
 , 郑磊1,2
, 郑磊1,2

1. 合肥工业大学 食品与生物工程学院,安徽 合肥 230009;
2. 安徽省现代农业产业技术体系 农业环境与食品安全研究室,安徽 合肥 230009
收稿日期:2020-08-16;接收日期:2020-10-20;网络出版时间:2020-12-22
基金项目:国家自然科学基金(No. 31972134),安徽省科技重大专项(No. 201903b06020003),合肥工业大学“黄山****”****基金(No. 407-037019) 资助
摘要:SlPAL5基因是酚类化合物代谢的关键基因。UV-C辐照可以有效提高番茄果实中酚类化合物的含量。因此研究调控SlPAL5基因表达的转录因子,对于进一步阐明UV-C诱导番茄果实酚类化合物合成的调控机制具有重要意义。文中通过构建番茄酵母单杂交文库,利用酵母单杂交技术筛选调控酚类化合物合成关键基因SlPAL5表达的转录因子。通过测序和Blast同源性分析得到转录因子SlERF7,并证实SlERF7可以与SlPAL5的启动子相互作用。另外,UV-C辐照可以显著提高SlERF7的表达水平。结果表明受UV-C辐照诱导的SlERF7可能参与了SlPAL5的转录调控,为研究UV-C诱导番茄果实酚类化合物合成的调控机制提供了基础。
关键词:SlPAL5番茄酵母单杂交转录因子UV-C辐照SlERF7
Screening for UV-C irradiation-enhanced transcription factors that regulate the metabolism of phenolic compounds in tomato fruit
Wenzhuo Hao1, Huanhuan Zheng1, Changhong Liu1

 , Lei Zheng1,2
, Lei Zheng1,2

1. School of Food and Biological Engineering, Hefei University of Technology, Hefei 230009, Anhui, China;
2. Research Laboratory of Agricultural Environment and Food Safety, Anhui Modern Agricultural Industry Technology System, Hefei 230009, Anhui, China
Received: August 16, 2020; Accepted: October 20, 2020; Published: December 22, 2020
Supported by: National Natural Science Foundation of China (No. 31972134), Key Science and Technology Specific Projects of Anhui Province, China (No. 201903b06020003), Funds for Huangshan Professorship of Hefei University of Technology (No. 407-037019)
Corresponding author: Changhong Liu. Tel: +86-551-62901516; E-mail: changhong22@hfut.edu.cn;
Lei Zheng. Tel: +86-551-62901516; E-mail: lzheng@hfut.edu.cn.
Abstract: Solanum lycopersicum phenylalanine ammonia-lyase 5 (SlPAL5) gene regulates the metabolism of phenolic compounds. The study of transcription factors that regulate the expression of SlPAL5 gene is of great significance to elucidate the regulatory mechanism underlying the biosynthesis of phenolic compounds in tomato fruit induced by UV-C irradiation. Here, yeast one-hybrid library of tomato fruit was constructed, and the yeast one-hybrid technology was used to screen the transcription factors that regulate the expression of SlPAL5, the key gene related to the synthesis of phenolic compounds in tomato fruit. As a result, a transcription factor, SlERF7, was obtained and sequenced, followed by the blast homology analysis. Further experiments confirmed that SlERF7 interacted with the promoter of SlPAL5 gene. In addition, UV-C irradiation significantly increased the expression level of SlERF7. These results indicate that SlERF7, which is regulated by UV-C irradiation, might be involved in regulating the transcription of SlPAL5, which provided foundations for further studying the regulation mechanism of the biosynthesis of phenolic compounds in tomato fruit induced by UV-C irradiation.
Keywords: SlPAL5tomatoyeast one-hybridtranscription factorUV-C irradiationSlERF7
IntroductionPlant growth, development and its response to the environment are regulated by gene expression. In addition, the gene expression is regulated by transcription factors[1]. Transcription factors could activate or inhibit transcription by identifying cis-acting elements in the promoter region of the target gene[2]. The metabolism of phenolic compounds is not only influenced by the related genes, but also related to the regulation of transcription factors[3]. Transcription factors can regulate the expression of genes encoding different enzymes in the metabolic pathways of phenolic compounds to regulate the metabolism of phenolic compounds[4-5]. Studies have shown that the MYB transcription factor family plays a crucial part in the regulation of plant phenolic metabolism[6]. Stracke et al[4] have also confirmed that the MYB transcription factors, MYB11, MYB12 and MYB111, of Arabidopsis thaliana can regulate the genes involved in the biosynthesis of phenolic compounds. In addition, UV-B irradiation can induce the expression of MYB1 transcription factor in carrot[7]. Other studies have reported that ERF and PAP1 can regulate the biosynthesis of phenolic acids[8-9]. In addition, the MYC2 transcription factor of Salvia miltiorrhiza could also induce the accumulation of phenolic compounds in Salvia miltiorrhiza. Yang et al[10] and Mohanty et al[11] have found that bHLH transcription factor is important in the synthesis of phenolic compounds. Furthermore, the transcription factor LBD50 of Salvia miltiorrhiza LBD can be induced by jasmonic acid to regulate the metabolism of phenolic compounds[12].
The wavelength range of UV-C is from 200 nm to 275 nm. UV-C irradiation can destroy DNA and produce pyrimidine dimers to interrupt the transcription and translation of DNA in microorganisms[13]. UV-C irradiation is an effective method to improve the quality of fruits and vegetables during storage[14]. UV-C irradiation can cause cell damage and DNA damage at high doses, while low dose treatment will bring a series of beneficial physiological effects. For example, UV-C irradiation can improve the sensory quality and nutritional quality of fruits and vegetables, and perfect the edible value of fruits and vegetables[15-17].
UV-C irradiation can cause DNA damage in many organisms. When plants are exposed to UV-C irradiation, they would induce the production of UV-absorbing phenolic compounds to avoid the harm of UV-C irradiation[18]. Phenylalanine ammonia-lyase (PAL) is a key enzyme in the synthesis of phenolic compounds and plays an important role in phenolic synthesis[19]. PAL5 is the most actively transcribed gene among various PAL genes[20], and our previous study has also shown that the expression of SlPAL5 gene was significantly up-regulated after UV-C irradiation. Especially, UV-C irradiation could promote the biosynthesis of phenolic compounds in tomato fruit via inducing the expression of several genes related to the biosynthesis of phenolic compounds, such as SlC4H, Sl4CL, SlCHS2, SlCHI, SlF3H and SlFLS[21]. Therefore, this study was aimed at further exploring the molecular mechanism underlying phenolic accumulation induced by UV-C irradiation in tomato fruit and at the identification of upstream transcription factors regulating SlPAL5 gene expression using the yeast one-hybrid method.
1 Materials and methods1.1 MaterialsTomato fruit (Solanum lycopersicum, cv. Wanza 15) were harvested at the pink stage from a commercial greenhouse in Hefei, Anhui province, China.
Vectors used in this study were as follows: pHIS2 vector, pGADT7 vector, pGAD53m vector, pHIS2-p53 vector, pDNOR222 vector.
Competent cells used in this study were as follows: E. coli DH5α competent cells, E. coli DH10B competent cells, yeast Y187 competent cells, yeast AH109 competent cells.
The information of main experimental reagents were as follows: LB (Luria-Bertani) broth, LB Nutrient Agar (Beijing Aoboxing Bio-tech Co., Ltd., China); SD/-Trp with agar, SD/-Trp-His with agar, SD/-Trp-Leu with agar, SD/-Trp-Leu-His with agar, 3-AT (3-amino-1, 2, 4-triazole) (Beijing Coolaber Technology Co., Ltd., China); Kanamycin (Beijing Solarbio Technology Co., Ltd., China); PEG, LiAc (Shanghai Weidi Biotechnology Co., Ltd., China); Agarose B, Low EEO (Sangon Biotech (Shanghai) Co., Ltd., China); DNA extraction kit, Gel recovery kit, TIANprep Mini Plasmid Kit (Tiangen Biotech (Beijing) Co., Ltd., China); DNA Ligation Kit (BioLion Technology Co., Ltd., China); CloneMinerTM II cDNA Library Construction Kit, FastTrackTM MAG Maxi mRNA Isolation Kit, PureLink? HiPure Plasmid Filter Midiprep Kit, BP Clonase?Ⅱ enzyme mix, LR ClonaseTM Ⅱ Enzyme Mix (Life Technologies); EcoR Ⅰ, Sac Ⅰ, TaKaRa MiniBEST Plant RNA Extraction Kit, PrimeScriptTMRT reagent Kit with gDNA Eraser (Perfect Real Time), SYBR? Premix Ex TaqTM (TaKaRa, Dalian, China).
1.2 Promoter sequence analysis of SlPAL5The 2.0 kb promoter sequence of the SlPAL5 (Gene ID: 101244220) was searched on NCBI. Then the primers were designed with two enzyme cutting sites of EcoR Ⅰ and Sac Ⅰ, and synthesized by Sangon Biotech Co., Ltd., (China). The SlPAL5 primer sequences were shown in Table 1.
Table 1 The primers used in this paper
| Number | Primer name | Forward/Reverse primer (5′–3′) |
| 1 | SlPAL5 | AAGAATTCTCCTTCTAGGGTTGGGTTGAGTTG |
| CCGAGCTCGACCAACAAAAAATGGTTTGATTTG | ||
| 2 | M13 | GTTGTAAAACGACGGCCAG |
| CAGGAAACAGCTATGAC | ||
| 3 | pDONR222 | GTAAAACGACGGCCAG |
| CAGGAAACAGCTATGAC | ||
| 4 | T7 | TAATACGACTCACTATAGGG |
| ADR | AGATGGTGCACGATGCACAG | |
| 5 | SlERF7 | GATAAGGTTCCGTGGAGTTCG |
| TGAAAGAGGAAGAAGCGATGT |
表选项
1.3 Construction of bait vectorsAccording to the SlPAL5 primer sequences, using tomato genomic DNA as a template, the core fragment of the SlPAL5 gene promoter region was obtained by PCR amplification. The genomic DNA of tomato fruit was extracted by plant DNA extraction kit. The PCR procedure was as follows: 98 ℃ for 5 min, followed by 35 cycles of 30 s at 98 ℃, 30 s at 55 ℃, and 57 s at 72 ℃, then 5 min at 72 ℃. After separated by 1% agarose with Sub-Cell? GT Agarose Gel Electrophoresis Systems (Sub-Cell? GT, Bio-Rad Laboratories, Inc.), the target fragments were recovered by the gel recovery kit. Then SlPAL5 gene promoter and pHIS2 vector were digested at 37 ℃ overnight with EcoR Ⅰ and Sac Ⅰ and ligated with DNA Ligation Kit at 16 ℃ for 1 h to form a recombinant bait vector pHIS2-SlPAL5. The recombinant bait pHIS2-SlPAL5 was transformed into E. coli DH5α competent cells with Electroporator (TX ECM 630) at 2.0 kV for 3 s, then the transformants were randomly picked and inoculated in LB broth which contained kanamycin. After incubated at 37 ℃ and 250 r/min for 16 h with constant temperature cultivation oscillator (HNY-200D, Tianjin Honour Instrument Co., Ltd., China), 1 μL of the bacterial solution was used for PCR with pHIS2 vector universal primers. The primer sequences were shown in Table 1.
Then the PCR products were identified by 1% agarose gel electrophoresis and the cloning of positive bands was sequenced. Then TIANprep Mini Plasmid Kit was used to extract pHIS2-SlPAL5.
1.4 Transform the bait vectors to Y187 yeast competent cellsAdd 5 μL of plasmid, 5 μL of ssDNA, 240 μL of 50% PEG and 36 μL of 1 mol/L LiAc to 50 μL of yeast competent cells, then vibrate vigorously with Vortex mixer (Vortex 3000, WIGGENS) for 1 min until completely mixed. Then the mixture was incubated in electric constant temperature water bath (HH-2, Jiangsu Jincheng Guosheng Experiment Instrument Factor) at 30 ℃ for 30 min and heat shock at 42 ℃ for 25 min, and finally resuscitated in a water bath at 30 ℃ for 1 h. After centrifuged with high speed refrigerated centrifuge (Sorvall Legend Micro 21R, Thermo Fisher Scientific) at 4 000× g for 5 min, the supernatant was discarded. The bacteria were suspended with 200 μL sterile water and incubated in SD/-Trp with agar medium at 30 ℃ for 3–4 d.
1.5 3-AT concentration screeningBecause the protein of yeast itself may weakly interact with the bait plasmid, resulting in the appearance of false positive clones, the inhibitor 3-AT should be used to inhibit this non-specific binding. Yeast containing bait plasmids which were verified correctly in 1.4 were plated and cultured on yeast SD/-Trp-His with agar medium with 3-AT concentrations of 0, 25, 50, 75 and 100 mmol/L, and the screening results were observed.
1.6 Construction of yeast one-hybrid libraryThe total RNA in tomato fruit was extracted using CTAB method, and then the mRNA in the extracted total RNA was isolated using FastTrackTM MAG Maxi mRNA Isolation Kit. The primary library was constructed using CloneMinerTMⅡ cDNA Library Construction Kit. The cDNA and pDONR222 vector were recombined with BP Clonase?Ⅱ enzyme mix, and transformed into E. coli DH10B competent cells, the transformed products were cultured, glycerol bacteria were preserved, and the quality of the primary library was identified.
Methods of identification of the primary library capacity were as follows: 10 μL of the transformed bacterial solution was diluted by 1 000 folds, 50 μL of it was taken out and coated on LB plate (containing corresponding resistance) and counted after cultured at 37 ℃ for 12–16 h. The library capacity was calculated using the following equations.
 | (1) |
 | (2) |
The PCR procedure was as follows: 94 ℃ for 5 min, followed by 35 cycles of 30 s at 94 ℃, 30 s at 58 ℃, and 2 min at 72 ℃, then 5 min at 72 ℃.
PureLink? HiPure Plasmid Filter Midiprep Kit was used to extract the plasmid in the primary library bacterial solution, and LR ClonaseTM Ⅱ Enzyme Mix was used to recombine the extracted plasmid with the pGADT7-Rec2-DEST vector and transform into AH109 yeast competent cells. After cultured, the glycerol bacteria were preserved, and the quality of the yeast one-hybrid library was identified.
Methods of identification of the yeast one-hybrid were same as the primary library. The T7 and ADR primer sequences were shown in Table 1.
The library quality was determined from three aspects: the library capacity, the average insert length and the recombination rate. The library capacity is the number of all positive clones contained in the library. For the cDNA library using E. coli as the host strain, the larger the library size, the greater the total number of cDNA fragments contained in the library. The library capacity is a significant index to measure the library quality, which reflects the integrity of the mRNA in the samples. The average inserted fragment length is the average of the fragment length of the products obtained by random colony PCR, which reflects the sequence integrity of the recombinant cDNA fragments in the library. The fragments are long enough to reflect the natural structure of the gene as much as possible, and it is easier to obtain the complete sequence and functional information of the target gene in the library. The recombination rate reflects the positive rate of all clones in the library, that is, the rate of clones containing recombination cDNA fragments.
1.7 Yeast one-hybrid screeningThe yeast transformants containing the correct pHIS2-SlPAL5 bait vector were used to prepare competent cells, and the yeast one-hybrid library plasmid pGADT7-HKL330 was transferred into it, and coated on SD/-Trp-Leu-His with agar plate containing the optimal concentration of 3-AT for 3–4 d. At the same time, the pGAD53m vector was transferred into the yeast containing pHIS2 vector as a negative control, and the pGAD53m vector was transferred into the yeast containing pHIS2-p53 vector as a positive control. In order to eliminate the interference of the background growth clones, at the third day of culture, the screen plate was photocopied and cleaned with flannelette and continued to culture for 7–14 d. The transformants were selected in batches for further detection. After photocopying and cleaning, the culture was continued for 7 d. 32 positive clones transformants were selected from the screen plate and cultured in SD/-Trp-Leu with agar medium for 2–3 d. Then the 32 positive clones were diluted with sterile water and cultured in the SD/-Trp-Leu medium and SD/-Trp-Leu-His with 100 mmol/L 3-AT, respectively. After incubated at 30 ℃ for 3–4 d, the results were observed.
1.8 Verification of interaction between SlPAL5 promoter fragment and SlERF7The pGADT7-SlERF7 vector was constructed and transferred into yeast competent cells containing the correct pHIS2-SlPAL5 vector as a verification group. Yeast cells containing only the bait vector were used as blank control, the positive and negative controls were the same as 1.4. After transformed, transformants were cultured in SD/-Trp-Leu. Afterwards, the transformants grown on the SD/-Trp-Leu plates were diluted with sterile water and seeded to SD/-Trp-Leu and SD/-Trp-Leu-His mediums containing 75 mmol/L 3-AT, respectively. Then the plates were cultured at 30 ℃ for 4 d, and the transformation results were observed.
1.9 Effect of UV-C irradiation on the expression level of SlERF7To find the expression of SlERF7 in tomato fruit after UV-C radiation, the breaker tomato fruit were irradiated with 4 kJ/m2 UV-C and stored at 14 ℃ and 95% relative humidity for 35 d. Samples were taken every 7 days to analyze the expression of SlERF7 by real-time quantitative PCR. Total RNA was extracted from breaker tomato fruit with TaKaRa MiniBEST Plant RNA Extraction Kit according to the instruction and cDNA was synthesized by PrimeScript RT reagent Kit. Real-time quantitative PCR was performed using the cDNA, and the SlERF7 primer sequences used were shown in Table 1.
Real-time quantitative PCR was performed using SYBR? Premix Ex TaqTM in Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA), and the methods of real-time quantitative PCR were according to Liu et al[22]. Tomato fruit without any treatment were used as the control group.
2 Results2.1 Construction of bait vectorsAfter transformed the recombinant bait pHIS2-SlPAL5 into E. coli DH5α competent cells, the transformants were randomly picked, inoculated in LB broth medium containing kanamycin and incubated at 37 ℃, 250 r/min for 16 h. 1 μL of the bacterial solution was used for PCR with pHIS2 plasmid primers, the agarose gel electrophoresis results showed that a fragment of about 2.0 kb was amplified (Fig. 1). The positive clone was confirmed to be correct after sequenced. The results showed that pHIS2-SlPAL5 was constructed correctly.
 |
| Fig. 1 Colony PCR of bait vector pHIS2-SlPAL5. Lane 1–6 were amplified products of bait vector pHIS2-SlPAL5 colonies selected randomly. "M" represents DNA marker. The gel electrophoresis results showed that the fragment size of pHIS2-SlPAL5 was about 2 kb. Therefore, the construction of bait vector pHIS2-PAL5 was correct, which can be used for follow-up experiments. |
| 图选项 |
2.2 Results of 3-AT concentration screening3-AT is a competitive inhibitor of yeast HIS3 protein synthesis, which is used to inhibit the leakage expression of HIS3 gene. As shown in Fig. 2, the growth value of the positive control was lower than that of the plate without 3-AT, and the growth rate would decrease as the concentration of 3-AT increased, but it was still significantly different from the negative control. Because the HIS3 reporter gene was not activated, the growth rate of negative control on the 3-AT supplemented plate was significantly reduced, and the higher the 3-AT concentration, the fewer the number of transformants. From the results of self-activation detection, the transformants containing bait vectors could not be inhibited significantly on 3-AT deficient plates with 25 mmol/L and 50 mmol/L respectively (the results were not shown), while the growth of transformants contained pHIS2-SlPAL5 bait was significantly inhibited on the plates containing 75 mmol/L and 100 mmol/L 3-AT, and the growth ratio was consistent with that of the negative control, which indicated that the HIS3 reporter gene was not activated. Therefore, the 3-AT concentration of the library screening was determined to be 75 mmol/L.
 |
| Fig. 2 Screening of 3-AT concentration. The positive control could grow normally on the plate with 3-AT inhibitor due to the activation of HIS3 reporter gene, and the number of transformants was the same as that without 3-AT, theoretically. However, the observed growth value of the positive control was about 10% lower than that of the plate without 3-AT, and the growth rate would decrease as the concentration of 3-AT increased, but it was still significantly different from the negative control. As the HIS3 reporter gene was not activated in the negative control, the growth of the negative control on the plate with 3-AT was significantly reduced, and the higher the concentration of 3-AT, the less the number of transformants. |
| 图选项 |
2.3 Construction and quality identification of primary libraryThe total RNA of tomato fruit that extracted by CTAB method was detected by agarose gel electrophoresis. The results showed that the bands of 28S rRNA and 18S rRNA were clear (Fig. 3A), and the brightness of 28S rRNA was about twice of 18S rRNA. The total RNA was of good quality and without degradation and contamination, so it could be used to construct cDNA library.
 |
| Fig. 3 Detection of total RNA by agarose gel electrophoresis. It was shown that the bands of 28S rRNA and 18S rRNA were clear, and the brightness of 28S rRNA was about twice of 18S rRNA (A). Identification of the primary library capacity. Coating amount of bacterial liquid was 50 μL and 150 clones were grown on LB plate. The total number of colonies was 1.2×107 according to the calculation formula (B). Identification of the insert length of primary library. 24 clones were randomly selected for colony PCR identification, the length of inserted fragments was about 800–3 000 bp and the recombination rate was 100% (C). |
| 图选项 |
The result of colony growth on LB plate was shown in Fig. 3B and 150 clones were grown. According to the calculation formula, the primary library capacity was above 106 CFU. Therefore, the primary library quality reached the requirements. A total of 24 clones were randomly selected for colony PCR identification, the length of inserted fragments was about 800–3 000 bp and the recombination rate was 100% (Fig. 3C).
2.4 Construction and quality identification of yeast one-hybrid libraryThe cDNA primary library plasmid was transferred into the pGADT7-Rec2-DEST vector and transformed into the AH109 yeast competent cells. A total of 10 μL of transformed yeast cells solution was diluted by 1 000 folds, and 50 μL of them was taken out and 180 clones were grown on LB medium (Fig. 4A). The yeast one-hybrid library capacity was more than 106 CFU and the results in Fig. 4B showed that the sizes of inserts were mainly distributed between 800 and 3 000 bp, and presented bright dispersive bands, indicating that the yeast one-hybrid library was successfully constructed.
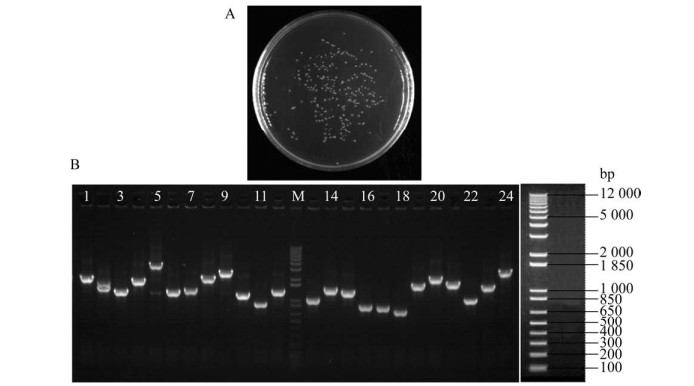 |
| Fig. 4 Identification of the yeast one-hybrid capacity. Coating amount of bacterial liquid was 50 μL and 180 clones were grown on LB plate. The total number of colonies was 1.44×107 according to the calculation formula (A). Identification of the insert length of yeast one-hybrid capacity. The results showed that the insert segments were mainly distributed between 800 and 3 000 bp, and presented bright dispersive bands (B). |
| 图选项 |
2.5 Yeast one-hybrid screeningThe results in Fig. 5 showed that the positive control could grow normally on the screening medium without 3-AT and with 3-AT, while the negative control could not activate the HIS3 reporter gene, so it could grow normally on the screening medium without 3-AT, but could not grow on the screening medium without histidine and with 3-AT. Therefore, among the 32 initial positive clones, the HIS3 reporter gene was activated, so the 32 clones could thrive on the screening medium with 3-AT. The above results showed that the 32 initial positive clones could activate HIS3 reporter gene.
 |
| Fig. 5 Detection of the activation of HIS3 reporter gene, "+" represents positive control and "–" represents negative control. The 32 positive clones were diluted with sterile water and cultured in the SD/-Trp-Leu medium and SD/-Trp-Leu-His with 100 mmol/L 3-AT, respectively. After incubated at 30 ℃ for 3–4 d, it was observed that 32 clones and the positive control could grow normally on the screening medium without 3-AT and with 3-AT, while the negative control could grow normally on the screening medium without 3-AT rather than on the screening medium without histidine and with 3-AT. |
| 图选项 |
In order to identify the genes of the 32 positive clones, all positive clones were cultured in liquid medium SD/-Trp-Leu, and the yeast plasmid was extracted thereafter. Then the extracted yeast plasmids were transformed into the top10 competent cells for amplification. The PCR products were sequenced and analyzed by blasting these sequences against the GenBank database. The results showed that the 32 positive clones encoded different proteins. The number and function of these gene fragments were shown in Table 2, among which two transcription factors SlERF7 and SlASR1 were speculated to bind the cis-acting elements of SlPAL5 gene.
Table 2 Proteins interacting with SlPAL5 gene in tomato fruit
| Number | NCBI accession | NCBI description |
| 1 | NM_001317970.1 | Solanum lycopersicum cytochrome b6-f complex iron-sulfur subunit, chloroplastic (LOC101243864) |
| 2 | XM_004232289.3 | PREDICTED: Solanum lycopersicum 60S ribosomal protein L11-1 (LOC101262882) |
| 3 | NM_001310077.1 | Solanum lycopersicum NEDD8-conjugating enzyme Ubc12 (LOC543810), mRNA |
| 4 | XM_004248147.3 | PREDICTED: Solanum lycopersicum 60S ribosomal protein L11-1 (LOC101246764) |
| 5 | XM_010314008.2 | PREDICTED: Solanum lycopersicum uncharacterized LOC101260185 (LOC101260185) |
| 6 | XM_010320190.2 | PREDICTED: Solanum lycopersicum threonine synthase, chloroplastic (LOC101260521), transcript variant X1 |
| 7 | NM_001252115.2 | Solanum lycopersicum ethylene responsive factor (ERF7) |
| 8 | XM_004246033.3 | PREDICTED: Solanum lycopersicum 40S ribosomal protein S6 (LOC101249290) |
| 9 | NM_001247015.2 | Solanum lycopersicum GTP-binding protein (ypt2), transcript variant 1 |
| 10 | XM_004235287.3 | PREDICTED: Solanum lycopersicum trans-resveratrol di-O-methyltransferase-like (LOC101263799) |
| 11 | XM_004245476.3 | PREDICTED: Solanum lycopersicum uncharacterized LOC101259182 (LOC101259182) |
| 12 | XM_004232364.3 | PREDICTED: Solanum lycopersicum protein N-lysine methyltransferase METTL21A (LOC101260984) |
| 13 | XM_004236103.3 | PREDICTED: Solanum lycopersicum pyridoxine/pyridoxamine 5′-phosphate oxidase 1, chloroplastic (LOC101259228), transcript variant X4 |
| 14 | XM_004229294.3 | PREDICTED: Solanum lycopersicum citrate synthase, mitochondrial (LOC101249011) |
| 15 | NM_001308186.1 | Solanum lycopersicum ACT domain-containing protein (LOC101267668), mRNA |
| 16 | BT013755.1 | Lycopersicon esculentum clone 132621F |
| 17 | XM_004245874.3 | PREDICTED: Solanum lycopersicum probable methyltransferase PMT21 (LOC101251208), transcript variant X4 |
| 18 | NM_001247208.2 | Solanum lycopersicum abscisic stress-ripening protein 1 (ASR1) |
| 19 | XM_004239709.3 | PREDICTED: Solanum lycopersicum serine/arginine-rich splicing factor RS2Z32 (LOC101253688), transcript variant X2 |
| 20 | XM_004245975.3 | PREDICTED: Solanum lycopersicum uncharacterized LOC101256209 (LOC101256209), transcript variant X1 |
| 21 | XM_004243154.3 | PREDICTED: Solanum lycopersicum protein MEI2-like 2 (LOC101247235) |
| 22 | XM_004231965.3 | PREDICTED: Solanum lycopersicum protein-lysine N-methyltransferase Mettl10 (LOC101263561) |
| 23 | NM_001306170.1 | Solanum lycopersicum stromal ascorbate peroxidase 7 (APX7) |
| 24 | XM_010328872.1 | PREDICTED: Solanum lycopersicum autophagy-related protein 8C (LOC101246095), transcript variant X1 |
| 25 | NM_001247385.2 | Solanum lycopersicum pathogenesis-related leaf protein 6 (PR1b1) |
| 26 | NM_001247686.2 | Solanum lycopersicum proteinase inhibitor Ⅰ (ER1) |
| 27 | XM_004244122.3 | PREDICTED: Solanum lycopersicum uncharacterized LOC101263228 (LOC101263228), transcript variant X2 |
| 28 | XM_010319984.2 | PREDICTED: Solanum lycopersicum uncharacterized LOC101255376 (LOC101255376) |
| 29 | NM_001347950.1 | Solanum lycopersicum THO complex subunit 4A (LOC101262878) |
| 30 | NM_001247474.2 | Solanum lycopersicum chitinase (CHI9) |
| 31 | XM_004237939.3 | PREDICTED: Solanum lycopersicum AT-hook motif nuclear-localized protein 17 (LOC101252483) |
| 32 | NM_001312890.1 | Solanum lycopersicum glucan endo-1, 3-beta-glucosidase B (LOC101261650) |
表选项
2.6 Verification of interaction between SlPAL5 promoter fragment and SlERF7ERFs (Ethylene responsive factors) are plant-specific transcription factors, which take a vital part in regulating plant growth and development and stress response[23]. Given that Severo et al[24] reported that UV-C irradiation regulated the production of ethylene by regulating the expression of ERF transcription factor. Therefore, the yeast-one hybrid was used to verify the interaction between the SlPAL5 gene promoter and transcription factor SlERF7. The results were shown in Fig. 6. In SD/-Trp-Leu medium, all the four transformants could grow normally. But in SD/-Trp-Leu-His medium containing 75 mmol/L 3-AT, only the positive control and the verification group could grow normally. It indicated that the target gene could be detected and the HIS3 reporter gene was activated in the verification group. Therefore, SlERF7 protein could interact with the promoter of SlPAL5 gene.
 |
| Fig. 6 Verification of the interaction between SlERF7 and SlPAL5 promoter, "1" represents blank control, "2" represents verification group, "+" represents positive control and "–" represents negative control. After transformed, transformants were cultured in SD/-Trp-Leu and SD/-Trp-Leu-His medium containing 75 mmol/L 3-AT, respectively, it was observed that all the four transformants could grow normally in SD/-Trp-Leu medium. But in SD/-Trp-Leu-His medium containing 75 mmol/L 3-AT, only the positive control and the verification group could grow normally. |
| 图选项 |
2.7 Effect of UV-C irradiation on the expression level of SlERF7The relative expression of transcription factor SlERF7 was examined in UV-C treated tomato fruit. It was shown that UV-C irradiation significantly increased the expression of SlERF7 on the 14th, 28th and 35th days of storage (P < 0.05), and at the end of storage, the expression of SlERF7 increased by 31.59% in comparison with the control (Fig. 7).
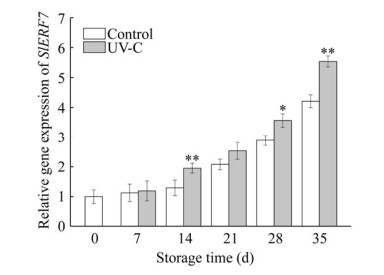 |
| Fig. 7 Effect of UV-C irradiation on expression of SlERF7 in postharvest tomato fruit. Results represent the expression level of SlERF7 in UV-C treated group and control group at different storage time. Error bars represent the standard deviation of the mean of three replicates. Statistical significance of the difference was confirmed according to Duncan's multiple range test at P < 0.05 (?:P < 0.05; ??: P < 0.01). |
| 图选项 |
3 DiscussionYeast one-hybrid technology is of great significance in the study of the interaction between DNA and protein[25]. The underlying principle is that a transcription factor can interact with a cis-acting element within the promoter of a target gene to activate the expression of the related reporter genes. The expression of the reporter gene often suggests the interaction of a transcription factor in question with the promoter of the targeted gene[26]. Therefore, through yeast one-hybrid, we can identify transcription factors that regulate the expression of related genes, and discover the molecular mechanisms underlying gene expression. Yeast one-hybrid technology is widely used to study the interaction between DNA and protein. Some transcription factors that regulate the expression of genes related to drought, low temperature, salinity and hormones have been isolated by yeast one-hybrid. For example, Tripathi et al[27] isolated transcription factors that have essential effect in drought signaling from the Arabidopsis thaliana. Wang et al[28] discovered transcription factors that play an important role in maize low temperature stress response. Tominaga-Wada et al[29] concluded that R3 MYB transcription factor regulated plant trichome and root-hair development of tomato (Solanum lycopersicum). Yoshida et al[30] found that AREB1, AREB2, and ABF3 were upregulated by ABA.
Tomato fruit contain many bioactive compounds beneficial to human health[31]. It is widely used as a model system for studying fruit development and maturation processes[32-33]. Furthermore, tomato fruit has been confirmed to contain higher levels of phenolic compounds[34-35]. UV-C irradiation is of great significance in the regulation of plant phenolic compounds metabolism[36-38]. PAL5 is a key gene in the process of phenolic compounds metabolism. Studies have shown that PAL5 gene plays an important regulatory role in plant abiotic stress response[39]. Screening the upstream transcriptional regulators of the key gene SlPAL5 involved in the phenolic compounds metabolism provided important information for studying the mechanism of phenolic compounds metabolism induced by UV-C. In this study, we discovered that SlERF7 transcription factor, which was upregulated by UV-C irradiation, was likely to regulate the transcription of SlPAL5 gene. This finding has contributed to the understanding of metabolism mechanism underlying the UV-C induced phenolic metabolism. Even so, the hypothesis still needs further experimental confirmation.
REFERENCES
| [1] | Yang HJ, Zhou Y, Zhang YN, et al. Identification of transcription factors of nitrate reductase gene promoters and NRE2 cis-element through yeast one-hybrid screening in Nicotiana tabacum. BMC Plant Biol, 2019, 19: 145. DOI:10.1186/s12870-019-1724-z |
| [2] | Zhang JZ. Overexpression analysis of plant transcription factors. Curr Opin Plant Biol, 2003, 6(5): 430-440. DOI:10.1016/S1369-5266(03)00081-5 |
| [3] | Ding K, Pei TL, Bai ZQ, et al. SmMYB36, a novel R2R3-MYB transcription factor, enhances tanshinone accumulation and decreases phenolic acid content in Salvia miltiorrhiza hairy roots. Sci Rep, 2017, 7: 5104. DOI:10.1038/s41598-017-04909-w |
| [4] | Stracke R, Ishihara H, Huep G, et al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J, 2007, 50(4): 660-677. DOI:10.1111/j.1365-313X.2007.03078.x |
| [5] | Ravaglia D, Espley RV, Henry-Kirk RA, et al. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol, 2013, 13: 68. DOI:10.1186/1471-2229-13-68 |
| [6] | Aharoni A, De Vos CHR, Wein M, et al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J, 2001, 28(3): 319-332. DOI:10.1046/j.1365-313X.2001.01154.x |
| [7] | Maeda K, Kimura S, Demura T, et al. DcMYB1 acts as a transcriptional activator of the carrot phenylalanine ammonia-lyase gene (DcPAL1) in response to elicitor treatment, UV-B irradiation and the dilution effect. Plant Mol Biol, 2005, 59(5): 739-752. DOI:10.1007/s11103-005-0910-6 |
| [8] | Huang Q, Sun MH, Yuan TP, et al. The AP2/ERF transcription factor SmERF1L1 regulates the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Food Chem, 2019, 274: 368-375. DOI:10.1016/j.foodchem.2018.08.119 |
| [9] | Zhang Y, Yan YP, Wang ZZ. The arabidopsis PAP1 transcription factor plays an important role in the enrichment of phenolic acids in Salvia miltiorrhiza. J Agric Food Chem, 2010, 58(23): 12168-12175. DOI:10.1021/jf103203e |
| [10] | Yang N, Zhou WP, Su J, et al. Overexpression of SmMYC2 increases the production of phenolic acids in Salvia miltiorrhiza. Front Plant Sci, 2017, 8: 1804. DOI:10.3389/fpls.2017.01804 |
| [11] | Mohanty B, Lakshmanan M, Lim SH, et al. Light-specific transcriptional regulation of the accumulation of carotenoids and phenolic compounds in rice leaves. Plant Signal Behav, 2016, 11(6): e1184808. DOI:10.1080/15592324.2016.1184808 |
| [12] | Lu XY, Liang XY, Li X, et al. Genome-wide characterisation and expression profiling of the LBD family in Salvia miltiorrhiza reveals the function of LBD50 in jasmonate signaling and phenolic biosynthesis. Ind Crop Prod, 2020, 144: 112006. DOI:10.1016/j.indcrop.2019.112006 |
| [13] | Franz CMAP, Specht I, Cho GS, et al. UV-C-inactivation of microorganisms in naturally cloudy apple juice using novel inactivation equipment based on dean vortex technology. Food Control, 2009, 20(12): 1103-1107. DOI:10.1016/j.foodcont.2009.02.010 |
| [14] | Liu CH, Cai LY, Lu XY, et al. Effect of postharvest UV-C irradiation on phenolic compound content and antioxidant activity of tomato fruit during storage. J Integr Agric, 2012, 11(1): 159-165. DOI:10.1016/S1671-2927(12)60794-9 |
| [15] | Jagadeesh SL, Charles MT, Gariepy Y, et al. Influence of postharvest UV-C hormesis on the bioactive components of tomato during post-treatment handling. Food Bioprocess Tech, 2009, 4(8): 1463-1472. |
| [16] | Bravo S, García-Alonso J, Martín-Pozuelo G, et al. The influence of post-harvest UV-C hormesis on lycopene, β-carotene, and phenolic content and antioxidant activity of breaker tomatoes. Food Res Int, 2012, 49(1): 296-302. DOI:10.1016/j.foodres.2012.07.018 |
| [17] | Urban L, Chabane Sari D, Orsal B, et al. UV-C light and pulsed light as alternatives to chemical and biological elicitors for stimulating plant natural defenses against fungal diseases. Sci Hortic, 2018, 235: 452-459. DOI:10.1016/j.scienta.2018.02.057 |
| [18] | Mazza CA, Boccalandro HE, Giordano CV, et al. Functional significance and induction by solar radiation of ultraviolet-absorbing sunscreens in field-grown soybean crops. Plant Physiol, 2000, 122(1): 117-126. DOI:10.1104/pp.122.1.117 |
| [19] | Ren SC, Sun JT. Changes in phenolic content, phenylalanine ammonia-lyase (PAL) activity, and antioxidant capacity of two buckwheat sprouts in relation to germination. J Funct Foods, 2014, 7: 298-304. DOI:10.1016/j.jff.2014.01.031 |
| [20] | Chang A, Lim MH, Lee SW, et al. Tomato phenylalanine ammonia-lyase gene family, highly redundant but strongly underutilized. J Biol Chem, 2008, 283(48): 33591-33601. DOI:10.1074/jbc.M804428200 |
| [21] | Liu CH, Zheng HH, Sheng KL, et al. Effects of postharvest UV-C irradiation on phenolic acids, flavonoids, and key phenylpropanoid pathway genes in tomato fruit. Sci Hortic, 2018, 241: 107-114. DOI:10.1016/j.scienta.2018.06.075 |
| [22] | Liu CH, Zheng HH, Sheng KL, et al. Effects of melatonin treatment on the postharvest quality of strawberry fruit. Postharvest Biol Tec, 2018, 139: 47-55. DOI:10.1016/j.postharvbio.2018.01.016 |
| [23] | Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta, 2012, 1819(2): 86-96. DOI:10.1016/j.bbagrm.2011.08.004 |
| [24] | Severo J, Tiecher A, Pirrello J, et al. UV-C radiation modifies the ripening and accumulation of ethylene response factor (ERF) transcripts in tomato fruit. Postharvest Biol Tec, 2015, 102: 9-16. DOI:10.1016/j.postharvbio.2015.02.001 |
| [25] | Hens K, Feuz JD, Deplancke B. A high-throughput gateway-compatible yeast one-hybrid screen to detect protein-DNA interactions//Deplancke B, Gheldof N, Eds. Gene Regulatory Networks. Totowa: Humana Press, 2012, 786: 335-355. |
| [26] | Yang HJ, Zhou Y, Zhang YN, Wang J, et al. Identification of transcription factors of nitrate reductase gene promoters and NRE2 cis-element through yeast one-hybrid screening in Nicotiana tabacum. BMC Plant Biol, 2019, 19: 145. DOI:10.1186/s12870-019-1724-z |
| [27] | Tripathi P, Rabara RC, Rushton PJ. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta, 2014, 239(2): 255-266. DOI:10.1007/s00425-013-1985-y |
| [28] | Wang L, Luo YZ, Zhang L, et al. Isolation and characterization of a C-repeat binding transcription factor from maize. J Integr Plant Biol, 2008, 50(8): 965-974. DOI:10.1111/j.1744-7909.2008.00683.x |
| [29] | Tominaga R, Nukumizu Y, Sato S, et al. Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS ONE, 2013, 8(1): e54019. DOI:10.1371/journal.pone.0054019 |
| [30] | Yoshida T, Fujita Y, Sayama H, et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J, 2010, 61(4): 672-685. DOI:10.1111/j.1365-313X.2009.04092.x |
| [31] | Wold AB, Rosenfeld HJ, Holte K, et al. Colour of post-harvest ripened and vine ripened tomatoes (Lycopersicon esculentum Mill.) as related to total antioxidant capacity and chemical composition. Int J Food Sci Technol, 2004, 39(3): 295-302. DOI:10.1111/j.1365-2621.2004.00784.x |
| [32] | Fagundes C, Moraes K, Pérez-Gago MB, et al. Effect of active modified atmosphere and cold storage on the postharvest quality of cherry tomatoes. Postharvest Biol Tec, 2015, 109: 73-81. DOI:10.1016/j.postharvbio.2015.05.017 |
| [33] | Aghdam MS, Luo ZS, Jannatizadeh A, et al. Employing exogenous melatonin applying confers chilling tolerance in tomato fruits by upregulating ZAT2/6/12 giving rise to promoting endogenous polyamines, proline, and nitric oxide accumulation by triggering arginine pathway activity. Food Chem, 2019, 275: 549-556. DOI:10.1016/j.foodchem.2018.09.157 |
| [34] | Martínez-Valverde V, Periago MJ, Provan G, et al. Phenolic compounds, lycopene and antioxidant activity in commercial varieties of tomato (Lycopersicum esculentum). J Sci Food Agric, 2002, 82(3): 323-330. DOI:10.1002/jsfa.1035 |
| [35] | Periago MJ, García-Alonso J, Jakob K, et al. Bioactive compounds, folates and antioxidant properties of tomatoes (Lycopersicum esculentum) during vine ripening. Int J Food Sci Nutr, 2009, 60(8): 694-708. DOI:10.3109/09637480701833457 |
| [36] | Tiecher A, De Paula LA, Chaves FC, et al. UV-C effect on ethylene, polyamines and the regulation of tomato fruit ripening. Postharvest Biol Tec, 2013, 86: 230-239. DOI:10.1016/j.postharvbio.2013.07.016 |
| [37] | El Ghaouth AE, Wilson CL, Callahan AM. Induction of chitinase, β-1, 3-glucanase, and phenylalanine ammonia lyase in peach fruit by UV-C treatment. Phytopathology, 2003, 93(3): 349-355. DOI:10.1094/PHYTO.2003.93.3.349 |
| [38] | Sheng KL, Zheng HH, Shui SS, et al. Comparison of postharvest UV-B and UV-C treatments on table grape: changes in phenolic compounds and their transcription of biosynthetic genes during storage. Postharvest Biol Tec, 2018, 138: 74-81. DOI:10.1016/j.postharvbio.2018.01.002 |
| [39] | Guo J, Wang MH. Characterization of the phenylalanine ammonia-lyase gene (SlPAL5) from tomato (Solanum lycopersicum L.). Mol Biol Rep, 2009, 36(6): 1579-1585. DOI:10.1007/s11033-008-9354-9 |
