王佳华1,2, 弋伊1,2, 徐俊1,2


1. 上海交通大学微生物代谢国家重点实验室, 上海 200240;
2. 上海交通大学生命科学技术学院, 上海 200240
收稿日期:2020-05-19;修回日期:2020-07-26;网络出版日期:2020-12-11
基金项目:国家重点基础研究发展计划(2014CB441503)
*通信作者:徐俊, Tel: +86-21-34207208; E-mail: xujunn@sjtu.edu.cn.
摘要:康氏菌科(Kangillaceae)是海洋螺杆菌目(Oceanosprillales)下的科级分类单元,包含康氏菌属(Kangiella)、异康氏菌属(Aliikangiella)和Pleionea三个属。在康氏菌属高度精简的基因组中存在异常丰富的胞外蛋白酶编码基因,暗示此类海洋细菌对海洋颗粒有机物中的蛋白质成分降解具有重要贡献。鉴于已分离鉴定的康氏菌属模式菌株专一性利用蛋白类营养物质,我们建议将该属译为“食朊菌属”。本文将结合本实验室的研究进展,对康氏菌科细菌的分离培养过程、系统发育关系、代谢特点和潜在生态功能进行综述,为研究此类海洋细菌的生理生态功能提供思路。
关键词:海洋螺杆菌目康氏菌属胞外蛋白酶颗粒有机物降解海洋细菌
Phylogeny and metabolic potential of marine bacteria in the family Kangillaceae
Jiahua Wang1,2, Yi Yi1,2, Jun Xu1,2


1. State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University, Shanghai 200240, China;
2. School of Life Sciences and Technology, Shanghai Jiao Tong University, Shanghai 200240, China
Received: 19 May 2020; Revised: 26 July 2020; Published online: 11 December 2020
*Corresponding author: Jun Xu, Tel: +86-21-34207208; E-mail: xujunn@sjtu.edu.cn.
Foundation item: Supported by the National Basic Research Program of China (2014CB441503)
Abstract: Kangillaceae is a family under the order Oceanosprillales that contains the genera Kangiella, Aliikangiella and Pleionea. The presence of abundant genes encoding extracellular proteases in the highly streamlined genome of the genus Kangiella suggests that it contributes significantly to the degradation of protein components in marine particulate organic matter. Considering that the type strains of Kangilla exclusively utilize protein as nutrient sources, we suggest to translate the genus as "Shi Ruan Jun" (protein degrading bacteria) in Chinese. In this paper, we review the research progress to identify novel species and phylogeny of Kangielleace. Based on the reconstituted metabolic networks of Kangiella, we reveal the potential ecological functions of this group of marine bacteria.
Keywords: OceanosprillalesKangiellaextracellular proteaseparticulate organic mattermarine bacteria
海洋螺杆菌目(Oceanospirillales)是基于16S rRNA基因系统发育关系建立的γ-变形菌纲中的重要分支,此类有机异养型细菌普遍表现出嗜盐或耐盐的特性,在海洋环境中广泛分布。目前,该目中已获得纯培养的科有11个:海洋螺菌科(Oceanospirillaceae)、食烷菌科(Alcanivoracaceae)、盐单胞菌科(Halomonadaceae)、河氏菌科(Hahellaceae)、嗜油菌科(Oleiphilaceae)、糖螺菌科(Saccharospirillaceae)、康氏菌科(Kangiellaceae)、Endozoicomonadaceae科、岸生菌科(Litoricolaceae)、Aestuariirhabdaceae科和Balneatrichaceae科,以及新增的Natronospirillaceae科[1]。
作为其中未知度较高的科级分类单元,康氏菌科包含了康氏菌属、异康氏菌属和Pleionea三属。当前已分离获得的15个模式菌株均为不产生孢子的革兰氏阴性杆菌,已测序的9个模式菌株的基因组大小范围为2.50-5.34 Mb,其中康氏菌属下的Kangiella geojedonensis YCS-5是海洋螺杆菌目中基因组最小的菌株。尽管多个康氏菌科细菌的基因组序列和基本培养特征已被报道,但其生理生态功能尚未被深入研究。本文综述了康氏菌科细菌的分离鉴定、系统发育关系和比较基因组学研究等方面的进展,以期为揭示此类海洋细菌的代谢潜能及其生态功能提供依据。
1 康氏菌科细菌的分离培养与系统发育分析 1.1 康氏菌科细菌的分离培养 2004年,康氏菌属K. koreensis SW-125和K. aquimarina SW-154由Yoon等首次报道分离自韩国黄海潮间带[2]。自2010年起,研究人员先后分离获得多个康氏菌属菌株,包括来源于日本海的3株K. japonica (KMM 3896、KMM 3897和KMM 3899)[3]、来源于美国佛罗里达群岛鸡肝海棉(Chondrilla nucula)的K. spongicola A79[4]、来源于韩国南岸水体的K. geojedonensis YCS-5[5]、来源于台湾北部浅滩区的K. taiwanensis KT1和K. marina KM1[6]、来源于韩国南海潮间带的K. sediminilitoris BB-Mw22[7]、来源于韩国济州岛的K. chungangensis CAU 1040[8]以及来源于西南印度洋2000 m沉积物的K. profundi FT102[9]。
2013年,Pleionea属首个纯培养菌株P. mediterranea MOLA115被分离自地中海巴纽尔斯海湾沿岸水体样品[10]。2016年,Wang等在海洋微藻培养基中分离获得首株异康氏菌属细菌A. marina GYP-15[11]。2019年底,Luo等报道分离另一Pleionea属菌株P. sediminis S1-5-21[12]。康氏菌科一般形态为非运动型、不产孢子的棒状细菌,各模式菌株的基本信息如表 1所示。
表 1. 康氏菌科现有的模式菌株的分离培养特征和基因组大小 Table 1. The main characteristics of the Kangillaceae type strains
| Species | Strain | Sampling site | Growth temperature/℃ | Genome size/Mb | GC content/% | Phenotype* |
| Kangiella koreensis | SW-125 | Tidal flat sediment of the Yellow Sea in Korea | 4-43 | 2.85 | 44.0 | Facultatively anaerobic |
| Kangiella aquimarina | SW-154 | Tidal flat sediment of the Yellow Sea in Korea | 10-48 | 2.68 | 44.0 | Facultatively anaerobic |
| Kangiella japonica | KMM-3896 KMM-3897 KMM-3899 | Coastal seawater and sediment of the Sea of Japan | 4-42 | - | 43.8- 45.8 | Facultatively anaerobic |
| Kangiella spongicola | A79 | Sponge Chondrilla nucula | 10-49 | 2.50 | 44.9 | Aerobic |
| Kangiella geojedonensis | YCS-5 | Seawater of the southern coast of Korea | 10-40 | 2.50 | 47.0 | Aerobic |
| Kangiella taiwanensis | KT1 | Shallow coastal regions of northern Taiwan | 15-40 | - | 43.9 | Facultatively anaerobic |
| Kangiella marina | KM1 | Shallow coastal regions of northern Taiwan | 15-40 | - | 46.3 | Facultatively anaerobic |
| Kangiella sediminilitoris | BB-Mw22 | Tidal flat sediment of the South Sea in South Korea | 10-40 | 2.50 | 48.9 | Facultatively anaerobic |
| Kangiella chungangensis | CAU 1040 | Marine sand from Jeju Island in South Korea | 20-40 | - | 45.3 | Aerobic |
| Kangiella profundi | FT102 | Deep-sea sediment in the south-west Indian Ocean | 10-50 | 2.65 | 45.0 | Facultatively anaerobic |
| Pleionea mediterranea | MOLA115 | Coastal water from the Mediterranean Sea | 15-37 | 4.04 | 44.5 | Aerobic |
| Pleionea sediminis | S1-5-21 | Coastal sediment from Quanzhou Bay in China | 15-40 | 5.19 | 40.1 | Facultatively anaerobic |
| Aliikangiella marina | GYP-15 | Culture broth of a marine microalga, Picochloruma sp. 122 | 15-37 | 5.34 | 44.7 | Aerobic |
| *: non-motile, non-spore-forming, rod-shaped. | ||||||
表选项
1.2 康氏菌科细菌的系统发育地位 最初,由于康氏菌属16S rRNA基因序列与当时数据库中食烷菌属(Alcanivorax)最为接近,故Yoon等将其划归入食烷菌科[2]。此后,Fagervold根据16S rRNA基因系统发育树建议将康氏菌属、Marinicella、Arenicella和Pleionea合并为一科[10],但该建议未被广泛采纳。直到2016年,Wang等将康氏菌属、异康氏菌属和Pleionea属最终划分在一个新科——康氏菌科中[11]。
除上述可培养菌株外,我们基于POGO-DB数据库[13]使用73个单拷贝持家基因编码的氨基酸序列对NCBI GenBank数据库中未分类的γ-变形菌纲基因组进行系统发育分析,发现其中3个来自于宏基因组的拼接基因组的分类位置属于康氏菌科。其中,GCA002746155.1为进化树长枝,或为康氏菌科新属;GCA002733465.1和GCA002401655.1可能为与异康氏菌属同源关系较近的新属(图 1)。
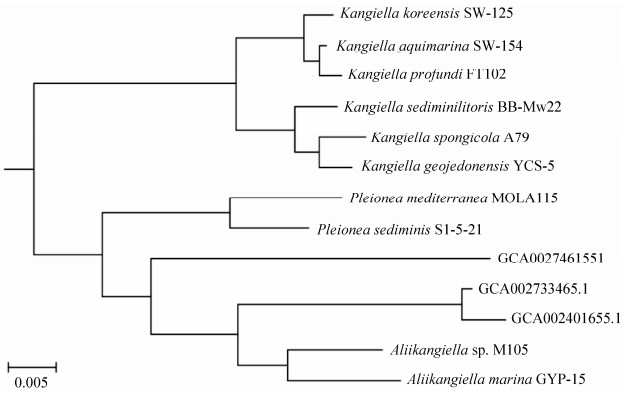 |
| 图 1 康氏菌科的系统发育树重建 Figure 1 The reconstructed phylogenetic tree of Kangillaceae. In this phylogenetic tree, the bootstraps of all nodes are 100; "*" means metagenome-derived genomes. Multiple sequences were aligned using Clustal Omega[14] and representative consensus Maximum-Likelihood tree were constructed using FastTree[15]. |
| 图选项 |
2 康氏菌科细菌的代谢潜能 2.1 康氏菌属细菌的代谢潜能 已分离培养的多个康氏菌属模式菌株皆无法使用单一性碳源(包括糖类、氨基酸、烷烃和羧酸等)实现培养[2, 4, 9]。我们的基因组学研究结果显示所有已知康氏菌属细菌的基因组中均缺乏糖类转运相关基因[16],且碳水化合物代谢相关基因(COG G)数目较海洋螺杆菌目其余微生物明显减少,甚至低于专职性烷烃降解模式菌株Alcanivorax borkumensis SK2[17](图 2),以上结果暗示糖类不是康氏菌属可利用的碳源。有论文推测盐沼中的康氏菌可能参与木质素降解[18],但该研究所用16S rRNA基因扩增子与康氏菌属16S rRNA基因序列相似性最高仅为91%。目前已公布康氏菌属基因组皆缺乏木质素降解相关基因,我们认为康氏菌属不可能参与木质素的降解过程。此外,康氏菌属的基因组天然精简,其中未发现潜在的次级代谢生物合成途径。
 |
| 图 2 海洋螺杆菌目细菌基因组中碳水化合物转运和代谢基因(COG G)数目统计 Figure 2 The distribution of carbohydrate transport and metabolism (COG category G) in the genomes of Oceanosprillales. |
| 图选项 |
而在氮源利用方面,绝大多数康氏菌属细菌缺乏氨转运体、硝酸盐还原酶基因,以及除氨基酸和短肽外的有机氮源转运基因[16],这提示蛋白氮或许是康氏菌属的主要氮源。比较基因组分析显示,康氏菌属细菌基因组中胞外蛋白酶数目、种类和比例均显著高于其他海洋螺杆菌目细菌[16]。基因组精简是康氏菌属细菌的典型特征,譬如在K. profundi FT102等5个康氏菌属细菌的核心基因组中仅1%的基因家族存在多拷贝。而这些多拷贝基因家族中胞外蛋白酶基因占比高达21%,这暗示降解胞外蛋白是康氏菌属细菌的重要生理功能[16]。实验证明,康氏菌属细菌能以混合氨基酸或蛋白质为唯一碳源生长,且其生物量与培养基中蛋白质含量、培养物中胞外蛋白酶粗酶活均呈正相关[16, 19]。因此,康氏菌属细菌很可能是一类基因组高度精简的专职性胞外蛋白降解细菌。
在氨基酸代谢方面,Neshich等在比较1748个细菌基因组中的赖氨酸代谢途径时发现K. koreensis极罕见地同时具有酵母氨酸(Saccharopine)和赖氨酸脱氢酶两条赖氨酸降解通路[20]。Augustus等在比较206个γ-变形菌纲微生物基因组的甲硫氨酸代谢相关通路时发现,K. koreensis的甲硫氨酸代谢基因簇结构十分罕见[21]。我们发现多个康氏菌属细菌基因组中都存在多种氨基酸合成途径的缺失,而海洋螺杆菌目其他科属细菌的基因组中均具有全部20种氨基酸的合成途径[16]。因此,可以推测康氏菌属细菌专职性胞外蛋白降解的营养特点或许部分依赖于氨基酸降解能力的增强,并导致了部分氨基酸合成能力的退化。
2.2 康氏菌科其他属细菌的代谢潜能 与康氏菌属明显不同,已分离的Pleionea属和异康氏菌菌株均能以糖类为唯一碳源生长[10-12]。P. mediterranea MOLA115基因组序列(Accession: GCF_003148745.1)中具有胞外蛋白酶和多种糖类(包括葡萄糖、果糖、麦芽糖、甘油蔗糖、葡萄糖酸和淀粉等)、氨基酸和寡肽的转运与代谢基因(图 3)。
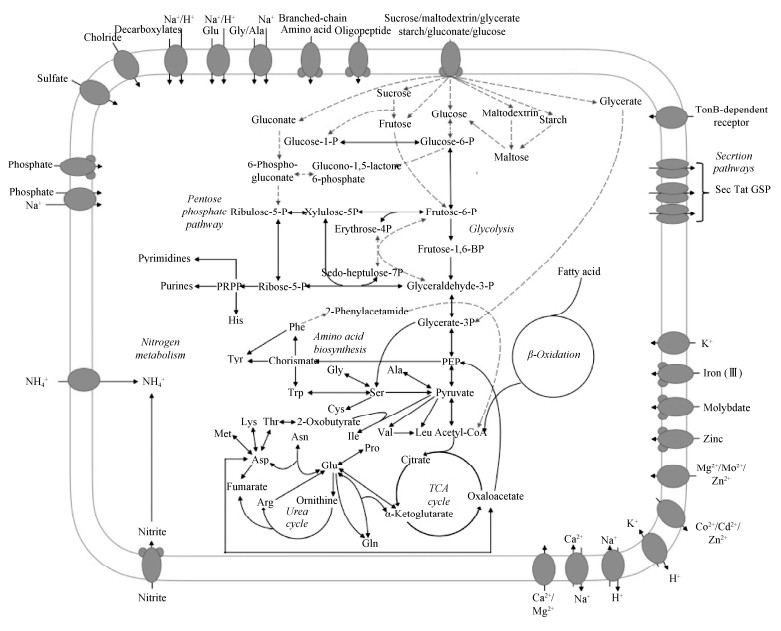 |
| 图 3 P. mediterranea MOLA115的代谢网络重建(未发表数据) Figure 3 The reconstructed metabolic pathways of P. mediterranea MOLA115 (unpublished data). The metabolic pathways in dash lines are not exist in any Kangilla genomes but are predicted to exist in Pleionea. |
| 图选项 |
与Pleionea属类似,康氏菌科其他非康氏属菌属(包括宏基因组bin)也具有糖类转运及胞外蛋白酶基因。其中,Aliikangiella marina GYP-15具有更多样的糖类(如岩藻糖和糊精)降解潜能、磷脂利用能力。总而言之,康氏菌科中的非康氏菌属细菌一般为兼性蛋白降解微生物。
3 康氏菌属微生物的环境分布与生态功能 利用16S rRNA基因扩增子测序发现,在深海的胡安·德富卡海岭熔岩流顶部的橘黄色絮状物质中康氏菌属细菌的丰度在γ-变形菌纲中位列第二(占比高达8.7%)[22],在人工饲养的南美白对虾(Litopenaeus vannamei)肠道中亦可发现康氏菌的存在[23]。Shi等发现康氏菌属N5菌株能够粘附并裂解水华过程中出现的肋骨条藻(Skeletonema costatum)[24]。我们对NCBI nr数据库中与康氏菌属16S rRNA基因序列高度相似的样本的采集地信息进行分析发现,在南极土壤、黄河三角洲土壤、马里亚纳海沟、高盐度海底微生物垫,甚至中国河西走廊盐碱地等环境中均有康氏菌属细菌存在,该结果显示康氏菌属细菌可以在海洋和陆地的高盐环境中大量分布(未发布结果)。
康氏属细菌在海洋生态系统中的作用迄今仅有两例相关的研究报告。Pelve等利用海洋沉积物捕获器和单细胞测序技术首次发现康氏菌属细菌存在于110 m水深的海洋颗粒有机物(particulate organic matter,POM)中[25]。随后,Boeuf等利用相同装置及宏转录组分析发现该属微生物在4000 m深海的POM中有活跃的基因表达[26]。我们依据康氏菌属细菌的胞外蛋白酶中含有胶原降解相关的PKD和PPC结构域[27-28],推测该属细菌可能参与海洋POM中难溶性蛋白的降解过程。
4 讨论和展望 康氏菌属细菌是一类在海洋环境中广泛分布的细菌,可能对海洋中蛋白类有机物作为生源要素的循环起到重要作用。虽然康氏菌属的多个模式种都是韩国科学家建立的,但我国的科研人员在确定此类海洋细菌的分类地位和系统发育关系,以及通过基因组学研究分析其代谢潜能和推断其生态功能方面则处于领先地位。与假单胞菌等许多已报道的兼性蛋白降解微生物相比,康氏菌具有高度的蛋白利用专一性,故而我们建议将该属译为“食朊菌属”。
我们发现康氏菌属细菌核心基因组中的绝大多数胞外蛋白酶基因在康氏菌科其余属中都存在直系同源基因,而其中仅有3个核心蛋白酶家族(M16B、S41A和M48C)是康氏菌属特有的(与非康氏菌属蛋白相似性低于50%)。这暗示绝大多数核心蛋白酶家族基因的获得时间早于康氏菌属的形成时间,同时也提示康氏菌属的分化及特化为专性蛋白营养型的过程可能存在其他机制。就胞外蛋白酶数目而言,在基因组高度精简背景下,康氏菌属部分胞外蛋白酶基因拷贝数的显著增加可能是康氏菌属专职化蛋白营养的重要基础。就功能而言,蛋白酶基因的局部重组也可能对其营养方式有所贡献。例如,康氏菌科中3个属的细菌基因组中均含有一个序列高度相似的、不含PKD结构域的S8A类胞外蛋白酶基因家族,但该家族仅在康氏菌属中连续扩增,且其中有个别成员5?端额外获得一个PKD结构域[16],并可能由此获得难溶性蛋白的降解功能。
此外,生理实验证明K. aquimarina和K. profundi均能利用硝酸盐进行厌氧生长[2, 16],但基因组分析未发现二者携带目前已知的硝酸盐还原酶基因序列,提示其中可能存在硝酸盐还原的新机制。近3年来,美国Delong研究小组利用沉积物捕获器、单细胞测序和宏转录组等技术手段开展的海洋原位观测实验证实了海洋康氏菌科细菌在海洋颗粒有机物中富集和活跃存在的现象[25-26]。因此,研究包括“食朊菌属”在内的康氏菌科(Kangiellaceae)细菌的生态作用对解释海洋氮不平衡问题具有积极的意义。
References
| [1] | Kevbrin V, Boltyanskaya Y, Grouzdev D, Koziaeva V, Park M, Cho JC. Natronospirillum operosum gen. nov., sp. nov., a haloalkaliphilic satellite isolated from decaying biomass of a laboratory culture of cyanobacterium Geitlerinema sp. and proposal of Natronospirillaceae fam. nov., Saccharospirillaceae fam. nov. and Gynuellaceae fam. nov.. International Journal of Systematic and Evolutionary Microbiology, 2020, 70(1): 511-521. DOI:10.1099/ijsem.0.003781 |
| [2] | Yoon JH, Oh TK, Park YH. Kangiella koreensis gen. nov., sp. nov. and Kangiella aquimarina sp. nov., isolated from a tidal flat of the Yellow Sea in Korea. International Journal of Systematic and Evolutionary Microbiology, 2004, 54(5): 1829-1835. DOI:10.1099/ijs.0.63156-0 |
| [3] | Romanenko LA, Tanaka N, Frolova GM, Mikhailov VV. Kangiella japonica sp. nov., isolated from a marine environment. International Journal of Systematic and Evolutionary Microbiology, 2010, 60(11): 2583-2586. DOI:10.1099/ijs.0.017087-0 |
| [4] | Ahn J, Park JW, McConnell JA, Ahn YB, Haggblom MM. Kangiella spongicola sp. nov., a halophilic marine bacterium isolated from the sponge Chondrilla nucula. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(4): 961-964. DOI:10.1099/ijs.0.021733-0 |
| [5] | Yoon JH, Kang SJ, Lee SY, Lee JS, Oh TK. Kangiella geojedonensis sp. nov., isolated from seawater. International Journal of Systematic and Evolutionary Microbiology, 2012, 62(3): 511-514. |
| [6] | Jean WD, Huang SP, Chen JS, Shieh WY. Kangiella taiwanensis sp. nov. and Kangiella marina sp. nov., marine bacteria isolated from shallow coastal water. International Journal of Systematic and Evolutionary Microbiology, 2012, 62(9): 2229-2234. |
| [7] | Lee SY, Park S, Oh TK, Yoon JH. Kangiella sediminilitoris sp. nov., isolated from a tidal flat sediment. International Journal of Systematic and Evolutionary Microbiology, 2013, 63(3): 1001-1006. |
| [8] | Kim JH, Ward AC, Kim W. Kangiella chungangensis sp. nov. isolated from a marine sand. Antonie Van Leeuwenhoek, 2015, 107(5): 1291-1298. DOI:10.1007/s10482-015-0423-5 |
| [9] | Xu FD, Li XG, Xiao X, Xu J. Kangiella profundi sp. nov., isolated from deep-sea sediment. International Journal of Systematic and Evolutionary Microbiology, 2015, 65(7): 2315-2319. |
| [10] | Fagervold SK, Urios L, Intertaglia L, Batailler N, Lebaron P, Suzuki MT. Pleionea mediterranea gen. nov., sp. nov., a gammaproteobacterium isolated from coastal seawater. International Journal of Systematic and Evolutionary Microbiology, 2013, 63(7): 2700-2705. |
| [11] | Wang GH, Tang MX, Wu HL, Dai SK, Li T, Chen CH, He H, Fan JW, Xiang WZ, Li X. Aliikangiella marina gen. nov., sp. nov., a marine bacterium from the culture broth of Picochlorum sp. 122, and proposal of Kangiellaceae fam. nov. in the order Oceanospirillales. International Journal of Systematic and Evolutionary Microbiology, 2015, 65(12): 4488-4494. |
| [12] | Luo YR, Lai QL, Yuan JJ, Huang ZB. Pleionea sediminis sp. nov., isolated from coastal sediment and emendation of the description of the genus Pleionea. International Journal of Systematic and Evolutionary Microbiology, 2019, 69(11): 3524-3528. DOI:10.1099/ijsem.0.003655 |
| [13] | Lan YM, Morrison JC, Hershberg R, Rosen GL. POGO-DB-a database of pairwise-comparisons of genomes and conserved orthologous genes. Nucleic Acids Research, 2014, 42(D1): D625-D632. DOI:10.1093/nar/gkt1094 |
| [14] | Sievers F, Higgins DG. Clustal omega, accurate alignment of very large numbers of sequences. Methods in Molecular Biology, 2014, 1079: 105-116. |
| [15] | Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One, 2010, 5(3): e9490. DOI:10.1371/journal.pone.0009490 |
| [16] | Wang JH, Lu Y, Nawaz MZ, Xu J. Comparative genomics reveals evidence of genome reduction and high extracellular protein degradation potential in Kangiella. Frontiers in Microbiology, 2018, 9: 1224. DOI:10.3389/fmicb.2018.01224 |
| [17] | Yakimov MM, Golyshin PN, Lang S, Moore ER, Abraham WR, Lünsdorf H, Timmis KN. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. International Journal of Systematic Bacteriology, 1998, 48(2): 339-348. DOI:10.1099/00207713-48-2-339 |
| [18] | Darjany LE, Whitcraft CR, Dillon JG. Lignocellulose-responsive bacteria in a southern california salt marsh identified by stable isotope probing. Frontiers in Microbiology, 2014, 5: 263. |
| [19] | Xu L, Wang JH, Lu Y, Xu J. Effect of major metal ions in sea water on growth and transcription of protease coding gene of 4Kangiella strains. Microbiology China, 2018, 45(4): 731-743. (in Chinese) 徐蕾, 王佳华, 卢烨, 徐俊. 海水中主要金属盐对4株Kangiella属细菌的生长及其蛋白酶相关基因表达的影响. 微生物学通报, 2018, 45(4): 731-743. |
| [20] | Neshich IA, Kiyota E, Arruda P. Genome-wide analysis of lysine catabolism in bacteria reveals new connections with osmotic stress resistance. The ISME Journal, 2013, 7(12): 2400-2410. DOI:10.1038/ismej.2013.123 |
| [21] | Augustus AM, Spicer LD. The MetJ regulon in gammaproteobacteria determined by comparative genomics methods. BMC Genomics, 2011, 12: 558. DOI:10.1186/1471-2164-12-558 |
| [22] | Meyer JL, Akerman NH, Proskurowski G, Huber JA. Microbiological characterization of post-eruption "snowblower" vents at Axial Seamount, Juan de Fuca Ridge. Frontiers in Microbiology, 2013, 4: 153. |
| [23] | Amoah K, Huang QC, Tan BP, Zhang S, Chi SY, Yang QH, Liu HY, Dong XH. Dietary supplementation of probiotic Bacillus coagulans ATCC 7050, improves the growth performance, intestinal morphology, microflora, immune response, and disease confrontation of pacific white shrimp, Litopenaeus vannamei. Fish & Shellfish Immunology, 2019, 87: 796-808. |
| [24] | Shi RJ, Huang HH, Qi ZH, Hu WA, Tian ZY, Dai M. Algicidal activity against Skeletonema costatum by marine bacteria isolated from a high frequency harmful algal blooms area in southern Chinese coast. World Journal of Microbiology and Biotechnology, 2013, 29(1): 153-162. DOI:10.1007/s11274-012-1168-1 |
| [25] | Pelve EA, Fontanez KM, DeLong EF. Bacterial succession on sinking particles in the ocean's interior. Frontiers in Microbiology, 2017, 8: 2269. DOI:10.3389/fmicb.2017.02269 |
| [26] | Boeuf D, Edwards BR, Eppley JM, Hu SK, Poff KE, Romano AE, Caron DA, Karl DM, DeLong EF. Biological composition and microbial dynamics of sinking particulate organic matter at abyssal depths in the oligotrophic open ocean. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(24): 11824-11832. |
| [27] | Wang YK, Zhao GY, Li Y, Chen XL, Xie BB, Su HN, Lv YH, He HL, Liu H, Hu J, Zhou BC, Zhang YZ. Mechanistic insight into the function of the C-terminal PKD domain of the collagenolytic serine protease deseasin MCP-01 from deep sea Pseudoalteromonas sp. SM9913:binding of the PKD domain to collagen results in collagen swelling but does not unwind the collagen triple helix. Journal of Biological and Chemical Sciences, 2010, 285(19): 14285-14291. |
| [28] | Huang JF, Wu RB, Liu D, Liao BQ, Lei M, Wang M, Huan R, Zhou MY, Ma CB, He HL. Mechanistic insight into the binding and swelling functions of prepeptidase C-Terminal (PPC) domains from various bacterial proteases. Applied and Environmental Microbiology, 2019, 85(14): e00611-19. |
