王雪1,2#, 赵佳男2, 刘智慧1,2, 张立敏3, 常晗4#, 秦姣2, 朱云芸5, 胡国成6, 胡诗佳2, 阳建春2, 贾仲昕1,2, 李俊保7, 鞠厚斌8, 王承民2, 季芳2

 , 秦建华1
, 秦建华1

1. 河北农业大学动物医学院, 河北 保定 071001;
2. 广东省科学院动物研究所, 广东省动物保护与资源利用重点实验室, 广东省野生动物保护与利用公共实验室, 广东 广州 510260;
3. 联勤保障部队北戴河康复疗养中心, 河北 秦皇岛 066100;
4. 中国科学院动物研究所, 北京 100101;
5. 北京动物园圈养野生动物技术北京市重点实验室, 北京 100044;
6. 生态环境部华南环境科学研究所, 广东 广州 510530;
7. 郑州市动物园, 河南 郑州 450008;
8. 上海市动物疫病预防控制中心, 上海 200336
收稿日期:2020-11-17;修回日期:2021-01-04;网络出版日期:2021-01-13
基金项目:河北省现代农业产业技术体系奶牛创新团队建设项目(HBCT2018120205);河北省重点研发项目(20326603D);广东省科学院人才专项(2016GDASRC-0205);广东省科学院科技发展专项(2018GDASCX-0107);北京动物园圈养野生动物技术北京市重点实验室开放课题(ZDK201909)
*通信作者:季芳, E-mail: 309533361@qq.com;
秦建华, E-mail: qjhqqq@126.com.
#并列第一作者。
摘要:[目的] 研究克雷伯氏菌与多复制子抗性质粒间的关系,分析细菌携带多复制子质粒对抗生素环境的响应机制。[方法] 以2018-2020年分离的56株不同来源克雷伯氏菌(Klebsiella sp.)分离株为研究对象,利用微量肉汤稀释法评估其多重耐药表型,对分离菌株进行全基因组测序(WGS),通过细菌全基因组关联分析(BGWAS)技术和比较基因组学方法深入解析多复制子抗性质粒形成的机制。[结果] 耐药表型分析发现野生动物来源的菌株具有更广的耐药谱系,总体Klebsiella sp.对氨苄西林表现出很高的耐药率(80.36%),尤其是马来穿山甲来源菌株对头孢类抗生素高度耐受,同时对氯霉素、左氧氟沙星和复方新诺明等药物耐受,基因组分析发现这些菌株携带了抗性质粒和更多的抗生素抗性基因。进一步对69个质粒序列分析,发现有28个质粒为多复制子质粒,主要携带blaCTX-M-15、blaCTX-M-14、blaCTX-M-55、blaOXA-1和blaTEM-1等β-内酰胺酶基因。细菌携带质粒类型分析认为Klebsiella pneumoniae可能是多复制子质粒的重要宿主,质粒骨架与结构分析发现多复制子质粒多由2个或2个以上单个质粒融合而成,携带此类质粒的菌株不仅获得了更广的耐药表型,而且在全球传播扩散分布逐年增加,因此产生对抗生素环境更强的适应性。[结论] 多重耐药性细菌呈现的表型与携带的多复制子质粒有关,相比较下多复制子质粒比非多复制子质粒有更强的抗性基因携带能力,或许是细菌在强大的抗生素压力下产生的重要响应机制。本研究对于未来探索细菌抗性基因的传播扩散机制具有重要意义。
关键词:多复制子质粒克雷伯氏菌抗生素抗性基因比较基因组学分析
Comparative genomic analysis of Klebsiella reveals extensive transmission of resistance genes mediated by multireplicon resistance plasmids
Xue Wang1,2#, Jianan Zhao2, Zhihui Liu1,2, Limin Zhang3, Han Chang4#, Jiao Qin2, Yunyun Zhu5, Guocheng Hu6, Shijia Hu2, Jianchun Yang2, Zhongxin Jia1,2, Junbao Li7, Houbin Ju8, Chengmin Wang2, Fang Ji2

 , Jianhua Qin1
, Jianhua Qin1

1. College of Veterinary Medicine, Hebei Agricultural University, Baoding 071001, Hebei Province, China;
2. Guangdong Key Laboratory of Animal Conservation and Resource Utilization, Guangdong Public Laboratory of Wild Animal Conservation and Utilization, Institute of Zoology, Guangdong Academy of Science, Guangzhou 510260, Guangdong Province, China;
3. Beidaihe Rehabilitation and Recuperation Center of Joint Logistics Support Force, Qinhuangdao 066100, Hebei Province, China;
4. Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China;
5. Beijing Key Laboratory of Captive Wildlife Technologies, Beijing Zoo, Beijing 100044, China;
6. South China Institute of Environmental Sciences, Ministry of Ecology and Environment, Guangzhou 510530, Guangdong Province, China;
7. Zhengzhou Zoo, Zhengzhou 450008, Henan Province, China;
8. Shanghai Animal Disease Prevention and Control Center, Shanghai 200336, China
Received: 17 November 2020; Revised: 4 January 2021; Published online: 13 January 2021
*Corresponding author: Fang Ji, E-mail: 309533361@qq.com;
Jianhua Qin, E-mail: qjhqqq@126.com.
Foundation item: Supported by the Earmarked Fund for Hebei Dairy Cattle Innovation Team of Modern Agro-industry Technology Research System (HBCT2018120205), by the Key R & D Projects of Hebei Province (20326603D), by the Introduction of Leading Talents Program of Guangdong Academy of Sciences (2016GDASRC-0205), by the GDAS Special Project of Science and Technology Development (2018GDASCX-0107) and by the Open Project of Beijing Key Laboratory of Captive Wildlife Technology in Beijing Zoo (ZDK201909)
#These authors contributed equally to this work.
Abstract: [Objective] To study the relationship between Klebsiella and multireplicon resistance plasmids, and to analyze the mechanism of multireplicon plasmid carrier in response to antibiotic pressure. [Methods] We collected fifty-six isolates of Klebsiella from different wild animals and environments during 2018 to 2020. We analyzed the multidrug resistance (MDR) phenotype by microbroth dilution method, sequenced and analyzed the representative isolates by bacterial genome-wide association study (BGWAS). [Results] Our results revealed that the isolates from non-human sources showed a more extensive drug resistance, and especially stronger resistance to ampicillin (reach 80.36%). In particular, the isolates from Malay pangolin were highly resistant to cephalosporins, chloramphenicol, levofloxacin and sulfamethoxazole. Genomic analysis showed that the resistance plasmids carried by these isolates were related to carry many antibiotic resistance genes. Further analysis of 69 plasmids demonstrated that 28 plasmids were multireplicon plasmids, mainly carrying beta-lactamase genes such as blaCTX-M-15, blaCTX-M-14, blaCTX-M-55, blaOXA-1 and blaTEM-1. According to the analysis of plasmids carried by different isolates, Klebsiella pneumoniae may be an important host of multireplicon plasmid. Plasmid skeleton and structure analysis showed that multireplicon plasmid was formed by the fusion of two or more single plasmids, which made the isolates strong adaptability to the antibiotic environment, and increased the ability of drug-resistant isolates to spread around the world year by year. [Conclusion] The phenotype of multidrug-resistant bacteria is related to the carrying multireplicon plasmid.Multireplicon plasmid has a stronger ability to carry resistance genes than non-multireplicon plasmid, which may be an important mechanism for bacteria response to adapt to stronger antibiotic pressure environment. This phenomenon is of great significance for exploring the transmission and diffusion mechanism of bacterial resistance genes in the future.
Keywords: multireplicon plasmidKlebsiellaantibiotic resistance genescomparative genomic analysis
抗性基因的广泛传播通常与质粒介导的基因水平转移有关,细菌的质粒接合可在不同的种属间发生,共轭质粒是革兰阴性细菌传播抗生素抗性基因簇的重要载体[1]。质粒类型多种多样,依据复制调控或分裂相关的基因元件之间的差异,质粒被定义为不同的不相容群[2],并且每个不相容群又细分为不同的不相容亚群[3]。在肠杆菌科细菌携带的质粒中,最常见的复制子类型包括(Inc)F、A/C、L/M、I1、HI2和N[4],而IncF、IncI、IncA/C、IncL(即IncL/M)、IncN和IncH经常被认为是携带更多种类抗性基因的质粒类型[5]。在肠杆菌科致病菌成员中,质粒呈现高度多样性,这为它们的宿主提供了获得新基因的机会[4]。IncF是在肠杆菌科中广泛流行的窄宿主谱质粒,可携带多种抗生素抗性基因(ARGs),并且在某些特异性抗性基因的扩散起主要作用[6]。大量研究认为IncF亚型质粒IncFIB/IncFII与ARGs间的关系密切[7-12],携带很多重要的抗性基因如头孢菌素酶基因(CTX-M等)、碳青霉烯酶基因(KPC等)、氨基糖苷乙酰化酶(aac6′-1b)[13]和粘菌素耐药基因mcr-1[14]。这些耐药质粒在环境中扩散,对公共卫生安全构成严重威胁。
近年来,一些具有复杂结构的多复制子质粒逐渐受到关注,这种多个复制子共存的情况在IncF不相容群组质粒中较为常见[3]。一项研究对质粒p721005-KPC、p504051-KPC和pA3295-KPC的分析显示,p721005-KPC/p504051-KPC的结构由IncR型骨架及IncFII型共轭移区、维持区和骨架共同组成,pA3295-KPC骨架是由IncFII型骨架区、维持区与IncN1型维持区、共轭转移区杂交而成[15];插入序列IS26介导了IncN1-F33:A-: B-质粒与携带mcr-1的噬菌体样质粒的融合,暗示其在质粒重组过程中发挥重要作用[16];另一项研究发现,携带IncFII家族pKPC-1k30/ pHN7A8复合质粒菌株CG258,在中国多家医院间发生克隆性传播[17]。Pesesky等[18]认为,转座子可能对质粒嵌合体的形成发挥主要作用,其在原核生物类群的基因组或质粒DNA中含量丰富且分布不均,证明这些质粒嵌合体是多种ARGs的有效载体。因此,深入研究在生态环境中的菌群携带质粒复制子类型与ARGs的关系,对于揭示细菌群体对抗生素环境的响应策略以及监测流行病学动态、建立特定质粒传播的干预方案具有重要意义。
在前期研究中,我们获得了来自马来穿山甲的6株多重耐药性肺炎克雷伯氏菌(Klebsiella pneumoniae),发现其对β-内酰胺类、氟喹诺酮类和氯霉素类药物高度耐受,并携带了多复制子复合质粒。为了评估多复制子质粒的流行扩散情况及其与ARGs间的关联,本研究进一步分析了源自多种野生动物和医院临床样本分离到的Klebsiella sp.菌株,利用细菌全基因组关联分析(BGWAS)方法,深入探讨多复制子质粒对菌株响应抗生素环境的生存策略的重要意义。
1 材料和方法 1.1 细菌菌株 本实验室收集和保存了2018–2020年期间分离的克雷伯氏菌(Klebsiella sp.)分离株,选取56株(其中动物来源21株,人类临床来源35株) Klebsiella sp.样本作为研究对象(附表 1)。将这些菌株冻存液重新划线于麦康凯琼脂平板(Beijing SanYao Science & Technology Development Co,Beijing,China),35 ℃下过夜复壮,次日挑取单菌落放入Mueller-Hinton Broth (MHB)培养基中,35 ℃过夜孵育富集,以用于后续实验。
附表 1. 菌株基本数据 Supplementary Table 1. Basic data of strains
| Strain name | Strain accession | Host | Strain | MLST | Genome size of strain/kb | Plasmid name | Plasmid accession | Plasmid Finder | Plasmid size of strain/kb |
| M164-1 | CP063992 | Manis javanica | Klebsiella pneumoniae | 147 | 5279.777 | N/A | N/A | N/A | N/A |
| M169-3 | CP063878 | Manis javanica | Klebsiella pneumoniae | 101 | 5487.893 | pM169-3.1 | CP063879 | IncFIB(K)/IncFII(pKP91) | 131.081 |
| pM169-3.2 | CP063880 | IncR | 37.458 | ||||||
| S161-2 | CP058544 | Manis javanica | Klebsiella pneumoniae | 1269 | 5334.536 | pS161-2.1 | CP058545 | IncFII(K) | 104.999 |
| pS161-2.2 | CP058546 | IncHI1B(pNDM-MAR)/repB | 178.411 | ||||||
| pS161-2.3 | CP058547 | IncFIB(pKPHS1) | 136.732 | ||||||
| S166-1 | CP063945 | Manis javanica | Klebsiella pneumoniae | 1910 | 5257.545 | pS166-1.1 | CP063946 | IncFII(K)/IncHI1B(pNDM-MAR)/IncR/IncR/repB | 433.968 |
| pS166-1.2 | CP063947 | novel | 27.799 | ||||||
| pS166-1.3 | CP063948 | IncFIA(HI1) | 22.895 | ||||||
| pS166-1.4 | CP063949 | IncN/IncN | 83.704 | ||||||
| S174-1 | CP063874 | Manis javanica | Klebsiella quasipneumoniae | 2354 | 5383.063 | pS174-1.1 | CP063875 | IncX1 | 51.856 |
| pS174-1.2 | CP063876 | IncFIA(HI1)/IncR | 56.886 | ||||||
| pS174-1.3 | CP063877 | IncFIB(K)(pCAV1099-114)/IncHI1B(pNDM-MAR)/IncQ1 | 159.310 | ||||||
| S165-1 | CP058548 | Manis javanica | Klebsiella pneumoniae | 231 | 5355.928 | pS165-1.1 | CP058549 | IncFIB(K)(pCAV1099-114)/IncHI1B(pNDM-MAR) | 169.990 |
| pS165-1.2 | CP058550 | IncFII(K) | 105.000 | ||||||
| pS165-1.3 | CP058551 | IncR | 56.644 | ||||||
| M1023-4Ar | CP063851 | Bos mutus | Klebsiella pneumoniae | 1 | 5141.853 | pM1023-4Ar.1 | CP063852 | IncFIB(K)/IncFII(K)/IncQ1 | 262.508 |
| pM1023-4Ar.2 | CP063853 | IncFIB(pKPHS1) | 110.270 | ||||||
| pM1023-4Ar.3 | CP063854 | novel | 77.050 | ||||||
| pM1023-4Ar.4 | CP063855 | IncFII(pHN7A8) | 89.958 | ||||||
| N1059-5At | CP063856 | Bos taurus | Klebsiella pneumoniae | 791 | 5222.128 | pN1059-5At | CP063857 | IncFII(K)/IncQ1 | 92.970 |
| M1026-3Ar | CP063858 | Cervus albirostris | Klebsiella pneumoniae | 1 | 5142.109 | pM1026-3Ar.1 | CP063859 | IncFIB(K)/IncFII(K)/IncQ1 | 262.519 |
| pM1026-3Ar.2 | CP063860 | novel | 77.053 | ||||||
| pM1026-3Ar.3 | CP063861 | IncFII(pHN7A8) | 89.965 | ||||||
| pM1026-3Ar.4 | CP063862 | IncFIB(pKPHS1) | 110.270 | ||||||
| M63-1 | CP063863 | Ailuropoda melanoleuca | Klebsiella pneumoniae | 628 | 5309.740 | pM63-1 | CP063864 | IncFIB(K)(pCAV1099-114) | 161.463 |
| M268-3 | CP064319 | Loxodonta africana | Klebsiella variicola | novel | 5521.406 | pM268-3.1 | CP064320 | novel | 26.136 |
| pM268-3.2 | CP064321 | novel | 28.989 | ||||||
| pM268-3.3 | CP064322 | IncFIB(K) | 31.972 | ||||||
| M297-1 | CP051490 | Macropus Rfus | Klebsiella pneumoniae | 290 | 5301.757 | pM297-1.1 | CP051491 | IncFIB(K)/IncFII(pKP91) | 222.864 |
| pM297-1.2 | CP051492 | IncFII(K)/IncQ1 | 225.763 | ||||||
| M142-3 | CP063867 | Psittacus erithacus | Klebsiella variicola | 3972 | 5615.590 | pM142-3 | CP063868 | IncFIB(K)(pCAV1099-114)/IncFII(K) | 180.701 |
| M911-1 | CP064129 | Aratinga solstitialis | Klebsiella pneumoniae | novel | 5211.192 | pM911-1.1 | CP064130 | novel | 75.711 |
| pM911-1.2 | CP064131 | IncR | 85.824 | ||||||
| pM911-1.3 | CP064132 | novel | 21.377 | ||||||
| S141 | CP063871 | Psittacula alexandri | Klebsiella pneumoniae | 1662 | 5383.698 | pS141.1 | CP063872 | IncFIB(K)(pCAV1099-114)/IncHI1B(pNDM-MAR) | 194.302 |
| pS141.2 | CP063873 | IncFIB(pKPHS1) | 112.160 | ||||||
| S15-2 | CP064046 | Eclectus roratus | Klebsiella quasipneumoniae | 2355 | 5330.587 | pS15-2 | CP064047 | IncFIB(K) | 163.717 |
| S129-1 | CP063954 | Sturnus nigricollis | Klebsiella variicola | novel | 5490.156 | N/A | N/A | N/A | N/A |
| S130-1 | CP063865 | Sturnus nigricollis | Klebsiella pneumoniae | 3753 | 5249.027 | pS130-1 | CP063866 | IncFIB(K) | 150.355 |
| S131-2 | CP063953 | Gracula religiosa | Klebsiella variicola | novel | 5490.142 | N/A | N/A | N/A | N/A |
| S90-2 | CP063881 | Alectoris chukar | Klebsiella pneumoniae | 629 | 5374.786 | pS90-2.1 | CP063882 | IncFIB(pKPHS1) | 110.388 |
| pS90-2.2 | CP063883 | IncFIA(HI1)/IncFII(K) | 109.675 | ||||||
| pS90-2.3 | CP063884 | IncR | 57.825 | ||||||
| M72-2-2 | CP063869 | Panthera tigris amoyensis | Klebsiella quasipneumoniae | 3864 | 5442.690 | pM72-2-2 | CP063870 | IncFIB(pKPHS1) | 108.143 |
| BS329-2 | CP063943 | Homo sapiens | Klebsiella pneumoniae | 1565 | 5244.603 | pBS329-2 | CP063944 | IncFII(K)/IncR | 104.835 |
| BS418 | CP063942 | Homo sapiens | Klebsiella quasipneumoniae | 2144 | 5184.470 | N/A | N/A | N/A | N/A |
| BM343 | CP063939 | Homo sapiens | Klebsiella pneumoniae | 133 | 5358.717 | pBM343.1 | CP063940 | IncFIB(K)/IncFII(K) | 189.015 |
| pBM343.2 | CP063941 | IncR | 68.142 | ||||||
| BS317-1 | CP063936 | Homo sapiens | Klebsiella pneumoniae | 1035 | 5058.265 | pBS317-1.1 | CP063937 | IncFIB(K)/IncFII(pKP91) | 182.144 |
| pBS317-1.2 | CP063938 | IncR | 62.783 | ||||||
| BS326-3 | CP063934 | Homo sapiens | Klebsiella pneumoniae | 1565 | 5244.341 | pBS326-3 | CP063935 | IncFII(K)/IncR | 104.836 |
| BS369-2 | CP063933 | Homo sapiens | Klebsiella variicola | 4115 | 5518.391 | N/A | N/A | N/A | N/A |
| BS375-3 | CP063932 | Homo sapiens | Klebsiella variicola | 4115 | 5517.087 | N/A | N/A | N/A | N/A |
| M186-2 | CP063930 | Homo sapiens | Klebsiella pneumoniae | 111 | 5237.027 | pM186-2 | CP063931 | IncFIB(K)(pCAV1099-114)/IncHI1B(pNDM-MAR) | 191.041 |
| S183-1 | CP063927 | Homo sapiens | Klebsiella pneumoniae | 2158 | 5167.418 | pS183-1.1 | CP063928 | IncFIB(K)(pCAV1099-114) | 208.376 |
| pS183-1.2 | CP063929 | IncFIA/IncFIB(AP001918)/IncFII/IncFII/IncFII(pHN7A8) | 190.929 | ||||||
| S187-1 | CP063926 | Homo sapiens | Klebsiella variicola | 4394 | 5484.033 | N/A | N/A | N/A | N/A |
| S210-3 | CP063925 | Homo sapiens | Klebsiella pneumoniae | 2668 | 5266.775 | N/A | N/A | N/A | N/A |
| BM338-1 | CP063922 | Homo sapiens | Klebsiella pneumoniae | 35 | 5384.549 | pBM338-1.1 | CP063923 | IncI1 | 82.097 |
| pBM338-1.2 | CP063924 | IncFII(K) | 116.734 | ||||||
| BM404-3-1 | CP064044 | Homo sapiens | Klebsiella quasipneumoniae | 2355 | 5330.587 | pBM404-3-1 | CP064045 | IncFIB(K) | 163.717 |
| BM366-1 | CP063921 | Homo sapiens | Klebsiella variicola | 4115 | 5519.299 | N/A | N/A | N/A | N/A |
| BM337-1 | CP063919 | Homo sapiens | Klebsiella pneumoniae | 1565 | 5292.492 | pBM337-1 | CP063920 | novel | 36.933 |
| BM374-1 | CP063917 | Homo sapiens | Klebsiella variicola | novel | 5540.484 | pBM374-1 | CP063918 | novel | 303.115 |
| BS327-2-1 | CP063916 | Homo sapiens | Klebsiella variicola | 4115 | 5518.238 | N/A | N/A | N/A | N/A |
| M186-1-2 | CP063915 | Homo sapiens | Klebsiella variicola | 4394 | 5484.339 | N/A | N/A | N/A | N/A |
| BM336-2-1 | CP063913 | Homo sapiens | Klebsiella pneumoniae | 36 | 5406.489 | pBM336-2-1 | CP063914 | IncFIB(K)/IncFII(K)/IncQ1 | 256.775 |
| BS359-2-1 | CP063912 | Homo sapiens | Klebsiella variicola | 919 | 5510.850 | N/A | N/A | N/A | N/A |
| M186-1 | CP063911 | Homo sapiens | Klebsiella variicola | 4394 | 5484.445 | N/A | N/A | N/A | N/A |
| BS325-2 | CP063910 | Homo sapiens | Klebsiella variicola | 4115 | 5519.050 | N/A | N/A | N/A | N/A |
| M212-2 | CP063908 | Homo sapiens | Klebsiella pneumoniae | 23 | 5419.625 | pM212-2 | CP063909 | IncHI1B(pNDM-MAR)/repB | 214.844 |
| BM327-1 | CP063906 | Homo sapiens | Klebsiella pneumoniae | 1565 | 5299.916 | pBM327-1 | CP063907 | IncFII(K)/IncR | 104.836 |
| BM334-2 | CP063904 | Homo sapiens | Klebsiella pneumoniae | 86 | 5409.887 | pBM334-2 | CP063905 | IncHI1B(pNDM-MAR)/repB | 226.993 |
| BS419-3 | CP063902 | Homo sapiens | Klebsiella quasipneumoniae | 2355 | 5330.355 | pBS419-3 | CP063903 | IncFIB(K) | 163.712 |
| BS324-2 | CP063900 | Homo sapiens | Klebsiella quasipneumoniae | 2558 | 5136.970 | pBS324-2 | CP063901 | IncFIB(K)(pCAV1099-114)/IncHI1B(pNDM-MAR) | 176.438 |
| BS326-1 | CP063898 | Homo sapiens | Klebsiella variicola | 347 | 5473.070 | pBS326-1 | CP063899 | IncFIB(K)(pCAV1099-114) | 139.497 |
| BS419-1 | CP063896 | Homo sapiens | Klebsiella quasipneumoniae | 2355 | 5329.905 | pBS419-1 | CP063897 | IncFIB(K) | 163.257 |
| BM378-2 | CP063893 | Homo sapiens | Klebsiella variicola | 1505 | 5581.862 | pBM378-2.1 | CP063894 | novel | 67.486 |
| pBM378-2.2 | CP063895 | novel | 33.451 | ||||||
| BS359-3 | CP063892 | Homo sapiens | Klebsiella variicola | 4115 | 5171.077 | N/A | N/A | N/A | N/A |
| BS433-2 | CP063890 | Homo sapiens | Klebsiella pneumoniae | 23 | 5467.009 | pBS433-2 | CP063891 | IncHI1B(pNDM-MAR)/repB | 227.749 |
| BS325-3-1 | CP063889 | Homo sapiens | Klebsiella variicola | 4115 | 5598.790 | N/A | N/A | N/A | N/A |
| BM419-3 | CP063887 | Homo sapiens | Klebsiella quasipneumoniae | 2355 | 5339.888 | pBM419-3 | CP063888 | IncFIB(K) | 163.710 |
| BS361-1 | CP063885 | Homo sapiens | Klebsiella pneumoniae | 1565 | 5301.662 | pBS361-1 | CP063886 | IncFII(K)/IncR | 104.834 |
| N/A: Not Applicable (No plasmid). | |||||||||
表选项
1.2 药敏试验 根据CLSI 2019年发布的《抗菌药物敏感性试验执行标准》[19],使用微生物药敏试剂盒(BIO-KONT,China)对所有Klebsiella sp.进行肉汤微量稀释法药敏(MIC)试验,检测氨苄西林、头孢呋辛钠、头孢唑林、头孢曲松、头孢吡肟、氨苄西林/舒巴坦、哌拉西林/他唑巴坦、美罗培南、庆大霉素、阿米卡星、氯霉素、左氧氟沙星、复方新诺明和替加环素(参照EUCAST标准)[20]等14种抗生素药物耐受情况,以评估其多重耐药表型。本实验用参考菌株E. coli ATCC25922作为质量控制菌株。
1.3 全基因组测序 选取56株Klebsiella sp.送测全基因组。具体方法如下:使用Nanopore测序平台[21-22]进行WGS (Biomarker Technologies,China),按照ONT提供的标准方法进行测序,采用NanoDrop、Qubit和0.35%琼脂糖凝胶电泳提取高质量的基因组DNA,检测其纯度、浓度和完整性。BluePippin核酸自动回收系统回收大片段DNA,连接测序试剂盒(SQK-LSK109 Ligation Sequence Kit,Oxford Nanopore Technologies,UK)构建文库,使用DNA损伤修复、末端修复和磁珠纯化等方法进行连接纯化,在机器上对Qubit文库进行定量和测序。提取菌株基因组16S rRNA数据绘制系统发育树,此项工作由NCBI在线BLAST工具和软件MEGA-X共同实现。
1.4 MLST分型和rMLST种属鉴定 从全基因组数据中提取MLST分型所需的7个保守的持家基因序列(rpoB、gapA、mdh、pgi、phoE、infB和tonB),使用Klebsiella sp. (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html)的多位点序列分型数据库,对菌株进行多位点序列分型(MLST),确定序列类型(ST)。同时使用核糖体MLST数据库(rMLST,https://pubmlst.org/species-id)进一步确定菌株种属信息。
1.5 多复制子质粒基因组DNA分析 使用PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/)数据库对质粒进行不相容群分型,利用质粒数据库数据比对测序质粒与已知质粒的相似程度(PLSDB,https://ccb-microbe.cs.uni-saarland.de/plsdb/,参数确定:Max.p-value=0,Max.distance=0.04,Per.Ident≥60%),确认相似质粒的不相容群组数据集,并依据PLSDB数据库确定相似质粒报道年份和全球出现频率。使用抗生素抗性基因专有数据库(CARD,https://card.mcmaster.ca/)对测序质粒进行基因注释,并依据基因组组分分析结果和功能注释(通用数据库Nr、GO、Uniprot、COG、SwissProt、Pfam和KEGG以及专有数据库ISfinder、INTEGRALL和TN Number Registry)对可移动元件和其他特征进行注释。使用软件SnapGene绘制了部分质粒的质粒图谱,选取pM1026-3Ar.1数据为代表质粒构建质粒融合模式图。
2 结果和分析 2.1 Klebsiella sp. 的耐药表型分析 药敏结果显示,56株Klebsiella sp.中有45株对氨苄西林表现出较强的耐药性,耐药率高达80.36%。马来穿山甲分离株对头孢类药物高度耐受,同时耐受药物还包括氯霉素、左氧氟沙星和复方新诺明,非马来穿山甲来源的动物分离株仅M1023-4Ar、N1059-5At、M1026-3Ar、M297-1、S141和S90-2对上述5类药物耐受性较强(分离自牦牛、奶牛、白唇鹿、赤袋鼠、绯胸鹦鹉和石鸡)。人类来源临床分离菌株耐药性相对较弱,仅BS375-3和S183-1对头孢呋辛钠、头孢唑林、头孢曲松和头孢吡肟显示出较强的耐药性(图 1)。值得注意的是,所有克雷伯氏菌主要对β-内酰胺类药物表现较强的耐受表型,但均对美罗培南和替加环素敏感。
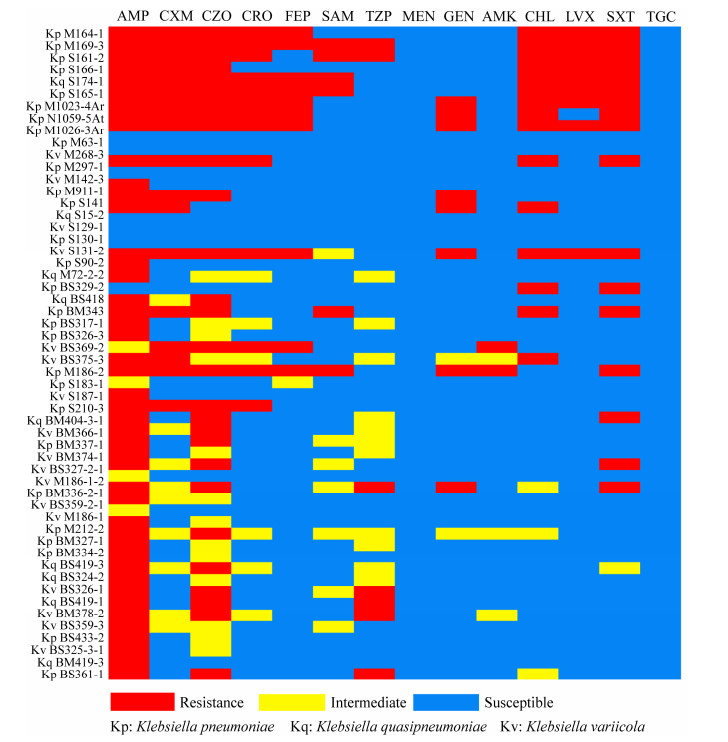 |
| 图 1 Klebsiella sp. 分离株的耐药表型 Figure 1 Drug resistance phenotype of Klebsiella sp. isolates. red: resistance; yellow: intermediate; blue: susceptible. AMP: ampicillin; CXM: cefuroxim; CZO: cefazolin; CRO: ceftriaxone; FEP: cefepime; SAM: ampicillin/sulbactam; TZP: piperacillin/tazobactam; MEM: meropenem; GEN: gentamicin; AMK: amikacin; CHL: chloramphenicol; LVX: levofloxacin; SXT: trimethoprim/sulfamethoxazole; TGC: tigecycline. |
| 图选项 |
2.2 多复制子质粒与宿主Klebsiella sp. 基因组的相关性分析 根据16S rRNA和rMLST鉴定结果,在56株Klebsiella sp.中,29株确定为肺炎克雷伯氏菌(K. pneumoniae),携带质粒菌株有27株(93.10%);9株确定为准肺炎克雷伯氏菌(K. quasipneumoniae),携带质粒菌株有8株(88.89%);18株确定为变栖克雷伯氏菌(K. variicola),携带质粒菌株有5株(27.78%)。MLST分型鉴定出33种不同ST型,其中16种ST型的菌株携带了多复制子质粒,包括ST1565型(4株),ST1和ST23型(各2株),ST36、ST86、ST101、ST111、ST133、ST231、ST290、ST791、ST1035、ST1662、ST1910、ST2558和ST3972型(各1株),研究发现多复制子质粒与菌株ST型并没有明显相关性(附表 1)。CARD数据库抗性基因注释结果表明,总体上看,Klebsiella sp.染色体DNA上通常携带β-内酰胺类药物固有抗性基因blaDHA、blaSHV、blaLEN和blaOKP-B,每个菌株至少携带两个β-内酰胺类药物抗性基因。此外,染色体DNA上还携带有ABC家族、MFS家族、SMR家族或MATE家族等耐药泵基因。
基于PlasmidFinder数据库,不相容群(Inc)分型是基于质粒DNA中复制子(rep)序列[23]。本研究中,对69个质粒DNA鉴定结果发现,30个质粒DNA属于单一复制子质粒,28个质粒DNA属于多复制子质粒以及11个质粒DNA属于未知复制子类型的质粒(unknown或novel) (附表 1)。进一步分析复制子类型与携带抗性基因的关系,发现多复制子质粒可携带的抗性基因数量明显多于非多复制子质粒(图 2),携带抗性基因数量≥5个的单一复制子型质粒比例仅为26.67%,novel质粒仅为9.09%,而多复制子质粒比例高达75.00%。当携带的抗性基因数量≥10个时,多复制子质粒占比为35.71%,且少数质粒携带抗性基因数量甚至大于25个,而单一复制子质粒只有1个,novel型质粒尚未检出。质粒携带抗性基因种类呈现多元化,多复制子质粒主要携带blaCTX-M-15、blaCTX-M-14、blaCTX-M-55、blaOXA-1和blaTEM-1等β-内酰胺酶基因及aac(6')-Ib-cr、aph(6)-Id和aph(3'')-Ib等氨基糖苷酶基因,部分质粒还携带磺胺类抗性基因sul1、sul2和sul3,四环素类抗性基因tetA、tetG,介导氟喹诺酮类抗性的qnr基因家族,以及介导氯霉素抗性的floR基因(附表 2)。综上所述,多复制子质粒携带抗生素抗性基因的能力要远远强于非多复制子质粒,Klebsiella sp.摄取并携带多复制子质粒或许是其应对抗生素环境压力的重要生存策略。
 |
| 图 2 质粒携带抗性基因数量的比较分析 Figure 2 Comparative analysis of drug resistance genes carried by plasmids. |
| 图选项 |
附表 2. 多复制子质粒基因组信息 Supplementary Table 2. Multireplicon plasmid genome information
| Name | Strain | Size/kb | G+C Content/% | Plasmid Finder | Number of Multi-drug resistance genes | Type of multi-drug resistance genes |
| pBM343.1 | Klebsiella pneumoniae | 189.015 | 52.68 | IncFIB(K)/IncFII(K) | 0 | |
| pM169-3.1 | Klebsiella pneumoniae | 131.081 | 50.89 | IncFIB(K)/IncFII(pKP91) | 11 | blaCTX-M-15, aac(6')-Ib-cr, tetG, aph(6)-Id, aph(3'')-Ib, sul2, blaOXA-1, arlR, mexD, oprM, mtrA |
| pBS317-1.1 | Klebsiella pneumoniae | 182.144 | 52.20 | IncFIB(K)/IncFII(pKP91) | 5 | mtrA, oprM, mexD, arlR, msrC |
| pM297-1.1 | Klebsiella pneumoniae | 222.864 | 51.76 | IncFIB(K)/IncFII(pKP91) | 9 | aph(3'')-Ib, aph(6)-Id, mtrA, oprM, mexD, arlR, blaCTX-M-14, blaTEM-191, qnrS1 |
| pM142-3 | Klebsiella variicola | 180.701 | 51.89 | IncFIB(K)(pCAV1099-114)/IncFII(K) | 7 | triC, triB, adeR, oprM, emrA, emrB, nmcR |
| pM1023-4Ar.1 | Klebsiella pneumoniae | 262.508 | 51.84 | IncFIB(K)/IncFII(K)/IncQ1 | 26 | mefB, sul3, qacH, aadA, cmlA1, aadA2, dfrA12, sul2, aph(3'')-Ib, aph(6)-Id, aph(3')-Ia, aac(3)-IIa, aac(6')-Ib-cr, blaOXA-1, catB3, arr-3, emrE, sul1, mrx, mphA, catII, nmcR, arlR, adeB, oprM, mtrA |
| pM1026-3Ar.1 | Klebsiella pneumoniae | 262.519 | 51.84 | IncFIB(K)/IncFII(K)/IncQ1 | 27 | mtrA, oprM, adeB, arlR, nmcR, tetG, catII, mphA, Mrx, sul1, emrE, arr-3, catB3, blaOXA-1, aac(6')-Ib-cr, aac(3)-IIa, aph(3')-Ia, aph(6)-Id, aph(3'')-Ib, sul2, dfrA12, aadA2, cmlA1, aadA, qacH, sul3, mefB |
| pBM336-2-1 | Klebsiella pneumoniae | 256.775 | 51.94 | IncFIB(K)/IncFII(K)/IncQ1 | 25 | nmcR, tetG, mphA, Mrx, sul1, emrE, arr-3, catB3, blaOXA-1, aac(6')-Ib-cr, aac(3)-IIa, adeB, oprM, mtrA, aph(6)-Id, aph(3'')-Ib, sul2, dfrA12, aadA2, cmlA1, aadA, qacH, sul3, mefB, arlR |
| pS141.1 | Klebsiella pneumoniae | 194.302 | 50.66 | IncFIB(K)(pCAV1099-114)/IncHI1B(pNDM-MAR) | 6 | ceoB, acrA, cmlv, mexT, vanTrL, fyuA |
| pM186-2 | Klebsiella pneumoniae | 191.041 | 50.21 | IncFIB(K)(pCAV1099-114)/IncHI1B(pNDM-MAR) | 5 | cmlv, mexT, vanTrL, fyuA, oleI |
| pBS324-2 | Klebsiella quasipneumoniae | 176.438 | 49.93 | IncFIB(K)(pCAV1099-114)/IncHI1B(pNDM-MAR) | 5 | nmcR, arlR, adeB, oprM, mtrA |
| pS165-1.1 | Klebsiella pneumoniae | 169.990 | 50.86 | IncFIB(K)(pCAV1099-114)/IncHI1B(pNDM-MAR) | 11 | nmcR, arlR, adeB, oprA, mtrA, sul2, floR, dfrA12, aadA2, aph(3')-Ia, tetG |
| pS174-1.3 | Klebsiella quasipneumoniae | 159.310 | 50.83 | IncFIB(K)(pCAV1099-114)/IncHI1B(pNDM-MAR)/IncQ1 | 8 | aph(6)-Id, aph(3'')-Ib, sul2, blaTEM-1, arlR, adeB, oprM, mtrA |
| pBS329-2 | Klebsiella pneumoniae | 104.835 | 52.83 | IncFII(K)/IncR | 2 | mexT, tetG |
| pBS326-3 | Klebsiella pneumoniae | 104.836 | 52.83 | IncFII(K)/IncR | 2 | tetG, mexT |
| pBM327-1 | Klebsiella pneumoniae | 104.836 | 52.83 | IncFII(K)/IncR | 2 | mexT, tetG |
| pBS361-1 | Klebsiella pneumoniae | 104.834 | 52.83 | IncFII(K)/IncR | 2 | mexT, tetG |
| pBM334-2 | Klebsiella pneumoniae | 226.993 | 50.24 | IncHI1B(pNDM-MAR)/repB | 8 | mtrA, oprM, mexD, arlR, mdtH, H-NS, oleI, mexS |
| pS161-2.2 | Klebsiella pneumoniae | 178.411 | 50.80 | IncHI1B(pNDM-MAR)/repB | 16 | aph(3')-Ia, mphA, mrx, sul1, nmcR, blaDHA-1, qnrB4, emrE, arr-3, catB3, blaOXA-1, aac(6')-Ib-cr, mtrA, oprM, mexD, arlR |
| pBS433-2 | Klebsiella pneumoniae | 227.749 | 50.03 | IncHI1B(pNDM-MAR)/repB | 8 | mtrA, oprM, mexD, arlR, H-NS, mdtH, oleI, mexS |
| pM212-2 | Klebsiella pneumoniae | 214.844 | 49.75 | IncHI1B(pNDM-MAR)/repB | 7 | mtrA, oprM, mexD, mdtH, H-NS, oleI, mexS |
| pN1059-5At | Klebsiella pneumoniae | 92.970 | 52.92 | IncFII(K)/IncQ1 | 18 | sul1, emrE, aadA16, dfrA5, arr-3, aac(6')-Ib-cr, aac(3)-IIa, aph(3')-Ia, aph(6)-Id, aph(3'')-Ib, sul2, qnrS1, blaCTX-M-3, blaTEM-1, floR, tetG, mphA, mrx |
| pM297-1.2 | Klebsiella pneumoniae | 225.763 | 52.59 | IncFII(K)/IncQ1 | 17 | sul2, aph(3')-Ia, aac(3)-IIa, floR, tetG, mphA, mrx, sul1, qnrB2, sul1, emrE, aadA16, arr-3, aac(6')-Ib-cr, blaTEM-1, blaCTX-M-3, qnrS1 |
| pS183-1.2 | Klebsiella pneumoniae | 190.929 | 52.40 | IncFIA/IncFIB(AP001918)/IncFII/IncFII/IncFII(pHN7A8) | 16 | aac(6')-Ib-cr, aac(3)-IIa, ermB, mphA, mrx, sul1, emrE, aadA5, blaCTX-M-55, fosA3, rmtB, blaTEM-1, sul1, emrE, arr-3, catB3 |
| pS90-2.2 | Klebsiella pneumoniae | 109.675 | 51.09 | IncFIA(HI1)/IncFII(K) | 2 | arlR, smeR |
| pS174-1.2 | Klebsiella quasipneumoniae | 56.886 | 52.95 | IncFIA(HI1)/IncR | 9 | aadA16, emrE, sul1, qnrB6, emrE, sul1, aac(6')-Ib-cr, arr-3, tetA |
| pS166-1.1 | Klebsiella pneumoniae | 433.968 | 51.99 | IncFII(K)/IncHI1B(pNDM-MAR)/IncR/IncR/repB | 26 | arlR, mexD, oprM, mtrA, aac(6')-Ib-cr, blaOXA-1, catB3, arr-3, emrE, sul1, qnrB4, blaDHA-1, nmcR, sul1, mrx, mphA, aph(3')-Ia, arlR, oprM, vanSN, sul2, floR, tetG, blaTEM-191, qnrS1, tetA |
| pS166-1.4 | Klebsiella pneumoniae | 83.704 | 51.45 | IncN/IncN | 4 | blaTEM-1, blaCTX-M-15, qepA, blaCTX-M-15 |
表选项
本研究又对K. pneumoniae、K. variicola和K. quasipneumoniae 3个种携带的质粒和耐药基因情况进行了比较分析,以评估不同种Klebsiella sp.与多复制子质粒间的关系。从整体上看,多复制子复合质粒分布广泛,与宿主菌株来源并无明显相关性,但发现多复制子复合质粒多由K. pneumoniae携带,而K. quasipneumoniae和K. variicola携带很少。利用16S rRNA构建了菌株系统发育树,发现明显分为5个分支,携带多复制子质粒的K. pneumoniae菌株明显在同一个进化分枝上,暗示这些菌株存在很近的同源关系(图 3)。值得注意的是,携带多复制子质粒的菌株多来源于野生动物,而并非人类(图 3),暗示野生动物可能是多重耐药菌的重要的保藏宿主。因此,K. pneumoniae被认为可能是多复制子质粒的重要宿主菌,野生动物又是多重耐药的K. pneumoniae的重要宿主。
 |
| 图 3 基于菌株16S rRNA序列的系统发育树 Figure 3 Contruction of Phylogenetic Tree based on 16S rRNA sequences. Inner layer 1: isolation source; inner layer 2: species; inner layer 3: isolates carrying plasmid; outer layer: plasmids carrying with β-lactam resistance genes and aminoglycoside resistance genes. |
| 图选项 |
2.3 多复制子质粒基因组成和骨架分析 本研究中,共发现28个多复制子质粒,其中IncFIB/IncFII/(IncQ1)型8个,IncFIB/IncHI1B/ (IncQ1)型5个,IncFII/IncR型4个,IncHI1B/repB型4个,IncFII/IncQ1型2个,IncFIA/IncFIB/ IncFII/IncFII/IncFII、IncFIA/IncFII、IncFIA/IncR、IncFII/IncHI1B/IncR/IncR/repB和IncN/IncN型各1个。除IncFII/IncR型和IncFIA/IncFII型质粒外,其余多复制子质粒都显示能够容纳更多的抗性基因。当质粒上的抗性基因数量增多时,质粒碱基数也明显增多,但质粒总体G+C含量并未明显增加,这些质粒可以将多种不同ST型K. pneumoniae作为宿主并被其稳定携带(附表 2)。
质粒骨架通常由复制调控区、分配系统、结合转移系统和质粒维持区组成[2, 24],选取28个多复制子质粒中的5种类型进行Blast分析,筛选出5个质粒骨架基因高同源性(同源性≥93%)同时携带耐药基因较多的质粒用于进一步分析质粒重组(表 1),质粒骨架基因包括rep基因、sop/par基因、tra/trb基因区域、抗生素抗性基因和可移动元件基因。通过质粒图谱发现,质粒pM1026- 3Ar.1 IncFIB (属于IncFIB/IncFII/(IncQ1)型)复制子的repB基因上下游基因结构为IS609 insQ- sopB-sopA-repB-intI-parD-ybdN-parB-IS110 tnp,IncFII复制子的repA基因上下游基因结构为Tn5393 tnpA-repA-repA2-pld-IS1 insB,IncQ1复制子的不完整repA基因被包含在18个抗性基因和5个可移动元件组成的复合结构之间,其上下游基因结构为repC-repA-IS431mec tnp-Tn21 tnpM-intI1;质粒pS174-1.3 IncFIB [属于IncFIB/IncHI1B/ (IncQ1)型]复制子的repB基因上下游结构为intM-int-repB-sopA-sopB,IncHI1B复制子的repA基因上下游基因结构为IS431mec tnp-other-repA- yadA-other-Tn903 tnp,IncQ1复制子的repA基因上下游基因结构为repC-repA-IS431mec tnp-Tn3 tnpR,它也被4个抗性基因和2个转座酶基因组成的复合结构包围在内;质粒pBS361-1 IncFII (属于IncFII/IncR型)复制子的repA1基因上下游基因结构为IS431mec tnp-yedK-repA1-pld-aer-Tn4653 tnpR-Tn1721 tnpA,IncR复制子的repB基因上下游基因结构为parM-parA-other-repB- other-resD,4个IncFII/IncR结构相似度很高(≥99%),暗示其可能为相同质粒的不同拷贝,携带的抗性基因数量都是2个,且均为mexT和tetG (附表 2)。
表 1. 代表性多复制子质粒基因组信息 Table 1. Plasmid genome information of representative multireplicon
| Name | Size/kb | G+C content/ % | Plasmid finder | Number of multi-drug resistance genes | Type of multi-drug resistance genes |
| pBS361-1 | 104.834 | 52.83 | IncFII(K)/IncR | 2 | mexT, tetG |
| pM297-1.2 | 225.763 | 52.59 | IncFII(K)/IncQ1 | 17 | sul2, aph(3')-Ia, aac(3)-IIa, floR, tetG, mphA, mrx, sul1, qnrB2, sul1, emrE, aadA16, arr-3, aac(6')-Ib-cr, blaTEM-1, blaCTX-M-3, qnrS1 |
| pM1026-3Ar.1 | 262.519 | 51.84 | IncFIB(K)/IncFII(K)/IncQ1 | 27 | mtrA, oprM, adeB, arlR, nmcR, tetG, catII, mphA, mrx, sul1, emrE, arr-3, catB3, blaOXA-1, aac(6')-Ib-cr, aac(3)-IIa, aph(3')-Ia, aph(6)-Id, aph(3'')-Ib, sul2, dfrA12, aadA2, cmlA1, aadA, qacH, sul3, mefB |
| pS161-2.2 | 178.411 | 50.80 | IncHI1B(pNDM-MAR)/repB | 16 | aph(3')-Ia, mphA, mrx, sul1, nmcR, blaDHA-1, qnrB4, emrE, arr-3, catB3, blaOXA-1, aac(6')-Ib-cr, mtrA, oprM, mexD, arlR |
| pS174-1.3 | 159.310 | 50.83 | IncFIB(K) (pCAV1099-114)/IncHI1B(pNDM-MAR)/IncQ1 | 8 | aph(6)-Id, aph(3'')-Ib, sul2, blaTEM-1, arlR, adeB, oprM, mtrA |
表选项
质粒pS161-2.2 IncHI1B (属于IncHI1B/repB型)复制子的repA基因上下游基因结构为Tn1721 tnpA-other-other-repA-other-other-Tn903 tnp,repB复制子的repA基因上下游基因结构为sopB-sopA- repA-int-intM,此类型质粒也具有较强的抗性基因携带能力,且拥有两套分配系统sopA-sopB/parA-parB;质粒pM297-1.2 IncFII (属于IncFII/ IncQ1型)复制子的repA基因与IncQ1复制子的repA基因物理距离十分接近(4038 bp),其结构为Tn903 tnp-other-repA(IncFII)-pld-aer-IS431mec tnp- other-repA(IncQ1)-repC。值得注意的是,IncQ1复制子的repA基因虽然没有处于抗性基因和可移动元件复合结构内,但repA-repC基因之后总是跟随一个磺胺类抗性基因sul2 (图 4)。质粒pM279-1.2、pM1026-3Ar.1和pS174-1.3均携带IncQ1复制子,且携带的抗性基因数量分别为17、27和8,对β-内酰胺类、氨基糖苷类和磺胺类抗性基因的携带能力较强(表 1),提示抗性基因携带能力可能与IncQ1型质粒有关。另外,质粒pM1026-3Ar.1、pBS361-1和pM297-1.2都存在共轭结合转移系统基因簇traABCDEFGHIGKLM NOPQRSTUVWXY/trbABCDEFGHIGKLMN-finO样结构,pM297-1.2甚至存在2套较为完整的转移系统。5个质粒都至少在其中1个rep基因的上下游发现了可移动元件(IS序列、转座子或整合酶基因),这可能是由于可移动元件能够作为融合关键位点,进而介导质粒片段融合而形成复合结构。
 |
| 图 4 多复制子质粒图谱 Figure 4 Multireplicon plasmid map. pM1026-3Ar.1, pS174-1.3, pBS361-1, pS161-2.2 and pM297-1.2 are selected as representatives, and different colors represent different functional region. |
| 图选项 |
多复制子质粒pM1026-3Ar.1基因组DNA上含有3个复制子结构IncFIB、IncFII和IncQ1,通过blast和PLSDB数据库查询到3个单一复制子质粒与其复制子高度同源(同源区域≥97%),其中IncFIB复制子区域(32883–96121 bp)与2018年在美国分离的人类宿主K. pneumoniae质粒p203同源性高(GenBank登录号:NZ_CP021166.1,Query Cover=40%),IncFII复制子区域(94997–1421 bp)与2018年在日本某污水处理厂分离的产KPC-2酶K. pneumoniae菌株质粒pGSU10-3-2同源性高(GenBank登录号:NZ_AP018673.1,Query Cover=31%),IncQ1复制子区域(451–5462 bp)与2012年分离的S. enterica亚种质粒pSRC15同源性高(GenBank登录号:NC_013104.1,Query Cover=1%),暗示多复制子质粒pM1026-3Ar.1可能由这3种质粒融合而成。根据这些质粒的结构信息绘制了可能的质粒融合模式图(图 5),质粒pM1026-3Ar.1全长262519 bp,推测p203样质粒作为基础的质粒结构提供了约63239 bp的基因片段,pGSU10-3-2样质粒提供了共轭转移区约40295 bp的基因片段,p203和pGSU10-3-2样质粒也为pM1026-3Ar.1提供了大量抗性基因和可移动元件(图 5-A)。而pSRC15样质粒提供了约5012 bp基因片段,包括aph(6)-Id、aph(3″)-Ib、sul2、blaOXA-1和aac(6′)-Ib-cr等抗生素抗性基因以及少量外排泵基因(图 5-B)。因此,在细菌面对外界抗生素的压力不断选择下,多个单复制子质粒向多复制子融合质粒的转变,为降低生存成本这种质粒融合模式或许是细菌对较强抗生素环境压力产生适应性进化的结果。
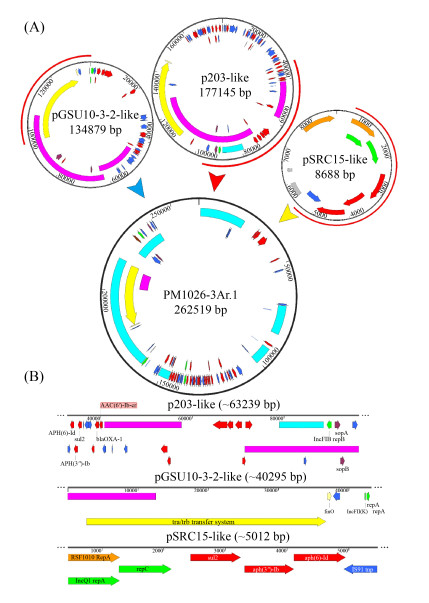 |
| Figure 5 The potential fusion process schematic diagram of plasmid pM1026-3Ar.1. A: The fusion process of plasmid P203, pGSU10-3-2 and pSRC15, the red semicircle on the outside of the plasmid indicates the fusion region; B: The specific structure and genetic composition of the fusion fragments provided by plasmids P203, pGSU10-3-2 and pSRC15. |
| 图选项 |
3 讨论 克雷伯氏菌属菌株Klebsiella sp.染色体DNA上大多仅携带固有抗性基因,而未携带质粒的菌株对抗生素耐受性较弱(图 1),因此固有抗性基因不足以赋予菌株对多种药物的耐受表型,质粒上携带的抗性基因是菌株多重耐药性表型的关键。我们研究发现,克雷伯氏菌属菌株携带的质粒复制子主要为IncFIB/IncFII/(IncQ1)型(无论其是否为多复制子质粒) (附表 1),相关研究表明,在中国四川、山东2省抽检的7个猪样品分离的肠沙门氏菌S. enterica分离株中,检测到与IncFIB/ IncFII型复制子相连锁的高水平甘氨酰环素类抗性基因tetX4[10],与转座子Tn1548相关的抗性基因qnrB2、aac(6′)-Ib-cr和blaCTX-M-3共定位于S. enterica亚种IncFII质粒上[25],IncFIB和IncFII型质粒是blaNDM、blaKPC、blaCTX-M和blaOXA等超广谱β-内酰胺酶(EBSLs)基因的适合载体[26-28],且具有容纳并稳定携带多种抗性基因的能力。
我们研究还发现K. pneumoniae携带的多复制子质粒主要由IncFIB、IncFII和IncQ1型复制子融合产生,IncQ1型质粒的宿主范围广泛,可以在细菌中稳定存在,是一种可被诱导结合的质粒类型,已知其能够编码抗性基因sul2、strAB、tetA和blaKPC-2,分别介导对磺胺甲恶唑、链霉素、四环素和亚胺培南等药物的抗性,证明该质粒与耐药性菌株的传播密切相关[29-30]。K. pneumoniae携带的pBS361-1、pM297-1.2、pM1026-3Ar.1、pS161-2.2和pS174-1.3均为多复制子质粒,单复制子质粒为其提供了rep基因和sop/par基因的框架区域、tra/trb-finO基因的结合转移区域、部分功能基因的维持区域以及各类抗生素抗性基因(图 4)。
多复制子质粒携带抗性基因的能力明显强于非多复制子质粒,研究中发现其携带抗性基因数量通常都多于5个,其质粒碱基数量随着携带抗性基因数量的增多而增多,当以细菌携带质粒的适应性成本与质粒编码抗生素抗性决定因素数量作为变量时,两变量呈负相关[31]。在我们的研究中发现,多复制子质粒并非单独与菌株共存,更多Klebsiella sp.在携带多复制子质粒的同时也会同时携带一个或数个单复制子质粒或未知类型质粒,这些质粒基因组大小和携带基因数量远远小于多复制子质粒(附表 1),甚至有些质粒似乎缺乏对宿主有用的基因[32]。有研究认为,肠杆菌科和芽孢杆菌中同时携带大质粒和小质粒时,小质粒的出现频率高于预期值,单个细菌携带多个无明显关联的可结合质粒,认为其携带成本比每个质粒单独携带成本更高,但实际结果却是携带两个结合型质粒的成本低于只携带单个质粒的成本[33]。
自然界中细菌菌株携带多个质粒现象十分常见,大质粒(100–400 kb)和小质粒(<25 kb)间的联系比预想的更为密切,质粒间的正上位性使质粒携带的成本负担最小化且有利于提高质粒稳定性[34]。我们研究发现多复制子质粒总G+C含量并未明显升高,少量质粒的G+C%甚至有所降低(附表 2),G+C含量是影响菌株携带质粒适应性成本的关键因素之一,当其降低时适应性成本也会随之降低[35]。多个复制子配合不同来源的维持接合区,使得多复制子质粒具备了稳定复制的能力[15],可能允许质粒以类似于分配系统的方式规避不相容性,或者提供一种改变质粒拷贝数以调节质粒基因表达的方式,从而达到拓宽窄宿主谱质粒宿主范围的目的[36],获得额外抗性质粒或抗性突变也会提高抗性菌株的适应性[37],这也解释了研究中发现的多复制子质粒类型为何如此复杂的同时仍然具备携带大量耐药基因并与菌株共存的现象。
本研究重点分析的5个质粒,rep基因上下游均含有包括插入序列、转座子和整合酶在内的大量可移动元件(图 4),表明多复制子质粒的形成与可移动元件间的联系密不可分。已有研究证明,多复制子质粒形成机制以指向插入序列(IS)介导的质粒融合重组事件为主,插入序列ISPa40介导子质粒pSa44-CRO、pSa44-CIP发生同源重组,整合为同时编码环丙沙星和头孢曲松耐药IncI1/ IncFIB型质粒pSa44-CIP-CRO,并在S. typhimurium中传播[38],IncFIB型质粒pBJ114-141与IncX3型质粒pBJ114-46的融合则可能由插入序列ISKpn19、IS3000和ISAba125间的转座事件共同介导[39],Leelaporn等[40]研究发现插入序列IS257元件可以通过未明确的复制转座机制介导质粒整合。IS26是在各种抗性质粒中经常被检测到的插入序列元件,它可介导IncN1-F33:A-: B-质粒与携带mcr-1的噬菌体样质粒的融合,也可同时介导毒力、抗性和高传播性的S. enteritidis的多复制子质粒pSE380T (IncHI2/IncFIA)的出现[16, 41]。本研究中,以质粒pM1026-3Ar.1为代表探讨质粒融合模式,认为p203样质粒提供了主要框架结构,pGSU10-3-2样质粒提供了共轭转移区,pSRC15样质粒提供了少量抗性基因(图 5),IS1、IS26、IS609和IS903B分布在这些结构两端,与插入序列介导质粒融合的报道相一致[16, 38-41],3个子质粒携带的IS元件可能是发生质粒融合的关键位点,在介导多复制子质粒形成的同时也为其提供了大量新的基因元件。
我们进一步研究了多复制子质粒与宿主菌株的关联性,认为多复制子质粒与K. pneumoniae具有良好的相容性,K. quasipneumoniae则具备携带质粒的潜力,而K. variicola并不是多复制子质粒的最佳宿主,这些菌株在以马来穿山甲为主的野生动物来源菌株中广泛分布(图 3)。多复制子质粒与菌株ST型并无明显相关性,在多种ST型中均可检出多复制子质粒(附表 1)。针对多复制子质粒pBS361-1、pM297-1.2、pM1026-3Ar.1、pS161-2.2和pS174-1.3的全球分布分析,发现这些质粒在中国及其周边国家广泛分布,检出频率逐年升高(图 6)。这些相似质粒中多复制子质粒占比分别为50.00%、37.10%、90.00%、92.86%和33.33%,暗示多复制子质粒正在扩散,由于其在具备良好的抗性基因携带能力同时也兼顾了低成本适应性优势,大量获得性革兰氏阴性多重耐药菌株的出现似乎与多复制子质粒广泛传播存在密切关联,或许是造成亚洲地区严重抗生素耐药的关键因素之一[42]。
 |
| 图 6 pBS361-1、pM297-1.2、pM1026-3Ar.1、pS161-2.2和pS174-1.3相似质粒的全球分布 Figure 6 Global distribution of similar plasmids pBS361-1, pM297-1.2, pM1026-3Ar.1, pS161-2.2 and pS174-1.3. |
| 图选项 |
总之,我们的研究证实,可移动元件介导质粒重组事件产生多复制子质粒,促进了细菌间各种抗性基因的积累,这些多复制子质粒具备容纳多种抗生素抗性基因的能力,携带该类质粒菌株的生存能力得以提升,但适应性成本并未因此而显著增加。因此,野生动物来源菌株与多复制子质粒间存在密切联系,作为其应对外界抗生素压力的重要生存手段,此现象使得菌株抗生素抗性机制变得更为复杂,从而造成不容忽视的公共健康隐患。
数据可用性声明 研究中所使用的测序样本序列资料,包括宿主、采集地和日期信息已提交至GenBank,具体登录号信息请见附表 1。
References
| [1] | Moran RA, Richardson IA, Hall RM. Analysis of two B/O plasmids, R805a from 1972 and pCERC6 from 2008, reveals extensive mosaicism in B/O plasmid backbones. Plasmid, 2019, 102: 62-70. DOI:10.1016/j.plasmid.2019.02.005 |
| [2] | Novick RP. Plasmid incompatibility. Microbiological Reviews, 1987, 51(4): 381-395. DOI:10.1128/mr.51.4.381-395.1987 |
| [3] | Villa L, García-Fernández A, Fortini D, Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. Journal of Antimicrobial Chemotherapy, 2010, 65(12): 2518-2529. DOI:10.1093/jac/dkq347 |
| [4] | Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrobial Agents and Chemotherapy, 2009, 53(6): 2227-2238. DOI:10.1128/AAC.01707-08 |
| [5] | Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. Journal of Antimicrobial Chemotherapy, 2018, 73(5): 1121-1137. |
| [6] | Carattoli A. Plasmids in Gram negatives: molecular typing of resistance plasmids. International Journal of Medical Microbiology, 2011, 301(8): 654-658. DOI:10.1016/j.ijmm.2011.09.003 |
| [7] | Wang QJ, Sun J, Li J, Ding YF, Li XP, Lin JX, Hassan B, Feng YJ. Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome, 2017, 5(1): 70. DOI:10.1186/s40168-017-0288-0 |
| [8] | Bougnom BP, Thiele-Bruhn S, Ricci V, Zongo C, Piddock LJV. Raw wastewater irrigation for urban agriculture in three African cities increases the abundance of transferable antibiotic resistance genes in soil, including those encoding extended spectrum β-lactamases (ESBLs). Science of the Total Environment, 2020, 698: 134201. DOI:10.1016/j.scitotenv.2019.134201 |
| [9] | Pérez-Vázquez M, Sola Campoy PJ, Ortega A, Bautista V, Monzón S, Ruiz-Carrascoso G, Mingorance J, González-Barberá EM, Gimeno C, Aracil B, Sáez D, Lara N, Fernández S, González-López JJ, Campos J, Kingsley RA, Dougan G, Oteo-Iglesias J, Spanish NDM Study Group. Emergence of NDM-producing Klebsiella pneumoniae and Escherichia coli in Spain: phylogeny, resistome, virulence and plasmids encoding blaNDM-like genes as determined by WGS. Journal of Antimicrobial Chemotherapy, 2019, 74(12): 3489-3496. DOI:10.1093/jac/dkz366 |
| [10] | Bai L, Du PC, Du YJ, Sun HH, Zhang P, Wan YP, Lin Q, Fanning S, Cui SH, Wu YN. Detection of plasmid-mediated tigecycline-resistant gene tet (X4) in Escherichia coli from pork, Sichuan and Shandong Provinces, China, February 2019. Eurosurveillance, 2019, 24(25): 1900340. |
| [11] | Wu WJ, Feng Y, Tang GM, Qiao F, McNally A, Zong ZY. NDM Metallo-β-lactamases and their bacterial producers in health care settings. Clinical Microbiology Reviews, 2019, 32(2): e00115-18. |
| [12] | Deng Y, Zeng Z, Chen S, He L, Liu Y, Wu C, Chen Z, Yao Q, Hou J, Yang T, Liu JH. Dissemination of IncFII plasmids carrying rmtB and qepA in Escherichia coli from pigs, farm workers and the environment. Clinical Microbiology and Infection, 2011, 17(11): 1740-1745. DOI:10.1111/j.1469-0691.2011.03472.x |
| [13] | Hawkey PM, Jones AM. The changing epidemiology of resistance. Journal of Antimicrobial Chemotherapy, 2009, 64(S1): i3-i10. |
| [14] | Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian GB, Dong BL, Huang XH, Yu LF, Gu DX, Ren HW, Chen XJ, Lv LC, He DD, Zhou HW, Liang ZS, Liu JH, Shen JZ. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious Diseases, 2016, 16(2): 161-168. DOI:10.1016/S1473-3099(15)00424-7 |
| [15] | Qu DF, Shen Y, Hu LF, Jiang XY, Yin Z, Gao B, Zhao YE, Yang WH, Yang HY, Han JZ, Zhou DS. Comparative analysis of KPC-2-encoding chimera plasmids with multi-replicon IncR: IncpA1763-KPC: IncN1 or IncFIIpHN7A8: IncpA1763-KPC: IncN1. Infection and Drug Resistance, 2019, 12: 285-296. DOI:10.2147/IDR.S189168 |
| [16] | He DD, Zhu YY, Li RC, Pan YS, Liu JH, Yuan L, Hu GZ. Emergence of a hybrid plasmid derived from IncN1-F33:A-: B-and mcr-1-bearing plasmids mediated by IS26. Journal of Antimicrobial Chemotherapy, 2019, 74(11): 3184-3189. DOI:10.1093/jac/dkz327 |
| [17] | Shi LN, Feng J, Zhan Z, Zhao YZ, Zhou HJ, Mao HF, Gao YJ, Zhang Y, Yin Z, Gao B, Tong YG, Luo YP, Zhang DF, Zhou DS. Comparative analysis of blaKPC-2- and rmtB-carrying IncFII-family pKPC-LK30/pHN7A8 hybrid plasmids from Klebsiella pneumonia CG258 strains disseminated among multiple Chinese hospitals. Infection and Drug Resistance, 2018, 11: 1783-1793. DOI:10.2147/IDR.S171953 |
| [18] | Pesesky MW, Tilley R, Beck DAC. Mosaic plasmids are abundant and unevenly distributed across prokaryotic taxa. Plasmid, 2019, 102: 10-18. DOI:10.1016/j.plasmid.2019.02.003 |
| [19] | CLSI. Performance standards for antimicrobial susceptibility testing. 29th ed. CLSI Supplement M100, Wayne, PA: Clinical and Laboratory Standards Institute, 2019. |
| [20] | The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0. [2020]. http://www.eucast.org. |
| [21] | Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nature Methods, 2015, 12(8): 733-735. DOI:10.1038/nmeth.3444 |
| [22] | Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S, Wain J, O'Grady J. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nature Biotechnology, 2015, 33(3): 296-300. DOI:10.1038/nbt.3103 |
| [23] | Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, M?ller Aarestrup F, Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrobial Agents and Chemotherapy, 2014, 58(7): 3895-3903. DOI:10.1128/AAC.02412-14 |
| [24] | Couturier M, Bex F, Bergquist PL, Maas WK. Identification and classification of bacterial plasmids. Microbiological Reviews, 1988, 52(3): 375-395. |
| [25] | Du XD, Li DX, Hu GZ, Wang Y, Shang YH, Wu CM, Liu HB, Li XS. Tn1548-associated armA is co-located with qnrB2, aac(6')-Ib-cr and blaCTX-M-3 on an IncFII plasmid in a Salmonella enterica subsp. enterica serovar Paratyphi B strain isolated from chickens in China. Journal of Antimicrobial Chemotherapy, 2012, 67(1): 246-248. DOI:10.1093/jac/dkr407 |
| [26] | Zhang RM, Li JY, Wang Y, Shen JZ, Shen ZQ, Wang SL. Wang SL. Presence of NDM in non-E. coli Enterobacteriaceae in the poultry production environment. Journal of Antimicrobial Chemotherapy, 2019, 74(8): 2209-2213. DOI:10.1093/jac/dkz193 |
| [27] | Liu Y, Long D, Xiang TX, Du FL, Wei DD, Wan LG, Deng Q, Cao XW, Zhang W. Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. Journal of Antimicrobial Chemotherapy, 2019, 74(5): 1233-1240. DOI:10.1093/jac/dkz023 |
| [28] | Musicha P, Msefula CL, Mather AE, Chaguza C, Cain AK, Peno C, Kallonen T, Khonga M, Denis B, Gray KJ, Heyderman RS, Thomson NR, Everett DB, Feasey NA. Genomic analysis of Klebsiella pneumoniae isolates from Malawi reveals acquisition of multiple ESBL determinants across diverse lineages. Journal of Antimicrobial Chemotherapy, 2019, 74(5): 1223-1232. DOI:10.1093/jac/dkz032 |
| [29] | Oliva M, Monno R, D'Addabbo P, Pesole G, Dionisi AM, Scrascia M, Chiara M, Horner DS, Manzari C, Luzzi I, Calia C, D'Erchia AM, Pazzani C. A novel group of IncQ1 plasmids conferring multidrug resistance. Plasmid, 2017, 89: 22-26. DOI:10.1016/j.plasmid.2016.11.005 |
| [30] | Martins WMBS, Nicolas MF, Yu Y, Li M, Dantas P, Sands K, Portal E, Almeida LGP, Vasconcelos ATR, Medeiros EA, Toleman MA, Walsh TR, Gales AC, Andrey DO. Clinical and molecular description of a high-copy IncQ1 KPC-2 plasmid harbored by the international ST15Klebsiella pneumoniae clone. mSphere, 2020, 5(5): e00756-20. |
| [31] | Vogwill T, MacLean RC. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evolutionary Applications, 2015, 8(3): 284-295. |
| [32] | Francino MP. Horizontal gene transfer in microorganisms. 3rd ed. Caister Academic Press, 2012. |
| [33] | Gama JA, Zilh?o R, Dionisio F. Impact of plasmid interactions with the chromosome and other plasmids on the spread of antibiotic resistance. Plasmid, 2018, 99: 82-88. |
| [34] | San Millan A, Heilbron K, MacLean RC. Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. The ISME Journal, 2014, 8(3): 601-612. |
| [35] | Yano H, Shintani M, Tomita M, Suzuki H, Oshima T. Reconsidering plasmid maintenance factors for computational plasmid design. Computational and Structural Biotechnology Journal, 2019, 17: 70-81. |
| [36] | Pilla G, Tang CM. Going around in circles: virulence plasmids in enteric pathogens. Nature Reviews Microbiology, 2018, 16(8): 484-495. |
| [37] | Silva RF, Mendon?a SCM, Carvalho LM, Reis AM, Gordo I, Trindade S, Dionisio F. Pervasive sign epistasis between conjugative plasmids and drug-resistance chromosomal mutations. PLoS Genetics, 2011, 7(7): e1002181. |
| [38] | Chen KC, Chan EWC, Chen S. Evolution and transmission of a conjugative plasmid encoding both ciprofloxacin and ceftriaxone resistance in Salmonella. Emerging Microbes & Infections, 2019, 8(1): 396-403. |
| [39] | Xie MM, Li RC, Liu ZH, Chan EWC, Chen S. Recombination of plasmids in a carbapenem-resistant NDM-5-producing clinical Escherichia coli isolate. Journal of Antimicrobial Chemotherapy, 2018, 73(5): 1230-1234. |
| [40] | Leelaporn A, Firth N, Paulsen IT, Skurray RA. IS257-mediated cointegration in the evolution of a family of staphylococcal trimethoprim resistance plasmids. Journal of Bacteriology, 1996, 178(20): 6070-6073. |
| [41] | Wong MHY, Chan EWC, Chen S. IS26-mediated formation of a virulence and resistance plasmid in Salmonella Enteritidis. Journal of Antimicrobial Chemotherapy, 2017, 72(10): 2750-2754. |
| [42] | Jean SS, Hsueh PR. High burden of antimicrobial resistance in Asia. International Journal of Antimicrobial Agents, 2011, 37(4): 291-295. |
