高源, 曹成亮

 , 李荣鹏, 蒋继宏
, 李荣鹏, 蒋继宏 江苏师范大学生命科学学院, 江苏省药用植物生物技术重点实验室, 江苏 徐州 221116
收稿日期:2020-10-07;修回日期:2021-02-09;网络出版日期:2021-06-08
基金项目:国家自然科学基金(41603078);江苏省研究生科研与实践创新计划(2020XKT476);徐州市科技计划(KC20049)
*通信作者:曹成亮, Tel/Fax: +86-516-83403515;E-mail: chengliangcao@jsnu.edu.cn.
摘要:伦茨菌属(Lentzea)由Yassin等于1995年建立,是一个经典的丝状稀有放线菌类群,目前共包含23个有效描述种。伦茨菌的细胞壁含有meso-二氨基庚二酸,优势甲基萘醌成分为MK-9(H4),磷酸类脂主要包括磷脂酰乙醇胺、双磷脂酸甘油、磷脂酰甘油和磷脂酰肌醇,基因组DNA的(G+C)含量为68.6 mol%–79.6 mol%。伦茨菌代谢产物具有显著的生物活性多样性,包括Ⅰ型人体免疫缺损病毒整合酶的抑制活性、抗肿瘤活性、抗结核活性等,在生物医药研究领域逐步显示出潜在的应用价值。本课题组在研究贵州石灰岩风化壳放线菌多样性时分离获得若干株伦茨菌,采用多相分类方法研究Lentzea xinjiangensis DHS C013T和Lentzea pudingi DHS C021T两株菌的分类学地位,证实均为伦茨菌属的新物种。在此研究基础上,引用有关文献和最新研究成果,对伦茨菌属的建立、分类学特征、生态位和物种分布、功能基因组、天然活性产物应用开发等方面的研究进展进行了综述。
关键词:伦茨菌属多相分类稀有放线菌天然活性产物
Recent advance on the genus Lentzea
Yuan Gao, Chengliang Cao

 , Rongpeng Li, Jihong Jiang
, Rongpeng Li, Jihong Jiang Key Laboratory for Biotechnology on Medicinal Plants of Jiangsu Province, School of Life Sciences, Jiangsu Normal University, Xuzhou 221116, Jiangsu Province, China
Received: 7 October 2020; Revised: 9 February 2021; Published online: 8 June 2021
*Corresponding author: Chengliang Cao, Tel/Fax: +86-516-83403515; E-mail: chengliangcao@jsnu.edu.cn.
Foundation item: Supported by the National Natural Science Foundation of China (41603078), by the Postgraduate Research & Practice Innovation Program of Jiangsu Province (2020XKT476) and by the Science and Technology Plan Project of Xuzhou City (KC20049)
Abstract: The genus Lentzea, established by Yassin et al in 1995, is a typical group of filamentous rare actinomycetes. This genus currently contains 23 valid species, and its typical taxonomic characteristics includes: meso-diaminopimelic acid in the cell wall; MK-9(H4) as the dominant menaquinone; the principal phosphate lipids include phosphatidylethanolamine, diphosphatidylglycerol, phosphotidylinositol and phosphatidylglycerol; the G+C content of genomic DNA is 68.6 mol%–79.6 mol%. Metabolites from strains of the genus Lentzea have significant diversity of biological activities, including human immunodeficiency virus type 1 integrase inhibitory activity, anti-tumor activity, anti-tuberculosis activity and so on, gradually showing the potential application value in biopharmaceutical research. Our research group isolated several strains of the genus Lentzea during the study on actinobacterial biodiversity of limestone crust in Guizhou province, and Lentzea xinjiangensis DHS C013T as well as Lentzea pudingi DHS C021T were proposed as novel species of the genus using the polyphasic taxonomy methods. On the basis of our study, the relevant literatures and the latest research findings, this paper reviews the establishment and taxonomic characteristics of the genus Lentzea, ecological diversity, functional genome and application of natural active metabolites.
Keywords: Lentzeapolyphasic taxonomyrare actinomycetesactive metabolites
微生物是天然活性产物的主要来源,其化合物种类繁多,得到广泛应用的主要有抗生素、抗肿瘤药物、免疫抑制剂、杀虫剂等。这些天然化合物或其先导化合物有50%以上是由放线菌类群产生的,长期以来放线菌在生物制药领域一直占据着重要地位[1]。近年来,从非链霉菌属的稀有放线菌(rare actinomycetes)获得新化合物的数量呈明显上升态势,表现出化学结构新颖、生物活性多样、细胞毒性低的特点。化合物分子结构类型涉及大环内酯、聚酮、蒽醌、联吡啶类、多烯等类型化合物[2],其中产生的一些抗菌药物如庆大霉素、红霉素、万古霉素和利福平等已成功应用于临床[3],这些新型化合物的发现为应对日益增长的耐药性病原菌种类和新致病菌的出现而再次得到了国内外****的高度关注。
伦茨菌属(Lentzea)是一类经典的丝状稀有放线菌,隶属于放线菌门(Actinobacteria)-放线菌纲(Actinobacteria)-假诺卡氏菌目(Pseudonocardiales)-假诺卡氏菌科(Pseudonocardiaceae)。随着分类学技术的发展,伦茨菌属的分类在建立之初发生过多次变动,重新修正伦茨菌属的分类地位,为后续新成员的分类研究提供科学依据。近年来,从伦茨菌分离获得越来越多的天然活性产物,愈发显示出该稀有放线菌类群在新药研发中的重要地位。此外,伦茨菌属产生的次级代谢产物具有多种生物活性,如环孢素A衍生物生物转化活性[4],抑菌抗癌活性的天然产物十字孢碱[5],蛋白质激酶的抑制剂活性等[6]。最引人关注的是伦茨菌Lentzea chajnantorensis H45T产生的代谢产物对Ⅰ型人体免疫缺损病毒(Human immunodeficiency virus type 1,HIV-1)整合酶具有显著的抑制活性[7],这一发现有力推动了新型抗HIV治疗药物的研发进程。
1 伦茨菌属的建立及研究现状 1.1 伦茨菌属的建立 1995年,Yassin等在研究病原放线菌时,从一位女性的患腹膜癌、结肠癌组织培养物中获得一株放线菌命名为IMMIB D-958。培养鉴定后发现该菌株与放线菌目的其他所有属的化学分类特征都不同,由此命名为白丝伦茨菌(Lentzea albidocapillata)[8],并建立了伦茨菌属。
2000年,Lee等发现紫色糖丝菌(Saccharothrix violacea)与白丝伦茨菌(L. albidocapillata)在进化树上形成一个分枝且化学特征相似,提议撤销伦茨菌属并将白丝伦茨菌转移到糖丝菌属(Saccharothrix)[9]。2001年Labeda等基于系统发育和化学分类学研究结果分析恢复伦茨菌属,并将新发表的2个菌种(Lentzea albida和Lentzea californiensis)以及糖丝菌属的2个菌种重新归类到伦茨菌属(L. albidocapillata和Lentzea waywayandensis)[10]。目前伦茨菌属共有23个有效描述种。另外两株Lentzea indica[11]和Lentzea isolaginshaensis[12]没有经过“List of Prokaryotic names with Standing in Nomenclature”的进一步确认,为非有效描述种。
大部分伦茨菌分离自土壤,也有的来自不同的极端环境如沙漠、酸性土壤及岩石风化土等。李文均团队报道从贵州喀斯特洞穴采集的样品中分离获得到伦茨菌[13]。Liu等[14]首次从地衣中分离得到伦茨菌。纯培养获得的伦茨菌新物种相对较少,而陆续报道的宏基因组学研究结果显示伦茨菌属放线菌类群在地球生态系统中分布十分广泛。Germida团队采用16S rRNA扩增测序技术研究油砂复垦区生长的一年生大麦和甜三叶草的根际菌群多样性,结果显示伦茨菌为优势属之一[15]。黄英团队综合免培养和纯培养分离方法系统分析了青藏高原土壤放线菌多样性特征,结果表明伦茨菌属丰度相对较高,并分离获得两株疑似新物种的伦茨菌[16]。Yang等研究短期施用氮肥对土壤分解几丁质的细菌群落(chitinolytic community,chiA)的影响,发现在丰度最高的前50个chiA操作分类单元(operational taxonomic units,OTU)多数归类为放线菌门中的链霉菌、伦茨菌和游动放线菌[17]。Hasyimi研究太平洋白虾Litopenaeus vannamei肠道微生物多样性组成,结果显示在属水平上伦茨菌属也具有较高的丰度[18]。
1.2 伦茨菌属与其近缘属的比较 伦茨菌属与邻近的束丝放线菌属(Actinosynnema)、动孢放线菌属(Actinokineospora)和糖丝菌属(Saccharothrix)同属于假诺卡氏菌科,各属的化学特征整体相似但是仍有明显差异(表 1)。伦茨菌能够形成良好的分枝状营养菌丝体,不产生孢囊结构,无游动孢子,而束丝放线菌成熟的气生菌丝形成孢子链,动孢放线菌的气生菌丝可产生游动孢子[19]。伦茨菌的菌落排列紧密,在ISP 2、ISP 3和ISP 4培养基上呈现黄色或褐色。在极性脂组成上,伦茨菌属放线菌没有羟基磷脂酰乙醇胺(hydroxy-phosphatidylethanolamine,OH-PE),而其3个近缘属均有检出。伦茨菌脂肪酸类型主要以iso-/anteiso-分支饱和脂肪酸为主。伦茨菌全细胞糖中不含阿拉伯糖,糖丝菌全细胞糖中有甘露糖和鼠李糖。相比于假诺卡氏菌科中的其他放线菌属,伦茨菌属有更高的基因组DNA (G+C) mol/%含量。
表 1. 伦茨菌属及相关菌属的分类学特征 Table 1. Taxonomy characteristics of the genus Lentzea and related genera
| Characteristics | Lentzea[8, 19] | Actinosynnema[20] | Actinokineospora[21-22] | Saccharothrix[23-24] |
| T/℃ | 10–37 | 28–30 | 10–45 | 10–45 |
| pH | 5–11 | 6.0–8.5 | 5–10 | 6–9 |
| NaCl tolerance/% | 0–4 | 0–2 | 0–5 | 0–3 |
| Colour of colony | Yellow | Pale greenish | Brown | Brown |
| Sporangia produced | None | Synnemata | None | None |
| Motile spores | No | Yes | Yes | No |
| Whole-cell sugars | Gal, Man, Rib | Gal, Man, Rha | Ara, Man, Rha | Gal, Man, Rha |
| Polar lipids | PE, DPG, PG, PI | PE, OH-PE, PI, PIM, DPG | PE, OH-PE | PE, OH-PE, PI, PIM, DPG, PG |
| Menaquinones | MK-9(H2) MK-9(H4) | MK-9(H4) MK-9(H6) | MK-9(H4) | MK-9(H4) MK-10(H4) |
| Cellular fatty acid | iso-C14:0 iso-C15:0 iso-C16:0 anteiso-C15:0 | C17:0 anteiso-C17:0 iso-C16:0 | iso-C16:0 iso-C15:0 iso-C16:02-OH | iso-C16:0 C17:1ω8c iso-C15:0 |
| DNA G+C content/mol% | 68.6–79.6 | 71–73 | 69.1–73.0 | 67–76 |
| Gal: galactose; Man: mannose; Rib: ribose; Ara: arabinose; Fru: fructose; Xyl: xylose; Rha: rhamnose; PE: phosphatidylethanolamine; PG: phosphatidylglycerol; PI: phosphatidylinositol; DPG: diphosphatidylglycerol; PIM: phosphatidylinositolmannoside; OH-PE: hydroxy-phosphatidylethanolamine. | ||||
表选项
1.3 伦茨菌属的多相分类特征
1.3.1 形态和生理生化特征: 伦茨菌为革兰氏阳性菌,需氧,不耐酸,细胞直径为0.5–0.7 μm,长度为2–3 μm。培养菌落紧密,基质菌丝发育良好,气生菌丝分枝不规则,成熟后断裂成杆状。在不同琼脂培养基上形成不同颜色的基质菌丝和气生菌丝,有淡黄色或黄褐色的基质菌丝以及白色或黄白色的气生菌丝,L. xinjiangensis能在改良的Bennett琼脂上形成白色到黄色的气生菌丝[25]。大多数菌种不产生可溶性色素,除了Lentzea kentuckyensis在蛋白胨-铁琼脂培养基上产生一种浅棕色可溶性色素[26],Lentzea aerocolonigenes在一些培养基上产生褐色的可溶性色素[27]。
该属大部分菌株生长温度范围一般为10–37 ℃,Lentzea terrae、L. xinjiangensis、Lentzea nigeriaca和Lentzea fradiae在45 ℃时依然能够存活[25, 28-30]。适宜生长的pH为5–11。除了Lentzea flaviverrucosa、Lentzea flava和L. terrae不能水解酪蛋白[27, 29, 31],多数菌种能够水解酪蛋白和淀粉,目前发现的菌株中,只有Lentzea cavernae不能水解淀粉[32];对于明胶的水解反应,L. albidocapillata、L. jiangxiensis、L. pudingi、Lentzea soli、Lentzea waywayandensis和Lentzea rhizosphaerae呈阴性[8, 33-37]。此外,只有L. flaviverrucosa、L. californiensis、L. waywayandensis和L. jiangxiensis的硝酸还原反应呈阳性[10, 31, 35, 37];能产生H2S气体的只有L. cavernae和Lentzea roselyniae。
1.3.2 化学分类特征: 伦茨菌属成员细胞壁类型为Ⅲ型(meso-DAP),全细胞糖由半乳糖、甘露糖、葡萄糖和核糖组成。L. cavernae的全细胞糖含有阿拉伯糖、果糖、甘露糖和木糖[32],而L. waywayandensis和L. aerocolonigenes只含有半乳糖以及鼠李糖。多数伦茨菌的主要甲基萘醌是MK-9(H4)和MK-9(H2)。除此之外,菌株L. soli的甲基萘醌还含有MK-9(H0),L. rhizosphaerae含有少量的MK-9(H6)。
脂肪酸结构包括直链饱和脂肪酸、不饱和脂肪酸和iso-/anteiso-类型的支链饱和脂肪酸。多数伦茨菌种的脂肪酸都含有iso-C16:0、iso-C15:0、C16:0、10Me-C16:0和anteiso-C15:0,L. cavernae中含有C14:0[32],L. terrae的脂肪酸中含有不饱和脂肪酸C16:1ω7c,L. albidocapillata的脂肪酸是3d型(10-甲基支链脂肪酸);该菌属部分成员的磷酸类脂组分有双磷脂酸甘油(diphosphatidylglycerol,DPG)、磷脂酰乙醇胺(phosphatidylethanolamine,PE)、磷脂酰肌醇甘露糖苷(phosphatidylinositolmannoside,PIM)、磷脂酰肌醇(phosphatidylinositol,PI)以及未鉴定的磷脂(unidentified lipid,UL)[26, 29, 33-35],菌株L. flava含有溶血磷脂酰乙醇胺(lysophosphatidylethanolamine,Lyso-PE)[38]。
1.3.3 分子分类特征: 伦茨菌属隶属于假诺卡氏菌科,在基于伦茨菌属及相关属代表菌株16S rRNA基因序列的系统发育进化树中(图 1),伦茨菌属各成员在假诺卡氏菌科内聚在一起形成一个稳定的分支。伦茨菌属与束丝放线菌属的关系最为接近,二者趋近聚合形成一个系统进化分支。本实验室从中国贵州省石灰岩风化壳样品中分离到的伦茨菌株L. guizhouensis DHS C013T与L. jiangxiensis FXJ1.034T位于一个亚分支内,16S rRNA基因序列相似性为98.7%[39]。另一株L. pudingi DHS C021T与L. albida CGMCC 4.1727T的16S rRNA基因序列相似性为98.8%[33]。伦茨菌属的特征性核苷酸位点是TCAA (617–620)、GCC (843–845)[40];伦茨菌属成员的基因组DNA (G+C)mol%含量范围是68.6%–79.6%。
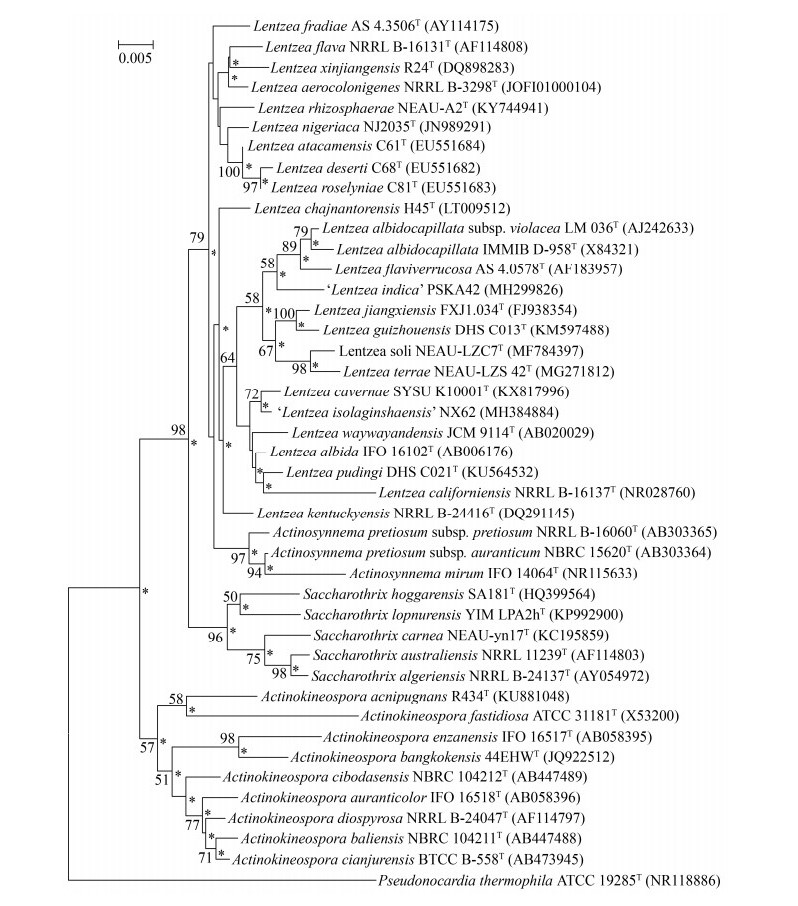 |
| 图 1 邻近法构建的基于伦茨菌属及相关属代表菌株16S rRNA基因序列的系统发育进化树 Figure 1 Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences of the representative members of the genera of Lentzea, Actinosynnema, Saccharothrix and Actinokineospora. Numbers at nodes indicate levels of bootstrap support based on a neighbour-joining analysis of 1000 resampled datasets, only values > 50% were given. Asterisks indicate branches that were also recovered using the maximum-parsimony and maximum-likelihood methods (Data were not presented in this paper). The sequences in bracket indicate the accession number in NCBI. Bar: 5 nucleotide substitution per 1000 nucleotides. |
| 图选项 |
1.4 基因组研究概况 截止2020年12月,NCBI和Ezbiocloud的Genome数据库共收录了19株伦茨菌的基因组序列。借助Type Strain Genome Server (https://tygs.dsmz.de/)对该19个菌种进行全基因组系统发育分析(图 2),并使用antiSMASH (5.2.0版本)注释预测。数据显示,伦茨菌基因组相对较大,约为8–10 Mb,拥有高达近万个蛋白编码基因(coding sequences,CDSs),聚酮化合物合成酶(polyketide synthase,PKS)和非核糖体肽合成酶(non-ribosomal peptide synthetase,NRPS)基因簇也相当丰富(表 2)。L. terrae NEAU-LZS 42T基因组含有9915个CDSs,L. waywayandensis DSM 44232T基因组含有39个次级代谢生物合成基因簇(biosynthetic gene clusters,BGCs),L. jiangxiensis CGMCC 4.6609T基因组含有22个PKS和NRPS基因簇。基因组研究表明,Lentzea sp. ATCC 31319基因组上存在聚酮类抗生素乳霉素(thiolactomycin,TLM)生物合成基因簇[41]。
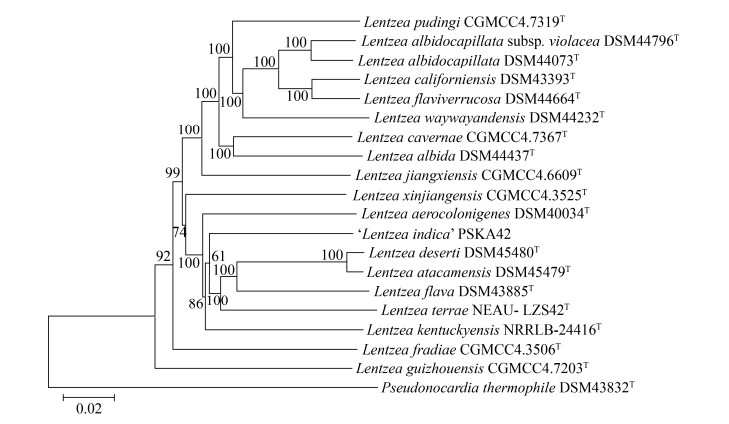 |
| 图 2 基于TYGS服务器中伦茨菌属基因组序列的系统发育树 Figure 2 Phylogenomic tree based on genome sequences of the Lentzea genus in the TYGS server. Tree inferred with FastME 2.1.6.1 from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of GBDP distance formula d5. The numbers above branches are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 94.6%. The tree was rooted at the midpoint. Bar: 2 nucleotide substitution per 100 nucleotides. |
| 图选项 |
表 2. 伦茨菌属基因组特征 Table 2. Features of lentzea genomes
| Species | Genome size/bp | CDSs | rRNAs | tRNAs | BGCs | PKS | NRPS |
| L. albida DSM 44437T | 9441135 | 8818 | 15 | 65 | 35 | 7 | 10 |
| L. terrae NEAU-LZS 42T | 10581732 | 9915 | 7 | 64 | 33 | 2 | 10 |
| L. albidocapillata DSM 44073T | 8639486 | 8185 | 14 | 62 | 28 | 2 | 9 |
| L. flaviverrucosa DSM 44664T | 9468454 | 8932 | 6 | 71 | 30 | 5 | 8 |
| L. albidocapillata subsp. violacea DSM 44796T | 8671075 | 8301 | 17 | 66 | 30 | 4 | 10 |
| L. waywayandensis DSM 44232T | 10153412 | 9339 | 15 | 67 | 39 | 4 | 13 |
| L. kentuckyensis NRRL B-24416T | 10210611 | 9374 | 5 | 63 | 35 | 10 | 9 |
| L. xinjiangensis CGMCC 4.3525T | 8684108 | 8199 | 14 | 62 | 37 | 7 | 14 |
| L. atacamensis DSM 45479T | 9306230 | 9058 | 8 | 68 | 26 | 4 | 8 |
| L. californiensis DSM 43393T | 8998498 | 8539 | 18 | 69 | 29 | 5 | 8 |
| L. deserti DSM 45480T | 9529573 | 9224 | 8 | 68 | 32 | 6 | 9 |
| L. flava DSM 43885T | 9746996 | 9203 | 16 | 68 | 28 | 3 | 8 |
| L. fradiae CGMCC 4.3506T | 8508028 | 8020 | 15 | 60 | 36 | 5 | 15 |
| L. guizhouensis DHS C013T | 9997872 | 9760 | 15 | 69 | 35 | 8 | 9 |
| L. jiangxiensis CGMCC 4.6609T | 8591279 | 8027 | 12 | 67 | 37 | 7 | 15 |
| L. aerocolonigenes NBRC 13195T | 10698154 | 9898 | 10 | 67 | 27 | 2 | 9 |
| L. indica PSKA42 | 9967419 | 8731 | 18 | 61 | 37 | 9 | 14 |
| L. pudingi CGMCC 4.7319T | 9209073 | 8539 | 5 | 69 | 26 | 4 | 7 |
| L. cavernae CGMCC 4.7367T | 9738430 | 9120 | 5 | 64 | 33 | 4 | 10 |
表选项
2 伦茨菌属的应用研究 2.1 HIV-1整合酶抑制活性 抗HIV治疗通常包括针对病毒复制周期不同阶段使用不同药物以及联合治疗,主要问题是病毒突变率高,导致耐药病毒的出现。为了克服这一问题,最初是让患者接受不同药物的联合治疗,以减少耐药突变株的选择,后来经过改进,开发出针对病毒复制周期内确定额外的药物靶点的新方法[42]。HIV-1整合酶是病毒复制周期中的关键酶之一,它负责将逆转录的病毒互补DNA (cDNA)整合到宿主细胞基因组中[43],该靶点对开发新的抗HIV治疗具有很强的选择特异性。
研究发现阿塔卡马沙漠中微生物类群也十分丰富[44-45],从中分离出许多放线菌新物种[46-47]。从其中L. chajnantorensis H45T菌株的天然代谢产物中分离出6种结构新颖的二烯和单烯糖苷化合物Lentzeaosides A–F,活性检测结果显示对HIV-1整合酶表现出不同水平的抑制活性[7]。抗病毒治疗以整合酶为靶点是对逆转录酶和蛋白酶抑制剂的宝贵补充[48],具有潜在的应用前景。化学全合成的雷特格韦(Raltegravir)是美国食品药物管理局(Food and Drug Administration,FDA)批准的第一个HIV-1整合酶抑制剂,多用于治疗对多种抗逆转录病毒药物耐药的患者[49]。
2.2 生物转化功能 环孢素A (cyclosporine A,CsA)是一种抑制细胞亲环素的环肽,用作器官移植的关键免疫抑制剂。CsA的抑制活性依赖于其与亲环素的结合,其复合物抑制钙调神经磷酸酶在T细胞中的作用[50]。Sasamura研究团队发现Lentzea sp. 7887 (与L. albidocapillata subsp. violacea LM 036T的16S rDNA相似性为99.2%)能够催化CsA衍生物FR901459多个位点发生羟基化反应,生物转化后表现出更好的免疫抑制活性[51]。同时发现底物分散剂大豆粉能有效提高该生物转化效率[52]。最新研究证实Lentzea sp. 7887的细胞色素P450可以催化抗真菌药物Sordaricin发生6-羟基化反应[53]。
2.3 伦茨菌的其他应用 1976年,****首次从菌株Streptomyces staurosporeus (现修正为L. albida)中分离出吲哚咔唑化合物十字孢碱(staurosporine)[54]。已知该化合物能和迄今发现的约90%人类激酶发生作用[55],具有良好的抗真菌、降血压作用[56]和抑制血小板聚集的特性[57],尤其是研究证实十字孢碱具有很好的抗癌活性。为减小其毒副作用,化学家已对其进行大量结构修饰,获得了多个高效低毒的十字孢碱衍生物[58]。另一伦茨菌L. aerocolonigenes sp.代谢产生的瑞贝霉素(rebeccamycin)也具有显著的抗肿瘤活性[59]。
从放线菌中分离得到的众多天然产物中许多都具有抗菌活性,很少有针对结核病原菌的研究。由结核分枝杆菌引起的结核病因耐药性问题,导致结核病难以短期治愈。最近研究首次发现L. albidocapillata subsp. violacea AS08对结核分枝杆菌Mycobacterium tuberculosis H37Rv表现出显著的拮抗作用,具有抗结核活性[60]。该伦茨菌是从印度喜马拉雅山脉西北部未开发地区的土壤中分离获得,早前研究证实L. albidocapillata subsp. violacea AS08产生的代谢产物具有多种生物活性,包括抗氧化、降胆固醇、杀虫等活性[61]。
聚乳酸(poly lactic acid,PLA)是一种生物可降解、低过敏性、抗菌、环保、多功能的高分子化合物[62-63],并且广泛应用于缝合线、支架、可吸收伤口闭合产品等生物医学领域[64]。Nimisha等研究发现菌株L. waywayandensis ATCC 51594能够显著提高聚乳酸的生物降解率[65];国内****堵国成团队进一步研究发现L. waywayandensis sp.代谢产生的胞外蛋白酶在降解PLA膜的过程中起到关键催化作用[66]。
3 展望 伦茨菌属发现和建立相对较晚,但是在过去的二十余年里从伦茨菌中陆续分离获得多种新颖的天然活性产物。这些天然产物通常具有显著的抗癌、生物降解、抗HIV等活性。尤其是HIV-1整合酶抑制活性的发现,在艾滋病治疗作为当今医学领域的热点和难点的大背景下,提供了HIV治疗药物研发新思路,突显出稀有放线菌伦茨菌属类群在生物医药领域的巨大价值。
截至目前有效发表的伦茨菌属物种还相对较少,可能仍然有许多新成员尚未被发现。在多样且复杂的地球极端环境中新物种资源挖掘依然存在很大空间,诸如海洋、极地、沙漠等特殊生境赋予了放线菌独特的基因型、特殊的生理机制以及丰富的代谢类型。据估计自然界中仍有90%以上的放线菌未被发现,而绝大多数微生物很难被培养,这就需要研究人员不断探索、改进培养基成分、创新分离技术来提高稀有放线菌的分离效率。已有研究表明在选择培养基中加入镧系等稀土元素可以培养出之前在实验室无法生长的微生物[67]。另外,利用组学技术在微生物群落研究中的优势,分析复杂环境中的微生物群落组成多样性及其与周围环境或宿主之间的关系,特别是能获得难以培养的微生物信息,有针对性地优化选择分离方法,促进难培养稀有放线资源的积累。同时,基于稀有放线菌基因组信息的新型基因簇的深度挖掘,针对沉默基因簇采用各种激活手段加强基因表达,增加结构新颖天然产物的产出,让稀有放线菌伦茨菌属类群成为新药研发的宝贵资源。
References
| [1] | Zheng QW. Advances have been made in the synthetic biology of natural products of actinomycetes in Shanghai Institute of Biotechnology. Pesticide Market News, 2017(5): 47-48. (in Chinese) 郑庆伟. 上海生科院在放线菌天然产物的合成生物学方面取得进展. 农药市场信息, 2017(5): 47-48. |
| [2] | Zhang JC, Yang XQ, Zhou H, Yang YB, Ding ZT. New natural products of rare actinomycetes from 2006 to 2018. Chinese Journal of Organic Chemistry, 2019, 39(4): 982-1012. (in Chinese) 张举成, 杨雪琼, 周皓, 杨亚滨, 丁中涛. 2006-2018年稀有放线菌中的新天然产物. 有机化学, 2019, 39(4): 982-1012. |
| [3] | Lazzarini A, Cavaletti L, Toppo G, Marinelli F. Rare genera of actinomycetes as potential producers of new antibiotics. Antonie Van Leeuwenhoek, 2000, 78(3/4): 399-405. DOI:10.1023/A:1010287600557 |
| [4] | Yabutani T, Tsujimoto M, Ohira S, Shimizu S, Nakano H. Strain improvement of LentZea sp. 7887 for higher yield per unit volume on hydroxylation of cyclosporine derivative FR901459. Bioscience, Biotechnology, and Biochemistry, 2017, 81(7): 1456-1459. DOI:10.1080/09168451.2017.1314759 |
| [5] | Nakano H, ōmura S. Chemical biology of natural indolocarbazole products: 30 years since the discovery of staurosporine. The Journal of Antibiotics, 2009, 62(1): 17-26. DOI:10.1038/ja.2008.4 |
| [6] | Tamaoki T, Nomoto H, Takahashi I, Kato Y, Morimoto M, Tomita F. Staurosporine, a potent inhibitor of phospholipid Ca+ dependent protein kinase. Biochemical and Biophysical Research Communications, 1986, 135(2): 397-402. DOI:10.1016/0006-291X(86)90008-2 |
| [7] | Wichner D, Idris H, Houssen WE, McEwan AR, Bull AT, Asenjo JA, Goodfellow M, Jaspars M, Ebel R, Rateb ME. Isolation and anti-HIV-1 integrase activity of lentzeosides A-F from extremotolerant LentZea sp. H45, a strain isolated from a high-altitude Atacama Desert soil. The Journal of Antibiotics, 2017, 70(4): 448-453. DOI:10.1038/ja.2016.78 |
| [8] | Yassin AF, Rainey FA, Brzezinka H, Jahnke KD, Schaal KP. Lentzea gen. nov., a New Genus of the Order Actinomycetales. International Journal of Systematic Bacteriology, 1995, 45(2): 357-363. DOI:10.1099/00207713-45-2-357 |
| [9] | Lee SD, Kim ES, Roe JH, Kim J, Kang SO, Hah YC. Saccharothrix violacea sp. nov., isolated from a gold mine cave, and Saccharothrix albidocapillata comb. nov.. International Journal of Systematic and Evolutionary Microbiology, 2000, 50(Pt 3): 1315-1323. |
| [10] | Labeda DP, Hatano K, Kroppenstedt RM, Tamura T. Revival of the genus LentZea and proposal for Lechevalieria gen. nov.. International Journal of Systematic and Evolutionary Microbiology, 2001, 51(3): 1045-1050. DOI:10.1099/00207713-51-3-1045 |
| [11] | Maiti PK, Mandal S. LentZea indica sp. nov., a novel actinobacteria isolated from Indian Himalayan-soil. Antonie Van Leeuwenhoek, 2020, 113(10): 1411-1423. DOI:10.1007/s10482-020-01449-8 |
| [12] | Wang LW, Li YM, Li YM. LentZea isolaginshaensis sp. nov., an actinomycete isolated from desert soil. Antonie Van Leeuwenhoek, 2019, 112(4): 633-639. DOI:10.1007/s10482-018-1193-7 |
| [13] | Zhang WQ, Fang BZ, Han MX, Li S, Dong L, Jiang HC, Li WJ. Diversity and antibacterial activity of culturable actinobacteria in Karst cave soil in Xingyi, Guizhou. Acta Microbiologica Sinica, 2020, 60(6): 1063-1073. (in Chinese) 张万芹, 房保柱, 韩明贤, 李帅, 董雷, 蒋宏忱, 李文均. 贵州兴义喀斯特洞穴可培养放线菌多样性及抗菌活性初筛. 微生物学报, 2020, 60(6): 1063-1073. |
| [14] | Liu CB, Jiang Y, Wang XY, Chen DB, Chen X, Wang LS, Han L, Huang XS, Jiang CL. Diversity, antimicrobial activity, and biosynthetic potential of cultivable actinomycetes associated with lichen symbiosis. Microbial Ecology, 2017, 74(3): 570-584. DOI:10.1007/s00248-017-0972-4 |
| [15] | Mitter EK, de Freitas JR, Germida JJ. Bacterial root microbiome of plants growing in oil sands reclamation covers. Frontiers in Microbiology, 2017, 8: 849. DOI:10.3389/fmicb.2017.00849 |
| [16] | Huang J, Yan BF, Huang Y. Diversity of culturable actinobacteria from soils collected in Ali, Naqu and Haixi Districts on the Qinghai-Tibet Plateau.. Acta Microbiologica Sinica, 2017, 57(9): 1342-1351. (in Chinese) 黄娇, 闫兵法, 黄英. 青藏高原阿里、那曲和海西地区土壤可培养放线菌的多样性. 微生物学报, 2017, 57(9): 1342-1351. |
| [17] | Ouyang Y, Norton JM. Short-term nitrogen fertilization affects microbial community composition and nitrogen mineralization functions in an agricultural soil. Applied and Environmental Microbiology, 2020, 86(5): e02278-19. |
| [18] | Hasyimi W, Widanarni W, Yuhana M. Growth performance and intestinal microbiota diversity in Pacific white shrimp Litopenaeus vannamei fed with a probiotic bacterium, honey prebiotic, and synbiotic. Current Microbiology, 2020, 77(10): 2982-2990. DOI:10.1007/s00284-020-02117-w |
| [19] | Goodfellow M, K?mpfer P, Busse HJ, Trujillo ME, Suzuki KI, Ludwig W, Whitman WB. Bergey's manual? of systematic bacteriology. New York, NY: Springer New York, 2012. |
| [20] | Labeda DP, Goodfellow M, Chun J, Zhi XY, Li WJ. Reassessment of the systematics of the suborder Pseudonocardineae: transfer of the genera within the family Actinosynnemataceae Labeda and Kroppenstedt 2000 emend. Zhi et al. 2009 into an emended family Pseudonocardiaceae Embley et al. 1989 emend. Zhi et al. 2009. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(6): 1259-1264. DOI:10.1099/ijs.0.024984-0 |
| [21] | Toru H. Actinokineospora: A new genus of the Actinomycetales. Actinomycetologica, 1988, 2(1): 31-45. DOI:10.3209/saj.2_31 |
| [22] | Lei YJ, Xia ZF, Luo XX, Zhang LL. Actinokineospora pegani sp. nov., an endophytic actinomycete isolated from the surface-sterilized root of Peganum harmala L.. International Journal of Systematic and Evolutionary Microbiology, 2020, 70(7): 4358-4363. DOI:10.1099/ijsem.0.004299 |
| [23] | Liu JJ, Sun Y, Liu JR, Wu YJ, Cao CL, Li RP, Jiang JH. Saccharothrix deserti sp. nov., an actinomycete isolated from desert soil. International Journal of Systematic and Evolutionary Microbiology, 2020, 70(3): 1882-1887. DOI:10.1099/ijsem.0.003989 |
| [24] | Labeda DP, Testa RT, Lechevalier MP, Lechevalier HA. Saccharothrix: a New Genus of the Actinomycetales Related to Nocardiopsis. International Journal of Systematic Bacteriology, 1984, 34(4): 426-431. DOI:10.1099/00207713-34-4-426 |
| [25] | Wang W, Zhang Z, Tang Q, Mao J, Wei D, Huang Y, Liu Z, Shi Y, Goodfellow M. Lechevalieria xinjiangensis sp. nov., a novel actinomycete isolated from radiation-polluted soil in China. International Journal of Systematic and Evolutionary Microbiology, 2007, 57(12): 2819-2822. DOI:10.1099/ijs.0.65134-0 |
| [26] | Labeda DP, Donahue JM, Sells SF, Kroppenstedt RM. Lentzea kentuckyensis sp. nov., of equine origin. International Journal of Systematic and Evolutionary Microbiology, 2007, 57(8): 1780-1783. DOI:10.1099/ijs.0.64245-0 |
| [27] | Labeda DP. Transfer of "Nocardia aerocolonigenes" (Shinobu and Kawato 1960) Pridham 1970 into the genus Saccharothrix Labeda, testa, Lechevalier, and Lechevalier 1984 as Saccharothrix aerocolonigenes sp. nov.. International Journal of Systematic Bacteriology, 1986, 36(1): 109-110. DOI:10.1099/00207713-36-1-109 |
| [28] | Zhang JL, Xie Q, Liu ZH, Goodfellow M. Lechevalieria fradiae sp. nov., a novel actinomycete isolated from soil in China. International Journal of Systematic and Evolutionary Microbiology, 2007, 57(4): 832-836. DOI:10.1099/ijs.0.64777-0 |
| [29] | Li D, Jiang H, Han L, Li Y, Zhao J, Jiang S, Wang X, Xiang W. Lentzea terrae sp. nov., isolated from soil and an emended description of Lentzea soli. International Journal of Systematic and Evolutionary Microbiology, 2018, 68(11): 3528-3533. DOI:10.1099/ijsem.0.003024 |
| [30] | Camas M, Veyisoglu A, Tatar D, Saygin H, Cetin D, Sazak A, Guven K, Sahin N. Lechevalieria nigeriaca sp. nov., isolated from arid soil. International Journal of Systematic and Evolutionary Microbiology, 2013, 63(10): 3750-3754. |
| [31] | Xie Q, Wang YM, Huang Y, Wu YL, Ba FS, Liu ZH. Description of Lentzea flaviverrucosa sp. nov. and transfer of the type strain of Saccharothrix aerocolonigenes subsp. staurosporea to Lentzea albida. International Journal of Systematic and Evolutionary Microbiology, 2002, 52(5): 1815-1820. |
| [32] | Fang BZ, Han MX, Liu L, Zhang ZT, Liu WL, Shen JT, Wang Y, Zhang WQ, Wei DQ, Li WJ. LentZea cavernae sp. nov., an actinobacterium isolated from a Karst cave sample, and emended description of the genus LentZea. International Journal of Systematic and Evolutionary Microbiology, 2017, 67(7): 2357-2362. DOI:10.1099/ijsem.0.001958 |
| [33] | Cao CL, Yuan B, Qin S, Jiang JH, Tao FX, Lian B. LentZea pudingi sp. nov., isolated from a weathered limestone sample in a Karst area. International Journal of Systematic and Evolutionary Microbiology, 2017, 67(11): 4873-4878. DOI:10.1099/ijsem.0.002400 |
| [34] | Li DM, Zheng WW, Zhao JW, Han LY, Zhao XL, Jiang H, Wang XJ, Xiang WS. LentZea soli sp. nov., an actinomycete isolated from soil. International Journal of Systematic and Evolutionary Microbiology, 2018, 68(5): 1496-1501. DOI:10.1099/ijsem.0.002698 |
| [35] | Li X, Zhang L, Ding Y, Gao Y, Ruan J, Huang Y. Lentzea jiangxiensis sp. nov., isolated from acidic soil. International Journal of Systematic and Evolutionary Microbiology, 2012, 62(10): 2342-2346. |
| [36] | Zhao JW, Li WC, Shi LL, Wang H, Wang XJ. Lechevalieria rhizosphaerae sp. nov., a novel actinomycete isolated from rhizosphere soil of wheat (Triticum aestivum L.) and emended description of the genus Lechevalieria. International Journal of Systematic and Evolutionary Microbiology, 2017, 67(11): 4655. DOI:10.1099/ijsem.0.002351 |
| [37] | Labeda DP, Lyons AJ. Saccharothrix texasensis sp. nov. and Saccharothrix waywayandensis sp. nov.. International Journal of Systematic Bacteriology, 1989, 39(3): 355-358. DOI:10.1099/00207713-39-3-355 |
| [38] | Grund E, Kroppenstedt RM. Transfer of Five Nocardiopsis Species to the Genus Saccharothrix Labeda et al. 1984. Systematic and Applied Microbiology, 1989, 12(3): 267-274. DOI:10.1016/S0723-2020(89)80073-6 |
| [39] | Cao CL, Zhou XQ, Qin S, Tao FX, Jiang JH, Lian B. LentZea guizhouensis sp. nov., a novel lithophilous actinobacterium isolated from limestone from the Karst area, Guizhou, China. Antonie Van Leeuwenhoek, 2015, 108(6): 1365-1372. DOI:10.1007/s10482-015-0589-x |
| [40] | 徐丽华, 李文均, 刘志恒. 放线菌系统学: 原理、方法及实践. 北京: 科学出版社, 2007. |
| [41] | Tao W, Yurkovich ME, Wen S, Lebe KE, Samborskyy M, Liu Y, Yang A, Liu Y, Ju Y, Deng Z, Tosin M, Sun Y, Leadlay PF. A genomics-led approach to deciphering the mechanism of thiotetronate antibiotic biosynthesis. Chemical Science, 2016, 7(1): 376-385. DOI:10.1039/C5SC03059E |
| [42] | Maes M, Loyter A, Friedler A. Peptides that inhibit HIV-1 integrase by blocking its protein-protein interactions. The FEBS Journal, 2012, 279(16): 2795-2809. DOI:10.1111/j.1742-4658.2012.08680.x |
| [43] | Sherman MP, Greene WC. Slipping through the door: HIV entry into the nucleus. Microbes and Infection, 2002, 4(1): 67-73. DOI:10.1016/S1286-4579(01)01511-8 |
| [44] | Crits-Christoph A, Robinson CK, Barnum T, Fricke WF, Davila AF, Jedynak B, McKay CP, Diruggiero J. Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome, 2013, 1(1): 28. DOI:10.1186/2049-2618-1-28 |
| [45] | Bull AT, Asenjo JA, Goodfellow M, Gómez-Silva B. The Atacama desert: technical resources and the growing importance of novel microbial diversity. Annual Review of Microbiology, 2016, 70: 215-234. DOI:10.1146/annurev-micro-102215-095236 |
| [46] | Okoro CK, Brown R, Jones AL, Andrews BA, Asenjo JA, Goodfellow M, Bull AT. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek, 2009, 95(2): 121-133. DOI:10.1007/s10482-008-9295-2 |
| [47] | Bull AT, Asenjo JA. Microbiology of hyper-arid environments: recent insights from the Atacama Desert, Chile. Antonie Van Leeuwenhoek, 2013, 103(6): 1173-1179. DOI:10.1007/s10482-013-9911-7 |
| [48] | Craigie R. HIV integrase, a brief overview from chemistry to therapeutics. The Journal of Biological Chemistry, 2001, 276(26): 23213-23216. DOI:10.1074/jbc.R100027200 |
| [49] | Hicks C, Gulick RM. Raltegravir: the first HIV type 1 integrase inhibitor. Clinical Infectious Diseases, 2009, 48(7): 931-939. DOI:10.1086/597290 |
| [50] | Ho S, Clipstone N, Timmermann L, Northrop J, Graef I, Fiorentino D, Nourse J, Crabtree GR. The mechanism of action of cyclosporin A and FK506. Clinical Immunology and Immunopathology, 1996, 80(3): S40-S45. DOI:10.1006/clin.1996.0140 |
| [51] | Sasamura S, Kobayashi M, Muramatsu H, Yoshimura S, Kinoshita T, Ohki H, Okada K, Deai Y, Yamagishi Y, Hashimoto M. Bioconversion of FR901459, a novel derivative of cyclosporin A, by LentZea sp. 7887. The Journal of Antibiotics, 2015, 68(8): 511-520. DOI:10.1038/ja.2015.19 |
| [52] | Yabutani T, Shimizu S, Nakano H. Pilot-scale whole-cell biocatalysis for the hydroxylation of cyclosporine derivative, FR901459, at higher concentrations by LentZea sp. 7887 using soybean flour as a novel substrate dispersant. Journal of Bioscience and Bioengineering, 2017, 123(1): 56-62. DOI:10.1016/j.jbiosc.2016.07.013 |
| [53] | Ueno M, Kobayashi M, Fujie A, Shibata T. Cloning and heterologous expression of P450Lent4B11, a novel bacterial P450 gene, for hydroxylation of an antifungal agent sordaricin. The Journal of Antibiotics, 2020, 73(9): 615-621. DOI:10.1038/s41429-020-0310-9 |
| [54] | ōmura S, Asami Y, Crump A. Staurosporine: new lease of life for parent compound of today's novel and highly successful anti-cancer drugs. The Journal of Antibiotics, 2018, 71(8): 688-701. DOI:10.1038/s41429-018-0029-z |
| [55] | Crespo A, Zhang X, Fernández A. Redesigning kinase inhibitors to enhance specificity. Journal of Medicinal Chemistry, 2008, 51(16): 4890-4898. DOI:10.1021/jm800453a |
| [56] | Hachisu M, Hiranuma T, Koyama M, Sezaki M. Antihypertensive compounds with potent protein kinases inhibitory activity. Life Sciences, 1989, 44(19): 1351-1362. DOI:10.1016/0024-3205(89)90392-5 |
| [57] | Sch?chtele C, Seifert R, Osswald H. Stimulus-dependent inhibition of platelet aggregation by the protein kinase C inhibitors polymyxin B, H-7 and staurosporine. Biochemical and Biophysical Research Communications, 1988, 151(1): 542-547. DOI:10.1016/0006-291X(88)90628-6 |
| [58] | Liu M, Jia H, Sha Y. Research progress in derivatization and structure-activity relationships of staurosporine. Journal of Shenyang Pharmaceutical University, 2014, 31(3): 224-240. (in Chinese) 刘敏, 贾号, 沙宇. 十字孢碱的衍生化及构效关系研究进展. 沈阳药科大学学报, 2014, 31(3): 224-240. |
| [59] | Bush JA, Long BH, Catino JJ, Bradner WT, Tomita K. Production and biological activity of rebeccamycin, a novel antitumor agent. The Journal of Antibiotics, 1987, 40(5): 668-678. DOI:10.7164/antibiotics.40.668 |
| [60] | Hussain A, Rather M, Shah A, Bhat Z, Shah A, Ahmad Z, Parvaiz Hassan Q. Antituberculotic activity of actinobacteria isolated from the rare habitats. Letters in Applied Microbiology, 2017, 65(3): 256-264. DOI:10.1111/lam.12773 |
| [61] | Selvin J, Shanmughapriya S, Gandhimathi R, Seghal Kiran G, Rajeetha Ravji T, Natarajaseenivasan K, Hema TA. Optimization and production of novel antimicrobial agents from sponge associated marine actinomycetes Nocardiopsis dassonvillei MAD08. Applied Microbiology and Biotechnology, 2009, 83(3): 435-445. DOI:10.1007/s00253-009-1878-y |
| [62] | Sedlarik V, Saha N, Sedlarikova J, Saha P. Biodegradation of blown films based on poly(lactic acid) under natural conditions. Macromolecular Symposia, 2008, 272(1): 100-103. DOI:10.1002/masy.200851214 |
| [63] | Lim LT, Auras R, Rubino M. Processing technologies for poly(lactic acid). Progress in Polymer Science, 2008, 33(8): 820-852. DOI:10.1016/j.progpolymsci.2008.05.004 |
| [64] | Zhao YM, Wang ZY, Wang J, Mai HZ, Yan B, Yang F. Direct synthesis of poly (D, L -lactic acid) by melt polycondensation and its application in drug delivery. Journal of Applied Polymer Science, 2004, 91(4): 2143-2150. DOI:10.1002/app.13354 |
| [65] | Nair NR, Nampoothiri KM, Pandey A. Preparation of poly(l-lactide) blends and biodegradation by LentZea waywayandensis. Biotechnology Letters, 2012, 34(11): 2031-2035. DOI:10.1007/s10529-012-1005-5 |
| [66] | Lin J, Zhou JW, Kang Z, Du GC, Chen J. Isolation, identification of poly lactic acid degrading microorganisms and optimization of the degradation process. Microbiology China, 2013, 40(9): 1560-1569. (in Chinese) 林娟, 周景文, 康振, 堵国成, 陈坚. 聚乳酸降解菌株筛选鉴定及降解过程优化. 微生物学通报, 2013, 40(9): 1560-1569. |
| [67] | Skovran E, Martinez-Gomez NC. Just add lanthanides. Science, 2015, 348(6237): 862-863. DOI:10.1126/science.aaa9091 |
