Yuping Wang#, Lingxin Kong#, Tao Zheng, Jinli Li, Yihua Sun, Delin You


State Key Laboratory of Microbial Metabolism, Joint International Research Laboratory of Metabolic and Developmental Sciences, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, Shanghai 200030, China
Received: 24 July 2018; Revised: 27 August 2018; Published online: 28 November 2018
Foundation item: Supported by the National Natural Science Foundation of China (31630002, 31470183, 21661140002) and by the Shanghai Pujiang Program from the Shanghai Municipal Council of Science and Technology
Corresponding author: Delin You, Tel: +86-21-62932943; E-mail: dlyou@sjtu.edu.cn.
#These authors contributed equally to this work
Abstract: [Objective] DNA phosphorothioate (PT) modification, in which the sulfur replaces a nonbridging oxygen, occurs naturally in diverse bacteria, archaea and human pathogens as a new kind of epigenetic modification. However, the regulatory mechanism of PT modification has not been fully characterized. In this study, the regulatory mechanism of spfB on DNA phosphorothioate (PT) modification in Pseudomonas fluorescens Pf0-1 was demonstrated.[Methods] Firstly, the spfB genetic interruption and complementary strains were constructed by homologous recombination and then tested the modification frequency in these strains by iodine cleavage. The operons within spf gene cluster were grouped by RT-PCR and the transcriptional level was analyzed in the ΔspfB mutant by quantitative real-time PCR. Finally, the possible regulatory region of SpfB on the spf operon was characterized by EMSA and DNase I footprinting assay.[Results] The inactivation of spfB led to more dispersed small fragments in genomic DNA of ΔspfB mutant and its complementation obviously restored the phenotype of wild type strain. Genes in spf gene cluster were assigned into one co-transcription unit, and the disruption of spfB directly up-regulated the transcription of the operon. In vitro SpfB directly protected two separate sequences within the spf promoter region from DNase I cleavage, and each protected sequence contained a direct repeat (5'-TGTTTGT-3').[Conclusion] SpfB in Pseudomonas fluorescens Pf0-1 was a negative regulator in DNA phosphorothioate modification.
Keywords: Pseudomonas fluorescensDNA phosphorothioationepigenetic modificationRT-PCRnegative regulator
假单胞菌Pseudomonas fluorescens Pf0-1中转录调控因子SpfB负调控DNA磷硫酰化修饰
王俞苹#, 孔令新#, 郑涛, 李金丽, 孙溢华, 由德林


上海交通大学生命科学技术学院, 微生物代谢国家重点实验室, 上海 200030
收稿日期:2018-07-24;修改日期:2018-08-27;网络出版日期:2018-11-28
基金项目:国家自然科学基金(31630002,31470183,21661140002);上海市浦江人才计划
通信作者:由德林, Tel: +86-21-62932943; E-mail: dlyou@sjtu.edu.cn.
#并列第一作者
摘要:[目的] DNA磷硫酰化修饰是DNA骨架上非桥接的氧原子以序列选择性和R-构型被硫取代的一种新型DNA修饰。目前,磷硫酰化修饰在多种细菌、古生菌以及人类致病菌中多有发现,但其分子调控机制尚不清楚。为了全面解析磷硫酰化修饰的调控机制,本文选择荧光假单胞菌Pf0-1为研究对象,开展了其DNA磷硫酰化修饰的调控机制研究。[方法] 首先,构建了spfB基因缺失和回补菌株,使用碘能特异性断裂磷硫酰化修饰DNA的方法,研究了该基因缺失对修饰表型的影响。利用cDNA在相邻同方向的基因间隔区进行PCR,确定了磷硫酰化修饰基因簇spfBCDE内的共转录单元。通过荧光定量RT-PCR,分析了spfB基因缺失突变株中磷硫酰化修饰基因的转录量。利用异源表达并纯化得到的重组蛋白SpfB进行了体外功能研究。通过EMSA实验,验证了SpfB蛋白具有与spfB启动子序列结合活性。通过DNase I footprinting实验,精确定位了SpfB蛋白与DNA结合序列。[结果] spfB基因的缺失加剧了磷硫酰化修饰DNA断裂所致电泳条带弥散的表型,spfB基因的回补能够恢复该表型,证明spfB基因负调控磷硫酰化修饰。鉴定了spf基因簇中只含有1个共转录单元,且该共转录单元在△spfB突变株中转录水平明显上升。通过EMSA和DNase I footprint实验,检测了SpfB蛋白与磷硫酰化修饰基因spfBCDE的启动子区域5'-TGTTTGT-3'相结合。[结论] SpfB作为转录调控因子负调控磷硫酰化修饰基因spfBCDE的表达,为解析磷硫酰化修饰的调控机制和全面理解基因组上的部分修饰特征奠定了基础。
关键词:假单胞菌DNA磷硫酰化修饰表观遗传修饰RT-PCR负调控蛋白
DNA phosphorothioate (PT) modification was originally characteristic of the DNA degradation (Dnd phenotype) during electrophoresis of genomic DNA from Gram-positive bacterium Streptomyces lividans 66[1] and many other distantly related bacteria[2]. The chemical nature of PT modification was found to be the incorporation of sulfur into DNA sugar-phosphate backbone for the replacement of the nonbridging oxygen by dnd (dndA–dndE) cluster in a sequence and stereospecific manner[3]. Since then, the five-member dnd cluster has been reported in various taxonomically unrelated bacteria, archaea[4] and even human pathogens[5]. Gene dndA can be found located either adjacent to clustered dndBCDE or elsewhere in the genome and the encoded DndA was an NifS-like cysteine desulfurase capable of assembling of DndC, which was a homologue of 3′-phosphoadenosine-5′-phosphosulfate (PAPS) reductase family proteins possessing ATP pyrophosphatase activity[6]. A DndD homologue in Pseudomonas fluorescens Pf0-1, SpfD, was proposed to be related to DNA structural alteration or nicking during sulfur incorporation with its ATPase activity[7]. Both of the canonical DndE[8] and DndEi with an additional DNA helicase domain[9] were assumed to bind the nicked double-stranded DNA during PT modifications. The biochemical study of PT modification revealed that DndA, C, D and E protein function as a large protein complex[10]. In some bacteria, PT-modifying genes dndACDE are in close proximity to restriction gene dndFGH, both of which form a novel host-specific restriction-modification (R-M) defense system for both of the host modification of conserved GpsAAC/GpsTTC sequences and the formation of double-stranded breaks in non-PT-protected foreign DNA[11]. Without PT modification, unrestrained restriction activity of DndFGH leads to double-stranded DNA damage which in turn triggered the SOS response, cell filamentation, and prophage induction[12]. Apart from that, PT modifications are dynamic and labile DNA modifications that exert effects on bacterial fitness in stressful environments, such as oxidative stress and exposure to peroxide and hypohalous acids[13]. The application of liquid chromatography-coupled tandem quadrupole mass spectrometry (LC-MS/MS), deep sequencing of iodine-induced cleavage at PT (ICDS), and single-molecule, real-time (SMRT) sequencing technology have revealed that PT was a partial, low frequency and dynamic genomic modification, which intriguing the question on the mechanism to maintain this low abundance modification suitable for cellular activities.
Recently, the transcriptional regulatory function of DptB in Salmonellaenterica serovar Cerro 87 has been reported. The DNA binding sequence of DptB was suggested to be conserved in other strains like Cedecea neteri strain ND14a and E.coli B7A[14]. As dptB homologs have been found in almost all the PT modification gene clusters including the spfBCDE gene cluster in P. fluorescens Pf0-1[7], which encoded proteins all exhibited high homology to that in S. lividans 1326. Especially, the work of the functional characterization of SpfD revealed for the first time the ATPase activity, contributing to the understanding of the biological PT modification. The whole genomic DNA of P.fluorescens Pf0-1 (GenBank ID: CP000094) has already been sequenced, interested by simple nutritional requirements but versatile metabolism and ability to function as a biological control agent[15]. Meanwhile, this strain has been served as good study material for PT modification. So, the regulatory study of PT modification in this strain will help expanding the knowledge of the various PT sustaining mechanism and benefit further biochemical characterization.
In this study, the regulatory role of spfB in genomic PT modifications was firstly investigated and the regulated region of spf cluster was finally identified. The reported data here demonstrated the negative regulatory role of SpfB for the maintaining of PT modification and revealed two pairs of direct repeats within the binding region of the regulated operon, which was different from the reported binding region of DptB in S.enterica serovar Cerro 87.
1 Materials and Methods 1.1 Bacterial strains and plasmids Bacteria and plasmids used in this study were listed in Table 1.
Table 1. Strains and plasmids
| Strains and plasmids | Characters | Source |
| Strains | ||
| E. coli DH10B | F-mcrA Δ(mrr-hsdRMS-mcrBC) Δ80d lacZΔM15 ΔlacX74 deoR recA1endA1araΔ139 D (ara, leu)7697 galU galK Δ-rpsL nupG | GIBCO BRL |
| E. coli BL21(DE3) | hsdS gal, chromosomal insertion of T7 pol | [23] |
| P. fluorescens Pf0-1 | Wild-type strain | Lab collection |
| E. coli S17-1 λpir | E. coli recA pro (RP4-2 Tet::Mu Kan::Tn7) | [24] |
| ΔspfB | P. fluorescens Pf0-1 mutant with disruption of spfB | This study |
| cΔspfB | The complementation strain of ΔspfB | This study |
| Plasmids | ||
| pET-28a | Expression vector, pBR322 replicon, PT7, His6 Tag, Kmr | Novagen |
| pPSUMO | Expression vector, pBR322 replicon, PT7, SUMO Tag, Kmr | [25] |
| pK18mobSacB | pMB1 ori, mob, Kmr, sacB, sacB | [26] |
| pBBR1MCS-2 | Complementary plasmid, a broad-host-range cloning vector, Kmr | [27] |
| pYP01 | Derivative plasmid of pK18mobSacB containing 1123-bp homologous fragment surrounding spfB | This study |
| pYP02 | Derivative plasmid of pPSUMO containing intact spfB for the expression of fused SUMO-SpfB | This study |
| pYP03 | pUC57 derivative containing 295-bp fragment of promoter region of the spfBCDE operon | This study |
| pYP04 | pBBR derivative complementary plasmid containing SUMO and spfB gene | This study |
表选项
1.2 General culture conditions The culture of Escherichia coli, P. fluorescens Pf0-1 and its derivate strains were conducted according to the protocol described before[7]. The wild type P. fluorescens Pf0-1 was used as original strain for construction of ΔspfB mutant. E. coli DH10B was used for routine gene cloning. Enzymes for DNA manipulation were purchased from Fermentas, genomic DNA isolation from P. fluorescens Pf0-1, ΔspfB and cΔspfB mutants were conducted using the TIANamp Bacteria DNA Kit (TIANGEN) and extraction of plasmids were performed using TIANprep Rapid Mini Plasmid Kit. E. coli BL21(DE3) was used as hetero-expression host for spfB. E. coli S17-1 λpir was used for conjugation. pPSUMO was used as protein expression vector.
1.3 Construction of the ΔspfB mutants The ΔspfB mutants were constructed by homologous recombination using the sucrose-sensitive plasmid pK18mobsacB. A 1.2 kb fragment containing partial fragment of spfB surrounding with homologous arms was amplified by two-step overlap extension PCR with Pfu DNA polymerase (Vazyme), using two pairs of primers spfB-LL/spfB-LR (5′-CG GAATTCTCCCTAGACGGCCTGCGAGT-3′/5′-GGC GACCATCAGCGGTTCGATGAGCCCGCAGCTCAGGCAT-3′) and spfB-RL/spfB-RR (5′-ATGCCTG AGCTGCGGGCTCATCGAACCGCTGATGGTCGCC-3′/5′-CGGGATCCGCCAGCGACGACTGAACCCA-3′). The pair of primer spfB-LL/spfB-RR carried a 40 nt overlap for the amplification of the fusion natural fragment with incorporation of EcoR I and BamH I restriction sites at the terminals. The resultant fragment was then digested with EcoR I and BamH I and cloned into plasmid pK18mobSacB for the construction of recombinant plasmid pYP01. Plasmid pYP01 was then transformed into S17-1 λpir and cultured with P. fluorescens Pf0-1 on NAN plate (peptone 5 g/L, yeast extract 1 g/L, beef extract 3 g/L, agar 15 g/L) for incubation of transformants. The mating and selection were conducted according to the procedure described before[16]. Mating bacterial colonies was firstly scratched and resuspend in 1 mL of PBS buffer. Then, 200 μL of each dilution (10×, 100×, 1000×) was plated on NAN medium supplied with ampicillin (100 mg/mL), chloramphenicol (25 mg/mL) and kanamycin (50 mg/mL) for selection of the single crossover intermediate strains. After that, 200 μL of each diluted colonies was plated on NAS medium (NA medium supplemented with 5% sucrose in the presence of ampicillin and chloramphenicol) at 30 ℃ for the final selection of the double crossover ΔspfB strain WYP01. The positive ΔspfB strains were verified through PCR with primers spfB-LL and spfB-RR.
1.4 The construction of complementary cΔspfB strain For the construction of ΔspfB complementary strain cΔspfB, the broad-host-range cloning vector pBBR1MCS-2 carrying SUMO tag encoding gene was used. The intact spfB gene with SUMO tag was amplified from pYP02 using primers spfB-csl/csr (5′-ATAAGCTTGATATCGAATTCATGGGCAGCAGCCATCATCA-3′/5′-GCTCTAGAACTAGTGGATCCTTAGCTTGTGGCCTGACGTG-3′). The PCR product was purified and ligated with pBBR1MCS-2 vector, resulting in the recombinant plasmid pYP04. The sequenced positive plasmid was then transformed into WYP01 by conjugation and the selected complementary strains cΔspfB were then verified by PCR using primers spfB-L/R (5′-ATACA TATGAACATGCTAAAATCCTCC-3′/5′-ATAGGATCCTTAGCTTGTGGCCTG-3′). In both cases, the corresponding strains containing vector pBBR1MCS-2 were used as control for RT-PCR or PT analysis.
1.5 Iodine cleavage at genomic PT sites PT-modified DNA can be cleaved by iodine at the modified sites. A 3 mmol/L iodine solution was freshly prepared. Reactions (total volume 20 μL) composed of 0.6 μg genomic DNA, 50 mmol/L Na2HPO4 (pH 9.0) and 3 mmol/L I2 were conducted in PCR tubes and incubated under 65 ℃ for 15 min. Then the systems were slowly cooled down to 4 ℃ with the rate of 0.1 ℃/s. PT modifications of gDNAs from P. fluorescens Pf0-1 (wild type), ΔspfB and cΔspfB were subjected to iodine cleavage. Then the samples were run on a 1% agarose gel buffered with 0.5×TAE buffer.
1.6 RNA preparation and quantitative real-time PCR (qPCR) The cells of P. fluorescens Pf0-1 (wild type), ΔspfB and cΔspfB were cultured following standard procedures until the OD600 reached 0.8–1.0. Total RNA was isolated with Qiagen RNeasy Protect Bacteria Mini Kit, following the manufacturer's protocol. To synthesize cDNA, 2 μg of purified total RNA was reverse transcribed using PrimeScriptRT reagent Kit with gDNA Eraser (TaKaRa) in a 20 μL reaction volume. The genome and cDNA of P. fluorescens Pf0-1 were respectively amplified with primers spfBCL and spfBCR (5′-CTTGGTTTGGGTT TGACGCC-3′/5′-GCCTCCGCTATAACCGACAA-3′) and spfBCR, spfCDL and spfCDR (5′-GCTTTGGA GTTTGCGAGAAGC-3′/5′-AAAATGACAGGTCGGCCAGG-3′), spfDEL and spfDER (5′-ATGAAAG TCGCCGTAGCGTC-3′/5′-GCAAGAGACATACAG AGCGCA-3′), which proved the co-transcription of spfBCDE. qPCR was operated using Maxima? SYBR Green/ROX qPCR Master Mix (Thermo) and an Applied Biosystms 7500 fast qPCR system with the cDNA (25 ng) as the template. The primers RT-spfCl and RT-spfCr (5′-CCTGCCCCACGAAAT AGGTT-3′/5′-ATCACGCGTGCTTCCACTAT-3′) were designed according to the spfC gene to quantify the transcription of the spf operon. The 16S rRNA was used as internal reference. mRNA levels were analyzed using the comparative threshold cycle (2–ΔΔCt) method. All qPCR assays were carried out triplicate using independent cultures.
1.7 Expression and purification of the N-terminal SUMO-tagged SpfB The spfB gene was PCR amplified with spfB-L/R and inserted into the N-terminal SUMO-tagged expression vector pPSUMO to construct pYP02. The spfB expression plasmid was transformed into E. coli BL21(DE3) and the resultant transformants were cultured at 37℃ and 220 r/min in LB medium supplemented with kanamycin (50 mg/mL) to OD600 reached 0.6–0.8. Isopropylthio-β-d-galactoside (IPTG) with final concentration 0.4 mmol/L was added into the culture to induce protein expression. The cells were further cultured at 22 ℃ for 10 h. Then the cells were harvested by centrifugation and resuspended in buffer A (50 mmol/L Tris-HCl, 200 mmol/L NaCl, pH 8.0) and then lysed by continuous high-pressure cell disrupter on 4 ℃. After centrifugation (10000×g for 60 min at 4 ℃), the supernatant was applied to a HiTrap chelating column charged with nickel. The impurity was eluted by buffer A with 20 mmol/L imidazole. The SUMO-tagged SpfB was eluted by buffer A with 200 mmol/L imidazole. Fractions containing SUMO-tagged labeled SpfB were pooled and concentrated to 2.5 mL with Amicon Ultra Centrifugal Filter 20000 MWCO (Millipore). PD-10 columns (GE healthcare) were then used for imidazole elimination and buffer exchange. The protein was stored in buffer A with 10% glycerol at –80 ℃. Protein concentration was determined with the Bradford assay using bovine serum albumin as a standard.
1.8 Electrophoretic mobility shift assay (EMSA) Putative DNA promoter regions were firstly amplified using primers spfB-proL/spfB-proR and then cloned into the plasmid pUC57 to construct pYP03. The resultant plasmids were used as template for the amplification of the 6-carboxyfluorescein (FAM)-labeled probes using primers M13F (FAM) and M13R (5′-TGTAAAACGACGGCCAGT-3′/5′-C AGGAAACAGCTATGACC-3′). FAM-labeled probes were purified by the Wizard? SV Gel and PCR Clean-Up System (Promega) and were quantified with NanoDrop 2000C (Thermo). The purified SUMO-tagged SpfB were incubated with labeled probes at room temperature in a total volume of 20 μL buffer comprising of 10 mmol/L Tris-HCl (pH 8.0), 25 mmol/L KCl, 2.5 mmol/L MgCl2, and 1.0 mmol/L dithiothreitol. To prevent nonspecific binding, sheared salmon sperm DNA was added to a final concentration of 100 ng/μL. After 20 min of incubation, the fragments were separated by a 2% agarose gel buffered with 0.5× Tris-borate-EDTA buffer. Gels were scanned with the ImageQuantTM LAS 4000 mini (GE Healthcare).
1.9 DNase I footprinting assay For preparation of fluorescent FAM labeled probes, the Dpx DNA polymerase (TOLO Biotech, Shanghai) was used for the amplification from pYP03. The following DNase I footprinting assays were performed according to the previously reported protocol [14].
For each assay, 350 ng probes were incubated with 14 μg of SpfB in a total volume of 40 μL. After incubation for 20 min at 30 ℃, 10 μL solution containing about 0.015 unit DNase I (Promega) and 100 nmol freshly prepared CaCl2 was added and further incubated for 1 min at 30 ℃. And then, the reaction was stopped by adding 140 μL DNase I stop solution (200 mmol/L unbuffered sodium acetate, 30 mmol/L EDTA and 0.15% SDS). Samples were firstly extracted with phenol/chloroform to remove the proteins, and then precipitated with ethanol and the resultant pellets were dissolved in 30 μL MiniQ water. The preparation of the DNA ladder, electrophoresis and data analysis were the same as described before[14], except that the GeneScan-LIZ600 size standard (Applied Biosystems) was used.
1.10 Phylogenetic analysis Multiple sequences were aligned using ClustalW[17-18] and the phylogenetic tree of SpfB was generated by MEGA 5[19] using neighbor-joining with Poisson correction and 500 replicate bootstrap analysis. The detailed information about the selected proteins listed designated by GenBank accession numbers as bellow. ABA72481 form P. fluorescens Pf0-1, AFM64477 from Pseudomonas aeruginosa DK2, ACB62514 from Burkholderia ambifaria MC40-6, AIV47941 from Burkholderia pseudomallei TSV 48, ACO34108 from Acidobacterium capsulatum ATCC 5119, ACL06476 from Desulfatibacillum alkenivorans AK-01, CCK76817 from Oleispira antarctica RB-8, AIJ12648 from Streptomyces lividans TK24, AIG73614 from Amycolatopsis japonica, ALM19267 from Mycobacterium abscessus, ATY86444 from Kyrpidia sp. EA-1, BAN34915 from Sulfuricella denitrificans skB26, ARB83643 from Yersinia sp. FDAARGOS_228, ADM97032 from Dickeya dadantii 3937, CP009459.1 from Cedecea neteri strain ND14a, AIF62361 from Escherichia coli B7A, APT80319 from S. enterica serovar Cerro 87, ABAM02000001 from Salmonella enterica SARA23, ABG30476 from Roseobacter denitrificans OCh 114, ALH94670 from Acinetobacter equi, ARO32310 from Rhizobium sp. NXC14.
2 Results and discussion 2.1 Disruption of spfB aggravates PT modification The interruption of individual dndCDE genes completely abolished DNA degradation, suggesting the essentiality of these genes for PT modification[20]; whereas, disruption of dndB in S. lividans and S. enterica serovar Cerro 87 led to highly degradation during electrophoresis. Especially, the study of DptB homologues in S. enterica serovar Cerro 87 has definitely proved the negative regulatory function in PT modification[14]. The analysis of similar binding short sequences in other PT-containing bacteria indicated the general regulatory mechanism by the corresponding DptB homologues. However, when we gave deep insight into the possible regulatory region in spfCDE for SpfB in P. fluorescens Pf0-1, no obvious similar sequence can be found. Moreover, sequence alignment of SpfB and DptB revealed 51% sequence identity and the same conserved DGQHR motif (Figure 1). In order to explore the possible regulatory function of SpfB, we constructed the in-frame scarless interruption of spfB (Figure 2-A) in P. fluorescens Pf0-1 and investigated DNA PT phenotype in ΔspfB mutant. Following the optimized iodine-induced PT-specific cleavage assay reported previously[21], more dispersed small fragments can be found in genomic DNA of ΔspfB mutant than that in wild-type strain, suggesting a similar significant increase of PT modification efficiencies in the genomic DNA of ΔspfB (Figure 2-B). To verify the increased modification frequency was related to the interruption of spfB, we constructed the complementary strain WYP02 and conducted the same iodine-induced PT-specific cleavage assay (Figure 2). Expectedly, the introduction of the intact spfB into ΔspfB mutant strain could restore the typical dnd phenotype to a certain extent. Taken together, SpfB plays a negative regulatory role for PT modification in P. fluorescens Pf0-1.
 |
| Figure 1 Multiple sequence alignment of SpfB and its homologues. Proteins used here were listed as follow with GenBank accession number: SpfB in P. fluorescens Pf0-1 (ABA72481), DptB in S. enterica Cerro 87 (ADN26581), DndB in S. lividans 1326 (AAZ29043), AJAP_03440 in Amycolatopsis japonica strain MG417-CF17 (AIG73614), HCH_07042 in Hahella chejuensis KCTC 2396 (ABC33659), Gura_2473 in Geobacter uraniireducens Rf4 (ABQ26651), Spea_2305 in Shewanella pealeana ATCC 700345 (ABV87625), PU1002_00508 in Candidatus Pelagibacter ubique HTCC1002 (EAS84156). The conserved DGQHR motif was marked with rectangle |
| 图选项 |
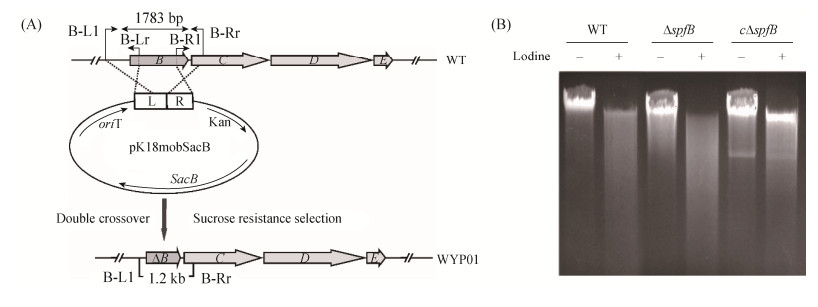 |
| Figure 2 The schematic construction of ΔspfB strain and characterization of Dnd phenotype. A: The construction of scarless genome deletion mutant (ΔspfB) in P. fluorescens Pf0-1. B: Iodine cleavage of genomic DNA from wild-type, ΔspfB and cΔspfB strain of P. fluorescens Pf0-1 |
| 图选项 |
2.2 SpfB negatively regulates expression of the spfBCDE operon Next, the reverse transcription polymerase chain reaction (RT-PCR) was conducted to validate SpfB as a negative regulator for regulating PT modification in P. fluorescens Pf0-1. Firstly, the number of transcripts within spfBCDE gene cluster was determined. To identify the operon organization in the spf cluster, one-step RT-PCR was performed to detect mRNA spanning different ORFs. All the intergenic gaps between neighboring genes with the same orientation were tested (Figure 3-A). The results of RT-PCR revealed that genes spfC, D and E were co-transcribed from the same promoter upstream of spfB, forming the spf operon. Next, qPCR was performed with RNAs isolated from the ΔspfB, wild type and cΔspfB strains grown in LB for 8 hours. From the data depicted in Figure 3-B, the transcription level of spf operon of ΔspfB mutant increased by 13-fold compared with that of wild type. Besides, the complementation of spfB contributed to the drop of the transcription to the similar level with that of wild type strain (Figure 3-B). These results obviously proved the negative regulatory function of SpfB during DNA PT modification in P. fluorescens Pf0-1.
 |
| Figure 3 Verification of the operon organization and qPCR analysis of the transcription of spf operon. A: Organization of the operon encoded by spf cluster. The position of primers used for RT-PCR of adjacent genes without overlap was showed by vertical solid arrows, and genes in an operon were marked with dashed arrows. M: 1 kb DNA ladder; lane 1: spfB-spfC in genome; lane 2: spfB-spfC in cDNA; lane 3: spfC-spfD in genome; lane 4: spfC-spfD in cDNA; lane 5: spfD-spfE in genome; lane 6: spfD-spfE in cDNA. B: QPCR analysis of transcription levels in ΔspfB mutants, cΔspfB mutants and wild type strain. The relative transcription levels of each gene were obtained after normalization against the internal reference 16S rRNA. Error bars showed the standard deviation of three independent experiments |
| 图选项 |
2.3 SpfB binding to the TGTTTGT motif upstream of spf operon To determine whether spfB directly regulates the spf operon, SpfB was next expressed for in vitro characterization analysis. However, His6-tagged SpfB protein was expressed in E. coli in the form of inclusion body, impeding further characterization. Fortunately, after the selection and optimization of protein expression, the SpfB could be soluble expressed as SUMO-tagged recombinant protein and the purified SpfB protein was detected by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Figure 4-A). Using the purified SpfB, the electrophoretic mobility shift assay (EMSA) experiment was performed according to the protocol described before[14]. The fragment of 295 bp upstream of spf operon was PCR amplified with primers spf-proL/R (5′-GGCACGGTAGGTAGCTG AGT-3′/5′-GTTTATGTGCAAATAAACAA-3′) for the construction of plasmid pYP03, and the resultant plasmid was then used as template for the preparation of fluorescent probe with primers M13F/R (5′-TGTAAAACGACGGCCAGT-3′/5′-CA GGAAACAGCTATGACC-3′). From Figure 4-B, the purified SpfB binding to the upstream of spfB and generated significantly shifted bands. DNase I footprinting assay with FAM-labeled primers uncovered two protected regions (Figure 4-C) and the two binding sites are: binding site I 5′-TGTT TGTGTTATCGA-3′ and binding site II 5′-TTGTTT GTTTATTTGC-3′ (Figure 4-D). The sequence analysis of these two sites revealed two direct repeats 5′-TGTTTGT-3′ in the promoter of spf operon (Figure 4-D). According to the typical regulatory mechanisms[22], it was proposed that SpfB might directly down-regulate the transcription of the spf operon by blocking the access of RNA polymerase to the two identified binding sites in this promoter region.
 |
| Figure 4 Binding characters of SpfB for controlling of the transcription of spf. A: Purified SpfB analyzed by SDS-PAGE. Lane 1, purified SUMO-tagged SpfB; M, molecular markers. B: EMSAs for binding of SpfB to the upstream region of spfB. The 295 bp FAM-labelled DNA fragment of the upstream region was incubated with increasing concentrations of SpfB protein (lanes 2–4; lanes contain 2.5, 7.0, 14 μg SpfB, respectively). Lane 1, negative control without SpfB. The shifted bands are indicated by arrows. C: Characterization of the direct binding site of SpfB by DNase I footprinting. Two protected regions were indicated. D: Nucleotide sequence of the SpfB-binding sites. The two SpfB-binding sites are underlined and the direct repeats are marked with gray rectangles. The bent arrows indicate the transcription start points and transcription orientation of spfB |
| 图选项 |
2.4 SpfB is the representative regulator among diverse Pseudomonas The transcriptional regulator DptB in S. enterica serovar Cerro 87 has been proved to repress the transcription of dptCDE and its own gene by binding to two regions (5′-ACGT/CAAN6ACGTAA-3′) in the upstream of dpt operon, each possessing a pair of imperfect 6 nt direct repeats (Figure 5)[14]. Considering the different binding sites of SpfB for the regulation of PT modification in P. fluorescens Pf0-1, we conducted the phylogenetic analysis of SpfB. As is shown in Figure 5, SpfB shared the same clade with other PT regulatory proteins like DndB from E. coli B7A and S. entarica S87, but further formed a small well-supported subclade with other homologous proteins from Pseudomonas. The sequence analysis of the binding site of SpfB revealed two direct repeats in the upstream region of sfpB, which was different from the recognized regulated region of DptB[14]. The identified two repeats for SpfB were firstly thought to generally exist in other Pseudomonas strains. However, after sequence analysis, the repeats could not be found in any of the other two Pseudomonas containing PT modification. These results here suggested that even with high sequence identity with DptB, SpfB might adjust the specificity to the host-specific DNA sequence for the regulatory role. Meanwhile, the absence of the same binding site in other Pseudomonas originated homologues suggested the more diverse regulatory mechanism.
 |
| Figure 5 Phylogenetic analysis of SpfB with its homologues. The identified direct repeats for SpfB binding was marked with solid square and the well-recognized sequence for other regulatory proteins represented by DndB from S. entarica S87 were marked with solid circles |
| 图选项 |
In conclusion, this work here revealed the negative regulatory protein SpfB encoded by spf cluster for the DNA PT modification in P. fluorescens Pf0-1. The identified binding sites of SpfB in the upstream region of the co-transcribed spfBCDE operon depicted the strict host-specific DNA-protein interaction. These findings pave the way for the study of other possible regulatory proteins in so many poorly characterized bacteria. Of course, the detailed regulatory mechanisms still need further exploration and may benefit the expanding of the knowledge of PT modifications.
Acknowledgement We thank Prof. Yawen He at Shanghai Jiao Tong University for providing the plasmid pK18mobSacB, pBBR1MCS-2 and strain E. coli S17-1 λpir for construction of ΔspfB mutant and cΔspfB strain.
References
| [1] | Zhou XF, He XY, Liang JD, Li AY, Xu TG, Kieser T, Helmann JD, Deng ZX. A novel DNA modification by sulphur. Molecular Microbiology, 2005, 57(5): 1428-1438. DOI:10.1111/j.1365-2958.2005.04764.x |
| [2] | He XY, Ou HY, Yu Q, Zhou XF, Wu J, Liang JD, Zhang W, Rajakumar K, Deng ZX. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Molecular Microbiology, 2007, 65(4): 1034-1048. DOI:10.1111/mmi.2007.65.issue-4 |
| [3] | Wang LR, Chen S, Xu TG, Taghizadeh K, Wishnok JS, Zhou XF, You DL, Deng ZX, Dedon PC. Phosphorothioation of DNA in bacteria by dnd genes. Nature Chemical Biology, 2007, 3(11): 709-710. DOI:10.1038/nchembio.2007.39 |
| [4] | Wang LR, Chen S, Vergin KL, Giovannoni SJ, Chan SW, DeMott MS, Taghizadeh K, Cordero OX, Cutler M, Timberlake S, Alm EJ, Polz MF, Pinhassi J, Deng ZX, Dedon PC. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(7): 2963-2968. DOI:10.1073/pnas.1017261108 |
| [5] | R?mling U, Tümmler B. Achieving 100% typeability of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. Journal of Clinical Microbiology, 2000, 38(1): 464-465. |
| [6] | You DL, Wang LR, Yao F, Zhou XF, Deng ZX. A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans. Biochemistry, 2007, 46(20): 6126-6133. DOI:10.1021/bi602615k |
| [7] | Yao F, Xu TG, Zhou XF, Deng ZX, You DL. Functional analysis of spfD gene involved in DNA phosphorothioation in Pseudomonas fluorescens Pf0-1. FEBS Letters, 2009, 583(4): 729-733. DOI:10.1016/j.febslet.2009.01.029 |
| [8] | Hu W, Wang CK, Liang JD, Zhang TL, Hu ZP, Wang ZJ, Lan WX, Li F, Wu HM, Ding JP, Wu G, Deng ZX, Cao CY. Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification. Cell Research, 2012, 22(7): 1203-1206. DOI:10.1038/cr.2012.66 |
| [9] | Zheng T, Jiang P, Cao B, Cheng QX, Kong LX, Zheng XQ, Hu QH, You DL. DndEi exhibits helicase activity essential for DNA phosphorothioate modification and ATPase activity strongly stimulated by DNA substrate with a GAAC/GTTC motif. The Journal of Biological Chemistry, 2016, 291(3): 1492-1500. DOI:10.1074/jbc.M115.694018 |
| [10] | Cao B, Zheng XQ, Cheng QX, Yao F, Zheng T, Ramesh Babu I, Zhou HC, Dedon P, You DL. In vitro analysis of phosphorothioate modification of DNA reveals substrate recognition by a multiprotein complex. Scientific Reports, 2015, 5: 12513. DOI:10.1038/srep12513 |
| [11] | Xu TG, Yao F, Zhou XF, Deng ZX, You DL. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Research, 2010, 38(20): 7133-7141. DOI:10.1093/nar/gkq610 |
| [12] | Cao B, Cheng QX, Gu C, Yao F, DeMott MS, Zheng XQ, Deng ZX, Dedon PC, You DL. Pathological phenotypes and in vivo DNA cleavage by unrestrained activity of a phosphorothioate-based restriction system in Salmonella. Molecular Microbiology, 2014, 93(4): 776-785. DOI:10.1111/mmi.2014.93.issue-4 |
| [13] | Kellner S, DeMott MS, Cheng CP, Russell BS, Cao B, You DL, Dedon PC. Oxidation of phosphorothioate DNA modifications leads to lethal genomic instability. Nature Chemical Biology, 2017, 13(8): 888-894. DOI:10.1038/nchembio.2407 |
| [14] | Cheng QX, Cao B, Yao F, Li JL, Deng ZX, You DL. Regulation of DNA phosphorothioate modifications by the transcriptional regulator DptB in Salmonella. Molecular Microbiology, 2015, 97(6): 1186-1194. DOI:10.1111/mmi.2015.97.issue-6 |
| [15] | Haas D, Blumer C, Keel C. Biocontrol ability of fluorescent pseudomonads genetically dissected: importance of positive feedback regulation. Current Opinion in Biotechnology, 2000, 11(3): 290-297. DOI:10.1016/S0958-1669(00)00098-7 |
| [16] | Schnider U, Keel C, Voisard C, Défago G, Haas D. Tn5-directed cloning of pqq genes from Pseudomonas fluorescens CHA0: mutational inactivation of the genes results in overproduction of the antibiotic pyoluteorin. Applied and Environmental Microbiology, 1995, 61(11): 3856-3864. |
| [17] | Zhang WK, Wang L, Kong LX, Wang T, Chu YW, Deng ZX, You DL. Unveiling the post-PKS redox tailoring steps in biosynthesis of the Type Ⅱ polyketide antitumor antibiotic xantholipin. Chemistry & Biology, 2012, 19(3): 422-432. |
| [18] | Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 1994, 22(22): 4673-4680. DOI:10.1093/nar/22.22.4673 |
| [19] | Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 2011, 28(10): 2731-2739. DOI:10.1093/molbev/msr121 |
| [20] | Xu TG, Liang JD, Chen S, Wang LR, He XY, You DL, Wang ZJ, Li AY, Xu ZL, Zhou XF, Deng ZX. DNA phosphorothioation in Streptomyces lividans: mutational analysis of the dnd locus. BMC Microbiology, 2009, 9: 41. DOI:10.1186/1471-2180-9-41 |
| [21] | Cao B, Chen C, DeMott MS, Cheng QX, Clark TA, Xiong XL, Zheng XQ, Butty V, Levine SS, Yuan G, Boitano M, Luong K, Song Y, Zhou XF, Deng ZX, Turner SW, Korlach J, You DL, Wang LR, Chen S, Dedon PC. Genomic mapping of phosphorothioates reveals partial modification of short consensus sequences. Nature Communications, 2014, 5: 3951. DOI:10.1038/ncomms4951 |
| [22] | Rojo F. Mechanisms of transcriptional repression. Current Opinion in Microbiology, 2001, 4(2): 145-151. DOI:10.1016/S1369-5274(00)00180-6 |
| [23] | Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods in Enzymology, 1990, 185: 60-89. DOI:10.1016/0076-6879(90)85008-C |
| [24] | Milton DL, Norqvist A, Wolf-Watz H. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. Journal of Bacteriology, 1992, 174(22): 7235-7244. DOI:10.1128/jb.174.22.7235-7244.1992 |
| [25] | Malakhov MP, Mattern MR, Malakhova OA, Drinker M, Weeks SD, Butt TR. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. Journal of Structural and Functional Genomics, 2004, 5(1/2): 75-86. DOI:10.1023/B:JSFG.0000029237.70316.52 |
| [26] | Sch?fer A, Tauch A, J?ger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene, 1994, 145(1): 69-73. |
| [27] | Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop Ⅱ RM, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene, 1995, 166(1): 175-176. DOI:10.1016/0378-1119(95)00584-1 |
