解过程 Lipid catabolic process | |
| BnaA05g25220D | 18.70 | GLIP4 | AT3G14225 | E-117 | 脂质分解过程 Lipid catabolic process | |
| BnaA05g26460D | 19.38 | DALL4 | AT1G06800 | 2E-27 | 脂质代谢过程 Lipid metabolic process | |
| A08 | BnaA08g09510D | 9.09 | OGOX2 | AT4G20840 | 0 | FAD-结合小檗碱家族蛋白 FAD-binding Berberine family protein |
| BnaA08g11130D | 10.19 | FAE1 | AT4G34520 | 0 | 3-酮酰辅酶A合成酶18 3-ketoacyl-CoA synthase 18 | |
| BnaA08g11140D | 10.19 | KCS17 | AT4G34510 | 0 | 3-酮酰辅酶A合成酶17 3-ketoacyl-CoA synthase 17 | |
| BnaA08g11440D | 10.39 | FAR3 | AT4G33790 | 8E-46 | 脂肪酸还原酶3 FATTY ACID REDUCTASE 3 | |
| BnaA08g11650D | 10.51 | MCCB | AT4G34030 | E-119 | 3-甲基巴豆酰辅酶A羧化酶 3-methylcrotonyl-CoA carboxylase | |
| BnaA08g12370D | 11.04 | WIN2 | AT4G31750 | 3E-71 | 编码HopW1-1-相互作用蛋白2 Encodes HopW1-1-Interacting protein 2 | |
| A09 | BnaA09g39290D | 27.87 | SLD1 | AT3G61580 | 0 | 脂肪酸去饱和酶 Fatty acid desaturase |
| A10 | BnaA10g10590D | 9.01 | 未知Unknown | AT5G56350 | 0 | 丙酮酸激酶家族蛋白 Pyruvate kinase family protein |
| BnaA10g10730D | 9.08 | END2 | AT5G56480 | 4E-62 | 脂质转移蛋白 Lipid-transfer protein | |
| C03 | BnaC03g63920D | 53.41 | OGOX2 | AT4G20840 | 0 | FAD-结合小檗碱家族蛋白 FAD-binding Berberine family protein |
| BnaC03g63930D | 53.42 | BBE22 | AT4G20860 | 5E-23 | FAD-结合小檗碱家族蛋白 FAD-binding Berberine family protein | |
| BnaC03g64130D | 53.56 | SPHK2 | AT4G21534 | 4E-55 | 二酰基甘油激酶家族蛋白 Diacylglycerol kinase family protein | |
| BnaC03g65980D | 55.68 | FAE1 | AT4G34520 | 0 | 3-酮酰辅酶A合成酶18 3-ketoacyl-CoA synthase 18 | |
| BnaC03g66040D | 55.81 | KCS17 | AT4G34510 | 0 | 3-酮酰辅酶A合成酶17 3-ketoacyl-CoA synthase 17 | |
| BnaC03g66380D | 56.21 | FAR3 | AT4G33790 | 3E-49 | 脂肪酸还原酶3 FATTY ACID REDUCTASE 3 | |
| BnaC03g67410D | 57.10 | WIN2 | AT4G31750 | 2E-73 | 编码HopW1-1-相互作用蛋白2 Encodes HopW1-1-Interacting protein 2 | |
新窗口打开|下载CSV
3 讨论
3.1 QTL定位结果的比较分析
油菜主要脂肪酸QTL定位研究前人已有报道,本研究的部分结果与已有研究结果一致性较高,但也发现了一些新的QTL区域。例如,本研究通过完备区间作图法发现了控制芥酸含量的3个QTL,其中位于A08和C03染色体上的2个主效QTL在2个环境下被重复检测到,第3个微效QTL也位于A08染色体,与前人将控制芥酸含量的QTL主要集中在A08和C03上的结果一致[23,30,35-36]。油酸的遗传比较复杂,本研究在2个环境中重复检测到位于A08和C03染色体上油酸含量的2个主效QTL,和芥酸的主效QTL重叠,加性效应相反,这与刘列钊等[30]的研究结果相同。不同的是,本研究亲本并非高油酸材料,却在2个环境中重复定位到位于A05染色体上油酸含量的1个微效QTL,该位点同时还是亚油酸的主效QTL位点,这在前人研究中未发现。本研究在该位点上筛选到候选基因FAD2,同时检测到该群体(RIL)中的4个高油酸材料的FAD2第722位碱基A突变成T导致酪氨酸变成苯丙氨酸,从而影响了蛋白质的空间结构,导致油酸含量提高,这与张宏军等[37]的研究结果一致。而亲本ZS11的FAD2正常,结合A05染色体上QTL位点的加性效应来源于亲本10D130,猜测FAD2的变异来自亲本10D130。亚麻酸含量是典型的数量性状,易受环境影响[30]。在17Cq环境中,亚麻酸含量检测到1个位于C07染色体上的主效QTL,解释表型变异的11.66%,而在18Cq环境中,检测到一个主效QTL位于A05染色体,解释亚麻酸含量表型变异的23.66%,这与前人主要将亚麻酸含量主效QTL定位在A04和C04染色体不同[12,24,38],可能的原因是受环境因素影响较大。本研究采用完备区间作图法对甘蓝型油菜RIL群体的一系列油菜籽脂肪酸含量进行QTL定位,一方面进一步验证了已有研究的可靠性,另一方面也说明这些能够被多次重复定位的主效区段和新发现的基因组区段均是控制油菜籽主要脂肪酸含量的重要区域,对剖析油菜籽脂肪酸含量的遗传具有重要意义。3.2 主要脂肪酸含量QTL之间的相关性
研究主要对甘蓝型油菜主要脂肪酸,包括硬脂酸、油酸、亚油酸、亚麻酸、廿碳烯酸及芥酸含量进行QTL定位分析。结果表明,控制这些脂肪酸含量的QTL之间存在明显的相关性,主要表现为A05、A08和C03染色体上出现QTL的“富集区”。在A05染色体的相同位置同时存在硬脂酸、油酸、亚油酸和亚麻酸4种脂肪酸的QTL,其中对于亚油酸和亚麻酸是主效QTL,且加性效应方向一致,可解释二者在2个环境中所表现的0.45和0.49的极显著正相关性。在A08和C03染色体上相同的置信区间内同时检测出油酸、廿碳烯酸和芥酸的主效QTL位点,即控制油酸含量的主效QTL同时也是控制廿碳烯酸和芥酸含量的主效QTL,油酸的2个主效位点加性效应值都为负,而廿碳烯酸和芥酸在此位点上的加性效应则表现为正值,充分解释了油酸和廿碳烯酸含量之间、油酸和芥酸含量之间在2个环境中都表现的极显著负相关性以及廿碳烯酸和芥酸含量在2个环境中的极显著正相关性。根据表型相关性分析和QTL相关性分析认为,油酸的2条代谢途径并不均衡,其“加碳”途径比“脱氢”途径更具优势。一方面表现在A08和C03上芥酸的2个主效QTL对于油酸也是主效位点,可解释油酸表型变异的28.28%-43.68%,而A05上亚油酸的主效QTL对于油酸是微效位点,只解释油酸表型变异的3.8%-5.7%;另一方面,A08染色体上芥酸的主效QTL同时也是亚油酸的QTL,可解释亚油酸表型变异的8.91%,而亚油酸在A05上的主效QTL并不能解释芥酸含量的表型变异,这就表明,“加碳”途径制约着亚油酸的含量,而“脱氢”途径则对芥酸的含量影响不大。在育种工作中,对亚油酸含量的提高,需要阻断油酸向芥酸转化的途径;而高油酸的获得,则需要对2条代谢途径同时进行抑制。所以针对这些区段的深入研究和挖掘为改善油菜脂肪酸组成奠定基础。
3.3 脂肪酸代谢候选基因
本研究在油菜籽脂肪酸组分QTL置信区间内发现22个脂肪酸代谢相关基因。这些候选基因主要集中在A05、A08和C03染色体上,其中位于A05染色体上的基因FAD2(BnaA05g26900D)编码脂肪酸脱氢酶,是调控多不饱和脂肪酸合成的关键基因[39]。其中坐落于A08和C03染色体上的同源基因FAE1(BnaA08g11130D和BnaC03g65980D)是参与长链脂肪酸合成的关键基因,已被很多****研究报道[40,41,42],这表明我们的QTL定位是可靠的。在A08和C03染色体上筛选的另外2个同源基因KCS17(BnaA08G11140D和BNAC03G66040D)和FAR3(BnaA08G11440D和BNAC03G66380D),通过各自编码一种醇形成的脂肪酰辅酶A还原酶参与饱和脂肪酸的生物合成[43,44]。位于A01染色体上的基因HAL3A(BnaA01g27100D)和HAL3B(BnaA01g27120D)参与辅酶A的生物合成,有研究[45]表明,辅酶A可与醋酸盐结合为脂肪酸从头合成的前体物质乙酰辅酶A。在A02染色体上的候选基因BnaA02g13270D与拟南芥基因LACS9同源,LACS9已被证实通过催化脂肪酸、ATP和辅酶A形成酰基辅酶A,参与拟南芥籽油的生物合成[46]。此外,本研究还在QTL置信区间内发现许多未知功能基因,可能存在油菜特有的脂肪酸代谢功能基因,有待进一步挖掘。4 结论
在2个环境中共检测到位于A01、A02、A05、A08、A09、A10、C03和C07染色体上的23个脂肪酸组分QTL位点,与硬脂酸、油酸、亚油酸、亚麻酸、廿碳烯酸和芥酸含量相关的QTL位点分别为6、3、4、5、2和3个,其中在A05、A08和C03染色体上发现多种脂肪酸含量的QTL“富集区”,可作为改善油菜籽脂肪酸组成的重要区段。通过与拟南芥脂肪酸代谢基因的比对分析,在QTL置信区间内筛选到22个脂肪酸代谢相关候选基因。(责任编辑 李莉)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
.
[本文引用: 2]
.
[本文引用: 1]
[本文引用: 1]
.
[本文引用: 1]
.
DOI:10.1097/WNQ.0b013e3181644e82URLPMID:20102727 [本文引用: 1]

Triacylglycerols (TAGs) constitute a highly efficient form of energy storage. In seeds of angiosperms, they can act as a reserve of carbon and energy allowing to fuel post-germinative seedling growth until photosynthesis becomes effective. They also constitute the economic value of seeds in many crops. In the past years, extensive tools allowing the molecular dissection of plant metabolism have been developed together with analytical and cytological procedures adapted for seed material. These tools have allowed gaining a comprehensive overview of the metabolic pathways leading to TAG synthesis. They have also unravelled factors limiting oil production such as metabolic bottlenecks and light or oxygen availability in seed tissues. Beyond these physiological aspects, accumulation of TAGs is developmentally regulated in seeds. The oil biosynthetic process is initiated at the onset of the maturation phase, once embryo morphogenesis is achieved. A wealth of recent studies has shed new lights on the intricate regulatory network controlling the seed maturation phase, including reserve deposition. This network involves a set of regulated transcription factors that crosstalk with physiological signaling. The knowledge thus acquired paves the way for the genetic engineering of oilseed crops dedicated to food applications or green chemistry.
[本文引用: 2]
DOI:10.1007/s00122-012-1849-zMagsci [本文引用: 1]

High oleic acid soybeans were produced by combining mutant FAD2-1A and FAD2-1B genes. Despite having a high oleic acid content, the linolenic acid content of these soybeans was in the range of 4-6 %, which may be high enough to cause oxidative instability of the oil. Therefore, a study was conducted to incorporate one or two mutant FAD3 genes into the high oleic acid background to further reduce the linolenic acid content. As a result, soybean lines with high oleic acid and low linolenic acid (HOLL) content were produced using different sources of mutant FAD2-1A genes. While oleic acid content of these HOLL lines was stable across two testing environments, the reduction of linolenic acid content varied depending on the number of mutant FAD3 genes combined with mutant FAD2-1 genes, on the severity of mutation in the FAD2-1A gene, and on the testing environment. Combination of two mutant FAD2-1 genes and one mutant FAD3 gene resulted in less than 2 % linolenic acid content in Portageville, Missouri (MO) while four mutant genes were needed to achieve the same linolenic acid in Columbia, MO. This study generated non-transgenic soybeans with the highest oleic acid content and lowest linolenic acid content reported to date, offering a unique alternative to produce a fatty acid profile similar to olive oil.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.3864/j.issn.0578-1752.2016.16.002Magsci [本文引用: 1]

【Objective】The present research aimed to explore the major QTL controlling the flowering time in European and Chinese rapeseed materials, to analyze the genetic influence of flowering time on QTL for 1000-seeds weight, and thus to provide available information for breeding purpose.【Method】The doubled haploid (DH) Sollux/Gaoyou population with 282 lines was used. The data set of flowering time was obtained from nine environments and over seven years. QTL identification of flowering time was performed using WinQTLCart 2.5. The candidate genes underlining QTLs were screened out by transcriptome analysis using RNA-Seq and alignment between QTL regions and Arabidopsis. Further, conditional QTL estimation was adopted to dissect the genetic relationships between flowering time and seed weight. Finally, using selected DH lines with extreme phenotypes of flowering time, an association evaluation between marker genotypes and phenotypes of flowering time was performed. 【Result】 Seven major QTLs were detected, which showing significant at least in three environments. Their additive effects ranged from 0.58-3.85 days and together accounted for around 84% of the phenotypic variation in population. The sum of eight pairs of epistatic loci (additive × additive) accounted for 41.8% of the total additive effects. QTL by environmental interactions were significant only in few environments with small amount of genetic effects. Four critical orthologous genes of Arabidopsis thaliana for flowering time were mapped in the peak positions of three most significant QTL regions (qFTA2, qFTC2, and qFTC6). It provides valuable information to anchor candidate genes underling QTL. The conditional QTL analysis revealed large impact of flowering time on seed weight in four QTLs (qSWA2, qSWA3, qSWA4, and qSWC2). This partly explained the significant negative correlation between flowering time and 1000-seed weight. While the most important two (qSWA7 and qSWC8) showed independent without being interfered. Six markers linked with three major QTLs showed good fitness between marker genotypes and trait phenotypes (70%-100%), indicating their potentials for breeding purpose. The results demonstrated that the combination of early flowering alleles from qFTA2, qFTC2 and qFTC6 by marker assistant selection of ZAAS548, DNAPL, ZAAS619sa, ZAAS616s, ZAAS846a and C6SGFLO-22 induced not only early flowering but also significantly increased 1000-seed weight, while the oil content and seeds per silique between two extreme flowering time groups showed almost the same.【Conclusion】All seven QTLs of flowering time showed Chinese parent Gaoyou induced early flowering. Four important candidate genes homologous to Arabidopsis controlling flowering time (FT, FLC, AP1, and FY) were physically aligned and mapped underlining the peak positions of the three major QTL qFTA2, qFTC2 and qFTC6. The results indicated that the four loci corresponding to seed weight were genetically influenced by flowering time, however, the most important two (qSWA7 and qSWC8) were independent. Six markers linked to the 3 major QTL were of potentials in the practical breeding program.
[本文引用: 1]
[本文引用: 1]
.
[本文引用: 1]
.
[本文引用: 1]
.
[本文引用: 1]
.
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
DOI:10.1007/s11032-007-9113-yMagsci [本文引用: 2]

<a name="Abs1"></a>Increasing oil content and improving the fatty acid composition in the seed oil are important breeding goals for rapeseed (<i>Brassica napus</i> L.). The objective of the study was to investigate a possible relationship between fatty acid composition and oil content in an oilseed rape doubled haploid (DH) population. The DH population was derived from a cross between the German cultivar Sollux and the Chinese cultivar Gaoyou, both having a high erucic acid and a very high oil content. In total, 282 DH lines were evaluated in replicated field experiments in four environments, two each in Germany and in China. Fatty acid composition of the seed oil was analyzed by gas liquid chromatography and oil content was determined by NIRS. Quantitative trait loci (QTL) for fatty acid contents were mapped and their additive main effects were determined by a mixed model approach using the program QTLMapper. For all fatty acids large and highly significant genetic variations among the genotypes were observed. High heritabilities were determined for oil content and for all fatty acids (<i>h</i> <sup>2</sup> = 0.82 to 0.94), except for stearic acid content (<i>h</i> <sup>2</sup>= 0.38). Significant correlations were found between the contents of all individual fatty acids and oil content. Closest genetic correlations were found between oil content and the sum of polyunsaturated fatty acids (18:2 + 18:3; <i>r</i> <sub>G </sub>= −0.46), the sum of monounsaturated fatty acids (18:1 + 20:1 + 22:1; <i>r</i> <sub>G </sub>= 0.46) and palmitic acid (16:0; <i>r</i> <sub>G </sub>= −0.34), respectively. Between one and eight QTL for the contents of the different fatty acids were detected. Together, their additive main effects explained between 28% and 65% of the genetic variance for the individual fatty acids. Ten QTL for fatty acid contents mapped within a distance of 0 to 10 cM to QTL for oil content, which were previously identified in this DH population. QTL mapped within this distance to each other are likely to be identical. The results indicate a close interrelationship between fatty acid composition and oil content, which should be considered when breeding for increased oil content or improved oil composition in rapeseed.
..
[本文引用: 1]
[本文引用: 1]
.
[本文引用: 2]
.
[本文引用: 1]
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
[本文引用: 4]
[本文引用: 4]
[本文引用: 4]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00122-006-0411-2Magsci [本文引用: 1]

<a name="Abs1"></a>We have developed a new DH mapping population for oilseed rape, named TNDH, using genetically and phenotypically diverse parental lines. We used the population in the construction of a high stringency genetic linkage map, consisting of 277 loci, for use in quantitative genetic analysis. A proportion of the markers had been used previously in the construction of linkage maps for <i>Brassica</i> species, thus permitting the alignment of maps. The map includes 68 newly developed Sequence Tagged Site (STS) markers targeted to the homologues of defined genes of <i>A. thaliana</i>. The use of these markers permits the alignment of our linkage map with the <i>A. thaliana</i> genome sequence. An additional 74 loci (31 newly developed STS markers and 43 loci defined by SSR and RFLP markers that had previously been used in published linkage maps) were added to the map. These markers increased the resolution of alignment of the newly constructed linkage map with existing <i>Brassica</i> linkage maps and the <i>A. thaliana</i> genome sequence. We conducted field trials with the TNDH population at two sites, and over 2 years, and identified reproducible QTL for seed oil content and erucic acid content. The results provide new insights into the genetic control of seed oil and erucic acid content in oilseed rape, and demonstrate the utility of the linkage map and population.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00122-007-0685-zMagsci [本文引用: 1]

<a name="Abs1"></a>The <i>fatty acid elongase 1</i> (<i>FAE1</i>) gene is a key gene in the erucic acid biosynthesis in rapeseed. The complete coding sequences of the <i>FAE1</i> gene were isolated separately from eight high and zero erucic acid rapeseed cultivars (<i>Brassica napus</i> L.). A four base pair deletion between T1366 and G1369 in the <i>FAE1</i> gene was found in a number of the cultivars, which leads to a frameshift mutation and a premature stop of the translation after the 466th amino acid residue. This deletion was predominantly found in the C-genome and rarely in the A-genome of <i>B. napus</i>. Expression of the gene isoforms with the four base pair deletion in a yeast system generated truncated proteins with no enzymatic activity and could not produce very long chain fatty acids as the control with an intact <i>FAE1</i> gene did in yeast cells. In the developing rape seeds the <i>FAE1</i> gene isoforms with the four base pair deletion were transcribed normally but failed to translate proteins to form a functional complex. The four base pair deletion proved to be a mutation responsible for the low erucic acid trait in rapeseed and independent from the point mutation reported by Han et al. (Plant Mol Biol 46:229–239, <cite>2001</cite>).
.
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
