
 , 洪伟1
, 洪伟1

1. 贵州医科大学 地方病与少数民族疾病教育部重点实验室,贵州 贵阳 550004;
2. 贵州医科大学 附属医院ICU,贵州 贵阳 550004;
3. 贵州医科大学 免疫学教研室,贵州 贵阳 550004;
4. 贵州医科大学 微生物学教研室,贵州 贵阳 550025
收稿日期:2019-12-09;接收日期:2020-02-26;网络出版时间:2020-03-16
基金项目:国家自然科学基金(Nos. 31560318,31760318),贵州省自然科学基金(Nos. [2020]1Z067,[2019]1441,[2018]1132,[2018]5779-17)资助
通讯作者:Xiaolan Qi. Tel: +86-851-86752814; E-mail: xiaolan76@163.com;
Wei Hong. Tel: +86-851-86752814; E-mail: hongwei@gmc.edu.cn.
摘要:构建基于TeI3c/4c嗜热二型内含子的温度诱导Targetron基因失活系统(Thermotargetron),并应用于中温微生物基因编辑。在大肠杆菌HMS174 (DE3)基因组中,选择Subunit of flagellum基因(fliC)和C4 dicarboxylate orotate:H+ symporter基因(dctA)为靶基因。根据TeI3c/4c DNA识别规则,在fliC和dctA基因中选择fliC489a、fliC828s、fliC1038s和dctA2a位点为基因打靶位点。使用重叠延伸PCR方法,基于pHK-TT1A质粒构建打靶载体。打靶载体转化HMS174菌株,对数期转化子培养液48 ℃热激1 h后涂布于氯霉素抗性LB平板上。使用菌落PCR和DNA测序检测突变株并计算基因失活效率。获得突变株后,通过琼脂穿刺和碳源代谢实验,鉴定ΔfliC、ΔdctA突变株表型变化。菌落PCR测序结果表明,TeI3c/4c插入到fliC和dctA基因设计位点,且打靶效率高达100%。突变株表型验证实验表明,ΔfliC突变株运动能力显著下降,ΔdctA突变株苹果酸代谢能力缺失。综上所述,文中建立了一套适用于嗜中温微生物的温度诱导型、高效基因失活系统,该系统可通过控制宿主菌在48 ℃保温时间实现高效、靶向、精准基因失活。
关键词:嗜热targetronⅡ型内含子温度诱导基因失活大肠杆菌fliCdctA
A temperature-inducible Targetron system for efficient gene inactivation in Escherichia coli
Xingxing Zhao1, Yumei Cheng2, Changxue Wu1, Wei Ren3, Fengqin Rao1, Qian Zhou1, Guzhen Cui4, Xiaolan Qi1

 , Wei Hong1
, Wei Hong1

1. Key Laboratory of Endemic and Ethnic Diseases, Ministry of Education, Guizhou Medical University, Guiyang 550004, Guizhou, China;
2. Department of ICU, The Affiliate Hospital of Guizhou Medical University, Guiyang 550004, Guizhou, China;
3. Department of Immunology, Guizhou Medical University, Guiyang 550004, Guizhou, China;
4. Department of Microbiology, Guizhou Medical University, Guiyang 550025, Guizhou, China
Received: December 9, 2019; Accepted: February 26, 2020; Published: March 16, 2020
Supported by: National Natural Science Foundation of China (Nos. 31560318, 31760318), Natural Science Foundation of Guizhou Province (Nos. [2020]1Z067, [2019]1441, [2018]1132, [2018]5779-17)
Abstract: To construct TeI3c/4c-based and temperature-inducible gene inactivation system (Thermotargetron) and to apply it to gene inactivation of mesophilic bacteria. The subunit of flagellum (fliC) and C4 dicarboxylate orotate:H+ symporter (dctA) genes were chosen as targets in the genome of Escherichia coli HMS174 (DE3) strain. According to recognition roles of TeI3c/4c intron, the fliC489a, fliC828s, fliC1038s and dctA2a sites were chosen as target sites. Gene-targeting plasmids were constructed based on pHK-TT1A by using overlap PCR method and transformed into HMS174 cells. An aliquot mid-log phase cultures of the transformants were shocked at 48 ℃ and plated on LB plate (containing chloramphenicol). Afterwards, gene mutants were screened by using colony PCR and DNA sequencing. After the mutants were obtained, the phenotypes of ΔfliC and ΔdctA gene mutants were characterized by using agar puncture and carbon metabolism experiments. Colony PCR and sequencing results show that TeI3c/4c intron was inserted in the designed sites of fliC and dctA genes. The gene-targeting efficiency of Thermotargetron system was 100%. Phenotype verification experiments of the mutants demonstrated that the cell motility of all ΔfliC mutants was damaged and the malate assimilation ability of ΔdctA mutant was deprived comparing to wild-type HMS174 strain. In our study, a temperature-inducible and high-efficiency gene inactivation system was established for mesophilic bacteria. This system could achieve high efficiency and precise gene inactivation by modulation of the incubation duration of the transformants at 48 ℃.
Keywords: Thermotargetrongroup Ⅱ intorntemperature induciablegene inactivationEscherichia colifliCdctA
可迁移二型内含子(Mobile group Ⅱ intron)是自我拼接内含子的一个亚类,具有在基因组DNA中自发迁移的能力,其迁移过程具有靶向性,并且这种靶向性可以通过突变二型内含子靶位点识别序列而改变,因此具有改造成为基因打靶系统的潜力[1-2]。2001年Karberg等利用来源于乳酸乳球菌Lactococcus lactis的二型内含子元件L1.LtrB,构建了一套靶向基因失活系统,实现了中温微生物基因的靶向失活[3-7]。2007年,Heap等进一步发展了这种基因靶向失活系统并将其命名为ClosTron[8]。ClosTron系统具有不依赖很高的外源DNA转化效率、基因失活效率高和操作简单等优点,适合用于外源DNA转化效率低的微生物基因编辑。然而,ClosTron系统也存在一定的局限性:(1)在中温微生物中使用ClosTron系统,需要首先构建适用于该中温微生物的诱导表达系统,以严格控制L1.LtrB元件的表达量(间接控制其活性),否则L1.LtrB元件会多拷贝插入靶基因组中,出现“脱靶”现象[9]。(2) ClosTron系统在高温条件下(48–60 ℃),因L1.LtrB元件失活,不能正常工作[10]。
为了克服ClosTron系统存在的不足,我们前期研究中使用来源于嗜热聚球藻Thermosynechococcus elongatus的TeI3c/4c二型内含子(ⅡB亚类)开发了温敏Targetron系统——Thermotargetron[10](图 1)。该系统由嗜热二型内含子RNA (TeI3c/4c)和逆转录酶(Reverse transcriptase,RT)组成[11]。其中,TeI3c/4c通过碱基互补配对原则识别基因组DNA双链上的靶位点,RT具有逆转录酶和脱氧核糖核酸内切酶的活性[12-13]。TeI3c/4c和RT共同组装成为核糖核蛋白复合体(RNP)。RNP通过“归巢(Retrohoming)”特异性插入靶基因组位点,其过程如下(图 1):(A) RNP中TeI3c/4c识别基因组DNA靶位点;(B) RT切割靶位点双链DNA并以内含子RNA为模板合成cDNA (逆转录酶);(C)将新合成TeI3c/4c的cDNA插入到靶位点。由于二型内含子编码序列中含有较多的终止密码子且转录成RNA后含有许多发夹结构,因此可以实现靶基因在翻译水平失活。
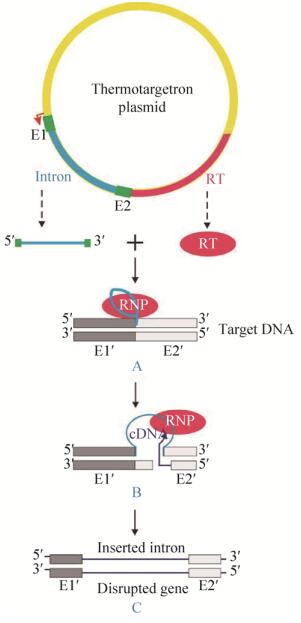 |
| 图 1 Thermotargetron基因“打靶”原理示意图 Fig. 1 Schematic diagram of the Thermotargetron system. (A) The mature ribonucleoprotein complex (RNP) was composed of the group Ⅱ intron RNA (TeI3c/4c) and its RT enzyme. RNP recognizes the target DNA by the principle of base complementary pairing (mainly by TeI3c/4c), and the recognition target is "5′-AAnnnnnnnnnnnnnA-3′" (16 bases in total, n=A, T, G or C). (B) The TeI3c/4c intron sequence was reverse transcribed into cDNA and inserted into the target site by the reverse splicing process. (C) DNA gaps were repaired by the host DNA repairing system. As the TeI3c/4c cDNA sequence contains stop codons and could easily form hairpins after transcription, thus the translation of the target gene is terminated at translation level. |
| 图选项 |
值得注意的是Thermotargetron核心元件TeI3c/4c二型内含子只在高温时(48–60 ℃)具有“归巢”活性。因此,Thermotargetron具有2个特征:(1)适用于嗜热微生物基因失活[14];(2)在可耐高温的中温微生物中,具有温度诱导特征。温度诱导属于物理诱导,相比ClosTron系统的化学诱导,具有易实现(无需从头构建诱导表达系统)、更精确(精确控制保温时间)和更直接(直接控制TeI3c/4c活性)等优点。除此之外Thermotargetron系统识别序列为“AAnnnnnnnnnnnnnA”,对于GC含量比较低的中温微生物具有更多的靶点可供选择,即其更适合低GC含量微生物的基因打靶。本研究利用来源于热纤梭菌groEL启动子表达Thermotargetron核心元件TeI3c/4c二型内含子,以HMS174 (DE3)为可耐高温的中温微生物模型,并以其fliC和dctA基因为靶基因,验证温度诱导型Thermotargetron基因失活系统的可行性,为中温微生物的基因编辑提供新的工具。
1 材料与方法1.1 材料1.1.1 主要试剂和培养基配方氯化钠(NaCl)、酵母提取物(Yeast extract)、胰蛋白胨(Tryptone)、琼脂粉(Agar power)、氯霉素(Chloramphenicol,Chl)、七水合磷酸氢二钠(Na2HPO4·7H2O)、氯化铵(NH4Cl)、磷酸二氢钾(KH2PO4)、硫酸镁(MgSO4)、氯化钙(CaCl2)、葡萄糖(Glucose)和L-苹果酸(L-malic acid)均购自上海阿拉丁生化科技股份有限公司(Aladdin,上海)。琼脂糖凝胶DNA回收试剂盒、质粒小提试剂盒购自天根生化科技(北京)有限公司(TIANGEN,北京);BsiWⅠ-HF (R3133L)和SpeⅠ-HF (R3553L)购自New England BioLabs (NEB,北京)。所有引物合成及测序均由生工生物工程(上海)股份有限公司(Sangon Biotech,上海)完成。
Luria-Bertani (LB)培养基:氯化钠10 g、酵母提取物5 g、胰蛋白胨10 g,定容至1 L。在转化子筛选时,向培养基中添加终浓度为10 μg/mL氯霉素。M9培养基:5×M9培养基母液(Na2HPO4·7H2O 12.8 g、NaCl 0.5 g、NH4Cl l g和KH2PO4 3 g),突变株表型鉴定时加入0.2% (W/V)葡萄糖或苹果酸,其中MgSO4、CaCl2、葡萄糖和L-苹果酸均溶解于ddH2O,使用0.22 μm滤膜过滤除菌。
1.1.2 菌株与培养条件大肠杆菌Escherichia coli NEBExpress高效感受态(NEB,北京)作为分子克隆和质粒构建的宿主细胞。Thermotargetron基因打靶实验在大肠杆菌HMS174 (DE3)细胞中进行(诺禾致源科技股份有限公司,Novogene,北京),以下简称HMS174。E. coli NEBExpress、E. coli HMS174菌株在LB培养基中,37 ℃好氧培养。LB培养基中加入氯霉素(10 μg/mL,LB-Chl)以筛选转化子。本研究用到的所有大肠杆菌菌株在7% (V/V)二甲基亚砜中长期冻存(?80 ℃)。
1.2 方法1.2.1 目标基因打靶引物的设计从National Center for Biotechnology Information (NCBI)数据库中获得HMS174菌株fliC和dctA基因序列。根据TeI3c/4c识别位点规律“5′-AAnnnnnnnnnnnnnA-3′”位点设计HMS174 fliC和dctA基因打靶载体引物(表 1)。每个打靶位点需要设计4条引物,即内含子结合位点12 (Intron-binding site 12,IBS12)、外显子正义链结合位点2 (Exon-binding site 2 sense,EBS2s)、外显子反义链结合位点1 (Exon-binding site 1 antisense,EBS1a)和通用引物(TeI3c-universal primer, TeI3c-UNV)。
表 1 文中所用引物Table 1 Primers used in this study
| Primer | Sequence (5′–3′) | Description |
| TeI3c-UNV | TAACGAGGCTTCTAGCG | Universal primer |
| fliC489aIBS12 | AGCCAAAGCAGGTTGACTAGTAAgagttttagcatcGTGCGACGCGAAAGCTAG | fliC489a targeting primers |
| fliC489aEBS2s | CGCTAGAAGCCTCGTTAaactcAGCAGGCCAAAGATGCTG | |
| fliC489aEBS1a | CGGAGTTGCTGTCCCCGTACGCTGAagcatcAGCAGCGtATCCAATCC | |
| fliC828sIBS12 | AGCCAAAGCAGGTTGACTAGTAAtactactaaagctGTGCGACGCGAAAGCTAG | fliC828s targeting primers |
| fliC828sEBS2s | CGCTAGAAGCCTCGTTAtagtaAGCAGGCCAAAGATGCTG | |
| fliC828sEBS1a | CGGAGTTGCTGTCCCCGTACGCTGAaaagctAGCAGCGTATCCAATCC | |
| fliC1038sIBS12 | AGCCAAAGCAGGTTGACTAGTAAaactattacctatGTGCGACGCGAAAGCTAG | fliC1038s targeting primers |
| fliC1038sEBS2s | CGCTAGAAGCCTCGTTAtagttAGCAGGCCAAAGATGCTG | |
| fliC1038sEBS1a | CGGAGTTGCTGTCCCCGTACGCTGAacctatAGCAGCGTATCCAATCC | |
| dctA2aIBS12 | AGCCAAAGCAGGTTGACTAGTAAcagagaggttttcGTGCGACGCGAAAGCTAG | dctA2a targeting primers |
| dctA2aEBS2s | CGCTAGAAGCCTCGTTActctgAGCAGGCCAAAGATGCTG | |
| dctA2aEBS1a | CGGAGTTGCTGTCCCCGTACGCTGAgttttcAGCAGCGtATCCAATCC | |
| DPfliC-F | AATTACAGTCAGGACGCGT | fliC gene detection primers |
| DPfliC-R | ATGTGACCGGGTTAGCC | |
| DPdctA-F | CCGCAGGTACCCCATAAC | dctA gene detection primers |
| DPdctA-R | CACTCGGGGAAGGGAGT | |
| DPjunction-U | GAGGAGGTGGGAAAAAGCGA | junction detection primers |
| DPjunction-D | TCGCTTTTTCCCACCTCCTC |
表选项
1.2.2 质粒构建打靶质粒构建过程需要进行两次PCR反应和一次连接反应。第一次PCR以pHK-TT1A质粒为模板(表 2),IBS12/TeI3c-UNV和EBS2s/EBS1a作引物分别进行扩增;第二次PCR反应以上一次PCR产物作为模板,IBS12与EBS1a作为引物,将第一次PCR产物连接成为一个携带与目标基因互补配对的基因打靶片段。基因打靶片段与BsiWⅠ-HF和SpeⅠ-HF线性化的pHK-TT1A载体,使用T5 exonuclease DNA assembly方法(TEDA)进行重组连接[15]。重组质粒送生工生物工程(上海)股份有限公司进行测序。
表 2 文中所用菌株及质粒Table 2 Strains and plasmids used in this study
| Strains and plasmids | Relevant features | Source or reference |
| Strains | ||
| NEB Express Competent E. coli (High efficiency) | fhuA2 [Ion] ompT gal sulA11 R(mcr-73::miniTn10--TetS)2[dcm] R(zgb-210::Tn10--TetS)endA1delta(mcrC-mrr)114::lS10 | NEB |
| E. coli HMS174(DE3) | F- recA1 hsdR(rK12-mK12+) (DE3) (RifR) | Novogene |
| ΔfliC489a | Derived from E. coli HMS174(DE3), ΔfliC489a | This work |
| ΔfliC828s | Derived from E. coli HMS174(DE3), ΔfliC828s | This work |
| ΔfliC1038s | Derived from E. coli HMS174(DE3), ΔfliC1038s | This work |
| ΔdctA2a | Derived from E. coli HMS174(DE3), ΔdctA2a | This work |
| Plasmids | ||
| pHK-TT1A | Targetron vector, GroEL promoter, CmR | [10] |
| pHK-TT1A-fliC489a | Derived from pHK-TT1A, targeting the antisense strand 489 site of fliC in HMS174(DE3) | This work |
| pHK-TT1A-fliC828s | Derived from pHK-TT1A, targeting the sense strand 828 site of fliC in HMS174(DE3) | This work |
| pHK-TT1A-fliC1038s | Derived from pHK-TT1A, targeting the sense strand 1038 site of fliC in HMS174(DE3) | This work |
| pHK-TT1A-dctA2a | Derived from pHK-TT1A, targeting the antisense strand 2 site of dctA in HMS174(DE3) | This work |
表选项
1.2.3 打靶质粒转化及突变株筛选测序验证正确的Thermotargetron打靶质粒使用化学热激法转化HMS174感受态细胞,并涂布于LB-Chl平板以筛选转化子。单个转化子挑取至LB-Chl液体培养基中培养过夜。培养过夜物稀释100倍后,置于1.5 mL无菌离心管中,37 ℃继续培养1 h,随后将培养温度提高到48 ℃,热激1 h (关键步骤,激活TeI3c/4c“归巢”活性)。热激后的培养液,稀释100倍后,均匀涂布于LB-Chl平板上,37 ℃过夜培养,平板上形成菌落待PCR验证。
上一步实验获得的菌落,使用fliC和dctA检测引物(表 1),PCR检测是否发生TeI3c/4c插入突变。使用Green Taq Mix试剂盒进行PCR (诺唯赞,南京),反应条件为:94 ℃ 5 min;94 ℃ 30 s,56 ℃ 30 s,72 ℃延伸150 s,30次循环;72 ℃ 5 min,4 ℃ 10 min。基因失活效率根据如下公式计算:
基因失活效率=突变株数量/检测菌株数量。
1.2.4 质粒丢失基因失活突变株通过连续在无抗性培养基中传代的方法丢失质粒,过程如下:吸取50 μL突变株培养液(OD600=0.8–1.2)到5 mL LB培养基中,37 ℃、200 r/min培养8 h,培养物在相同培养基中连续转接10次,最后划线于LB平板上。待平板长出单克隆后,使用影印平板法[16],将同一克隆挑取至LB-Chl和LB平板上,并置于恒温培养箱37 ℃培养24 h。质粒丢失效率使用如下公式计算:丢失氯霉素抗性克隆数/总检测克隆数。
1.2.5 突变株表型检测突变株运动能力检测方法如下:将野生型HMS174与ΔfliC突变株穿刺接种于含有0.8% (W/V)琼脂的LB半固体培养基的试管中,37 ℃恒温培养过夜,穿刺培养管使用佳能G11相机拍照记录。苹果酸代谢能力检测方法:野生型HMS174与ΔdctA突变株经LB培养基活化,以1%接种量(V/V)接种于含0.2%葡萄糖的M9液体培养基中,培养至对数生长中期(OD600=0.5– 0.8)以去除LB培养基中痕量苹果酸[17]。培养物转接至含0.2%苹果酸作为唯一碳源的M9液体培养基中,200 r/min、37 ℃培养24 h,定时取样,测定OD600值。
2 结果与分析2.1 基因打靶位点设计HMS174 fliC 基因(ECHMS174_01916)和dctA基因(ECHMS174_03796)全长分别为1 497 bp和1 287 bp (图 2A)。在fliC基因中选择位点符合TeI3c/4c识别规律的fliC489a (5′-AA gagttttagcatcA-3′)、fliC828s (5′-AAtactactaaagct A-3′)和fliC1038s (5′-AAaactattacctatA-3′)位点。在dctA基因中选择dctA2a (5′-AAcagagaggttttc A-3′)位点。使用PCR突变野生型TeI3c/4c IBS1、IBS2、EBS1和EBS2位点,构建针对fliC489a、828s、1038s和dctA2a四个识别位点的打靶质粒pHK-TT1A-fliC489a、pHK-TT1A-fliC828s、pHK- TT1A-fliC1038s和pHK-TT1A-dctA2a (图 2B)。
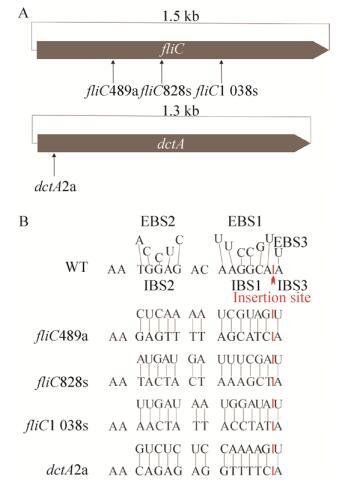 |
| 图 2 HMS174 fliC与dctA基因中的Thermotargetron打靶识别位点 Fig. 2 Gene targeting sites in fliC and dctA genes. (A) Gene targeting sites in fliC and dctA genes. (B) The bases composition of fliC489a, fliC828s, fliC1038s and dctA2a targeting sites. |
| 图选项 |
2.2 Thermotargetron打靶质粒构建以pHK-TT1A-fliC489a打靶载体构建过程为例(图 3),详述Thermotargetron打靶质粒的构建过程如下:(1)以pHK-TT1A为载体模板,使用fliC489aIBS12/TeI3c-UNV,fliC489aEBS2s/ fliC489aEBS1a引物(表 1),PCR扩增获得IBS1/2和EBS1/2突变片段,其长度分别为296 bp (IBS1/2)和114 bp (EBS1/2) (图 4A)。第二次通过重叠延伸PCR反应,以fliC489aIBS12/ fliC489aEBS1a为引物,上一步PCR获得的IBS1/2和EBS1/2突变片段为模板,重叠延伸PCR扩增获得打靶片段(393 bp) (图 4B)。打靶片段与线性化的pHK-TT1A载体(BsiWⅠ-SpeⅠ双酶切),使用TEDA法重组连接[15],获得pHK-TT1A-fliC489a打靶质粒。使用相同的方法构建pHK-TT1A- fliC828s、pHK-TT1A-fliC1038s和pHK-TT1A- dctA2a打靶质粒,测序结果表明4个打靶质粒均构建成功。
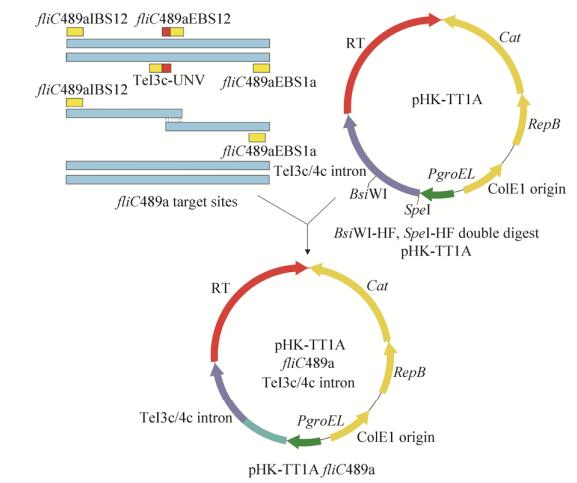 |
| 图 3 Thermotargetron载体构建 Fig. 3 The construction of Thermotargetron gene-targeting vectors. Two rounds of PCRs were carried to construct pHK-TT1A-fliC489a plasmid, which targeting fliC gene. Firstly, fliC489aIBS12/TeI3c-UNV and fliC489aEBS2s/fliC489aEBS1a primer pairs were used to generate mutations in IBS1/2, EBS1/2 sites of wild-type TeI3c/4c intron, which produce 296 bp and 114 bp amplicons. Secondly, the 296 bp and 114 bp amplicons were assembled together by using overlap PCR, in which fliC489aIBS12/fliC489aEBS1a primers were used. The overlap PCR produce 393 bp amplicon, which containing desire mutations for targeting fliC gene. Finally, 393 bp amplicon were assembled with BsiWⅠ-SpeⅠ linerized pHK-TT1A plasmid by using TEDA method and the resultant plasmid was denoted pHK-TT1A-fliC489a. Same work flow was carried out to construct pHK-TT1A-fliC828s, pHK-TT1A-fliC1038s and pHK-TT1A-dctA2a plasmids. |
| 图选项 |
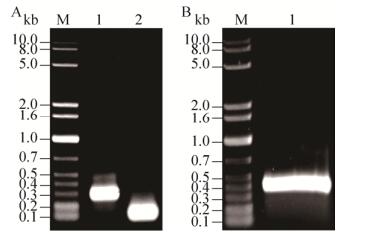 |
| 图 4 两轮PCR突变TeI3c/4c靶序列识别位点 Fig. 4 Mutate TeI3c/4c gene-targeting sites by two- rounds PCRs. (A) Amplicons containing mutated IBS1/2 (296 bp) and EBS1/2 (114 bp) sites. (B) Assemble 296 bp and 114 bp amplicons, containing mutated IBS1/2 and EBS1/2 sites, by using overlap PCR, which produce the gene-targeting fragment. |
| 图选项 |
2.3 突变株筛选使用HMS174 fliC和dctA两侧检测引物(DPfliC-F/DPfliC-R和DPdctA-F/DPdctA-R,表 1) PCR鉴定经48 ℃热激诱导1 h后的转化子,突变株PCR扩增条带比野生型菌株增加839 bp,检测结果表明TeI3c/4c二型内含子成功失活fliC (1 558 vs. 2 397bp)和dctA基因(1 376 vs. 2 215 bp),且4个打靶质粒基因失活效率均为100% (表 3,每个打靶位点检测15个克隆)。测序结果表明,TeI3c/ 4c二型内含子插入到HMS174 ΔfliC489a (图 5A)、ΔfliC828s (图 5B)、ΔfliC1038s (图 5C)和ΔdctA2a (图 5D)突变株基因组中的设计位点(图 5)。
表 3 Thermotargetron基因失活效率Table 3 Gene inactivation efficiency of Thermotargetron system
| Mutant strain | Target sites | Gene inactivation efficiency (%) |
| ΔfliC489a | GAGTTTTAGCATC | 100 |
| ΔfliC828s | TACTACTAAAGCT | 100 |
| ΔfliC1038s | AACTATTACCTAT | 100 |
| ΔdctA2a | CAGAGAGGTTTTC | 100 |
表选项
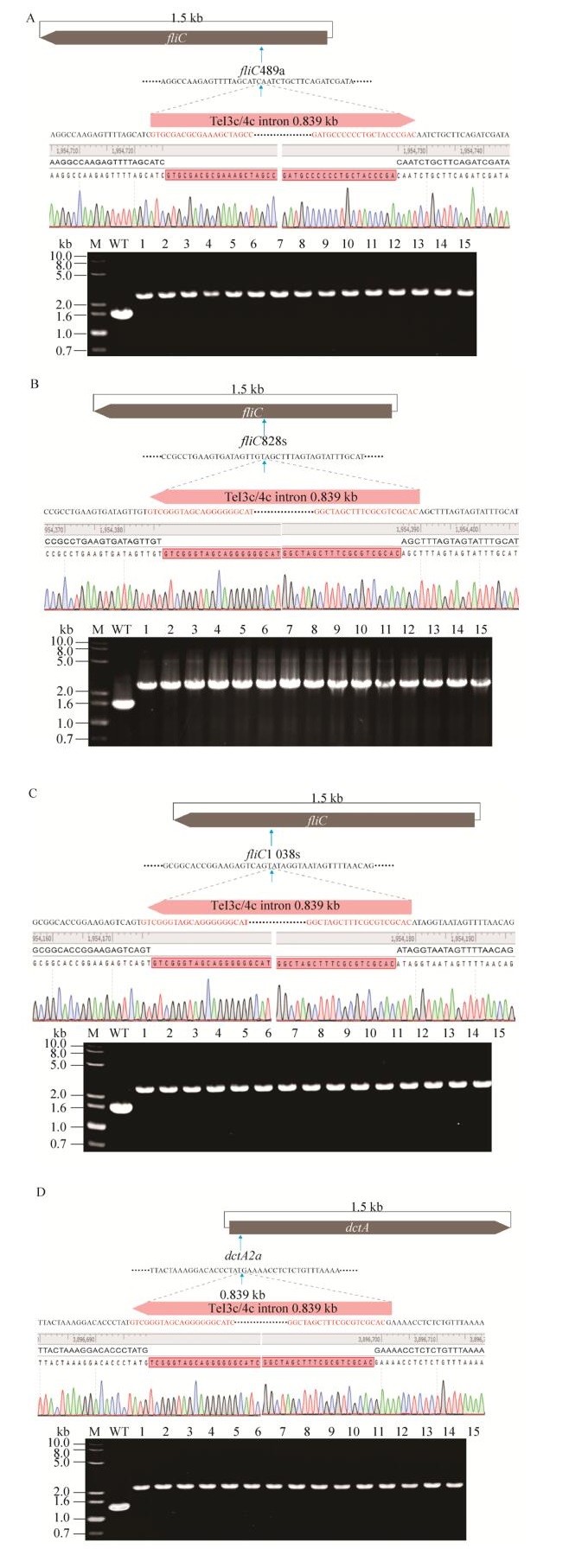 |
| 图 5 突变株筛选及测序验证 Fig. 5 Screen mutants by using PCR and DNA sequencing. (A) Screen E. coli HMS174 ΔfliC489a mutants by using PCR. M: DNA marker; WT: wild type; lane 1–15: E. coli HMS174 ΔfliC489a mutants. (B) Screen E. coli HMS174 ΔfliC828s mutants by using PCR. M: DNA marker; WT: wild type; lane 1–15: E. coli HMS174 ΔfliC828s mutants. (C) Screen E. coli HMS174 ΔfliC1038s mutants by using PCR. M: DNA marker; WT: wild type; Lane 1–15: E. coli HMS174 ΔfliC1038s mutants. (D) Screen E. coli HMS174 ΔdctA2a mutants by using PCR. M: DNA marker; WT: wild type; lane 1–15: E. coli HMS174 ΔdctA2a mutants. The TeI3c/4c insertion sites in E. coli HMS174 genome are indicated as blue arrows. TeI3c/4c intron is depicted in red, E. coli HMS174 genomic DNA was depict in black. The junctions between TeI3c/4c intron DNA and E. coli HMS 174 genomic DNA are sequenced by using primers DPjunction-U/DPjunction-D. Sequencing data are visualized using the SnapGene software (GSL Biotech; available at snapgene.com). |
| 图选项 |
2.4 质粒丢失筛选获得的大肠杆菌HMS174 ΔfliC489a、ΔfliC828s、ΔfliC1038s和ΔdctA2a突变株仍携带其基因打靶质粒(分别为pHK-TT1A-fliC489a、pHK-TT1A-fliC828s、pHK-TT1A-fliC1038s和pHK-TT1A-dctA2a)。该突变株连续10次无抗性培养传代后,使用克隆平板法筛选在有抗性的平板上不能生长,而在无抗性平板上能生长的为质粒丢失单克隆。如图 6所示,经过连续10次无抗性培养传代后,大肠杆菌HMS174 ΔfliC489a、ΔfliC828s、ΔfliC1038s和ΔdctA2a突变株质粒成功丢失(质粒丢失效率为100%,每个突变株检测15–24个克隆)。
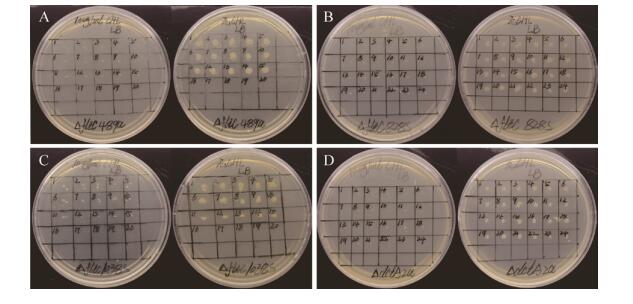 |
| 图 6 克隆平板实验验证基因失活质粒丢失情况 Fig. 6 Detection of plasmids curing by using replica plate method. (A) Replica plate of E. coli ΔfliC489a mutants. (B) Replica plate of E. coli ΔfliC828s mutants. (C) Replica plate of E. coli ΔfliC1038s mutants. (D) Replica plate of E. coli ΔdctA2a mutants; Colonies only formed on LB plate but not on LB-Chl plate, this result indicate that gene targeting plasmids were cured in all mutants and the plasmids curing efficiency was 100%. |
| 图选项 |
2.5 HMS174 ΔfliC突变株运动能力变化检测通过穿刺培养法对比野生型HMS174菌株与ΔfliC489a、ΔfliC828s和ΔfliC1038s突变株运动能力变化。野生型菌株经过12 h培养后可以均匀扩散至培养管中,ΔfliC突变株只局限于接种针接触培养基的位置生长(图 7),提示ΔfliC突变株在琼脂中运动能力明显下降。
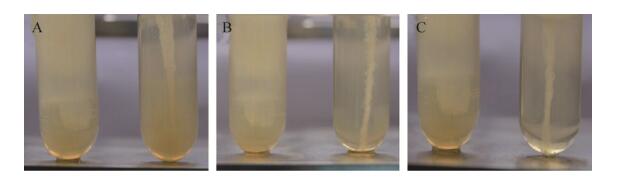 |
| 图 7 野生型HMS174与ΔfliC突变株运动能力对比 Fig. 7 Comparison motility of HMS174 strains. (A) Comparison of motility E. coli HMS174 wild-type strain and E. coli HMS174 ΔfliC489a mutant. (B) Comparison of E. coli HMS174 wild-type strain and E. coli HMS174 ΔfliC828s mutant. (C) Comparison of E. coli HMS174 wild-type strain and E. coli HMS174 ΔfliC1038s mutant. Wild-type E. coli HMS174 strain (left), E. coli HMS174 ΔfliC mutants (right). |
| 图选项 |
2.6 HMS174 ΔdctA突变株苹果酸代谢能力测定为测试HMS174 ΔdctA2a基因失活突变株的苹果酸代谢能力变化,首先将野生型HMS174菌株和HMS174 ΔdctA2a突变株培养于以0.2%葡萄糖作为唯一碳源的M9液体培养基中,再转接至以0.2%苹果酸作为唯一碳源的M9液体培养基。野生型大肠杆菌HMS174在以上两种碳源的M9培养基中均能正常生长,ΔdctA2a则只能在0.2%葡萄糖作为唯一碳源的M9液体培养基中生长,不能在0.2%苹果酸作为唯一碳源的M9培养基中生长(图 8)。提示dctA基因是大肠杆菌苹果酸利用的关键基因。
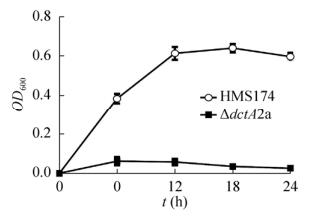 |
| 图 8 对比HMS174野生型与ΔdctA2a突变株苹果酸利用能力 Fig. 8 Comparision of L-malic acid utilization ability of wild-type E. coli HMS174 and E. coli HMS174 ΔdctA2a mutant. |
| 图选项 |
3 讨论Thermotargetron是基于嗜热聚球藻Thermosynechococcus elongatus TeI3c/4c嗜热二型内含子建立的靶向基因失活系统[10]。TeI3c/4c最大的特点是只在高温(48–60 ℃)时具有活性,这种特性是实现温度诱导的关键。对于可耐受高温的中温微生物(如Clostridium属和Escherichia属的大多数微生物),将Thermotargetron打靶质粒转入宿主细胞,经48 ℃高温诱导,即可激活TeI3c/4c的“归巢”活性,从而实现靶向基因失活。温度诱导相比化学诱导(如脱水四环素[18]、乳糖诱导[19])具有诱导条件简单、易控(控制保温时间)、打靶元件活力易控制(恢复至37 ℃打靶元件即失活)等优点。不仅如此,Thermotargetron还具有基因失活效率高的优点,在本研究中设计的4个打靶位点基因失活效率高达100%,明显高于化学诱导系统[10]。可见,Thermotargetron系统具有广泛应用于耐高温中温菌的潜力,可在中温菌中实现高效、精确的基因失活。
在前期的研究中,T7启动子被用于控制TeI3c/4c的表达,打靶效率在50%左右[10]。但是T7启动子具有以下两个缺点:(1) T7启动子为强启动子,启动表达效率很高,导致诱导试剂的加入量比较难控制,容易造成TeI3c/4c表达过量,过多的TeI3c/4c表达量是造成其脱靶的主要原因;(2) T7启动子并不能在嗜热菌中工作。为了改善以上问题,本研究在打靶质粒的设计上作出了两点改进:(1)将打靶质粒中的T7启动子改为热纤梭菌中的groEL启动子,该启动子表达量适中,易于在打靶高效和严谨性上找到平衡点;(2)将化学诱导改为通过温度诱导(控制48 ℃保温时间,控制过程直接、精确)[10]。改进后的Thermotargetron系统打靶效率高达100%,明显优于T7启动子介导的化学诱导系统。
使用大肠杆菌作为Thermotargetron基因打靶的模式生物主要有两个原因:(1)大肠杆菌外源基因转化方法成熟且基因组背景清楚;(2)大肠杆菌作为革兰氏阴性菌的代表,对高温的耐受性低于细胞壁结构复杂的革兰氏阳性菌。以热耐受性低的微生物为起点,温度诱导更易推广。除此之外,化学法热激转化大肠杆菌是大肠杆菌的常规实验操作,学界普遍认为热激不会改变大肠杆菌基因背景[20]。为了最大限度保证热激前、后大肠杆菌的基因背景的一致性,所有突变株均在无抗性的LB培养基中37 ℃连续转接10次,此步骤有两个目的:(1)使突变株从热应激状态恢复为正常状态;(2)将突变株中的Thermotargetron打靶质粒丢失。
HMS174中fliC基因编码的鞭毛蛋白是鞭毛丝的主要结构亚基,参与细菌的一系列致病过程[21]。fliC参与形成的鞭毛结构是多种细菌最主要的运动器官,主要介导细菌的运动、趋化和黏附,帮助细菌附着于宿主体内并迁移到营养物质丰富的位置去,在细菌的感染与免疫过程中发挥着重要的作用:(1)鞭毛在生物膜的扩大阶段介导菌体-菌体之间的相互作用,从而产生较强的毒性与抗生素耐药性[22];(2)鞭毛介导的运动性对于沙门氏菌、大肠杆菌、李氏杆菌和耶尔森氏菌等微生物的生物膜的形成和成熟起着关键作用[23]。通过对大肠杆菌HMS174 ΔfliC突变株和野生型菌株运动能力的分析,证明了fliC基因编码产物对于鞭毛的运动功能至关重要。
在无氧条件下,大肠杆菌3个独立二羧酸摄取(Dcu)系统DcuA、DcuB和DcuC共同介导对四碳-二羧酸和L-天冬氨酸的摄取、交换和外排[24]。在有氧条件下,dctA基因编码2-羟基羧酸摄取的转运蛋白(2-hydroxycarboxylic acid transporter,2-HCT)以Na+或H+作为共转运阳离子驱动柠檬酸、苹果酸或乳酸等的转运[25],该蛋白也介导对四碳-二羧酸的摄取[26]。苹果酸是四碳- 2-羟基羧酸的代表,常用作2-HCT转运蛋白功能鉴定底物。Jiang等报道了dctA基因参与了苹果酸的摄入过程[17]。本研究通过dctA基因的靶向失活,验证了dctA是大肠杆菌中苹果酸利用的必需基因。
综上所述,本研究通过温度诱导控制TeI3c/4c的活力,高效失活了4个靶基因位点(效率均为100%),并在基因和表型两个不同层次,验证了Thermotargetron可用于嗜中温(可耐高温的)微生物基因靶向失活。相比于前期研究中使用的化学诱导方式,温度诱导对TeI3c/4c“归巢”活力的控制比化学诱导更加直接和便捷,因此,温度诱导Targetron系统的构建为中温微生物的基因编辑提供了可靠、高效和便捷的新方法。
参考文献
| [1] | Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harbor Perspect Biol, 2011, 3(8): a003616. |
| [2] | Lambowitz AM, Zimmerly S. Mobile group II introns. Ann Rev Genet, 2004, 38: 1-35. DOI:10.1146/annurev.genet.38.072902.091600 |
| [3] | Karberg M, Guo HT, Zhong J, et al. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotechnol, 2001, 19(12): 1162-1167. DOI:10.1038/nbt1201-1162 |
| [4] | McNeil BA, Semper C, Zimmerly S. Group II introns: versatile ribozymes and retroelements. Wiley Interdiscipl Rev: RNA, 2016, 7(3): 341-355. DOI:10.1002/wrna.1339 |
| [5] | Zimmerly S, Semper C. Evolution of group II introns. Mobile DNA, 2015, 6: 7. DOI:10.1186/s13100-015-0037-5 |
| [6] | Qu GS, Kaushal PS, Wang J, et al. Structure of a group II intron in complex with its reverse transcriptase. Nat Struct Mol Biol, 2016, 23(6): 549-557. DOI:10.1038/nsmb.3220 |
| [7] | Novikova O, Belfort M. Mobile group II introns as ancestral eukaryotic elements. Trends Genet, 2017, 33(11): 773-783. DOI:10.1016/j.tig.2017.07.009 |
| [8] | Heap JT, Pennington OJ, Cartman ST, et al. The ClosTron: A universal gene knock-out system for the genus Clostridium. J Microbiol Methods, 2007, 70(3): 452-464. DOI:10.1016/j.mimet.2007.05.021 |
| [9] | Zhang J, Liu YJ, Cui GZ, et al. A novel arabinose-inducible genetic operation system developed for Clostridium cellulolyticum. Biotechnol Biof, 2015, 8: 36. DOI:10.1186/s13068-015-0214-2 |
| [10] | Mohr G, Hong W, Zhang J, et al. A targetron system for gene targeting in thermophiles and its application in Clostridium thermocellum. PLoS ONE, 2013, 8(7). |
| [11] | Zhao C, Pyle AM. The group II intron maturase: a reverse transcriptase and splicing factor go hand in hand. Curr Opin Struct Biol, 2017, 47: 30-39. DOI:10.1016/j.sbi.2017.05.002 |
| [12] | Liu YJ, Zhang J, Cui GZ, et al. Current progress of targetron technology: Development, improvement and application in metabolic engineering. Biotechnol J, 2015, 10(6): 855-865. DOI:10.1002/biot.201400716 |
| [13] | Zimmerly S, Guo HT, Perlman PS, et al. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell, 1995, 82(4): 545-554. DOI:10.1016/0092-8674(95)90027-6 |
| [14] | Hong W, Zhang J, Feng YG, et al. The contribution of cellulosomal scaffoldins to cellulose hydrolysis by Clostridium thermocellum analyzed by using thermotargetrons. Biotechnol Biof, 2014, 7: 80. DOI:10.1186/1754-6834-7-80 |
| [15] | Xia YZ, Li K, Li JJ, et al. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res, 2018, 47(3): e15. |
| [16] | Hong W, Zhang J, Cui GZ, et al. Multiplexed CRISPR-Cpf1-mediated genome editing in Clostridium difficile toward the understanding of pathogenesis of C. difficile infection. ACS Synth Biol, 2018, 7(6): 1588-1600. DOI:10.1021/acssynbio.8b00087 |
| [17] | Jiang JQ, Zhang ZL, Xu T, et al. Knock-out of dctA gene from Escherichia coli K12 and characterization of its function for malic acid transportation. J Northeast Agric Univ, 2018, 49(5): 45-52 (in Chinese). 姜巨全, 张正来, 徐桐, 等. 大肠杆菌中dctA基因敲除及其苹果酸摄取功能鉴定. 东北农业大学学报, 2018, 49(5): 45-52. DOI:10.3969/j.issn.1005-9369.2018.05.006 |
| [18] | Stebbins MJ, Urlinger S, Byrne G, et al. Tetracycline-inducible systems for Drosophila. Proc Natl Acad Sci USA, 2001, 98(19): 10775-10780. DOI:10.1073/pnas.121186498 |
| [19] | Banerjee A, Leang C, Ueki T, et al. Lactose-inducible system for metabolic engineering of Clostridium ljungdahlii. Appl & Environ Microbiol, 2014, 80(8): 2410-2416. |
| [20] | Murano EA, Pierson MD. Effect of heat shock and incubation atmosphere on injury and recovery of Escherichia coli O157:H7. J Food Prot, 1993, 56(7): 568-572. DOI:10.4315/0362-028X-56.7.568 |
| [21] | Yu HY, Ma Y. Influence of FliC knock-out Escherichia coli on the biofilm formation. Chin J Vet Sci, 2016, 36(8): 1301-1306 (in Chinese). 喻华英, 马燕. 大肠杆菌FliC基因敲除后对大肠杆菌生物膜形成的影响. 中国兽医学报, 2016, 36(8): 1301-1306. |
| [22] | Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol, 1998, 30(2): 285-293. DOI:10.1046/j.1365-2958.1998.01061.x |
| [23] | Wolska KI, Grudniak AM, Rudnicka Z, et al. Genetic control of bacterial biofilms. J Appl Genet, 2016, 57(2): 225-238. DOI:10.1007/s13353-015-0309-2 |
| [24] | Golby P, Davies S, Kelly DJ, et al. Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J Bacteriol, 1999, 181(4): 1238-1248. DOI:10.1128/JB.181.4.1238-1248.1999 |
| [25] | Sobczak I, Lolkema JS. The 2-hydroxycarboxylate transporter family: physiology, structure, and mechanism. Microbiol Mol Biol Rev, 2005, 69(4): 665-695. |
| [26] | Davies SJ, Golby P, Omrani D, et al. Inactivation and regulation of the aerobic C4-dicarboxylate transport (dctA) gene of Escherichia coli. J Bacteriol, 1999, 181(18): 5624-5635. DOI:10.1128/JB.181.18.5624-5635.1999 |
