刘兰1, 明语真1, 吕爱萍1, 焦建宇1, 李文均1,2


1. 中山大学生命科学学院, 有害生物控制与资源利用国家重点实验室, 广东 广州 510275;
2. 中国科学院新疆生态与地理研究所, 荒漠与绿洲生态国家重点实验室, 新疆 乌鲁木齐 830011
收稿日期:2020-12-10;修回日期:2021-02-19;网络出版日期:2020-03-11
基金项目:广州市科技计划(201803030030);广东省基础与应用基础研究基金联合基金(2019A1515110227);中国博士后科学基金(2019M653156)
作者简介:刘兰, 中山大学生命科学学院, 有害生物控制与资源利用国家重点实验室博士后。主要研究方向为热泉微生物资源与生态, 重点关注热泉厌氧微生物资源与生态功能的研究。在高温厌氧微生物的富集、分离、鉴定及多组学研究方面积累了丰富的经验和相关成果。在National Science Review、The ISME Journal、Systematic and Applied Microbiology、Microbial Ecology、Anaerobe、Internatinal Journal of Systematic and Evolutionary Microbiology及Antonie van Leeuwenhoek等期刊参与发表学术论文60余篇, 其中以第一作者发表学术论文8篇, 以并列第一作者(第二位)或第二作者发表学术论文8篇, 谷歌学术总引用428次.
*通信作者:李文均。Tel/Fax: +86-20-84111727;E-mail: liwenjun3@mail.sysu.edu.cn.
摘要:厌氧氨氧化是指微生物在无氧条件下,以NO2-为电子受体,将NH4+氧化成N2的过程,该过程主要由浮霉菌门下的厌氧氨氧化细菌参与。厌氧氨氧化细菌广泛存在于海洋生态系统、淡水生态系统、陆地生态系统及其他一些特殊生境中,其在废水生物脱氮和地球氮循环中扮演着重要角色。本文从厌氧氨氧化细菌的发现历程、种类、特性、代谢途径、分布、检测方法及应用上进行了较为全面的总结;最后对厌氧氨氧化细菌研究前沿问题和未来发展方向进行了探讨与展望。
关键词:厌氧氨氧化细菌种类特征分布代谢途径
Recent advance on the anaerobic ammonium oxidation bacteria
Lan Liu1, Yuzhen Ming1, Aiping Lv1, Jianyu Jiao1, Wenjun Li1,2


1. State Key Laboratory of Biocontrol, School of Life Sciences, Sun Yat-Sen University, Guangzhou 510275, Guangdong Province, China;
2. State Key Laboratory of Desert and Oasis Ecology, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Urumqi 830011, Xinjiang Uygur Autonomous Region, China
Received: 10 December 2020; Revised: 19 February 2021; Published online: 11 March 2020
*Corresponding author: Wenjun Li. Tel/Fax: +86-20-84111727; E-mail: liwenjun3@mail.sysu.edu.cn.
Foundation item: Supported by the Science and Technology Program of Guangzhou, China (201803030030), by the Guangdong Basic and Applied Basic Research Foundation (2019A1515110227) and by the China Postdoctoral Science Foundation (2019M653156)
Abstract: The anaerobic ammonia oxidation (anammox) reaction with nitrite as an electron acceptor and nitrogen as a product is mediated by bacteria which belong to Planctomycetales. AnAOB are widely present in marine, freshwater and terrestrial ecosystem, it also can be found in some other extreme environments. In this study, we reviewed the most new research advance on taxonomy, characteristics, metabolism, distribution, biotechniques used for the analysis of AnAOB and application. Finally, we discussed some questions exiting in this field and prospected future research of anaerobic ammonium oxidation microorganisms.
Keywords: anaerobic ammonium oxidation bacteriaspeciescharacteristicsdistributionmetabolism pathway
1 厌氧氨氧化细菌的发现历程 1977年,根据自由能的变化,Engelbert Broda最先预言自然界中存在着以NO2–和NO3–为电子受体氧化NH4+的过程[1];10年后,Arnold Mulder发现在反硝化处理过程中,随着NO3–的减少,NH4+会损失,N2会增加[2];1995年,Arnold Mulder在脱氮流化床反应器内证实了上述现象,并将这一过程称为厌氧氨氧化(anaerobic ammonium oxidation,anammox)[3],但其并不清楚这一现象是自发的化学反应还是由微生物参与的反应;随后van de Graaf通过实验确认了厌氧氨氧化是微生物参与的氧化还原反应[4];1999年,Strous等[5]利用梯度密度离心法,从生物膜上获得了纯度高达99.6%的具有厌氧氨氧化功能的微生物,系统发育分析显示,这类微生物属于浮霉菌,是浮霉菌门起源比较早的分支,至此,厌氧氨氧化细菌(anaerobic ammonium-oxidizing bacteria,AnAOB)被首次确认。
厌氧氨氧化细菌的发现经历了漫长的历程,这类微生物会在无氧的条件下,以NH4+为电子供体,NO2–为电子受体,产生N2,但最新的研究表明,厌氧氨氧化细菌可以直接以NO为电子受体将NH4+氧化为N2,且N2是唯一的最终产物,没有N2O和NO3–的产生[6]。厌氧氨氧化细菌是介导氮循环的重要微生物类群,它的发现颠覆了我们对氮循环的认识[7],且其所具有的脱氮功能,在水体净化以及环境污染治理方面具有重要的意义。
2 厌氧氨氧化细菌的种类 厌氧氨氧化细菌属于浮霉菌门下的类群,伯杰氏手册中收录的厌氧氨氧化细菌为5个属,但时至今日,厌氧氨氧化细菌共有6个属23个种(表 1和图 1),且都未获得纯培养。这6个属分别是Candidatus Brocadia (B. anammoxidans、B. fulgida、B. sinica、B. brasiliensis、B. caroliniensis和B. sapporoensis),Candidatus Kuenenia (K. stuttgartiensis),Candidatus Jettenia (J. asiatica、J. ecosi、J. moscovienalis和J. caeni),Candidatus Scalindua (S. sorokinii、S. brodae、S. wagneri、S. arabica、S. sinooilfield、S. rubra、S. japonica、S. profunda、S. marina和S. richardsii), Candidatus Anammoxoglobus (A. propionicus)及Candidatus Brasilis (B. concordiensis)。在上述微生物中,Candidatus Brasilis、Candidatus Kuenenia、Candidatus Brocadia、Candidatus Anammoxoglobus和Candidatus Jettenia均最先发现于生物反应器中,而Candidatus Scalindua主要存在于海洋和一些低氧的环境内。Candidatus ‘Brocadia anammoxidans’是首个被富集确定的厌氧氨氧化细菌,为该类群的模式种[5];Candidatus‘Brocadia fulgida’是目前发现的厌氧氨氧化细菌中唯一一个具有发光性能的微生物[8];Candidatus‘Kuenenia stuttgartiensis’是该类群中第二个被富集到[9],且是首个获得全基因组序列的物种[10];Candidatus Anammoxoglobus能氧化甲酸和丙酸,有利于厌氧氨氧化细菌在生活污水脱氮上的应用[11];与其他属相比,Candidatus Jettenia对亚硝酸盐的耐受性较高,多存在于淡水中[12];Candidatus Scalindua包含10个种,其中7个发现于海洋中,多为嗜盐菌[13-14],2个发现于反应器内[15],另外一个则存在于油藏环境中[16]。
表 1. 已有的厌氧氨氧化细菌的种类 Table 1. The species of anaerobic ammonium oxidation bacteria
| Generic names | Species and genus names | Publication time | Sources | References |
| Candidatus Brocadia | B. anammoxidans | 1999 | Wastewater | [5] |
| B. fulgida | 2008 | Bioreactor | [17] | |
| B. sinica | 2010 | Bioreactor | [18] | |
| B. brasiliensis | 2011 | Wastewater | [19] | |
| B. caroliniensis | 2013 | Wastewater | [20] | |
| B. sapporoensis | 2017 | Bioreactor | [21] | |
| Candidatus Scalindua | S. brodae | 2003 | Wastewater | [15] |
| S. wagneri | 2003 | Wastewater | [15] | |
| S. sorokinii | 2003 | Seawater | [22] | |
| S. arabica | 2008 | Seawater | [23] | |
| S. sinooilfield | 2010 | Oil reservoirs | [16] | |
| S. marina | 2011 | Marine sediment | [24] | |
| S. richardsii | 2012 | Black sea suboxic zone | [25] | |
| S. profunda | 2013 | Marine sediment | [26] | |
| S. rubra | 2017 | Seawater | [27] | |
| S. japonica | 2017 | Bay sediment | [28] | |
| Candidatus Jettenia | J. asiatica | 2008 | Bioreactor | [29] |
| J. caeni | 2014 | Bioreactor | [30] | |
| J. ecosi | 2018 | Bioreactor | [31] | |
| J. moscovienalis | 2015 | Bioreactor | [32] | |
| Candidatus Kuenenia | K. stuttgartiensis | 2000 | Bioreactor biofilm | [9] |
| Candidatus Anammoxoglobus | A. Propionicus | 2007 | Bioreactor | [33] |
| Candidatus Brasilis | B. concordiensis | 2011 | Bioreactor | [34] |
表选项
 |
| 图 1 基于16S rRNA基因序列所构建的厌氧氨氧化细菌进化树 Figure 1 Phylogenetic tree of AnAOB based on 16S rRNA gene sequences. |
| 图选项 |
3 厌氧氨氧化细菌的性质与特殊结构 厌氧氨氧化细菌是能以二氧化碳为唯一碳源的自养类群[35],部分厌氧氨氧化细菌可以氧化乙酸和丙酸等有机物质,如在反应器中添加乙酸和丙酸可分别富集到Candidatus‘Brocadia fulgida’[8]和Candidatus‘Anammoxoglobus propionicus’[33],但这些小分子有机酸不能被这2种微生物用作碳源来合成细胞物质,而最新的研究却表明Candidatus ‘Kuenenia stuttgartiensis’能直接以甲酸为碳源,是混合营养型微生物[36]。厌氧氨氧化细菌大多嗜中温,最适生长温度为15–40 ℃,最佳生长pH为6.7–8.3,是严格厌氧微生物,其在不同环境中的群落结构组成和活性受到盐浓度[37-38]、温度[39]、有机碳[40]及无机氮[41]等的影响。厌氧氨氧化细菌含有丰富的细胞色素, 富集后呈红色,俗称“红菌”,其细胞成不规则的球状或者卵状,为革兰氏阴性菌,细胞内拥有特殊的细胞器,该细胞器被称为厌氧氨氧化体。厌氧氨氧化体最早于2001年被提出[42],其占细胞总体积的50%–80%,是进行厌氧氨氧化过程的场所,它的存在使厌氧氨氧化微生物的细胞物质被细胞质膜、胞浆内膜、厌氧氨氧化体膜分隔成3个部分,从外到内分别为外室细胞质、核糖细胞质和厌氧氨氧化体[43]。Damsté等[44]用气质联用仪(GC-MS)和高场核磁共振仪对厌氧氨氧化体的膜结构进行了分析,发现其膜上含有一种特殊的成分-梯烷,梯烷是一种由3–5个线性串联的环丁烷组成的梯状结构。厌氧氨氧化体的膜主要由梯烷通过烷基链与甘油分子结合而成,甘油有sn-1、sn-2、sn-3共3个连接位点,其中sn-1和sn-2位点连接烷烃支链,sn-3位与一个磷脂分子结合,磷脂分子是整个梯烷脂的首基部分,磷脂酰胆碱(PC)和磷脂酰乙醇胺(PE)是主要的首基,sn-3位失去极性首基,则被称为核心梯烷脂,含有极性首基则被称为完整梯烷脂[45],而梯烷脂的多样性主要由sn-1位上连接的烷烃种类来决定。梯烷结构致密,渗透性差,可阻止厌氧氨氧化过程中产生的有毒物质外泄,避免对厌氧氨氧化细菌自身的毒害作用,其是厌氧氨氧化细菌的特有成分,目前未见于其他细菌中[35]。
4 厌氧氨氧化细菌的代谢途径 4.1 氮代谢途径 最初认为,在厌氧氨氧化反应中,NO3–是电子受体[3],随后的研究证实NO2–更适合做电子受体[4, 46];van de Graaf使用同位素标记法表明,在厌氧氨氧化过程中有NH2OH的产生,其由NO2–转化而来,当系统中有过量NH2OH和NH4+时,N2H4会有短暂的积累,随后转化为N2,N2H4氧化过程释放的4个电子被用于NO2–的还原[47];Schalk等认为N2H4的生成是由细胞质内一种被膜包裹的复杂酶催化而成[48](后被证实这种酶是联氨合成酶),且部分(近20%)NO2–会转化成NO3–;研究者在对Candidatus‘Brocadia anammoxidans’进行研究时,通过透射电镜发现厌氧氨氧化体是进行厌氧氨氧化代谢的细胞器,是N2H4生成的主要场所[49]。至此,研究者认为厌氧氨氧化可能的代谢途径是NO2–转化为NH2OH,NH2OH与NH4+转化为N2H4,最后N2H4再转化为N2,且部分NO2–会转化为NO3–,N2H4氧化过程释放的4个电子被用于NO2–的还原。2006年,随着测序技术的发展,研究者得到了Candidatus ‘Kuenenia stuttgartiensis’的基因组序列[10],从Candidatus‘Kuenenia stuttgartiensis’的基因组数据中,第二种厌氧氨氧化细菌的代谢途径被提出,该途径认为NO2–还原产生的中间产物并非NH2OH,而是NO,即NO2–在亚硝酸盐还原酶(NIR)的作用下转化为NO,NO和NH4+在联氨合成酶(HZS)的作用下转化为N2H4,随后N2H4在羟胺氧化还原酶(HAO)或者联氨氧化还原酶(HZO)的作用下生成N2,N2H4转化过程中释放4个电子,这4个电子分别传递给NIR和HZS,其中3个电子传递给NIR,1个电子传递给HZS。伴随电子传递,质子被排放至厌氧氨氧化体膜外侧,该膜两侧形成质子梯度,驱动ATP和NADPH的合成[50]。2019年,Kartal博士发现厌氧氨氧化细菌可以直接以NO为电子受体,将NH4+转化为N2,且不生成NO3–和N2O等产物[6],减少了温室气体的排放,这一发现进一步拓宽了我们对地球氮循环的认识。因此,综合上述研究结果,现被提出的厌氧氨氧化细菌参与的氮循环共有3条途径(图 2)。
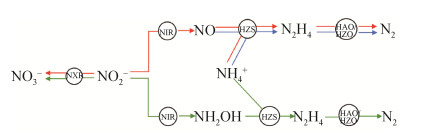 |
| 图 2 厌氧氨氧化细菌的氮代谢途径 Figure 2 Nitrogen metabolism of anammox bacteria. Different colors represent different pathways of nitrogen metabolism. |
| 图选项 |
厌氧氨氧化细菌介导的氮循环过程需要多种酶的参与,这些酶包括羟胺氧化还原酶(HAO),亚硝酸盐还原酶(NIR),亚硝酸氧化还原酶(NXR),联氨合成酶(HZS),联氨氧化还原酶(HZO)。HAO可以同时作用于NH2OH和N2H4,而HZO只作用于N2H4,HAO和HZO作用于N2H4时,其活性受到NH2OH浓度的影响,当NH2OH浓度过高时,HZO活性被抑制,HAO发挥作用,当NH2OH浓度较低时,HZO和HAO可同时发挥作用[51-52]。同时,HZS和HZO是厌氧氨氧化细菌所特有的酶,而其他几种酶在好氧氨氧化细菌和反硝化细菌中都有发现。由于厌氧氨氧化细菌会分泌大量的胞外聚合物,这使得对其代谢酶的研究有很大难度,到目前为止,研究较为深入的酶是HAO和HZO两种。
4.2 碳代谢途径 厌氧氨氧化细菌属化能自养型微生物,主要碳源是CO2或HCO3–。厌氧氨氧化细菌的碳代谢是指通过碳固定途径将CO2、碳酸盐或者碳酸氢盐等无机碳源还原为有机化合物。目前已知的微生物固碳途径主要有7条:卡尔文循环、还原型三羧酸循环、还原乙酰辅酶A途径、3-羟基丙酸双循环、3-羟基丙酸/4-羟基丁酸循环、二羧酸/4-羟基丁酸循环和反向甘氨酸裂解途径[53-54],反向甘氨酸裂解途径是最近提出的第7种固碳途径[53]。最初,人们通过同位素示踪法和酶活法推断厌氧氨氧化细菌的固碳方式应该为卡尔文循环或者还原性乙酰辅酶A途径[55],但始终未能确认到底是哪一条途径。2006年,Strous等采用宏基因组技术对Candidatus‘Kuenenia stuttgartiensis’的序列进行分析,发现其基因组中存在实现还原性乙酰辅酶A途径所需的所有关键基因,而卡尔文循环中的关键基因有所缺失,无法形成完整的途径[56],这一证据表明了厌氧氨氧化细菌对CO2的固定方式是还原性乙酰辅酶A途径。同时,在Candidatus ‘K. stuttgartiensis’基因组中检出了8个编码甲酸脱氢酶(Formate dehydrogenase,FDH)的基因和5个编码一氧化碳脱氢酶(Carbon monoxide dehydrogense,CODH)的基因,并检测出了FDH和CODH酶活。
4.3 其他代谢途径 最初的研究认为,厌氧氨氧化细菌是专性化能自养型微生物[57],只能利用NH4+作为电子供,NO2–作为电子受体。随着对厌氧氨氧化过程及厌氧氨氧化细菌研究的深入,人们发现厌氧氨氧化细菌还具有一些独特的生理功能。Strous等的研究表明,在Candidatus K. stuttgartiensis的基因组中,有多达200个基因负责指导合成与代谢和呼吸作用有关的酶,厌氧氨氧化细菌可能具有代谢多样性,能够利用有机物、金属离子(Fe2+、Fe3+和Mn2+)等充当其电子供体和受体。同时,相关研究表明,金属离子的添加,能促进厌氧氨氧化细菌混培物的生长,促进反应器对氮的去除[58-60]。
过去的观点认为有机物的存在会对厌氧氨氧化细菌产生不利影响,一方面是因为某些有机物对厌氧氨氧化细菌有毒性抑制作用[61],另一方面是因为有机物对厌氧氨氧化细菌的基质竞争性抑制作用[62]。但随着研究的深入,研究者发现在某些有机碳源条件下厌氧氨氧化细菌仍然能够保持一定活性,和反硝化细菌竞争底物。有研究表明,甲烷[63]和甲酸盐[64]均已被证实能够作为某些厌氧微生物的电子供体来还原NO2–/NO3–;乙酸盐和丙酸盐不仅不影响厌氧氨氧化细菌的生长,还能被厌氧氨氧化细菌利用。2008年,研究者通过密度梯度离心的方法获得了高纯度的厌氧氨氧化细菌,并利用短期和长期运行实验证明该细菌能直接利用NH4+为电子供体,SO42–为电子受体,实现NH4+和SO42–的同时去除[65]。
5 厌氧氨氧化细菌的分布 厌氧氨氧化过程最早发现于生物废水处理反应器中,因此早期对厌氧氨氧化的研究主要集中在废水处理系统上,现在已知的厌氧氨氧化物种大部分都发现于生物反应器中(表 1)。2002年,Thamdrup等首次在海洋中发现了厌氧氨氧化细菌[66],其研究表明在波罗海大陆架沉积物中24%–67%氮气的生成与厌氧氨氧化有关,这一发现拉开了海洋生境厌氧氨氧化细菌研究的序幕。2003年,Kuypers等通过荧光原位杂交,同位素标记法和梯烷膜脂的检测,证实在世界最大的厌氧盆地黑海中存在着厌氧氨氧化细菌[22]。Dalsgaard等利用15N稳定性同位素示踪的方法发现在哥斯达黎加的Golfo Dulce海湾中有19%–35%的氮气的生成与厌氧氨氧化过程有关[67]。随后又在其他很多海洋系统中检测到了厌氧氨氧化细菌,如在鄂霍茨克海的深海沉积物中[68],中国南海红树林沉积物中[69],东海郁陵岛流域中[41]及张江河口红树林湿地[70]等等。在这些海洋生态系统中,Candidatus Scalindua都是主要的厌氧氨氧化细菌类群。
除了存在于反应器和海洋生态系统中外,厌氧氨氧化细菌还存在淡水生态系统中。2006年,Schubert等通过荧光原位杂交实验及膜脂成分分析,第一次从世界第二大湖泊Tanganyika中检测到了厌氧氨氧化细菌,基于16S rRNA基因进行的系统发育分析显示该湖泊中的厌氧微生物与Candidatus‘Scalindua brodae’的相似性为95.7%,同位素标记实验表明该湖泊中13%的氮气生成与厌氧氨氧化细菌有关[71]。在Superior湖内,存在着独特的厌氧氨氧化细菌类群,其对氮气的排放有着重要的贡献[72]。城市河水系统中,厌氧氨氧化细菌的多样性丰富,NH4+对厌氧氨氧化细菌的群落结构有着重要影响[73]。除此之外,厌氧氨氧化细菌还存在于地下水、蓄水层及池塘[74-76]等多种淡水生态系统中。
与海洋和淡水生态系统相比较,陆地生态系统的厌氧氨氧化细菌的发现较晚。2010年,Humbert等首次从沼泽、湖岸、含水土层、冻土及农业土壤中检测到了厌氧氨氧化细菌,与海洋生态系统相比较,Candidatus Brocadia和Candidatus Kunenenia在上述陆地生态系统中普遍存在,表明陆地生态系统中厌氧氨氧化细菌的多样性较海洋生态系统的丰富,可能原因是陆地生态系统中适合厌氧氨氧化细菌生长的生态位更丰富[77]。2011年,胡宝兰等通过富集,从氮含量较高的泥碳土中得到了占比为40%–50%的厌氧氨氧化细菌的富集物[78]。2015年,研究者在蔬菜地检测到了厌氧氨氧化细菌,且其在蔬菜地中的氮气排放率占到5.9%–20.5%[79]。随后,在根际[80]及水稻田土壤[81]中也发现了厌氧氨氧化细菌的踪迹。
厌氧氨氧化细菌还存在于一些特殊的环境中。2009年,研究者在高达52 ℃的California和Nevada热泉中,检测到了Candidatus‘Brocadia fulgida’,Candidatus‘Brocadia anammoxidans’和Candidatus‘Kuenenia stuttgartiensis’[82]。通过分子生态学手段,对17个高温油藏中的厌氧氨氧化细菌进行了分析,其中有9个油藏环境中都有厌氧氨氧化细菌,基于16S rRNA基因检测的物种与已知的Candidatus Brocadia、Candidatus Kuenenia、Candidatus Scalindua和Candidatus Jettenia密切相关,而基于HZO基因检测的物种与Candidatus Anammoxoglobus、Candidatus Kuenenia、Candidatus Scalindua和Candidatus Jettenia密切相关[16]。Byrne等在对大西洋中脊的3次海洋考察期间,从5个深度为750到3650米的热液口采集了样品,16S rRNA基因序列的扩增,脂质分析和同位素检测结果表明,新分支的厌氧氨氧化类群可能在这些热的栖息地中具有活性[83]。厌氧氨氧化细菌同样存在于海绵这样的古老动物中,主要类群为Candidatus Scalindua和Candidatus Brocadia[84-85],活性检测显示,其对氮气产生的贡献率为1.25%[84]。
6 厌氧氨氧化细菌的研究手段 迄今为止,厌氧氨氧化类群未获得纯培养物,因此分子生物学手段成为了研究厌氧氨氧化细菌的重要手段,包括种类检测、数量检测、分布检测及活性检测。种类检测方面,主要是根据16S rRNA基因或者其他功能基因(表 2)对环境样品的DNA进行特异性扩增,结合变形梯度凝胶电泳技术或者克隆文库的构建,对扩增产物进行测序比对分析,建立系统发育树,从而鉴定其种类[86-88]。数量的检测方面主要是借助于qPCR技术,采用特异性引物对DNA进行扩增,获得其拷贝数;同时也可以检测样品中总细菌的拷贝数,根据厌氧氨氧化细菌与总细菌拷贝数的比值来确定厌氧氨氧化细菌在样品中所占的比例[89-91]。厌氧氨氧化细菌拥有特异的16S rRNA基因或其他功能基因,以荧光素标记针对上述基因的特有探针并与上述基因特异性结合,然后在荧光显微镜下观察,分析厌氧氨氧化细菌的空间分布[92]。同位素标记法广泛用于检测环境样品中厌氧氨氧化细菌活性,用15NH4+和14NO3–作为基质,收集产生的N2,其中29N2为厌氧氨氧化细菌产物,由29N2产生速度即可推断厌氧氨氧化细菌的活性及其在氮素循环中的贡献率[81, 93]。厌氧氨氧化细菌具有梯烷这一特殊结构,因此也有不少研究者通过多梯烷的分析来展开对厌氧氨氧化细菌的研究[94-95]。
表 2. AnAOB检测常用引物 Table 2. PCR primer for detecting of AnAOB
| Primers | Amplified fragment | Specificity |
| Common primers for 16S rRNA gene amplification | ||
| A438f-A684r | 250 | All AnAOB |
| Amx368F-Amx820R | 470 | Except Scalindua |
| Amx368F-BS820R | 470 | Scalindua |
| AMXU368F-AMXU820R | 470 | All AnAOB |
| Brod541F-Amx820R | 279 | Scalindua |
| Amx368F-1390R | 1040 | All AnAOB (Nested PCR) |
| An7F-An1388R | 1360 | All AnAOB (Nested PCR) |
| Pla46F-1390R | 1360 | All AnAOB (Nested PCR) |
| Common primers for functional gene amplification | ||
| hzocl1F1-hzocl1R2 | 470 | All AnAOB |
| Ana-hzo1F-Ana-hzo2R | 1030 | Jettenia, Kuenenia |
| Scnir372F-Scnir845R | 470 | Scalindua |
| AnnirS379F-AnnirS821R | 440 | Except Scalindua |
| Common primers for qPCR | ||
| Amx808f-Amx1040R | 232 | All AnAOB |
| hzsA-1597f/hzsA-1857r | 260 | All AnAOB |
表选项
近年来,随着测序技术的发展,扩增子及宏基因组数据为厌氧氨氧化细菌的研究提供了新的手段。罗氏454测序结果显示,在扬子江内存着Candidatus Scalindua和Candidatus Brocadia两个属内的菌株,且TOC和TN对其的丰度有着显著影响[90]。青海和西藏高原的Hiseq测序结果显示,Candidatus Brocadia,Candidatus Jettenia和Candidatus Kuenenia是土壤中的主要厌氧氨氧化类群,海拔的不同对其多样性、群落组成和丰度不构成影响[96]。在中国南部的酸性土壤中,Candidatus Brocadia是主要的厌氧氨氧化类群,所占比例为93.03%,氮气的排放速率为0.01–0.59 nmol/(g·h),对氮气排放的贡献率为16.67%–53.27%。此外,研究者借助高通量数据还对云南淡水湖[91]、北京污水处理厂的反应器[97]及南海沉积物中厌氧氨氧化细菌进行了研究[98]。
7 厌氧氨氧化细菌的应用 厌氧氨氧化细菌的应用主要体现在工业污水处理上。随着城市人口的增多和工业化水平的进步,我国水资源污染问题日渐突出,水体富营养化问题加剧,污水处理问题已成为当下热点。氨态氮是水体富营养化的一种重要污染物质,经济有效控制含氨氮废水污染成为环境工作所面临的重大问题。废水中氨氮的去除方法主要有物化法和生物法,生物脱氮法主要有消化、反硝化、短程消化与反硝化、同时消化与反硝化及厌氧氨氧化,与其他生物脱氮相比,厌氧氨氧化工艺是在无氧的条件下将NH4+和NO2–转化为N2,在好氧阶段只需将NH4+转化为NO2–,省略后续NO2–氧化为NO3–,可节约62.5%的曝气量;厌氧氨氧化细菌是无机营养型,所以无需额外添加碳源;该过程N2O和NO的排放量减少,减少了温室气体对环境的污染;同时该技术污泥产生量也减少了90%[99]。因此,厌氧氨氧化处理工艺不仅展现出更好的脱氮性能,而且可以节约成本,减少二次污染,避免温室气体的排放,减少实验所需的占地空间,具有广阔的应用前景。
厌氧氨氧化工艺最早是在一个中试规模的反硝化流化床中被发现[3],随后研究者对此工艺的研究主要集中在反应机理、微生物特征及控制条件等方面。2002年,荷兰研究者根据前期的研究成果,通过数学模型模拟设计出了世界上第一个生产性规模的厌氧氨氧化反应器,并于2002年6月投入运行,主要用于污泥消化液的脱氮处理[100]。目前该工艺主要用于垃圾渗滤液[101],污泥消化液[102],食品加工厂废水[103]及制药厂废水[104]的处理上,据统计,全世界现有110个全规模的厌氧氨氧化装置[105]。国外关于厌氧氨氧化工艺的研究比较成熟,且取得了一定成果。2002年,荷兰设计并启动了第一套全规模厌氧氨氧化系统,每天的脱氮净重为750 kgN[100],为厌氧氨氧化技术在废水脱氮领域的处理奠定了基础。在亚洲,第一个大型全规模应用在经历了60 d的启动后在日本投入使用[106]。国内关于厌氧氨氧化工艺的研究虽然较国外起步晚,2009年,厌氧氨氧化工艺才首次在台湾应用于垃圾渗滤液的脱氮全规模应用中[107],但国内关于厌氧氨氧工艺的应用研究也相对成熟且取得了一定的研究成果[108-110]。
要想通过厌氧氨氧化过程来去除水体中的氨氮,则必须提供亚硝态氮,但污水中通常不含有亚硝态氮,必须将水体中的部分氨氮通过消化作用转化为硝态氮,因此集合短程的消化作用和厌氧氨氧化过程开发了系列的工艺,如SHARON- ANAMMOX (亚硝酸型硝化-厌氧氨氧化联合工艺)[111-112]、CANON (全程自养脱氮工艺)[113-114]、OLAND (限氧自养反硝化工艺)[115]及DEMON (好氧反氨化工艺)[116]。与此同时,为保证该工艺的正常运行,不同类型的反应器也相继问世。如续批式反应器[117]、膜反应器[118]、移动床生物膜反应器[119]及整合固定膜活性污泥反应器[120]等。
厌氧氨氧化细菌生长缓慢,倍增时间长达10– 15 d[56],细胞产率低,厌氧氨氧化细菌在细胞密度高达1010个/mL以上时,才能显现出厌氧氨氧化活性,这导致厌氧氨氧化工艺启动时间较长,因此,提高厌氧氨氧化细菌的相对丰度和活性,加快厌氧氨氧化启动进程才能提高工艺脱氮效率。研究表明添加外加物质(金属离子、无机碳、导电性材料及厌氧氨氧化中间产物),增大前期厌氧氨氧化种泥接种量或增加后期厌氧氨氧化细菌流加频率[121-125],可加快厌氧氨氧化细菌的富集,提高相对丰度和活性,缩短厌氧氨氧化启动周期,快速启动厌氧氨氧化工艺。同时厌氧氨氧化细菌对温度、溶解氧、pH值、有机物、基质浓度、磷酸盐、盐度、硫化物、重金属以及光照等环境条件异常敏感[126-131],导致厌氧氨氧化工艺不易启动,容易失稳,失稳后难以恢复,厌氧氨氧化细菌的这些缺陷限制了其工艺的广泛运用。
8 展望 厌氧氨氧化细菌是介导氮循环的重要微生物类群,具有脱氮功能,在水体净化以及环境污染治理方面具有重要的意义,是一类重要的微生物资源。近年来厌氧氨氧化细菌成为了研究的热点,具有很多重大的发现,这些深化了我们对厌氧氨氧化微生物及其所介导的氮循环的认识,但关于厌氧氨氧化微生物现在仍有很多盲点,还有诸多问题需要进一步探究。
(1)近几十年来,无数研究者致力于厌氧氨氧化细菌的富集及分离,但仅通过富集得到了丰度更高的该类群,却始终未获得纯培养菌株,因此,结合目前的研究结果,根据厌氧氨氧化微生物的特性(如生长缓慢,细胞产率和密度低;大多嗜中温,最适生长温度为15–40 ℃;受无机氮,有机碳和盐浓度的影响等)优化分离培养条件,开创新的分离培养方法来获得厌氧氨氧化细菌的纯培养物是后续研究的重点。
(2)酶学研究和基因组分析揭示了厌氧氨氧化细菌的代谢方式,但目前对厌氧氨氧化细菌的代谢途径的理解大多停留在对于基因组数据分析的推测层面,一些关键基因的功能及其功能酶的生物化学特性仍有待进一步确认和验证;代谢途径中的很多酶类还没有分离获得,代谢途径还存在很多疑点;目前关于代谢途径的研究仅局限在Candidatus‘Brocadia anammoxidans’和Candidatus‘Kunenenia stuttgartiensis’上,对其他20多个厌氧氨氧化细菌的代谢途径并没有深入研究。因此,关键酶类的体外解析是否可以帮助完善这类细菌的代谢通路是我们需要探讨的内容;纯培养菌株的获得及以更多的厌氧氨氧化细菌为研究对象,结合多组学研究方法,对其代谢途径进行研究,以获得完整明确的代谢通路是急需开展的工作。
(3)随着测序技术的发展,高通量数据为我们提供更全面更详细的信息,让我们对环境中厌氧氨氧化细菌的组成与分布有了更清楚的认识,了解到环境中还存在很多未知的厌氧氨氧化类群,因此采用合适的技术方法来挖掘这些新的厌氧氨氧化物种是今后厌氧氨氧化细菌研究中需要长期进行的研究。
(4)基于16S rRNA基因和功能基因的研究为认识环境中厌氧氨氧化微生物的组成与分布提供了重要信息,目前针对这些基因的引物种类繁多,且这些基因扩增引物的特异性、代表性和有效性存在争议。因此,功能基因与16S rRNA基因所代表的微生物之间的聚类关系如何对应,针对厌氧氨氧化微生物的扩增有没有一个统一的标准是我们需要去解答的问题。
(5)目前已知的6个属的厌氧氨氧化微生物都属于浮霉菌门,但在其他细菌类群甚至古菌中是否存在具有厌氧氨氧化功能的微生物,是后续研究中我们需要关注的问题。
(6)以更多的生境为研究对象,结合多种检测手段,完善厌氧氨氧化微生物在自然界中的分布情况,明确其重要贡献。如厌氧氨氧化细菌虽存在于多种环境中,但目前鲜见关于热泉环境厌氧氨氧化细菌的报道,因此,本课题组对云南热泉环境的厌氧氨氧化微生物进行了研究,宏基因组数据及16S rRNA基因的扩增子数据表明在云南热泉环境中存在厌氧氨氧化细菌,主要类群为Candidatus Brocadia和Candidatus Kuenenia。后期会对相关样点的厌氧氨氧化细菌进行富集,比较富集前后类群和丰度的差异,并对其氮气产生的贡献率进行检测,相关研究可为全面认识云南热泉生态系统中厌氧氨氧化细菌提供重要的理论基础,为掌握其生态学分布及对云南热泉生境中氮循环的贡献率提供重要的理论依据。
References
| [1] | Broda E. Two kinds of lithotrophs missing in nature. Zeitschrift für Allgemeine Mikrobiologie, 1977, 17(6): 491-493. DOI:10.1002/jobm.19770170611 |
| [2] | Kuenen JG. Anammox bacteria: from discovery to application. Nature Reviews: Microbiology, 2008, 6(4): 320-326. DOI:10.1038/nrmicro1857 |
| [3] | Mulder A, Van de Graaf AA, Robertson L, Kuenen J. Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiology Ecology, 1995, 16(3): 177-183. DOI:10.1111/j.1574-6941.1995.tb00281.x |
| [4] | Van de Graaf AA, Mulder A, de Bruijn P, Jetten M, Robertson L, Kuenen JG. Anaerobic oxidation of ammonium is a biologically mediated process. Applied and Environmental Microbiology, 1995, 61(4): 1246-1251. DOI:10.1128/AEM.61.4.1246-1251.1995 |
| [5] | Strous M, Fuerst JA, Kramer EH, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MS. Missing lithotroph identified as new planctomycete. Nature, 1999, 400(6743): 446-449. DOI:10.1038/22749 |
| [6] | Hu Z, Wessels H, van Alen T, Jetten MSM, Kartal B. Nitric oxide-dependent anaerobic ammonium oxidation. Nature Communications, 2019, 10(1): 1244. DOI:10.1038/s41467-019-09268-w |
| [7] | Thamdrup B. New Pathways and Processes in the Global Nitrogen Cycle. Annual Review of Ecology, Evolution, and Systematics, 2012, 43(1): 407-428. DOI:10.1146/annurev-ecolsys-102710-145048 |
| [8] | Kartal B, van Niftrik L, Rattray J, van de Vossenberg JL, Schmid MC, Sinninghe Damste J, Jetten MS, Strous M. Candidatus 'Brocadia fulgida': an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiology Ecology, 2008, 63(1): 46-55. DOI:10.1111/j.1574-6941.2007.00408.x |
| [9] | Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M, Metzger JW, Schleifer KH, Wagner M. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Systematic and Applied Microbiology, 2000, 23(1): 93-106. DOI:10.1016/S0723-2020(00)80050-8 |
| [10] | Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Medigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJ, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes HW, Weissenbach J, Jetten MS, Wagner M, Le Paslier D. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature, 2006, 440(7085): 790-794. DOI:10.1038/nature04647 |
| [11] | Kartal B, Rattray J, van Niftrik LA, van de Vossenberg J, Schmid MC, Webb RI, Schouten S, Fuerst JA, Damste JS, Jetten MS, Strous M. Candidatus "Anammoxoglobus propionicus" a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Systematic and Applied Microbiology, 2007, 30(1): 39-49. DOI:10.1016/j.syapm.2006.03.004 |
| [12] | Quan ZX, Rhee SK, Zuo JE, Yang Y, Bae JW, Park JR, Lee ST, Park YH. Diversity of ammonium-oxidizing bacteria in a granular sludge anaerobic ammonium-oxidizing (anammox) reactor. Environmental Microbiology, 2008, 10(11): 3130-3139. DOI:10.1111/j.1462-2920.2008.01642.x |
| [13] | van de Vossenberg J, Woebken D, Maalcke WJ, Wessels HJ, Dutilh BE, Kartal B, Janssen-Megens EM, Roeselers G, Yan J, Speth D, Gloerich J, Geerts W, van der Biezen E, Pluk W, Francoijs KJ, Russ L, Lam P, Malfatti SA, Tringe SG, Haaijer SC, Op den Camp HJ, Stunnenberg HG, Amann R, Kuypers MM, Jetten MS. The metagenome of the marine anammox bacterium 'Candidatus Scalindua profunda' illustrates the versatility of this globally important nitrogen cycle bacterium. Environmental Microbiology, 2013, 15(5): 1275-1289. DOI:10.1111/j.1462-2920.2012.02774.x |
| [14] | Brandsma J, van de Vossenberg J, Risgaard-Petersen N, Schmid MC, Engstrom P, Eurenius K, Hulth S, Jaeschke A, Abbas B, Hopmans EC, Strous M, Schouten S, Jetten MS, Damste JS. A multi-proxy study of anaerobic ammonium oxidation in marine sediments of the Gullmar Fjord, Sweden. Environmental Microbiology Reports, 2011, 3(3): 360-366. DOI:10.1111/j.1758-2229.2010.00233.x |
| [15] | Schmid M, Walsh K, Webb R, Rijpstra WI, van de Pas-Schoonen K, Verbruggen MJ, Hill T, Moffett B, Fuerst J, Schouten S. Candidatus "Scalindua brodae", sp. nov., Candidatus "Scalindua wagneri", sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Systematic and Applied Microbiology, 2003, 26(4): 529-538. DOI:10.1078/072320203770865837 |
| [16] | Li H, Chen S, Mu B-Z, Gu J-D. Molecular detection of anaerobic ammonium-oxidizing (anammox) bacteria in high-temperature petroleum reservoirs. Microbial Ecology, 2010, 60(4): 771-783. DOI:10.1007/s00248-010-9733-3 |
| [17] | Kartal B, Van Niftrik L, Rattray J, Van De Vossenberg JL, Schmid MC, Sinninghe Damsté J, Jetten MS, Strous M. Candidatus 'Brocadia fulgida': an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiology Ecology, 2008, 63(1): 46-55. DOI:10.1111/j.1574-6941.2007.00408.x |
| [18] | Hu BL, Zheng P, Tang CJ, Chen JW, van der Biezen E, Zhang L, Ni BJ, Jetten MS, Yan J, Yu HQ. Identification and quantification of anammox bacteria in eight nitrogen removal reactors. Water Research, 2010, 44(17): 5014-5020. DOI:10.1016/j.watres.2010.07.021 |
| [19] | Araujo J, Campos A, Correa M, Silva E, Matte M, Matte G, Von Sperling M, Chernicharo C. Anammox bacteria enrichment and characterization from municipal activated sludge. Water Science and Technology, 2011, 64(7): 1428-1434. DOI:10.2166/wst.2011.632 |
| [20] | Vanotti MB, Szogi AA, Rothrock MJ. Anammox bacterium isolate. Google Patents; 2013. |
| [21] | Narita Y, Zhang L, Kimura ZI, Ali M, Fujii T, Okabe S. Enrichment and physiological characterization of an anaerobic ammonium-oxidizing bacterium 'Candidatus Brocadia sapporoensis'. Systematic and Applied Microbiology, 2017, 40(7): 448-457. DOI:10.1016/j.syapm.2017.07.004 |
| [22] | Kuypers MM, Sliekers AO, Lavik G, Schmid M, J?rgensen BB, Kuenen JG, Damsté JSS, Strous M, Jetten MS. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature, 2003, 422(6932): 608-611. DOI:10.1038/nature01472 |
| [23] | Woebken D, Lam P, Kuypers MM, Naqvi SWA, Kartal B, Strous M, Jetten MS, Fuchs BM, Amann R. A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindua phylotype in marine oxygen minimum zones. Environmental Microbiology, 2008, 10(11): 3106-3119. DOI:10.1111/j.1462-2920.2008.01640.x |
| [24] | Brandsma J, van de Vossenberg J, Risgaard-Petersen N, Schmid MC, Engstr?m P, Eurenius K, Hulth S, Jaeschke A, Abbas B, Hopmans EC. A multi-proxy study of anaerobic ammonium oxidation in marine sediments of the Gullmar Fjord, Sweden. Environmental Microbiology Reports, 2011, 3(3): 360-366. DOI:10.1111/j.1758-2229.2010.00233.x |
| [25] | Fuchsman CA, Staley JT, Oakley BB, Kirkpatrick JB, Murray JW. Free-living and aggregate-associated Planctomycetes in the Black Sea. FEMS Microbiology Ecology, 2012, 80(2): 402-416. DOI:10.1111/j.1574-6941.2012.01306.x |
| [26] | van de Vossenberg J, Rattray JE, Geerts W, Kartal B, van Niftrik L, van Donselaar EG, Sinninghe Damste JS, Strous M, Jetten MS. Enrichment and characterization of marine anammox bacteria associated with global nitrogen gas production. Environmental Microbiology, 2008, 10(11): 3120-3129. DOI:10.1111/j.1462-2920.2008.01643.x |
| [27] | Speth DR, Lagkouvardos I, Wang Y, Qian PY, Dutilh BE, Jetten MS. Draft genome of Scalindua rubra, obtained from the interface above the discovery deep brine in the Red Sea, sheds light on potential salt adaptation strategies in anammox bacteria. Microbial Ecology, 2017, 74(1): 1-5. DOI:10.1007/s00248-017-0929-7 |
| [28] | Oshiki M, Mizuto K, Kimura ZI, Kindaichi T, Satoh H, Okabe S. Genetic diversity of marine anaerobic ammonium-oxidizing bacteria as revealed by genomic and proteomic analyses of 'Candidatus Scalindua japonica'. Environmental Microbiology Reports, 2017, 9(5): 550-561. DOI:10.1111/1758-2229.12586 |
| [29] | Quan ZX, Rhee SK, Zuo JE, Yang Y, Bae JW, Park JR, Lee ST, Park YH. Diversity of ammonium-oxidizing bacteria in a granular sludge anaerobic ammonium-oxidizing (anammox) reactor. Environmental Microbiology, 2008, 10(11): 3130-3139. DOI:10.1111/j.1462-2920.2008.01642.x |
| [30] | Ali M, Oshiki M, Awata T, Isobe K, Kimura Z, Yoshikawa H, Hira D, Kindaichi T, Satoh H, Fujii T. Physiological characterization of anaerobic ammonium oxidizing bacterium 'Candidatus Jettenia caeni'. Environmental Microbiology, 2015, 17(6): 2172-2189. DOI:10.1111/1462-2920.12674 |
| [31] | Botchkova E, Litti YV, Novikov A, Grouzdev D, Bochkareva E, Beskorovayny A, Kuznetsov B, Nozhevnikova A. Description of "Candidatus jettenia ecosi" sp. nov., a new species of anammox bacteria. Microbiology, 2018, 87(6): 766-776. DOI:10.1134/S002626171806005X |
| [32] | Nikolaev YA, Kozlov M, Kevbrina M, Dorofeev A, Pimenov N, Kallistova AY, Grachev V, Kazakova E, Zharkov A, Kuznetsov B. Candidatus "Jettenia moscovienalis" sp. nov., a new species of bacteria carrying out anaerobic ammonium oxidation. Microbiology, 2015, 84(2): 256-262. DOI:10.1134/S0026261715020101 |
| [33] | Kartal B, Rattray J, van Niftrik LA, van de Vossenberg J, Schmid MC, Webb RI, Schouten S, Fuerst JA, Damsté JS, Jetten MS. Candidatus "Anammoxoglobus propionicus" a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Systematic and Applied Microbiology, 2007, 30(1): 39-49. DOI:10.1016/j.syapm.2006.03.004 |
| [34] | Viancelli A, Kunz A, Esteves PA, Bauermann FV, Furukawa K, Fujii T, Ant?nio RV, Vanotti M. Bacterial biodiversity from an anaerobic up flow bioreactor with ANAMMOX activity inoculated with swine sludge. Brazilian Archives of Biology and Technology, 2011, 54(5): 1035-1041. DOI:10.1590/S1516-89132011000500022 |
| [35] | Kartal B, de Almeida NM, Maalcke WJ, Op den Camp HJ, Jetten MS, Keltjens JT. How to make a living from anaerobic ammonium oxidation. FEMS Microbiology Reviews, 2013, 37(3): 428-461. DOI:10.1111/1574-6976.12014 |
| [36] | Lawson CE, Nuijten GHL, de Graaf RM, Jacobson TB, Pabst M, Stevenson DM, Jetten MSM, Noguera DR, McMahon KD, Amador-Noguez D, Lücker S. Autotrophic and mixotrophic metabolism of an anammox bacterium revealed by in vivo 13C and 2H metabolic network mapping. The ISME Journal, 2021, 15: 673-687. DOI:10.1038/s41396-020-00805-w |
| [37] | Wu D, Li GF, Shi ZJ, Zhang Q, Huang BC, Fan NS, Jin RC. Co-inhibition of salinity and Ni(Ⅱ) in the anammox-UASB reactor. Science of the Total Environment, 2019, 669: 70-82. DOI:10.1016/j.scitotenv.2019.03.130 |
| [38] | Meng Y, Yin C, Zhou Z, Meng F. Increased salinity triggers significant changes in the functional proteins of ANAMMOX bacteria within a biofilm community. Chemosphere, 2018, 207: 655-664. DOI:10.1016/j.chemosphere.2018.05.076 |
| [39] | Yang XR, Weng BS, Li H, Marshall CW, Li H, Chen YS, Yu S, Zhu GB, Zhu YG. An overlooked nitrogen loss linked to anaerobic ammonium oxidation in estuarine sediments in China. Journal of Soils and Sediments, 2017, 17(10): 2537-2546. DOI:10.1007/s11368-017-1728-y |
| [40] | Lu X, Bade DL, Leff LG, Mou XJ. The relative importance of anammox and denitrification to total N2 production in Lake Erie. Journal of Great Lakes Research, 2018, 44(3): 428-435. DOI:10.1016/j.jglr.2018.03.008 |
| [41] | Na T, Thamdrup B, Kim B, Kim SH, Vandieken V, Kang DJ, Hyun JH. N2 production through denitrification and anammox across the continental margin (shelf-slope-rise) of the Ulleung Basin, East Sea. Limnology and Oceanography, 2018, 63(S1): S410-S424. DOI:10.1002/lno.10750 |
| [42] | Jetten Mike SM WM, Fuerst J, van Loosdrecht M, Kuenen GJ, Strous M. Microbiology and application of the anaerobic ammonium oxidation ('anammox') process. Current Opinion in Biotechnology, 2001, 12: 283-288. DOI:10.1016/S0958-1669(00)00211-1 |
| [43] | Oshiki M, Shimokawa M, Fujii N, Satoh H, Okabe S. Physiological characteristics of the anaerobic ammonium-oxidizing bacterium 'Candidatus Brocadia sinica'. Microbiology, 2011, 157(6): 1706-1713. DOI:10.1099/mic.0.048595-0 |
| [44] | Damsté JSS, Strous M, Rijpstra WIC, Hopmans EC, Geenevasen JA, van Duin AC, Van Niftrik LA, Jetten MS. Linearly concatenated cyclobutane lipids form a dense bacterial membrane. Nature, 2002, 419(6908): 708-712. DOI:10.1038/nature01128 |
| [45] | Boumann HA, Hopmans EC, Van De Leemput I, Op den Camp HJ, Van De Vossenberg J, Strous M, Jetten MS, Sinninghe Damsté JS, Schouten S. Ladderane phospholipids in anammox bacteria comprise phosphocholine and phosphoethanolamine headgroups. FEMS Microbiology Letters, 2006, 258(2): 297-304. DOI:10.1111/j.1574-6968.2006.00233.x |
| [46] | Van de Graaf AA, de Bruijn P, Robertson LA, Jetten MS, Kuenen JG. Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiology, 1996, 142(8): 2187-2196. DOI:10.1099/13500872-142-8-2187 |
| [47] | Van De Graaf AA, De Bruijn P, Robertson LA, Jetten MS, Kuenen JG. Metabolic pathway of anaerobic ammonium oxidation on the basis of 15N studies in a fluidized bed reactor. Microbiology, 1997, 143(7): 2415-2421. DOI:10.1099/00221287-143-7-2415 |
| [48] | Schalk J, Oustad H, Kuenen JG, Jetten MS. The anaerobic oxidation of hydrazine: a novel reaction in microbial nitrogen metabolism. FEMS Microbiology Letters, 1998, 158(1): 61-67. DOI:10.1111/j.1574-6968.1998.tb12801.x |
| [49] | Jetten MS, Wagner M, Fuerst J, van Loosdrecht M, Kuenen G, Strous M. Microbiology and application of the anaerobic ammonium oxidation ('anammox') process. Current Opinion in Biotechnology, 2001, 12(3): 283-288. DOI:10.1016/S0958-1669(00)00211-1 |
| [50] | Jetten MS, Niftrik L, Strous M, Kartal B, Keltjens JT, Op den Camp HJ. Biochemistry and molecular biology of anammox bacteria. Critical Reviews in Biochemistry and Molecular Biology, 2009, 44(2/3): 65-84. |
| [51] | Shimamura M, Nishiyama T, Shigetomo H, Toyomoto T, Kawahara Y, Furukawa K, Fujii T. Isolation of a multiheme protein with features of a hydrazine-oxidizing enzyme from an anaerobic ammonium-oxidizing enrichment culture. Applied and Environmental Microbiology, 2007, 73(4): 1065-1072. DOI:10.1128/AEM.01978-06 |
| [52] | Shimamura M, Nishiyama T, Shinya K, Kawahara Y, Furukawa K, Fujii T. Another multiheme protein, hydroxylamine oxidoreductase, abundantly produced in an anammox bacterium besides the hydrazine-oxidizing enzyme. Journal of Bioscience and Bioengineering, 2008, 105(3): 243-248. DOI:10.1263/jbb.105.243 |
| [53] | Figueroa IA, Barnum TP, Somasekhar PY, Carlstr?m CI, Engelbrektson AL, Coates JD. Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proceedings of the National Academy of Sciences of the Unites States of America, 2018, 115(1): E92-E101. DOI:10.1073/pnas.1715549114 |
| [54] | Liu YY, Wang S, Li SZ, Deng Y. Advances in molecular ecology on microbial functional genes of carbon cycle. Microbiology China, 2017, 44(7): 1676-1689. (in Chinese) 刘洋荧, 王尚, 厉舒祯, 邓晔. 基于功能基因的微生物碳循环分子生态学研究进展. 微生物学通报, 2017, 44(7): 1676-1689. |
| [55] | Schouten S, Strous M, Kuypers MM, Rijpstra WI, Baas M, Schubert CJ, Jetten MS, Sinninghe Damsté JS. Stable carbon isotopic fractionations associated with inorganic carbon fixation by anaerobic ammonium-oxidizing bacteria. Applied and Environmental Microbiology, 2004, 70(6): 3785-3788. DOI:10.1128/AEM.70.6.3785-3788.2004 |
| [56] | Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature, 2006, 440(7085): 790-794. DOI:10.1038/nature04647 |
| [57] | Fuerst JA. Intracellular compartmentation in planctomycetes. Annual Review of Microbiology, 2005, 59: 299-328. DOI:10.1146/annurev.micro.59.030804.121258 |
| [58] | Shu D, He Y, Yue H, Yang S. Effects of Fe (Ⅱ) on microbial communities, nitrogen transformation pathways and iron cycling in the anammox process: kinetics, quantitative molecular mechanism and metagenomic analysis. RSC Advances, 2016, 6(72): 68005-68016. DOI:10.1039/C6RA09209H |
| [59] | Chen H, Yu J-J, Jia X-Y, Jin R-C. Enhancement of anammox performance by Cu(Ⅱ), Ni(Ⅱ) and Fe(Ⅲ) supplementation. Chemosphere, 2014, 117: 610-616. DOI:10.1016/j.chemosphere.2014.09.047 |
| [60] | Huang X, Gao D, Peng S, Tao Y. Effects of ferrous and manganese ions on anammox process in sequencing batch biofilm reactors. Journal of Environmental Sciences, 2014, 26(5): 1034-1039. DOI:10.1016/S1001-0742(13)60531-8 |
| [61] | Güven D, Dapena A, Kartal B, Schmid MC, Maas B, van de Pas-Schoonen K, Sozen S, Mendez R, den Camp HJO, Jetten MS. Propionate oxidation by and methanol inhibition of anaerobic ammonium-oxidizing bacteria. Applied and Environmental Microbiology, 2005, 71(2): 1066-1071. DOI:10.1128/AEM.71.2.1066-1071.2005 |
| [62] | Chamchoi N, Nitisoravut S, Schmidt JE. Inactivation of ANAMMOX communities under concurrent operation of anaerobic ammonium oxidation (ANAMMOX) and denitrification. Bioresource Technology, 2008, 99(9): 3331-3336. DOI:10.1016/j.biortech.2007.08.029 |
| [63] | Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, Schreiber F, Dutilh BE, Zedelius J, de Beer D. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature, 2010, 464(7288): 543-548. DOI:10.1038/nature08883 |
| [64] | Van De Vossenberg J, Rattray JE, Geerts W, Kartal B, Van Niftrik L, Van Donselaar EG, Sinninghe Damsté JS, Strous M, Jetten MS. Enrichment and characterization of marine anammox bacteria associated with global nitrogen gas production. Environmental Microbiology, 2008, 10(11): 3120-3129. DOI:10.1111/j.1462-2920.2008.01643.x |
| [65] | Liu S, Yang F, Gong Z, Meng F, Chen H, Xue Y, Furukawa K. Application of anaerobic ammonium-oxidizing consortium to achieve completely autotrophic ammonium and sulfate removal. Bioresource Technology, 2008, 99(15): 6817-6825. DOI:10.1016/j.biortech.2008.01.054 |
| [66] | Thamdrup B, Dalsgaard T. Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Applied and Environmental Microbiology, 2002, 68(3): 1312-1318. DOI:10.1128/AEM.68.3.1312-1318.2002 |
| [67] | Dalsgaard T, Canfield DE, Petersen J, Thamdrup B, Acu?a-González J. N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature, 2003, 422(6932): 606-608. DOI:10.1038/nature01526 |
| [68] | Shao S, Luan X, Dang H, Zhou H, Zhao Y, Liu H, Zhang Y, Dai L, Ye Y, Klotz MG. Deep-sea methane seep sediments in the Okhotsk Sea sustain diverse and abundant anammox bacteria. FEMS Microbiology Ecology, 2014, 87(2): 503-516. DOI:10.1111/1574-6941.12241 |
| [69] | Cao W, Guan Q, Li Y, Wang M, Liu B. The contribution of denitrification and anaerobic ammonium oxidation to N2 production in mangrove sediments in Southeast China. Journal of Soils and Sediments, 2017, 17(6): 1767-1776. DOI:10.1007/s11368-017-1653-0 |
| [70] | Zhang M, Dai P, Lin X, Lin L, Hetharua B, Zhang Y, Tian Y. Nitrogen loss by anaerobic ammonium oxidation in a mangrove wetland of the Zhangjiang Estuary, China. Science of the Total Environment, 2020, 698: 134291. DOI:10.1016/j.scitotenv.2019.134291 |
| [71] | Schubert CJ, Durisch-Kaiser E, Wehrli B, Thamdrup B, Lam P, Kuypers MM. Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika). Environmental Microbiology, 2006, 8(10): 1857-1863. DOI:10.1111/j.1462-2920.2006.01074.x |
| [72] | Crowe SA, Treusch AH, Forth M, Li J, Magen C, Canfield DE, Thamdrup B, Katsev S. Novel anammox bacteria and nitrogen loss from Lake Superior. Scientific Reports, 2017, 7(1): 13757. DOI:10.1038/s41598-017-12270-1 |
| [73] | Zheng Y, Hou L, Liu M, Yin G. Dynamics and environmental importance of anaerobic ammonium oxidation (anammox) bacteria in urban river networks. Environmental Pollution, 2019, 254(Pt A): 112998. |
| [74] | Moore TA, Xing Y, Lazenby B, Lynch MD, Schiff S, Robertson WD, Timlin R, Lanza S, Ryan MC, Aravena R. Prevalence of anaerobic ammonium-oxidizing bacteria in contaminated groundwater. Environmental Science & Technology, 2011, 45(17): 7217-7225. |
| [75] | Smith RL, Bohlke J, Song B, Tobias CR. Role of anaerobic ammonium oxidation (anammox) in nitrogen removal from a freshwater aquifer. Environmental Science & Technology, 2015, 49(20): 12169-12177. |
| [76] | Shen LD, Wu HS, Gao ZQ, Ruan YJ, Xu XH, Li J, Ma SJ, Zheng PH. Evidence for anaerobic ammonium oxidation process in freshwater sediments of aquaculture ponds. Environmental Science and Pollution Research International, 2016, 23(2): 1344-1352. DOI:10.1007/s11356-015-5356-z |
| [77] | Humbert S, Tarnawski S, Fromin N, Mallet M-P, Aragno M, Zopfi J. Molecular detection of anammox bacteria in terrestrial ecosystems: distribution and diversity. The ISME Journal, 2010, 4(3): 450-454. DOI:10.1038/ismej.2009.125 |
| [78] | Hu B-l, Rush D, van der Biezen E, Zheng P, van Mullekom M, Schouten S, Damsté JSS, Smolders AJ, Jetten MS, Kartal B. New anaerobic, ammonium-oxidizing community enriched from peat soil. Applied and Environmental Microbiology, 2011, 77(3): 966-971. DOI:10.1128/AEM.02402-10 |
| [79] | Shen LD, Wu HS, Gao ZQ, Xu XH, Chen TX, Liu S, Cheng HX. Occurrence and importance of anaerobic ammonium-oxidising bacteria in vegetable soils. Applied Microbiology and Biotechnology, 2015, 99(13): 5709-5718. DOI:10.1007/s00253-015-6454-z |
| [80] | Zhou X, Zhang J, Wen C. Community composition and abundance of anammox bacteria in cattail rhizosphere sediments at three phenological stages. Current Microbiology, 2017, 74(11): 1349-1357. DOI:10.1007/s00284-017-1324-9 |
| [81] | Ding B, Zhang H, Luo W, Sun S, Cheng F, Li Z. Nitrogen loss through denitrification, anammox and Feammox in a paddy soil. Science of the Total Environment, 2021, 773: 145601. DOI:10.1016/j.scitotenv.2021.145601 |
| [82] | Jaeschke A, Op den Camp HJ, Harhangi H, Klimiuk A, Hopmans EC, Jetten MS, Schouten S, Sinninghe Damsté JS. 16S rRNA gene and lipid biomarker evidence for anaerobic ammonium-oxidizing bacteria (anammox) in California and Nevada hot springs. FEMS Microbiology Ecology, 2009, 67(3): 343-350. DOI:10.1111/j.1574-6941.2008.00640.x |
| [83] | Byrne N, Strous M, Crépeau V, Kartal B, Birrien J-L, Schmid M, Lesongeur F, Schouten S, Jaeschke A, Jetten M. Presence and activity of anaerobic ammonium-oxidizing bacteria at deep-sea hydrothermal vents. The ISME Journal, 2009, 3(1): 117-123. DOI:10.1038/ismej.2008.72 |
| [84] | Hoffmann F, Radax R, Woebken D, Holtappels M, Lavik G, Rapp HT, Schl?ppy ML, Schleper C, Kuypers MM. Complex nitrogen cycling in the sponge Geodia barretti. Environmental Microbiology, 2009, 11(9): 2228-2243. DOI:10.1111/j.1462-2920.2009.01944.x |
| [85] | Mohamed NM, Saito K, Tal Y, Hill RT. Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. The ISME Journal, 2010, 4(1): 38-48. DOI:10.1038/ismej.2009.84 |
| [86] | Yang Y, Li M, Li H, Li XY, Lin JG, Denecke M, Gu JD. Specific and effective detection of anammox bacteria using PCR primers targeting the 16S rRNA gene and functional genes. Science of the Total Environment, 2020, 734: 139387. DOI:10.1016/j.scitotenv.2020.139387 |
| [87] | Wang S, Hong Y, Wu J, Xu XR, Bin L, Pan Y, Guan F, Wen J. Comparative analysis of two 16S rRNA gene-based PCR primer sets provides insight into the diversity distribution patterns of anammox bacteria in different environments. Applied Microbiology and Biotechnology, 2015, 99(19): 8163-8176. DOI:10.1007/s00253-015-6814-8 |
| [88] | Li M, Gu JD. The diversity and distribution of anammox bacteria in the marine aquaculture zones. Applied Microbiology and Biotechnology, 2016, 100(20): 8943-8953. DOI:10.1007/s00253-016-7690-6 |
| [89] | Wu J, Hong Y, He X, Jiao L, Wen X, Chen S, Chen G, Li Y, Huang T, Hu YJFim. Anaerobic ammonium oxidation in acidic red soils. Frontiers in Microbiology, 2018, 9: 2142. DOI:10.3389/fmicb.2018.02142 |
| [90] | Fu L, Chen Y, Li S, He H, Mi T, Zhen Y, Yu Z. Shifts in the anammox bacterial community structure and abundance in sediments from the Changjiang Estuary and its adjacent area. Systematic and Applied Microbiology, 2019, 42(3): 383-396. DOI:10.1016/j.syapm.2018.12.008 |
| [91] | Yang Y, Dai Y, Li N, Li B, Xie S, Liu Y. Temporal and spatial dynamics of sediment anaerobic ammonium oxidation (Anammox) bacteria in freshwater lakes. Microbial Ecology, 2017, 73(2): 285-295. DOI:10.1007/s00248-016-0872-z |
| [92] | Stultiens K, van Kessel M, Frank J, Fischer P, Pelzer C, van Alen TA, Kartal B, Op den Camp HJM, Jetten MSM. Diversity, enrichment, and genomic potential of anaerobic methane- and ammonium-oxidizing microorganisms from a brewery wastewater treatment plant. Applied Microbiology and Biotechnology, 2020, 104(16): 7201-7212. DOI:10.1007/s00253-020-10748-z |
| [93] | Wu J, Hong Y, He X, Jiao L, Wen X, Chen S, Chen G, Li Y, Huang T, Hu Y, Liu X. Anaerobic ammonium oxidation in acidic red soils. Frontiers Microbiology, 2018, 9: 2142. DOI:10.3389/fmicb.2018.02142 |
| [94] | Moss FR, 3 rd, Shuken SR, Mercer JAM, Cohen CM, Weiss TM, Boxer SG, Burns NZ. Ladderane phospholipids form a densely packed membrane with normal hydrazine and anomalously low proton/hydroxide permeability. Proceedings of the National Academy of Sciences of the Unites States of America, 2018, 115(37): 9098-9103. DOI:10.1073/pnas.1810706115 |
| [95] | Zhao Z, Cao Y, Fan Y, Yang H, Feng X, Li L, Zhang H, Xing L, Zhao M. Ladderane records over the last century in the East China sea: Proxies for anammox and eutrophication changes. Water Research, 2019, 156: 297-304. DOI:10.1016/j.watres.2019.03.046 |
| [96] | Zhao S, Zhuang L, Wang C, Li Y, Wang S, Zhu G. High-throughput analysis of anammox bacteria in wetland and dryland soils along the altitudinal gradient in Qinghai- Tibet Plateau. Microbiologyopen, 2018, 7(2): e00556. DOI:10.1002/mbo3.556 |
| [97] | Guo J, Peng Y, Fan L, Zhang L, Ni BJ, Kartal B, Feng X, Jetten MS, Yuan Z. Metagenomic analysis of anammox communities in three different microbial aggregates. Environmental Microbiology, 2016, 18(9): 2979-2993. DOI:10.1111/1462-2920.13132 |
| [98] | Wu J, Hong Y, Chang X, Jiao L, Li Y, Liu X, Xie H, Gu JD. Unexpectedly high diversity of anammox bacteria detected in deep-sea surface sediments of the South China Sea. FEMS Microbiology Ecology, 2019, 95(3): fiz013. |
| [99] | Liu Y, Niu Q, Wang S, Ji J, Zhang Y, Yang M, Hojo T, Li YY. Upgrading of the symbiosis of Nitrosomanas and anammox bacteria in a novel single-stage partial nitritation-anammox system: Nitrogen removal potential and Microbial characterization. Bioresource Technology, 2017, 244(Pt 1): 463-472. |
| [100] | Toh SK, Webb RI, Ashbolt NJ. Enrichment of autotrophic anaerobic ammonium-oxidizing consortia from various wastewaters. Microbial Ecology, 2002, 43(1): 154-167. DOI:10.1007/s00248-001-0033-9 |
| [101] | Wang Z, Peng Y, Miao L, Cao T, Zhang F, Wang S, Han J. Continuous-flow combined process of nitritation and ANAMMOX for treatment of landfill leachate. Bioresource Technology, 2016, 214: 514-519. DOI:10.1016/j.biortech.2016.04.118 |
| [102] | Zhang L, Zhang S, Peng Y, Han X, Gan Y. Nitrogen removal performance and microbial distribution in pilot- and full-scale integrated fixed-biofilm activated sludge reactors based on nitritation-anammox process. Bioresource Technology, 2015, 196: 448-453. DOI:10.1016/j.biortech.2015.07.090 |
| [103] | Li-dong S, An-hui H, Ren-cun J, Dong-qing C, Ping Z, Xiang-yang X, Bao-lan H. Enrichment of anammox bacteria from three sludge sources for the startup of monosodium glutamate industrial wastewater treatment system. Journal of Hazardous Materials, 2012, 199-200: 193-199. DOI:10.1016/j.jhazmat.2011.10.081 |
| [104] | Tang CJ, Zheng P, Chen TT, Zhang JQ, Mahmood Q, Ding S, Chen XG, Chen JW, Wu DT. Enhanced nitrogen removal from pharmaceutical wastewater using SBA-ANAMMOX process. Water Research, 2011, 45(1): 201-210. DOI:10.1016/j.watres.2010.08.036 |
| [105] | Lackner S, Gilbert EM, Vlaeminck SE, Joss A, Horn H, van Loosdrecht MC. Full-scale partial nitritation/anammox experiences——an application survey. Water Research, 2014, 55: 292-303. DOI:10.1016/j.watres.2014.02.032 |
| [106] | Ni SQ, Zhang J. Anaerobic ammonium oxidation: from laboratory to full-scale application. Biomed Research International, 2013, 2013: 469360. |
| [107] | Wang CC, Lee PH, Kumar M, Huang YT, Sung S, Lin JG. Simultaneous partial nitrification, anaerobic ammonium oxidation and denitrification (SNAD) in a full-scale landfill-leachate treatment plant. Journal of Hazardous Materials, 2010, 175(1-3): 622-628. DOI:10.1016/j.jhazmat.2009.10.052 |
| [108] | Yang ZL, Tian FR, Lin HY, Wang KC, Dong ZB, Wang KY. Test and study on quick start-up of anaerobic ammonium oxidation process. Guangzhou Chemical Industry, 2018, 46(22): 50-52. (in Chinese) 杨志林, 田凤蓉, 林浩宇, 王开春, 董自斌, 王克云. 厌氧氨氧化工艺的快速启动试验研究. 广州化工, 2018, 46(22): 50-52. DOI:10.3969/j.issn.1001-9677.2018.22.022 |
| [109] | Deng K, Tang L, Li J, Meng J, Li J. Practicing anammox in a novel hybrid anaerobic-aerobic baffled reactor for treating high-strength ammonium piggery wastewater with low COD/TN ratio. Bioresource Technology, 2019, 294: 122193. DOI:10.1016/j.biortech.2019.122193 |
| [110] | Li X, Zhang J, Zhang X, Li J, Liu F, Chen Y. Start-up and nitrogen removal performance of CANON and SNAD processes in a pilot-scale oxidation ditch reactor. Process Biochemistry, 2019, 84: 134-142. DOI:10.1016/j.procbio.2019.06.010 |
| [111] | van Dongen U, Jetten MS, van Loosdrecht MC. The SHARON-Anammox process for treatment of ammonium rich wastewater. Water Science & Technology, 2001, 44(1): 153-160. |
| [112] | Sri Shalini S, Joseph K. Combined SHARON and ANAMMOX processes for ammoniacal nitrogen stabilisation in landfill bioreactors. Bioresource Technology, 2018, 250: 723-732. DOI:10.1016/j.biortech.2017.10.077 |
| [113] | Gonzalez-Martinez A, Osorio F, Morillo JA, Rodriguez-Sanchez A, Gonzalez-Lopez J, Abbas BA, van Loosdrecht MC. Comparison of bacterial diversity in full scale anammox bioreactors operated under different conditions. Biotechnology Progress, 2015, 31(6): 1464-1472. DOI:10.1002/btpr.2151 |
| [114] | Zhang FZ, Wang SY, Peng YZ, Miao L, Cao TH, Wang Z. Start-up and characterization of nitrogen and COD removal from mature landfill leachate via CANON process. Journal of Chemical Industry and Engineering: China, 2016, 67(9): 3910-3918. (in Chinese) 张方斋, 王淑莹, 彭永臻, 苗蕾, 曹天昊, 王众. CANON工艺处理实际晚期垃圾渗滤液的启动实验. 2016, 67(9): 3910-3918. |
| [115] | Wett B. Development and implementation of a robust deammonification process. Water Science & Technology, 2007, 56(7): 81-88. |
| [116] | Vlaeminck SE, Terada A, Smets BF, van der Linden D, Boon N, Verstraete W, Carballa M. Nitrogen removal from digested black water by one-stage partial nitritation and anammox. Environmental Science & Technology, 2009, 43(13): 5035-5041. |
| [117] | Yang Y, Zhang L, Han X, Zhang S, Li B, Peng Y. Determine the operational boundary of a pilot-scale single-stage partial nitritation/anammox system with granular sludge. Water Science & Technology, 2016, 73(9): 2085-2092. |
| [118] | Tao Y, Gao DW, Fu Y, Wu WM, Ren NQ. Impact of reactor configuration on anammox process start-up: MBR versus SBR. Bioresource Technology, 2012, 104: 73-80. DOI:10.1016/j.biortech.2011.10.052 |
| [119] | Gilbert EM, Agrawal S, Karst SM, Horn H, Nielsen PH, Lackner S. Low temperature partial nitritation/anammox in a moving bed biofilm reactor treating low strength wastewater. Environmental Science & Technology, 2014, 48(15): 8784-8792. |
| [120] | Zhang X, Zhang H, Ye C, Wei M, Du J. Effect of COD/N ratio on nitrogen removal and microbial communities of CANON process in membrane bioreactors. Bioresource Technology, 2015, 189: 302-308. DOI:10.1016/j.biortech.2015.04.006 |
| [121] | Yao L, Zhang JY, Wang HY, Wang YY, Wei YS. Comparisons of enhancement of low abundance anammox bacteria through different additives. Chinese Journal of Environmental Engineering, 2018, 12(5): 1490-1500. (in Chinese) 姚丽, 张俊亚, 王红艳, 王元月, 魏源送. 不同添加剂强化低丰度厌氧氨氧化菌群的比较. 环境工程学报, 2018, 12(5): 1490-1500. |
| [122] | Wei CY, Zhang J, Ren LL, Peng SC, Lu PL, Zhang DJ. Recent advances in activity inhibition, recovery and enhancement, and application of the anaerobic ammonium oxidation process. Chinese Journal of Applied and Environmental Biology, 2018, 24(3): 671-680. (in Chinese) 魏彩莹, 张静, 任露露, 彭淑婵, 卢培利, 张代钧. 厌氧氨氧化活性抑制、恢复与强化及工艺应用研究进展. 应用与环境生物学报, 2018, 24(3): 671-680. |
| [123] | Lv Gang, Li Tian, Xu Lezhong, Shen Yaoliang, Wu Peng, Zhang Ting, Thomas S. Quick start-up performance of combined ANAMMOX reactor based on different inoculated sludge types. Environmental Science, 2017, 38(10): 4324-4331. (in Chinese) 闾刚, 李田, 徐乐中, 沈耀良, 吴鹏, 张婷, Thomas S. 基于不同接种污泥复合型厌氧氨氧化反应器的快速启动特征. 环境科学, 2017, 38(10): 4324-4331. |
| [124] | Ali M, Oshiki M, Rathnayake L, Ishii S, Satoh H, Okabe S. Rapid and successful start-up of anammox process by immobilizing the minimal quantity of biomass in PVA-SA gel beads. Water Research, 2015, 79: 147-157. DOI:10.1016/j.watres.2015.04.024 |
| [125] | Yao Z, Lu P, Zhang D, Wan X, Li Y, Peng S. Stoichiometry and kinetics of the anaerobic ammonium oxidation (Anammox) with trace hydrazine addition. Bioresource Technology, 2015, 198: 70-76. DOI:10.1016/j.biortech.2015.08.098 |
| [126] | Wang YY, Li L, Ma X, Lin XM, Pan ML, Dai XH. Bio-characteristics of anammox bacteria and CANON anammox process. Acta Scientiae Circumstantiae, 2014, 34(6): 1362-1374. (in Chinese) 王亚宜, 黎力, 马骁, 林喜茂, 潘绵立, 戴晓虎. 厌氧氨氧化菌的生物特性及CANON厌氧氨氧化工艺. 环境科学学报, 2014, 34(6): 1362-1374. |
| [127] | Chen YL, Sui QW, Jiang LA, Yao L, Chen MX, Wei YS. Quick enrichment of ANAMMOX bacteria and microbial community mechanism analysis. China Water & Wastewater, 2018, 34(13): 26-31. (in Chinese) 陈彦霖, 隋倩雯, 姜黎安, 姚丽, 陈梅雪, 魏源送. 厌氧氨氧化菌快速富集培养及微生物机制解析. 中国给水排水, 2018, 34(13): 26-31. |
| [128] | Magrí A, Béline F, Dabert P. Feasibility and interest of the anammox process as treatment alternative for anaerobic digester supernatants in manure processing——an overview. Journal of Environmental Management, 2013, 131: 170-184. |
| [129] | Jin RC, Yang GF, Zhang QQ, Ma C, Yu JJ, Xing BS. The effect of sulfide inhibition on the ANAMMOX process. Water Research, 2013, 47(3): 1459-1469. DOI:10.1016/j.watres.2012.12.018 |
| [130] | Anjali G, Sabumon PC. Unprecedented development of anammox in presence of organic carbon using seed biomass from a tannery Common Effluent Treatment Plant (CETP). Bioresource Technology, 2014, 153: 30-38. DOI:10.1016/j.biortech.2013.11.061 |
| [131] | Persson F, Sultana R, Suarez M, Hermansson M, Plaza E, Wilén BM. Structure and composition of biofilm communities in a moving bed biofilm reactor for nitritation-anammox at low temperatures. Bioresource Technology, 2014, 154: 267-273. DOI:10.1016/j.biortech.2013.12.062 |
