任世恩1, 南小宁1

 , 许淼1, 邹垚1, 梁南楠1, 石建宁2, 韩崇选1
, 许淼1, 邹垚1, 梁南楠1, 石建宁2, 韩崇选1

1. 西北农林科技大学西部森林生物灾害治理国家林业和草原局重点实验室, 陕西 杨凌 712100;
2. 宁夏森林病虫害防治检疫总站, 宁夏 银川 750001
收稿日期:2019-07-29;修回日期:2019-09-05;网络出版日期:2019-09-11
基金项目:林木重大生物灾害扩散流行的生态适应与分子基础(2017YFD0600100)
*通信作者:南小宁. E-mail:nxn@nwsuaf.edu.cn;
韩崇选. E-mail:sendakingcat@nwsuaf.edu.cn.
摘要:[目的] 肠道微生物在宿主代谢和健康方面发挥着重要的作用,已有报道表明甘肃鼢鼠肠道菌群和地域、性别以及季节等因素有关,但对于野生和人工饲喂条件下,取食不同食物的甘肃鼢鼠肠道菌群是否存在差异尚不明确。[方法] 本研究选取苜蓿、樟子松根、油松根和云杉根对甘肃鼢鼠进行分组饲喂,同时以野生个体作为对照,采用16S rRNA V3-V4高通量测序技术对野生和人工饲喂甘肃鼢鼠肠道细菌进行比较。[结果] 野生和人工饲喂条件下,优势菌门均为Firmicutes和Bacteroidetes,但含量却存在显著性差异。在人工饲喂条件下,樟子松根、油松根和云杉根饲喂的三组甘肃鼢鼠肠道细菌群落结构相似度较高,而与苜蓿组和野生组之间均存在较大差异,樟子松组、油松组和云杉组甘肃鼢鼠肠道细菌多样性高于苜蓿组和野生组。[结论] 野生和人工饲喂条件下甘肃鼢鼠肠道菌群组成和结构存在显著性差异。
关键词:甘肃鼢鼠肠道细菌高通量测序16S rRNA
Comparison of intestinal bacterial diversity of Gansu zokor under wild and artificial feeding conditions
Shien Ren1, Xiaoning Nan1

 , Miao Xu1, Yao Zou1, Nannan Liang1, Jianning Shi2, Chongxuan Han1
, Miao Xu1, Yao Zou1, Nannan Liang1, Jianning Shi2, Chongxuan Han1

1. Key Laboratory of National Forestry and Grassland Administration on Management of Western Forest Bio-Disaster, Northwest A&F University, Yangling 712100, Shaanxi Province, China;
2. Ningxia Hui Autonomous Region Forest Disease and Pest Control Quarantine Station, Yinchuan 750001, Ningxia Hui Autonomous Region, China
Received: 29 July 2019; Revised: 5 September 2019; Published online: 11 September 2019
*Corresponding author: Nan Xiaoning, E-mail:nxn@nwsuaf.edu.cn;
Han Chongxuan, E-mail:sendakingcat@nwsuaf.edu.cn.
Foundation item: Supported by the Ecological Adaptation and Molecular Basis of the Spread of Major Biological Disasters in Forest (2017YFD0600100)
Abstract: [Objective] Intestinal bacteria play an important role in host metabolism and health. It has been reported that the intestinal flora of Gansu zokor is related to regional, gender and seasonal factors, however, it is not clear whether there is a difference in the intestinal bacterial of Gansu zokor feeding with different feeds under wild and artificial feeding conditions. [Methods] In this study, we selected alfalfa, the roots of Mongolian, Chinese pine and spruce for feeding groups, wild Gansu zokors captured in the same habitat as controls. Bacterial diversity of digestive tract contents in Gansu zokor was analyzed by 16S rRNA V3-V4 high-throughput sequencing technology. [Results] Under the conditions of wild and artificial feeding, the dominant bacteria were both Firmicutes and Bacteroidetes, but the content was significantly different. Under artificial feeding conditions, the intestinal bacterial community structure of Mongolian pine, Chinese pine and spruce was similar, they all had significant difference with alfalfa and wild groups. The intestinal bacterial diversity of Mongolian pine, Chinese pine, spruce were higher than alfalfa and wild groups. [Conclusion] The results showed that there were significant differences in the composition and structure of intestinal bacterial of Gansu zokor under wild and artificial feeding conditions.
Keywords: Gansu zokor (Myospalax cansus)intestinal bacteriahigh-throughput sequencing16S rRNA
动物肠道微生物组已被誉为动物的“第二套基因组”,其数量庞大,事实上,动物就生活在一个充满微生物的世界里[1]。宿主和微生物之间的相互作用对宿主来说具有重要意义,其中肠道微生物又在宿主代谢和健康方面发挥至关重要的作用[2-4],因此肠道微生物的组成、结构及其影响因素是人们重点关注的内容。研究表明,肠道微生物群落结构受饮食、遗传和地理等因素影响[5-9],并且已经得到广泛认可的是宿主饮食在肠道中产生强烈的选择压力,其形成肠道微生物的群落结构[10-12]。
甘肃鼢鼠主要分布在中国西北地区,是一种典型的地下啮齿动物[13],其终生生活在封闭的洞道环境中[14],以草本和木本植物的根系为食。就甘肃鼢鼠的肠道细菌而言,以往的研究主要评估了甘肃鼢鼠肠道细菌群落与地理、性别和季节因素之间的关系[15],而忽略了食物因素对肠道菌群的影响。其次,在宁夏六盘山地区退耕还林工程建设中,樟子松、油松、云杉均为主要造林树种,但由于甘肃鼢鼠取食危害造林树种的根部,导致新造林苗木成活率较低,之前的研究表明甘肃鼢鼠对樟子松、油松、云杉三年累计危害率分别为43.5%、55.2%、10.1%[16]。因此,本研究选择甘肃鼢鼠危害率不同的造林树种对其进行饲喂,以同域分布的野生个体作为对照,采用高通量测序技术对甘肃鼢鼠盲肠内容物中细菌群落结构及多样性进行分析,研究野生和人工饲喂条件下甘肃鼢鼠肠道细菌群落差异,探讨人工饲喂条件下甘肃鼢鼠肠道菌群结构及其多样性变化,为揭示不同类型食物与动物肠道菌群结构之间的关系提供依据。
1 材料和方法 1.1 样品采集与保存 本研究所用甘肃鼢鼠均为在宁夏六盘山地区使用接地式活体捕鼠器捕捉的健康成年个体(表 1),于当地在室内条件下单笼饲养。首先对甘肃鼢鼠饲喂苜蓿统一驯养3 d,驯养结束后选择体重均一个体随机分为4组,每组10只,分别饲喂苜蓿(A组)、苜蓿+樟子松(M组)、苜蓿+油松(C组)和苜蓿+云杉(S组),持续饲喂15 d。以相同栖息地中捕获的野生个体作为野生对照组(W组)。用樟子松、油松和云杉的根部以及苜蓿的整株去饲喂甘肃鼢鼠。其中,苜蓿与樟子松根、油松根和云杉根的比例为1︰2,分别在每天10:00和18:00各饲喂1次,饲喂的食物均为新鲜采集,并在每次饲喂前打扫笼子。不同分组中食物成分含量见表 2。
表 1. 甘肃鼢鼠分组情况 Table 1. Information of Gansu zokor samples
| Groups | Samples name |
| A | A1、A2、A3、A4 |
| M | M1、M2、M3、M4、M5 |
| C | C1、C2、C3、C4、C5、C6 |
| S | S1、S2、S3、S4、S5、S6 |
| W | W1、W2、W3、W4 |
| A: alfalfa group; M: Mongolian pine group; C: Chinese pine group; S: spruce group; W: wild group. | |
表选项
表 2. 不同分组中食物营养成分含量[17-20] Table 2. Food nutrition contents in different groups[17-20]
| Groups | Crude fiber/% | Crude fat/% | Crude protein/% |
| A | 28.94 | 1.77 | 17.40 |
| M | 45.97 | 3.98 | 7.53 |
| C | 43.55 | 3.85 | 6.87 |
| S | 43.63 | / | 6.62 |
| W | / | / | / |
| A: alfalfa group; M: Mongolian pine group; C: Chinese pine group; S: spruce group; W: wild group. | |||
表选项
持续饲喂15 d后将甘肃鼢鼠放入装有乙酸乙酯的塑料密封袋中进行安乐死,死后立即收集盲肠内容物。采集的盲肠样品及时装入冻存管置于液氮罐保存,带回实验室冻存在–80 ℃中。本研究一共采集了5组26个盲肠样品。
1.2 DNA提取和测序 使用TIANamp Stool DNA Kit (TIANGEN, China)对甘肃鼢鼠的盲肠内容物总基因组DNA进行提取,具体步骤见试剂盒说明书。使用通用引物338F-806R (338F:5′-ACTCCTACGGGAGGCA GCAG-3′,806R:5′-GGACTACHVGGGTWTCTAA T-3′)对16S rRNA基因V3-V4区进行扩增[21]。20 μL反应体系如下:模板DNA 10 ng,5×FastPfu缓冲液4 μL,dNTPs (2.5 mmol/L) 2 μL,FastPfu聚合酶0.4 μL,BSA 0.2 μL,正反引物(5 μmol/L)均为0.8 μL,添加ddH2O补足反应体系。然后使用以下条件扩增DNA:95 ℃ 3 min;95 ℃ 30 s,55 ℃ 30 s,72 ℃ 45 s,循环25次;72 ℃ 10 min。在Illumina MiSeq平台上对纯化的PCR产物进行测序。
1.3 序列处理和分析 使用QIIME Pipeline Version 1.7.0分析原始序列[22],基于独特的条形码对序列进行分离。在去除低质量序列和嵌合体后,以97%的序列相似度为阈值对非重复序列进行OTUs (operational taxonomic units)的聚类。采用RDP classifier贝叶斯算法对OTU代表序列在97%相似度水平上进行分类学分析,将每个16S rRNA基因序列以70%的置信阈值和Silva (Release128 http://www.arb- silva.de)数据库进行比对[23-24]。
绘制稀疏曲线以证明每个样品的群落丰度和测序数据[25]。使用mothur软件计算Alpha多样性指数,包括群落丰富度指数(Chao 1,ACE)、群落多样性指数(Shannon)和群落覆盖度指数(Good’s coverage)[26]。使用Kruskal-Wallis秩和检验计算组间存在差异的细菌种类,使用R软件进行非度量多维尺度分析(NMDS)。
2 结果和分析 2.1 测序结果 从26个样品中获得了1405165条原始16S rRNA基因序列。计算并且绘制稀疏曲线。随着数据大小的增加稀疏曲线逐渐变得平缓,表明测序深度已经达到要求,数据可以进行后续分析。总共获得了658216条高质量序列,平均每个样品具有25316条序列。根据97%的序列相似性鉴定出1294个核心OTUs,苜蓿组平均每个样品564个OTUs,樟子松组平均每个样品656个OTUs,油松组平均每个样品629个OTUs,云杉组平均每个样品670个OTUs,野生组平均每个样品607个OTUs。
2.2 肠道细菌群落组成分析
2.2.1 细菌门分类水平比较: 从26个甘肃鼢鼠盲肠样本中共检测出11个门、17个纲、27个目、47个科和118个属。在细菌门水平上(图 1),所有样本中相对含量大于1%的细菌门共有6个,分别为厚壁菌门(Firmicutes)、拟杆菌门(Bacteroidetes)、变形菌门(Proteobacteria)、蓝细菌门(Cyanobacteria)、软壁菌门(Tenericutes)和疣微菌门(Verrucomicrobia)。其中厚壁菌门为所有样品中的优势菌,其次是拟杆菌门。在野生和人工饲喂条件下两个菌门所占的比例分别为:苜蓿组74.48%和21.11%,樟子松组73.53%和19.30%,油松组86.54%和9.45%,云杉组80.34%和16.09%,野生组75.64%和20.04%。
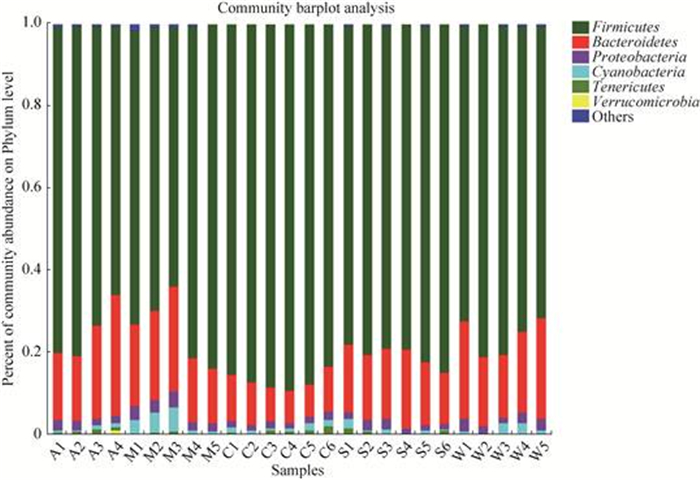 |
| 图 1 甘肃鼢鼠肠道样品门分类水平物种组成 Figure 1 Gansu zokor intestinal bacterial compositions at phylum. A: alfalfa group; M: Mongolian pine group; C: Chinese pine group; S: spruce group; W: wild group. |
| 图选项 |
2.2.2 细菌属分类水平比较: 在细菌属水平上(图 2),所有样本中相对含量大于1%的细菌属共有31个,其中丰度最高的前15个细菌属分别是unclassified_f_Lachnospiraceae、Lachnospiraceae_ NK4A136_group、Bacteroidales_S24-7_group、norank_f_Ruminococcaceae、norank_f_ Lachnospiraceae、Ruminococcus_1、unclassified_f_ Ruminococcaceae、Ruminiclostridium_6、Ruminococcaceae_NK4A214_group、Clostridiales_ vadinBB60_group、[Eubacterium]_ruminantium_ group、颤杆菌属(Oscillibacter)、Ruminiclostridium、norank_o_Gastranaerophilales、脱硫弧菌属(Desulfovibrio)。野生和人工饲喂条件下属水平含量占前三的细菌属分别是unclassified_ f_Lachnospiraceae、Lachnospiraceae_NK4A136_ group、Bacteroidales_S24-7_group。
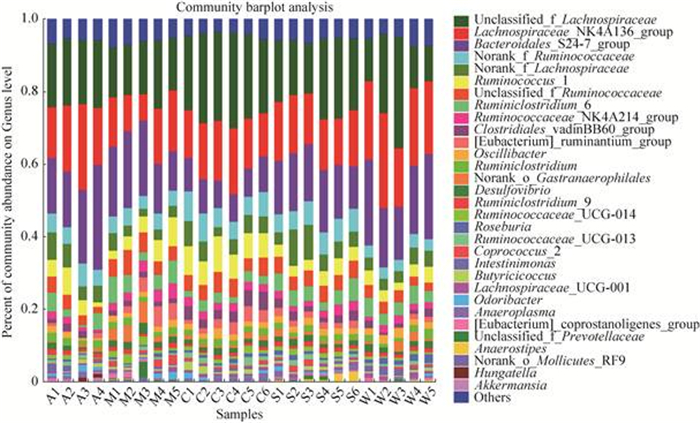 |
| 图 2 甘肃鼢鼠肠道样品属分类水平物种组成 Figure 2 Gansu zokor intestinal bacterial compositions at genus. A: alfalfa group; M: Mongolian pine group; C: Chinese pine group; S: spruce group; W: wild group. |
| 图选项 |
2.3 肠道细菌群落多样性分析
2.3.1 Alpha多样性分析: 野生和人工饲喂条件下甘肃鼢鼠肠道细菌Alpha多样性指数展示在表 3中,Good’s coverage分析表明每个分组中测得了99%的细菌种类。LSD分析显示Simpson指数在苜蓿组和云杉组间存在显著性差异,说明甘肃鼢鼠肠道细菌多样性云杉组显著高于苜蓿组;Shannon指数在苜蓿组与樟子松组、云杉组2组间,油松组与云杉组间存在显著性差异,说明樟子松组和云杉组甘肃鼢鼠肠道细菌群落均匀度显著高于苜蓿组,同时云杉组显著高于油松组;Chao 1和Ace指数苜蓿组和野生组分别与樟子松组和云杉组间存在显著性差异,说明樟子松组和云杉组甘肃鼢鼠肠道细菌群落丰富度显著高于苜蓿组和野生组,此外Ace指数在苜蓿组和油松组间也存在显著性差异,说明油松组甘肃鼢鼠肠道细菌群落均匀度显著高于苜蓿组(表 4)。
表 3. 甘肃鼢鼠肠道样品Alpha多样性指数表 Table 3. Number of alpha diversity indices of Gansu zokor intestinal samples
| Groups | Simpson/(mean±S.E) | Shannon/(mean±S.E) | Chao 1/(mean±S.E) | ACE/(mean±S.E) | Good’s coverage |
| A | 0.016±0.003 | 5.14±0.16 | 902±57 | 881±50 | 0.99 |
| M | 0.010±0.001 | 5.45±0.06 | 1015±12 | 1008±9 | 0.99 |
| C | 0.013±0.001 | 5.25±0.07 | 973±17 | 964±17 | 0.99 |
| S | 0.009±0.000 | 5.50±0.03 | 1008±18 | 990±13 | 0.99 |
| W | 0.012±0.003 | 5.37±0.08 | 928±10 | 923±9 | 0.99 |
| A: alfalfa group; M: Mongolian pine group; C: Chinese pine group; S: spruce group; W: wild group. | |||||
表选项
表 4. 甘肃鼢鼠肠道样品Alpha多样性指数LSD分析表 Table 4. Multiple comparisons of Alpha diversity index of Gansu zokor intestinal samples
| Alpha | Mean (i-j) (P) | W (j) | S (j) | C (j) | M (j) |
| Simpson | A (i) | 0.004 (0.22) | 0.007 (0.03) | 0.003 (0.31) | 0.006 (0.06) |
| M (i) | –0.002 (0.43) | 0.001 (0.75) | –0.003 (0.28) | ||
| C (i) | 0.001 (0.78) | 0.004 (0.15) | |||
| S (i) | –0.003 (0.26) | ||||
| Shannon | A (i) | –0.23 (0.07) | –0.37 (0.01) | –0.11 (0.34) | –0.31 (0.02) |
| M (i) | 0.08 (0.49) | –0.06 (0.60) | 0.19 (0.09) | ||
| C (i) | –0.11 (0.30) | –0.25 (0.02) | |||
| S (i) | 0.14 (0.22) | ||||
| Chao 1 | A (i) | –26 (0.49) | –106 (0.01) | –71 (0.06) | –113 (0.01) |
| M (i) | 87 (0.02) | 7 (0.83) | 42 (0.22) | ||
| C (i) | 45 (0.19) | –35 (0.29) | |||
| S (i) | 79 (0.03) | ||||
| Ace | A (i) | –42 (0.20) | –109 (0.00) | –83 (0.01) | –127 (0.00) |
| M (i) | 85 (0.01) | 18 (0.54) | 44 (0.14) | ||
| C (i) | 41 (0.17) | –26 (0.36) | |||
| S (i) | 67 (0.03) | ||||
| A: alfalfa group; M: Mongolian pine group; C: Chinese pine group; S: spruce group; W: wild group. | |||||
表选项
2.3.2 Beta多样性分析: 使用非度量多维尺度分析(NMDS)检验野生和人工饲喂条件下甘肃鼢鼠肠道细菌群落结构差异(图 3)。在NMDS图中,不同颜色或形状的点代表不同分组的样本,通过对样本距离进行等级排序,使样本在低维空间中的排序反映彼此之间的距离远近关系,NMDS分析中两样本点之间的距离越近,表明两个样本之间的细菌群落结构相似度越高,差异越小。NMDS的结果和聚类分析的结果一致,不同分组的样品单独聚集在一起,说明组内相似度高。樟子松组、油松组和云杉组3组间样本距离接近,其次是野生组,最后是苜蓿组,说明樟子松、油松组和云杉组这3组样本的肠道细菌群落结构较为相似,而与苜蓿组之间存在较大差异。
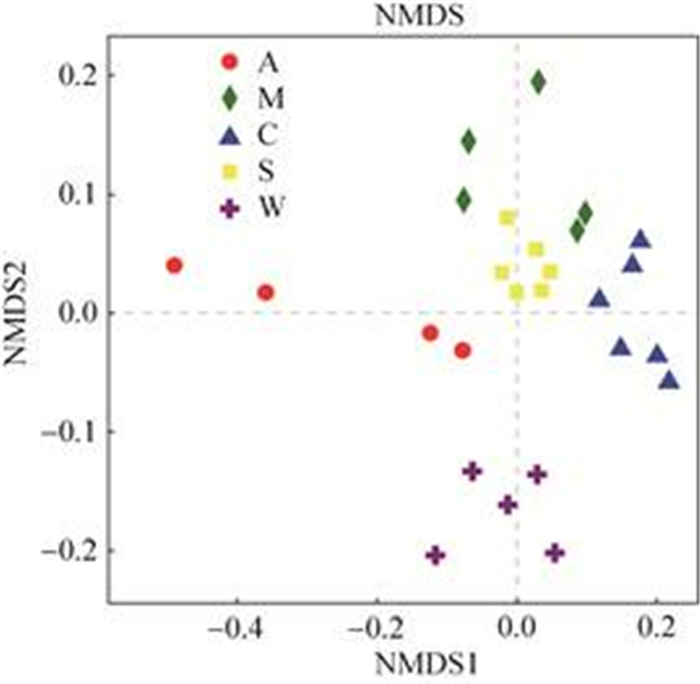 |
| 图 3 甘肃鼢鼠肠道细菌群落NMDS图 Figure 3 NMDS of Gansu zokor intestinal bacterial population structures. A: alfalfa group; M: Mongolian pine group; C: Chinese pine group; S: spruce group; W: wild group. |
| 图选项 |
为研究不同样本物种组成结构的相似性和差异关系,使用未加权的unifrac距离对野生和人工饲喂条件下5个分组中26个盲肠样品肠道细菌之间的关系进行分析,其结果通过树状图呈现(图 4)。树上的每个分支代表一个肠道细菌样本,樟子松组和云杉组的甘肃鼢鼠的肠道样本混在一起,而苜蓿组、油松组和野生组的甘肃鼢鼠肠道样品则位于不同的分支上。樟子松组、油松组和云杉组3组之间样本距离较近,其次是野生组,与苜蓿组之间距离最远。说明樟子松组、油松组和云杉组这3组与苜蓿组之间肠道样品物种组成结构相似性较小,差异较大。
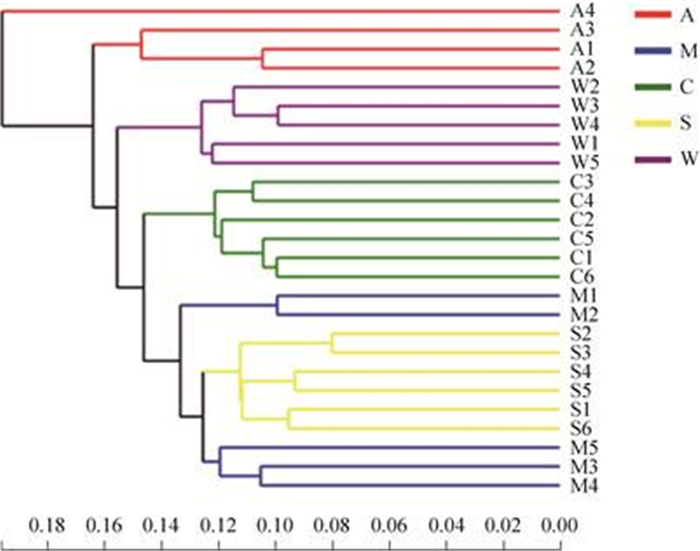 |
| 图 4 甘肃鼢鼠肠道样品聚类分析图 Figure 4 Clustering analysis of Gansu zokor intestinal samples. A: alfalfa group; M: Mongolian pine group; C: Chinese pine group; S: spruce group; W: wild group. |
| 图选项 |
2.4 组间差异分析 组间差异分析是运用严格的统计学方法,依据得到的群落丰度数据,对不同分组(或样本)微生物群落之间的物种进行假设检验,评估物种丰度差异水平,从而获得分组(或样本)间存在显著性差异的物种。Kruskal-Wallis秩和检验结果表明,在门水平上(图 5-A),厚壁菌门(Firmicutes)、拟杆菌门(Bacteroidetes)、软壁菌门(Tenericutes)、Saccharibacteria、迷踪菌门(Elusimicrobia)和疣微菌门(Verrucomicrobia)的相对含量在5个分组间具有显著性差异。和野生组对比发现,苜蓿组中,软壁菌门的含量增加。樟子松组中,厚壁菌门、软壁菌门和迷踪菌门的含量增加,而Saccharibacteria的含量降低。油松组中,厚壁菌门和软壁菌门的含量增加,而拟杆菌门、Saccharibacteria和迷踪菌门的含量降低。云杉组中,厚壁菌门和软壁菌门的含量增加,而拟杆菌门和Saccharibacteria的含量降低。值得注意的是,只有苜蓿组中检测到了疣微菌门。在属水平上(图 5-B),unclassified_f__ Lachnospiraceae、Lachnospiraceae_NK4A136_ group、Bacteroidales_S24-7_group、norank_f_ Ruminococcaceae、Ruminococcus_1、unclassified_f_ Ruminococcaceae、Ruminococcaceae_NK4A214_ group和Clostridiales_vadinBB60_group的相对含量在5个分组间具有显著性差异。和野生组对比发现,苜蓿组中,unclassified_f__Lachnospiraceae、norank_f__Ruminococcaceae和Clostridiales_ vadinBB60_group的含量增加,而Lachnospiraceae_ NK4A136_group和Ruminococcaceae_NK4A214_ group的含量降低。樟子松组中,norank_f_ Ruminococcaceae、unclassified_f_Ruminococcaceae、Ruminococcaceae_NK4A214_group和Clostridiales_ vadinBB60_group的含量增加,而unclassified_f_ Lachnospiraceae和Lachnospiraceae_NK4A136_ group的含量降低。油松组中,unclassified_f_ Lachnospiraceae、norank_f__Ruminococcaceae、Ruminococcus_1、unclassified_f_Ruminococcaceae、Ruminococcaceae_NK4A214_group和Clostridiales_ vadinBB60_group的含量增加,而Lachnospiraceae_ NK4A136_group和Bacteroidales_S24-7_group的含量降低。云杉组中,unclassified_f_ Lachnospiraceae、norank_f_Ruminococcaceae、unclassified_f_Ruminococcaceae、Ruminococcaceae_ NK4A214_group和Clostridiales_vadinBB60_group的含量增加,而Lachnospiraceae_NK4A136_group和Bacteroidales_S24-7_group的含量降低。
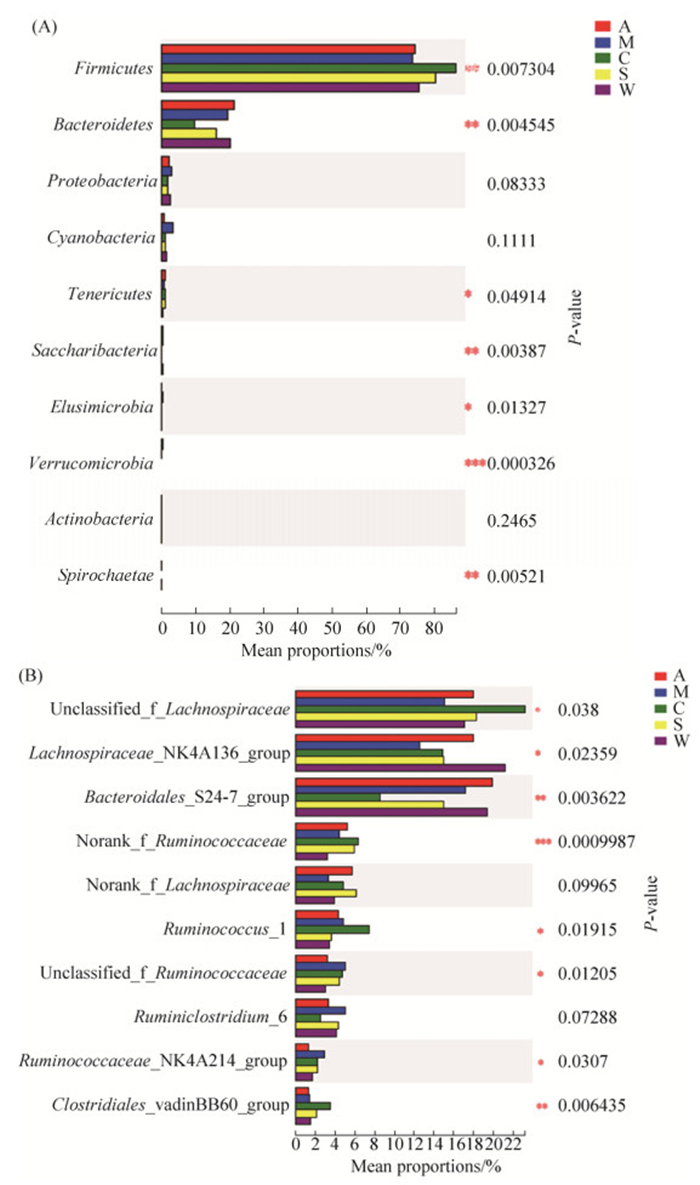 |
| 图 5 甘肃鼢鼠肠道细菌门水平(A)和属水平(B)差异检验 Figure 5 Statistical comparison of Gansu zokor intestinal bacteria at phylum (A) and genus (B) level. * 0.01 < P≤0.05, ** 0.001 < P≤0.01, *** P≤0.001. A: alfalfa group; M: Mongolian pine group; C: Chinese pine group; S: spruce group; W: wild group. |
| 图选项 |
3 讨论 本研究采用Illumina MiSeq技术首次对野生环境和人工饲喂条件下甘肃鼢鼠肠道细菌群落进行高通量测序。对5个分组中甘肃鼢鼠盲肠内容物细菌种类组成进行分析,结果表明不同分组中甘肃鼢鼠肠道门和属水平的优势菌具有一致性。在门分类水平上,来自5组中的优势细菌门均为厚壁菌门(Firmicutes)和拟杆菌门(Bacteroidetes),这与其他哺乳动物如马[27]和梅花鹿[28]的研究结果一致。但厚壁菌门和拟杆菌门的含量在不同分组间却存在显著性差异,以野生组作为对照,厚壁菌门在樟子松、油松和云杉组中的含量增加,拟杆菌门在油松和云杉组中的含量降低。大量研究表明厚壁菌门和拟杆菌门的比例(F/B)是衡量肥胖的一个指标[29-30],并且肥胖人群肠道中F/B的比例高于清瘦个体[31]。与苜蓿组相比,樟子松、油松和云杉组中粗纤维含量较高而粗蛋白的含量较低,因此,F/B比例的升高可能有助于甘肃鼢鼠从食物中获取更多的能量。此外,软壁菌门(Tenericutes)在人工饲喂组中的含量增加,Saccharibacteria在樟子松、油松和云杉组中的含量降低。可以看出,门水平细菌含量在樟子松、油松和云杉组中的变化是一致的,而苜蓿组的细菌组成和含量与野生组较为相似。
在属分类水平上,来自5组中的优势细菌属均为unclassified_f__Lachnospiraceae、Lachnospiraceae_ NK4A136_group和Bacteroidales_S24-7_group。Lachnospiraceae未分类菌和Lachnospiraceae_ NK4A136_group是甘肃鼢鼠肠道细菌群落的主要成分,两者都属于梭菌目并且普遍存在于人类和其他哺乳动物的肠道细菌群落中[32]。梭菌有利于消化纤维素和基于半纤维素的植物食物资源[33]。其次在属水平含量较高的是Bacteroidales_S24-7_ group、norank Ruminococcaceae、Ruminococcus_1,Ruminococcus的主要成员是Ruminococcus albus和Ruminococcus flavefaciens,它们是纤维降解过程中被研究最多的细菌,也是纤维素分解的主要细菌[34-35],这两种细菌在纤维和半纤维素的降解中是非常重要的,其分泌具有高活性的纤维素酶并且能够溶解微晶纤维素。因此,甘肃鼢鼠肠道中门和属水平的优势菌均表明了其对植食性的高度适应。但unclassified_f__Lachnospiraceae、Lachnospiraceae_NK4A136_group和Bacteroidales_ S24-7_group的含量在不同分组间却存在显著性差异,以野生组作为对照,unclassified_f__ Lachnospiraceae在人工饲喂组中的含量增加,Lachnospiraceae_NK4A136_group在人工饲喂组中的含量降低,Bacteroidales_S24-7_group在油松组中的含量降低。此外,norank_f__Ruminococcaceae在人工饲喂组中的含量增加,unclassified_f_ Ruminococcaceae、Ruminococcaceae_NK4A214_ group和Clostridiales_vadinBB60_group在樟子松、油松和云杉组中的含量增加。可以看出,属水平细菌含量在樟子松、油松和云杉组中的变化是一致的,这可能和它们3组中食物营养成分含量较为相似有关。而苜蓿组的细菌组成和含量与野生组较为相似,这可能和夏季甘肃鼢鼠取食的植物种类中草本类占的比例较大有关。
在NMDS图中,野生组甘肃鼢鼠肠道细菌群落结构与人工饲喂的4组之间存在明显差异,尽管野生环境中甘肃鼢鼠取食植物种类较多,但野生组中的肠道样品仍聚集在一起,表明野生环境条件下甘肃鼢鼠在长期进化过程中肠道菌群形成了一定的定植模式,导致了其肠道菌群的相似性。此外,苜蓿组和野生组与樟子松组、油松组和云杉组饲喂的甘肃鼢鼠肠道细菌群落存在明显差异,并且层级聚类树中的距离更远。在试验开始前,这5个分组中的个体均具有相似的地理条件和宿主遗传背景。然而,本研究表明它们各组的肠道细菌群落结构却并不相同,樟子松组、油松组和云杉组具有比苜蓿组和野生组更相似的群落结构和更高的物种丰富度。研究表明,多种因素都会对宿主肠道细菌群落产生影响,但饮食是其中最重要的因素之一[36-37],同时具有相同饮食结构的物种之间其肠道菌群结构会更为相似[38]。因此,野生和人工饲喂条件下甘肃鼢鼠肠道细菌群落结构存在显著性差异可能有两种原因,一方面,野生和人工饲喂条件下甘肃鼢鼠的食物不同,食物的不同会导致肠道细菌群落结构的差异[39];另一方面,野生和人工饲喂条件下甘肃鼢鼠生活的环境条件不同,例如环境微生物和氧含量的不同可能会影响肠道细菌群落结构。
本研究首次进行了野生和人工饲喂条件下甘肃鼢鼠肠道细菌多样性比较,研究发现野生环境和人工饲喂条件下甘肃鼢鼠肠道细菌群落组成和结构均存在显著性差异,人工饲喂显著改变了甘肃鼢鼠肠道细菌多样性和群落结构,并且饲喂木本植物的分组间甘肃鼢鼠肠道细菌群落结构更为相似。
致谢
感谢宁夏回族自治区森防总站及其固原地区泾源县林木病虫害检疫站、隆德县林木病虫害检疫站、原州区林木病虫害检疫站、彭阳县林木病虫害检疫站和西吉县林木病虫害检疫站在样品采集过程中的合作与支持。特别感谢学弟徐达、王鸿飞、张恒、苏嘉亮、燕宇航、郭晋泽和学妹侯佳钰在采集树根及饲喂鼢鼠过程中提供的帮助。
References
| [1] | McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lo?o T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(9): 3229-3236. |
| [2] | Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 2006, 444(7122): 1027-1031. |
| [3] | Leser TD, M lbak L. Better living through microbial action:the benefits of the mammalian gastrointestinal microbiota on the host. Environmental Microbiology, 2009, 11(9): 2194-2206. |
| [4] | Buddington RK, Sangild PT. Companion animals symposium:development of the mammalian gastrointestinal tract, the resident microbiota, and the role of diet in early life. Journal of Animal Science, 2011, 89(5): 1506-1519. |
| [5] | Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science, 2011, 332(6032): 970-974. |
| [6] | Godoy-Vitorino F, Goldfarb KC, Karaoz U, Leal S, Garcia-Amado MA, Hugenholtz P, Tringe SG, Brodie EL, Dominguez-Bello MG. Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. The ISME Journal, 2012, 6(3): 531-541. |
| [7] | Phillips CD, Phelan G, Dowd SE, McDonough MM, Ferguson AW, Delton Hanson J, Siles L, Ordó ez-Garza N, San Francisco M, Baker RJ. Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Molecular Ecology, 2012, 21(11): 2617-2627. |
| [8] | Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature, 2012, 486(7402): 222-227. |
| [9] | Kong FL, Zhao JC, Han SS, Zeng B, Yang JD, Si XH, Yang BQ, Yang MY, Xu HL, Li Y. Characterization of the gut microbiota in the red panda (Ailurus fulgens). PLoS One, 2014, 9(2): e87885. |
| [10] | Duncan SH, Scott KP, Ramsay AG, Harmsen HJM, Welling GW, Stewart CS, Flint HJ. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Applied and Environmental Microbiology, 2003, 69(2): 1136-1142. |
| [11] | Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome:a metagenomic analysis in humanized gnotobiotic mice. Science Translational Medicine, 2009, 1(6): 6ra14. |
| [12] | Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li HZ, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science, 2011, 334(6052): 105-108. |
| [13] | Northwest Institute of Plateau Biology, Chinese Academy of Sciences. Economic animal fauna of Qinghai. Xining: Qinghai People's Publishing House, 1989: 681-682. (in Chinese) 中国科学院西北高原生物研究所.青海经济动物志.西宁: 青海人民出版社, 1989: 681-682. |
| [14] | Nevo E. Mosaic evolution of subterranean mammals: regression, progression and global convergence. New York: Oxford University Press, 1999. |
| [15] | Yang J, Nan XN, Zou Y, Zhang FR, Shi JN, Han CX. Effects of the three factors on intestinal bacterial diversity of Eospalax cansus in the region of Liupan Mountains. Acta Microbiologica Sinica, 2018, 58(8): 1382-1396. (in Chinese) 杨静, 南小宁, 邹垚, 张斐然, 石建宁, 韩崇选. 不同因素对六盘山地区甘肃鼢鼠肠道细菌多样性的影响. 微生物学报, 2018, 58(8): 1382-1396. |
| [16] | Han CX, Xin XH, Zhang F, Wang MC, Yang QE, Zhang HL. Zokor resistance of main tree species on Loess Plateau. Journal of Northwest Forestry University, 2009, 24(1): 116-121. (in Chinese) 韩崇选, 辛晓辉, 张放, 王明春, 杨清娥, 张宏利. 黄土高原主要造林树种对鼢鼠危害的抗性研究. 西北林学院学报, 2009, 24(1): 116-121. |
| [17] | 王清华, 武娜, 王婉婉, 郭会茹, 韩雪林, 郑爱荣, 贺永惠, 牛岩, 王跃先, 张晓霞. 国产苜蓿干草营养成分分析与质量分级研究. 中国畜牧杂志, 2019. DOI:10.19556/j.0258-7033.20190218-06 |
| [18] | Chen XD, Hu ZL, Wang MC, Han CX, Yang XJ, Cao F. Study on the feeding habits and appetite of Gansu sokhor in the forested land of Northern Shaanxi. Shaanxi Forest Science and Technology, 1994(1): 40-45. (in Chinese) 陈孝达, 胡忠朗, 王明春, 韩崇选, 杨学军, 曹锋. 陕北林区甘肃鼢鼠的食性及食量研究. 陕西林业科技, 1994(1): 40-45. |
| [19] | Wan SM, Hu SL, Zhang B, Huang QH, Wang LC. The study on hay yield and nutrition of alfalfa cultivars. Acta Agriculturae Boreali-occidentalis Sinica, 2004, 13(1): 14-17. (in Chinese) 万素梅, 胡守林, 张波, 黄庆辉, 王龙昌. 不同紫花苜蓿品种产草量及营养成分研究. 西北农业学报, 2004, 13(1): 14-17. |
| [20] | 濮玲.腐朽对木材化学成分及活性炭性能的影响.青海民族大学硕士学位论文, 2016. |
| [21] | Xu N, Tan GC, Wang HY, Gai XP. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. European Journal of Soil Biology, 2016, 74: 1-8. |
| [22] | Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature Methods, 2013, 10(1): 57-59. |
| [23] | Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The ribosomal database project:improved alignments and new tools for rRNA analysis. Nucleic Acids Research, 2009, 37(S1): D141-D145. |
| [24] | Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Gl ckner FO. The SILVA ribosomal RNA gene database project:improved data processing and web-based tools. Nucleic Acids Research, 2013, 41(D1): D590-D596. |
| [25] | Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Gaskins HR, Stumpf RM, Yildirim S, Torralba M, Gillis M, Wilson BA, Nelson KE, White BA, Leigh SR. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. The ISME Journal, 2013, 7(7): 1344-1353. |
| [26] | Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One, 2011, 6(12): e27310. |
| [27] | Costa MC, Arroyo LG, Allen-Vercoe E, Stampfli HR, Kim PT, Sturgeon A, Weese JS. Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene. PLoS One, 2012, 7(7): e41484. |
| [28] | Guan Y, Yang HT, Han SY, Feng LM, Wang TM, Ge JP. Comparison of the gut microbiota composition between wild and captive sika deer (Cervus nippon hortulorum) from feces by high-throughput sequencing. AMB Express, 2017, 7(1): 212. |
| [29] | Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature, 2011, 469(7331): 543-547. |
| [30] | Li ST, Zhang C, Gu YY, Chen L, Ou SY, Wang Y, Peng XC. Lean rats gained more body weight than obese ones from a high-fibre diet. British Journal of Nutrition, 2015, 114(8): 1188-1194. |
| [31] | Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature, 2006, 444(7122): 1022-1023. |
| [32] | Kanki P, Grimes DJ. Infectious diseases: selected entries from the encyclopedia of sustainability science and technology. New York: Springer, 2013. |
| [33] | van Dyke MI, McCarthy AJ. Molecular biological detection and characterization of Clostridium populations in municipal landfill sites. Applied and Environmental Microbiology, 2002, 68(4): 2049-2053. |
| [34] | Koike S, Kobayashi Y. Development and use of competitive PCR assays for the rumen cellulolytic bacteria:Fibrobacter succinogenes, Ruminococus albus and Ruminococcus flavefaciens. FEMS Microbiology Letters, 2001, 204(2): 361-366. |
| [35] | Kohl KD, Miller AW, Marvin JE, Mackie R, Dearing MD. Herbivorous rodents (Neotoma spp.) harbour abundant and active foregut microbiota. Environmental Microbiology, 2014, 16(9): 2869-2878. |
| [36] | Zhang CH, Zhang MH, Wang SY, Han RJ, Cao YF, Hua WY, Mao YJ, Zhang XJ, Pang XY, Wei CC, Zhao GP, Chen Y, Zhao LP. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. The ISME Journal, 2010, 4(2): 232-241. |
| [37] | De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(33): 14691-14696. |
| [38] | Li H, Li TT, Beasley DE, Heděnec P, Xiao ZS, Zhang SH, Li JB, Lin Q, Li XZ. Diet diversity is associated with beta but not alpha diversity of Pika Gut Microbiota. Frontiers in Microbiology, 2016, 7: 1169. |
| [39] | Li ZP, Wright ADG, Liu HL, Bao K, Zhang TT, Wang KY, Cui XZ, Yang FH, Zhang ZG, Li GY. Bacterial community composition and fermentation patterns in the rumen of sika deer (Cervus nippon) fed three different diets. Microbial Ecology, 2015, 69(2): 307-318. |
