郭淑华1, 易现峰2

1. 潍坊学院生物与农业工程学院, 山东 潍坊 261061;
2. 江西师范大学生命科学学院, 江西 南昌 330022
收稿日期:2018-06-01;修回日期:2018-09-13;网络出版日期:2019-11-20
基金项目:江西省自然科学基金(20161ACB20017, 20171BBF60073), 国家自然科学基金(31760156)
作者简介:易现峰。Tel/Fax:+86-791-88120390, E-mail:yxfeng1975@126.com.
摘要:[目的] 象甲是栎属植物橡子中主要的寄生昆虫,但其适应高单宁食物(橡子)的肠道微生物基础尚待揭示。本研究分析了蒙古栎和辽东栎橡子中两种柞栎象(Curculio arakawai和C.dentipes)幼虫的肠道菌群结构和多样性,试图阐明柞栎象幼虫适应高单宁食物的肠道微生物基础。[方法] 分别提取蒙古栎和辽东栎橡子中象甲幼虫各50头的肠道DNA,利用Illumina MiSeq技术对肠道菌群的V3-V4区序列进行16S rRNA测序,统计样品操作分类单元(OTU)数量,分析物种组成丰度、α多样性和β多样性。[结果] 结果表明,可用于物种分类的OTU分别有2054和2308个,C.arakawai和C.dentipes共有的OUT 481个。在两种柞栎象C.arakawai和C.dentipes肠道菌群中,共注释到的主要分类阶元有27个门、145个科和274个属。变形菌门(Proteobacteria)、拟杆菌门(Bacteroidetes)和厚壁菌门(Firmicutes)在两种象甲肠道菌群中占优势;假单胞菌属Pseudomonas(63.8%)、沙雷氏菌属Serratia(6%)和不动杆菌属Acinetobacter(5.2%)是C.arakawai肠道中的主要类群,而沙雷氏菌属Serratia(32%)、拉恩菌属Rahnella(24.2%)、气单胞菌属Aeromonas(6.8%)和立克次体属Rickettsia(6.6%)在C.dentipes肠道菌群中占主导优势。C.arakawai和C.dentipes肠道菌群α多样性无显著差异,β多样性则差异显著。具有单宁酶活性的肠道细菌,如粘质沙雷菌Serratia marcescens、乳球菌Lactococcus lactis、假单胞菌Pseudomonasspp.在C.arakawai和C.dentipes之间差异不显著。[结论] 寄生在蒙古栎和辽东栎橡子中的C.arakawai和C.dentipes肠道菌群组成迥异,这可能与遗传因素和食物特点有关。具有单宁酶活性的粘质沙雷氏菌Serratia marcescens和乳球菌Lactococcus lactis等菌类可能是两种象甲幼虫适应高单宁食物的肠道微生物基础。
关键词:蒙古栎辽东栎柞栎象甲16S rRNA肠道菌群群落多样性单宁菌
Gut bacterial composition of two Curculio species and their adaptation to high-tannin food
Shuhua Guo1, Xianfeng Yi2

1. School of Biological and Agricultural Engineering, Weifang University, Weifang 261061, Shandong Province, China;
2. College of Life Sciences, Jiangxi Normal University, Nanchang 330022, Jiangxi Province, China
Foundation item: Supported by the Natural Science Foundation of Jiangxi Province (20161ACB20017, 20171BBF60073) and by the National Natural Science Foundation of China (31760156)
Abstract: [Objective] Curculio beetles are major parasitic insects of oak acorns, but the mechanism of their adaptation to high-tannin food (e.g., acorns) is not clear. In this study, we compared the structure and diversity of gut bacteria of the larvae of Curculio arakawai and C. dentipes infested in Mongolian oak (Quercus mongolica) and Liaodong oak (Q. liaotungensis), to elucidate the adaptive mechanisms of weevil larvae to high-tannin food. [Methods] We collected gut samples of weevil larvae for DNA extraction. V3-V4 areas were sequenced using the Illumina MiSeq technology 16S rRNA sequencing. We carried out sequence statistics of Operational Taxonomic Units (OTUs), analysis of species abundance, a and b diversity. [Results] There were 2054 OTUs and 2308 OTUs of the high quality sequences obtained from the guts of the 2 weevil species, in which 481 OTUs were shared. In the gut bacterial community of C. arakawai and C. dentipes, 27 phyla, 145 families and 274 genera were identified as the major taxonomic groups. At the phylum level, Proteobacteria, Bacteroidetes and Firmicutes were dominant in the guts of C. arakawai and C. dentipes. At the genus level, Pseudomonas (63.8%), Serratia (6%) and Acinetobacter (5.2%) were the main groups in the gut of C. arakawai, while Serratia (32%), Rahnella (24.2%), Aeromonas (6.8%) and Rickettsia (6.6%) were dormant in the gut of C. dentipes. Our a diversity analyses showed no significant differences between the gut bacteria of C. arakawai and C. dentipes; however, b diversity showed significant difference between the gut bacteria of C. arakawai and C. dentipes. [Conclusion] Differences in gut bacterial community may be related to differences in species of weevils and host tannin content. We also found several bacteria related to the degradation of tannins, e.g., Serratia marcescens and Lactococcus lactis, which may reflect the adaptive basis for two weevil species to use high tannin food.
Keywords: Quercus mongolicaQ. liaotungensisCurculio weevil16S rRNAgut bacteriacommunity diversitytannin-degrading bacteria
栎属(Quercus)植物在全世界约有450余种,中国约有51种[1],广泛分布于北温带至热带山地[2]。由于栎属植物的橡子个体较大且营养丰富,因而成为许多动物(如鼠类、鸟类和昆虫等)捕食的对象。象甲(Curculio)是寄生在栎属植物橡子中一类常见的植食性昆虫,其幼虫的取食活动显著降低了橡子的活力,已成为栎林天然更新的重要限制因素。与此同时,栎属植物橡子进化出了高单宁的化学防御机制抵御象甲幼虫的蛀食。在长期的协同进化过程中,植食性昆虫又发展出丰富的肠道微生物类群。这些微生物直接或间接对宿主发挥着重要的作用,对许多植物次生代谢物质具有较强的抵抗作用[3-9],从而维持昆虫的生存和繁衍。
蒙古栎(Quercus mongolica)为我国落叶栎中分布最北的一个种,同时也是我国东北和华北地区针阔叶混交林的主要建群种。辽东栎(Q. liaotungensis)是我国暖温带落叶阔叶林主要优势种之一,遍布于黄河流域和辽东半岛等广大地区[10]。研究表明,两种植物橡子中象甲幼虫维持较高的寄生率[11-12]。柞栎象Curculio arakawai是蛀食蒙古栎橡子的主要昆虫种类[11],而C. dentipes是寄生辽东栎橡子的重要类群[13]。作为植物重要的化学防御物质之一,单宁在栎属植物橡子内的含量高达3%-10%。但是,以橡子为单一食物的象甲幼虫适应高单宁食物的肠道微生物基础还有待于揭示。近年来,随着高通量测序技术的发展,许多昆虫的肠道微生物作用机制得到阐明。因此,本研究采用高通量测序技术16S rRNA对蒙古栎和辽东栎象甲幼虫肠道菌群的多样性和组成进行分析,试图阐明两种象甲幼虫适应高单宁食物的肠道微生物基础。
1 材料和方法 1.1 象甲幼虫采集 于2016年9月,分别在辽宁省抚顺市清原县以及北京东灵山小龙门采集蒙古栎和辽东栎橡子各2000粒。将橡子放置在室温下,使内部的象甲幼虫自由逃出。每天收集逃出的活体象甲幼虫,液氮冷冻后保存于?80 ℃冰箱内。待收集的象甲幼虫数量分别达到50头时,在无菌条件下解剖并挑取C. arakawai和C. dentipes幼虫的肠容物,分别混合形成3个待测样本,干冰封装后送到上海伯豪生物技术有限公司进行测序分析。
1.2 肠道细菌16S rRNA高通量测序 象甲肠道菌群DNA以QIAGEN QIAamp DNA Stool Mini Kit进行提取,用AxyPrep DNA纯化试剂盒(Axygen Biosciences,Union City,CA,U.S.)纯化。用0.8%琼脂糖凝胶电泳定性检测和质控后(QuantiFluor?-ST,Promega,U.S.),每个样品用总DNA作为模板,进行16S rDNA的V3-V4区PCR扩增,引物为338F (5′-barcode- ACTCCTAC GGGAGGCAGCAG-3′)和806R (5′-GGACTACHV GGGTWTCTAAT-3′)。PCR反应体系:4 μL 5× FastPfu缓冲液,2 μL 2.5 mmol/L dNTPs,上下游引物各0.8 μL (5 μmol/L), 0.4 μL FastPfu聚合酶,10 ng模板DNA。扩增条件:95 ℃ 2 min;95 ℃ 30 s,55 ℃ 30 s,72 ℃ 30 s,25个循环;72 ℃ 5 min,4 ℃保温。PCH产物经2%凝胶电泳检测合格,将所提取的16S rRNA V3-V4扩增产物按照Miseq (Illumina公司)建库说明完成建库。经Qubit定量,将样品等量混合后,完成Miseq双末端测序。
1.3 序列数据分析 原始数据在QIIME (version 1.17)中去噪,删除质量差(低于平均质量20)和短序列(< 50 bp),并使用USEARCH (v1.8.0)工具去除嵌合体。用UQIIME (version 7.1 http://drive5.com/uparse/)在97%的相似水平下划分分类操作单元(operational taxonomic units,OTU),将得到的干净序列用UCHIME筛选嵌合体并去除嵌合体序列。每个操作分类单元中丰度最高的序列被选为OTU的代表序列,代表序列使用RDP数据库(http://rdp.cme.msu.edu/)进行注释和分类。将OTU和物种注释结合,从而得到样品的OTU和分类谱系的基本分析结果。再对OTU进行丰度、α和β多样性指数等分析,最后对物种注释到各个分类水平上并统计各样本中的群落组成。
1.4 肠道细菌群群落多样性分析 用α多样性分析单个样本内部的细菌群落多样性,按照QIIME (Quantitative Insights Into Microbial Ecology)的分析流程进行物种分类地位的确定,并进行稀疏曲线(rarefaction curve)的绘制、计算测定物种指数(Chao 1指数、Shannon指数、observed_species以及goods_coverage)等。计算结果用R (Version 3.1.3)进行作图。根据差异OTU的丰度利用R(Version 3.1.3)进行丰度聚类热图的绘制。用α多样性分析样本间的细菌群落组成差异,做PCoA (principal co-ordinates analysis)图。用R语言Vegan软件包分析不同样本OTU (97%相似性)组成,反映样本间的差异和距离。
1.5 统计分析 组间各分类水平上的差异使用Metastats进行显著性检验;单因素方差ANOVA用α多样性的差异检验;群落相似性检验(analysis of similarities ANOSIM)采用R语言的ANOSIM函数进行比较。
2 结果和分析 两种柞栎象C. arakawai和C. dentipes肠道菌群的OTU数分别为2054和2308个,二者共有OTU为481个(图 1)。所有OTU可归入27个门(图 2),其中变形菌门(Proteobacteria)是两种柞栎象肠道中的优势菌门,分别占到了总序列数的96.7%和90.4%。拟杆菌门(Bacteroidetes)次之,分别占0.9%和3.2%。硬壁菌门(Firmicutes)、放线菌门(Actinobacteria)和蓝细菌门(Cyanobacteria)等比例均较低。
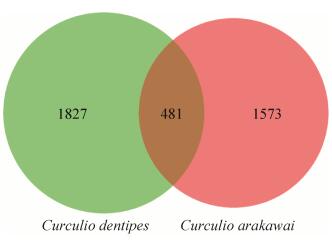 |
| 图 1 柞栎象C. arakawai和C. dentipes肠道菌群的韦恩图 Figure 1 Venn diagram of gut bacteria of C. arakawai and C. dentipes. |
| 图选项 |
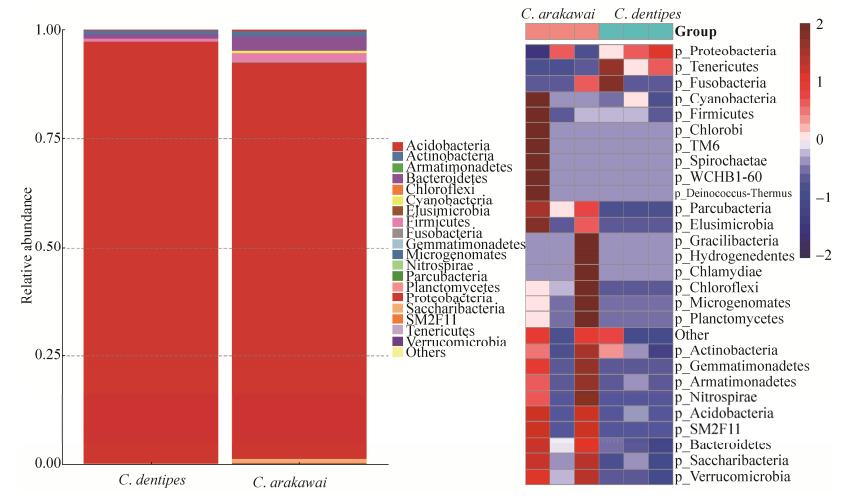 |
| 图 2 柞栎象C. arakawai和C. dentipes肠道菌群门水平上的比例分布和热图 Figure 2 Proportion and heat map of gut bacteria of C. arakawai and C. dentipes at phylum level. |
| 图选项 |
在C. arakawai和C. dentipes肠道菌群已知属信息的OTU中,按丰度排序的前20个属分布比例如图 3所示,二者组成差异明显。在C. arakawai的肠道中,占优势地位的为假单胞菌属Pseudomonas(63.8%)、沙雷氏菌属Serratia (6.0%)、不动杆菌属Acinetobacter (5.2%)、拉恩菌属Rahnella (4.8%)和寡养单胞菌属Stenotrophomonas (1.6%)。在C. dentipes的肠道菌群中,占优势为沙雷氏菌属Serratia (32.0%)、拉恩菌属Rahnella (24.2%)、气单胞菌属Aeromonas (6.8%)、立克次体属Rickettsia (6.6%)、Curculioniphilus (5.0%)、假单胞菌属Pseudomonas (4.7%)、不动杆菌属Acinetobacter (1.0%)、肠杆菌属Enterobacter (1.0%)和水杆菌属Aquabacterium (1.0%)。C. arakawai肠道菌群内假单胞菌属Pseudomonas的比例显著高于C. dentipes (P=0.014),但沙雷氏菌属Serratia、拉恩菌属Rahnella和立克次体属Rickettsia显著低于C. dentipes (P=0.003,P=0.008,P=0.033)。两种柞栎象C. arakawai和C. dentipes肠道菌群的差异还可从属水平上的聚类热图上看出(图 3)。C. arakawai和C. dentipes肠道菌群样本分别有7.7%和10.3%的OTU无法确定到属的水平。
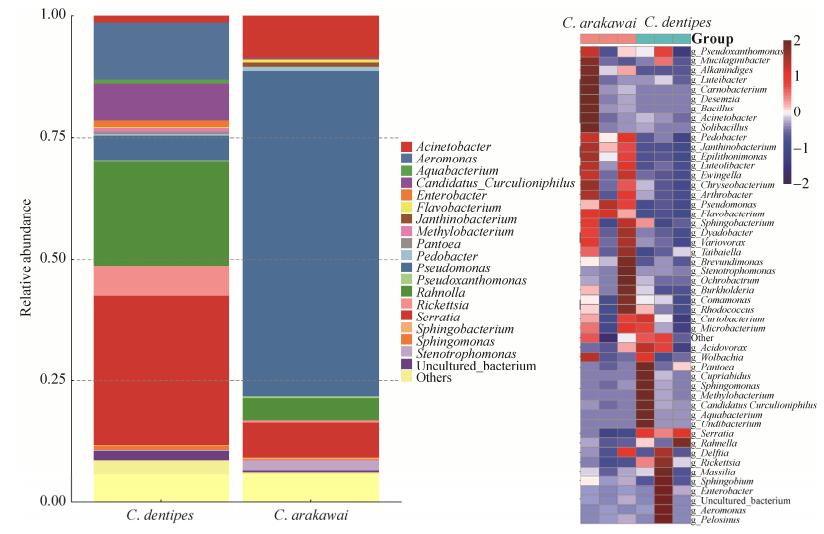 |
| 图 3 柞栎象C. arakawai和C. dentipes肠道菌群属水平上的比例分布和热图 Figure 3 Proportion and heat map of gut bacteria of C. arakawai and C. dentipes at genus level. |
| 图选项 |
图 4所示为两种柞栎象C. arakawai和C. dentipes肠道菌群中按丰度排序的前20个种分布比例,二者组成差异明显。C. arakawai的肠道中丁香假单胞菌Pseudomonas syringae、黄杆菌Flavobacterium rivuli、氧化葡萄糖酸杆菌Gluconobacter oxydans、肠球菌Enterococcus gallinarum、哈氏噬纤维菌Cytophaga hutchinsonii ATCC_33406、粘液杆菌Mucilaginibacter sp.、粘土杆菌Pelobacteraceae bacterium、麦芽香肉杆菌Carnobacterium maltaromaticum的比例均显著高于C. dentipes (P < 0.05)。柞栎象C. arakawai和C. dentipes肠道菌群的差异还可从种水平上的热图看出(图 4)。通过与已有文献比对,我们在C. arakawai和C. dentipes肠道中发现具有单宁酶活性的肠道菌,如粘质沙雷菌Serratia marcescens (0.0024%和0.0550%)、乳球菌Lactococcus lactis (0.0380%和0.0024%)、假单胞菌Pseudomonas_ spp. (0.014%和0.014%)等。尽管寄主有所不同,但C. arakawai和C. dentipes肠道中单宁菌的丰度差异不显著(P > 0.05)(图 4)。另外,我们还在2种柞栎象肠道中发现具有固氮能力的水拉恩氏菌Rahnella aquatilis以及栎属植物的专性寄生菌Gibbsiella quercinecans,且比例较高(图 4)。
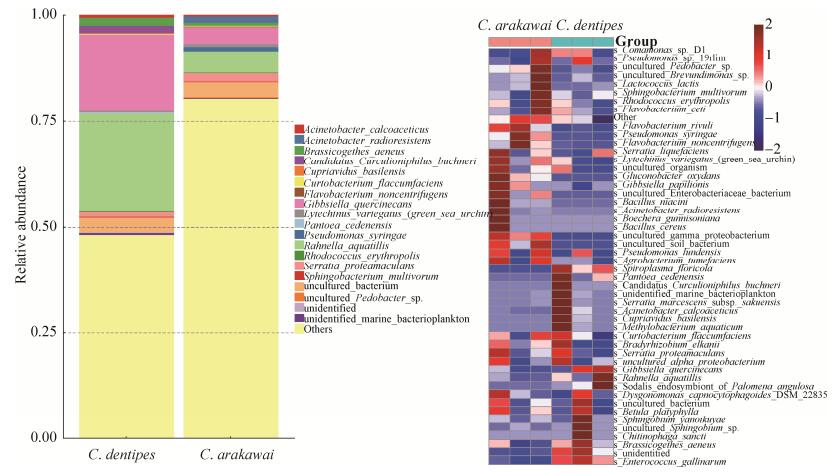 |
| 图 4 柞栎象C. arakawai和C. dentipes肠道菌群种水平上的比例分布和热图 Figure 4 Proportion and heat map of gut bacteria of C. arakawai and C. dentipes at species level. |
| 图选项 |
尽管C. arakawai和C. dentipes肠道菌群组成在属和种的水平上存在明显差异,但经Wicox检验后,α多样性指数之间并无明显差异:Chao 1指数(P=0.3827)、observed_species (P=0.1904)、Shannon指数(P=1.0)和goods_coverage (P=0.6625) (图 5)。β多样性分析表明,C. arakawai和C. dentipes肠道菌群在群落组成上存在明显差异(ANOSIM,P < 0.05)(图 6)。
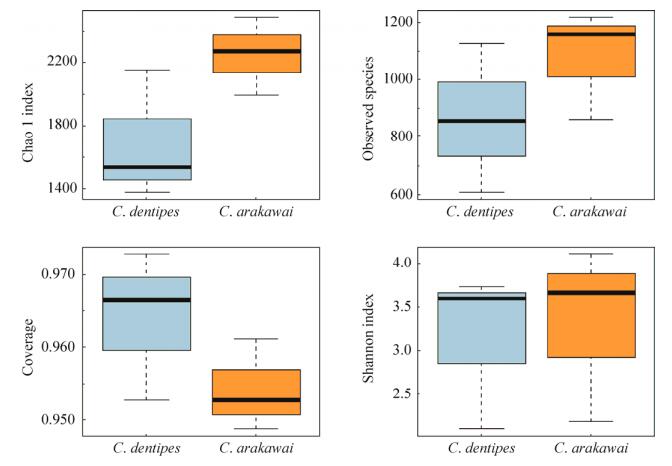 |
| 图 5 柞栎象C. arakawai和C. dentipes肠道菌群的α多样性比较 Figure 5 Alpha diversity of gut bacteria of C. arakawai and C. dentipes. |
| 图选项 |
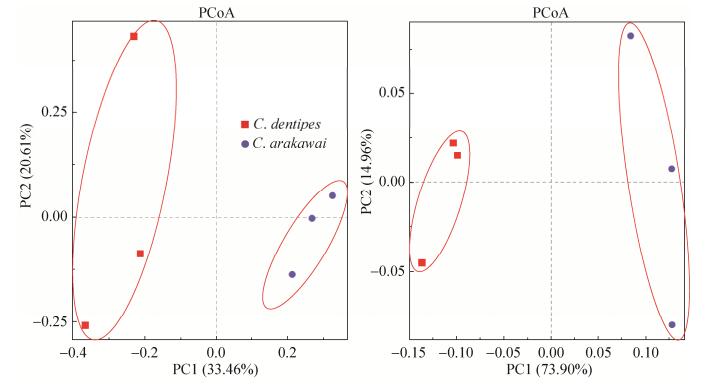 |
| 图 6 柞栎象C. arakawai和C. dentipes肠道菌群的unweighted和weighted UniFrac聚类图 Figure 6 Principal coordinates analysis of unweighted and weighted UniFrac distances between the gut bacteria of C. arakawai and C. dentipes. |
| 图选项 |
3 讨论 大量研究表明,植食性昆虫肠道内的微生物具有一系列重要的生理功能[5, 7]。譬如,Rinke et al.[6]从甘蔗象甲Sphenophorus levis幼虫体内分离出具有降解纤维素能力的肠道微生物。Berasategui et al.[3-4]对松树象甲Hylobius abietis进行宏基因组测定,发现了分解萜类化合物的肠杆菌科的种类。国内****Xu等[8-9]发现红脂大小蠹Dendroctonus valens肠道微生物具有催化分解α-蒎烯的能力。近年来,一些研究对象甲属Curculio昆虫进行了肠道微生物组成分析,发现了一些重要的微生物类群(如伴蝇菌属Sodalis,沃巴赫体属Wolbachia,立克次体属Rickettsia,螺原体属Spiroplasma和沙雷氏菌属Serratia)[14-17],但尚未将肠道微生物组成与其生物学功能联系起来进行分析。尽管2种柞栎象C. arakawai和C. dentipes的单一食物橡子富含单宁,但其肠道微生物组成在门水平上与其他植食性昆虫差异不大,均以变形菌门(Proteobacteria)、拟杆菌门(Bacteroidetes)和硬壁菌门(Firmicutes)为主[3-4, 6-9]。本研究还发现,C. arakawai和C. dentipes肠道微生物组成与已报到的象甲属昆虫具有较大差别[14-17]。如在属的水平上,C. arakawai和C. dentipes肠道微生物分别以假单胞菌属Pseudomonas (63.8%)和沙雷氏菌属Serratia (32.0%)为主。假单胞菌属和沙雷氏菌属内通常包括众多具有单宁降解活性的菌群。另外,我们在C. arakawai和C. dentipes肠道中发现Curculio属昆虫专有的共生菌属Curculioniphilus[15]。在种的分类水平上,我们还在C. arakawai和C. dentipes肠道中鉴定出具有固氮能力的水生拉恩氏菌Rahnella aquatilis。由于单宁可与蛋白质发生缩合反应产生收敛性,水生拉恩氏菌的存在可能与象甲虫单一的食物来源有关,可补充氮源的不足[18]。C. arakawai和C. dentipes幼虫肠道中还存在引起栎属植物感染导致种群退化的种类栎类赤霉菌Gibbsiella quercinecans[19-20]。因此,象甲虫寄生不仅造成橡子营养和生活力的下降,还可能引起致病微生物在栎属植物种群内部的扩散。
大量研究表明,许多肠道微生物可产生单宁酶,具有降解单宁的能力。比如乳球菌属Lactococcus[21]、假单胞菌属Pseudomonas[22-23]、链球菌Streptococcus spp.[24]、肠杆菌Enterobacter spp.[25]、葡萄球菌Staphylococcus lugdunensis[26]、沙雷氏菌Serratia marcescens[27]以及乳杆菌属Lactobacillus[28-29]。本研究中,我们在C. arakawai和C. dentipes肠道中发现几种与单宁降解有关的菌群,如沙雷氏菌Serratia marcescens和乳球菌Lactococcus lactis。尽管我们未对它们进行分离提纯和功能鉴定,但先前的研究表明这两种肠道菌具有较强的分解单宁的能力,有助于动物对高单宁食物的摄取[21, 30-33]。由于单宁广泛存在于栎属植物的橡子中[11-12, 34],且具有生理毒害作用[35-39],肠道中分解单宁的菌群对于C. arakawai和C. dentipes取食高单宁的橡子具有积极意义,有助于象甲幼虫在橡子内的取食和生存。
尽管蒙古栎和辽东栎属于近源种,橡子化学组成相近[40],但寄生在橡子中的C. arakawai和C. dentipes的肠道微生物组成存在较大差异。先前的研究分析了多种寄生于栎属植物橡子中的象甲的肠道微生物组成,也发现明显的种间差异[14-17]。这种差异可能与遗传因素以及寄主(橡子)的营养成分和单宁含量有关[40]。尽管两种象甲的肠道微生物组成存在较大差异,具有分解单宁功能的沙雷氏菌Serratia marcescens和乳球菌Lactococcus lactis在C. arakawai和C. dentipes之间差异不大。这些共有单宁菌的存在有助于象甲幼虫取食和利用高单宁食物——栎属植物橡子,是象甲幼虫适应高单宁食物的基础。由此看来,象甲幼虫与栎属植物橡子之间的博弈与肠道微生物组成和功能息息相关。
References
| [1] | Fu LG, Chen TQ, Lang KY, Hong T, Lin Q, Li Y. Higher Plants in China (Vol. 1). Qingdao: Qingdao Publishers, 2012. (in Chinese) 傅立国, 陈潭清, 郎楷永, 洪涛, 林祁, 李勇. 中国高等植物(第一卷). 青岛: 青岛出版社, 2012. |
| [2] | Li WY, Gu WC. Study on phenotypic diversity of natural population in Quercus mongolica. Scientia Silvae Sinicae, 2005, 41(1): 49-56. (in Chinese) 李文英, 顾万春. 蒙古栎天然群体表型多样性研究. 林业科学, 2005, 41(1): 49-56. DOI:10.3321/j.issn:1001-7488.2005.01.010 |
| [3] | Berasategui A, Axelsson K, Nordlander G, Schmidt A, Borg-Karlson AK, Gershenzon J, Terenius O, Kaltenpoth M. The gut microbiota of the pine weevil is similar across Europe and resembles that of other conifer-feeding beetles. Molecular Ecology, 2016, 25(16): 4014-4031. DOI:10.1111/mec.13702 |
| [4] | Berasategui A, Salem H, Paetz C, Santoro M, Gershenzon J, Kaltenpoth M, Schmidt A. Gut microbiota of the pine weevil degrades conifer diterpenes and increases insect fitness. Molecular Ecology, 2017, 26(15): 4099-4110. DOI:10.1111/mec.2017.26.issue-15 |
| [5] | Morales-Jiménez J, Zú?iga G, Villa-Tanaca L, Hernández-Rodríguez C. Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae). Microbial Ecology, 2009, 58(4): 879-891. DOI:10.1007/s00248-009-9548-2 |
| [6] | Rinke R, Costa AS, Fonseca FPP, Almeida LC, Delalibera I, Henrique-Silva F. Microbial diversity in the larval gut of field and laboratory populations of the sugarcane weevil Sphenophorus levis (Coleoptera, Curculionidae). Genetics and Molecular Research, 2011, 10(4): 2679-2691. DOI:10.4238/2011.November.1.1 |
| [7] | Xu LT, Lou QZ, Cheng CH, Lu M, Sun JH. Gut-associated bacteria of Dendroctonus valens and their involvement in verbenone production. Microbial Ecology, 2015, 70(4): 1012-1023. DOI:10.1007/s00248-015-0625-4 |
| [8] | Xu LT, Lu M, Xu DD, Chen L, Sun JH. Sexual variation of bacterial microbiota of Dendroctonus valens guts and frass in relation to verbenone production. Journal of Insect Physiology, 2016, 95: 110-117. DOI:10.1016/j.jinsphys.2016.09.014 |
| [9] | Xu LT, Shi ZH, Wang B, Lu M, Sun JH. Pine defensive monoterpene α-pinene influences the feeding behavior of Dendroctonus valens and its gut bacterial community structure. International Journal of Molecular Sciences, 2016, 17(11): 1734. DOI:10.3390/ijms17111734 |
| [10] | Sun SC, Chen LZ. Seed demography of Quercus liaotungensis in Dongling mountain region. Acta Phytoecologica Sinica, 2000, 24(2): 215-221. (in Chinese) 孙书存, 陈灵芝. 东灵山地区辽东栎种子库统计. 植物生态学报, 2000, 24(2): 215-221. DOI:10.3321/j.issn:1005-264X.2000.02.016 |
| [11] | Wang X, Xiao ZS, Zhang ZB, Pan HC. Insect seed predation and its relationships with seed crop and seed size of Quercus mongolica. Acta Theriologica Sinica, 2008, 51(2): 161-165. (in Chinese) 王学, 肖治术, 张知彬, 潘红春. 昆虫种子捕食与蒙古栎种子产量和种子大小的关系. 昆虫学报, 2008, 51(2): 161-165. DOI:10.3321/j.issn:0454-6296.2008.02.009 |
| [12] | Yu XD, Zhou HZ, Luo TH. Spatial and temporal variations in insect-infested acorn fall in a Quercus liaotungensis forest in north China. Ecological Research, 2003, 18(2): 155-164. DOI:10.1046/j.1440-1703.2003.00543.x |
| [13] | Yu XD, Zhou HZ, Luo TH, He JJ, Zhang ZB. Insect infestation and acorn fate in Quercus liaotungensis. Acta Entomologica Sinica, 2001, 44(4): 518-524. (in Chinese) 于晓东, 周红章, 罗天宏, 何君舰, 张知彬. 昆虫寄生对辽东栎种子命运的影响. 昆虫学报, 2001, 44(4): 518-524. DOI:10.3321/j.issn:0454-6296.2001.04.020 |
| [14] | Merville A, Venner S, Henri H, Vallier A, Menu F, Vavre F, Heddi A, Bel-Venner MC. Endosymbiont diversity among sibling weevil species competing for the same resource. BMC Evolutionary Biology, 2013, 13: 28. DOI:10.1186/1471-2148-13-28 |
| [15] | Toju H, Hosokawa T, Koga R, Nikoh N, Meng XY, Kimura N, Fukatsu T. "Candidatus Curculioniphilus buchneri, " a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Applied and Environmental Microbiology, 2010, 76(1): 275-282. DOI:10.1128/AEM.02154-09 |
| [16] | Toju H, Tanabe AS, Notsu Y, Sota T, Fukatsu T. Diversification of endosymbiosis: Replacements, co-speciation and promiscuity of bacteriocyte symbionts in weevils. ISME Journal, 2013, 7(7): 1378-1390. DOI:10.1038/ismej.2013.27 |
| [17] | Toju H, Fukatsu T. Diversity and infection prevalence of endosymbionts in natural populations of the chestnut weevil: Relevance of local climate and host plants. Molecular Ecology, 2011, 20(4): 853-868. DOI:10.1111/mec.2011.20.issue-4 |
| [18] | Berge O, Heulin T, Achouak W, Richard C, Bally R, Balandreau J. Rahnella aquatilis, a nitrogen-fixing enteric bacterium associated with the rhizosphere of wheat and maize. Canadian Journal of Microbiology, 1991, 37(3): 195-203. DOI:10.1139/m91-030 |
| [19] | Brady C, Denman S, Kirk S, Venter S, Rodríguez-Palenzuela P, Coutinho T. Description of Gibbsiella quercinecans gen. nov., sp. nov., associated with acute oak decline. Systematic and Applied Microbiology, 2010, 33(8): 444-450. DOI:10.1016/j.syapm.2010.08.006 |
| [20] | Bull CT, de Boer SH, Denny TP, Firrao G, Fischer-Le Saux M, Saddler GS, Scortichini M, Stead DE, Takikawa Y. List of new names of plant pathogenic bacteria (2008-2010). Journal of Plant Pathology, 2012, 94: 21-27. |
| [21] | Varsha, Mukherjee A, Bhanwar S, Ganguli A. Characterization of tannase production by Lactococcus lactis subsp. lactis and its potential in enhancing nutritional value of a composite sourdough. International Journal of Genetic Engineering and Biotechnology, 2014, 5(1): 77-84. |
| [22] | Belur PD, Mugeraya G. Microbial production of tannase: State of the art. Research Journal of Microbiology, 2011, 6(1): 25-40. DOI:10.3923/jm.2011.25.40 |
| [23] | Selwal MK, Yadav A, Selwal KK, Aggarwal NK, Gupta R, Gautam SK. Optimization of cultural conditions for tannase production by Pseudomonas aeruginosa IIIB 8914 under submerged fermentation. World Journal of Microbiology and Biotechnology, 2010, 26(4): 599-605. DOI:10.1007/s11274-009-0209-x |
| [24] | Osawa R, Mitsuoka T. Selective medium for enumeration of tannin-protein complex-degrading Streptococcus spp. in feces of koalas. Applied and Environmental Microbiology, 1990, 56(11): 3609-3611. |
| [25] | Sharma KP, John PJ. Purification and characterization of tannase and tannase gene from Enterobacter sp.. Process Biochemistry, 2011, 46(1): 240-244. DOI:10.1016/j.procbio.2010.08.016 |
| [26] | Noguchi N, Ohashi T, Shiratori T, Narui K, Hagiwara T, Ko M, Watanabe K, Miyahara T, Taira S, Moriyasu F, Sasatsu M. Association of tannase-producing Staphylococcus lugdunensis with colon cancer and characterization of a novel tannase gene. Journal of Gastroenterology, 2007, 42(5): 346-351. DOI:10.1007/s00535-007-2012-5 |
| [27] | Pepi M, Lampariello LR, Altieri R, Esposito A, Perra G, Renzi M, Lobianco A, Feola A, Gasperini S, Focardi SE. Tannic acid degradation by bacterial strains Serratia spp. and Pantoea sp. isolated from olive mill waste mixtures. International Biodeterioration and Biodegradation, 2010, 64(1): 73-80. DOI:10.1016/j.ibiod.2009.10.009 |
| [28] | Sasaki E, Shimada T, Osawa R, Nishitani Y, Spring S, Lang E. Isolation of tannin-degrading bacteria isolated from feces of the Japanese large wood mouse, Apodemus speciosus, feeding on tannin-rich acorns. Systematic and Applied Microbiology, 2005, 28(4): 358-365. DOI:10.1016/j.syapm.2005.01.005 |
| [29] | Osawa R, Fujisawa T, Pukall R. Lactobacillus apodemi sp. nov., a tannase-producing species isolated from wild mouse faeces. International Journal of Systematic & Evolutionary Microbiology, 2006, 56: 1693-1696. |
| [30] | Kohl KD, Weiss RB, Dale C, Dearing MD. Diversity and novelty of the gut microbial community of an herbivorous rodent (Neotoma bryanti). Symbiosis, 2011, 54(1): 47-54. DOI:10.1007/s13199-011-0125-3 |
| [31] | Kohl KD, Dearing MD. Experience matters: Prior exposure to plant toxins enhances diversity of gut microbes in herbivores. Ecology Letters, 2012, 15(9): 1008-1015. DOI:10.1111/j.1461-0248.2012.01822.x |
| [32] | Osawa R, Kuroiso K, Goto S, Shimizu A. Isolation of tannin-degrading Lactobacilli from humans and fermented foods. Applied and Environmental Microbiology, 2000, 66(7): 3093-3097. DOI:10.1128/AEM.66.7.3093-3097.2000 |
| [33] | Shimada T, Saitoh T, Sasaki E, Nishitani Y, Osawa R. Role of tannin-binding salivary proteins and tannase-producing bacteria in the acclimation of the Japanese wood mouse to acorn tannins. Journal of Chemical Ecology, 2006, 32(6): 1165-1180. DOI:10.1007/s10886-006-9078-z |
| [34] | Steele MA, Hadj-Chikh LZ, Hazeltine J. Caching and feeding decisions by Sciurus carolinensis: Responses to weevil-infested acorns. Journal of Mammalogy, 1996, 77(2): 305-314. DOI:10.2307/1382802 |
| [35] | Roehrig NE, Capinera JL. Behavioural and developmental responses of range caterpillar larvae, Hemileuca oliviae, to condensed tannin. Journal of Insect Physiology, 1983, 29(12): 901-906. DOI:10.1016/0022-1910(83)90053-7 |
| [36] | Zhu J, Filippich LJ, Alsalami MT. Tannic acid intoxication in sheep and mice. Research in Veterinary Science, 1992, 53(3): 280-292. DOI:10.1016/0034-5288(92)90128-O |
| [37] | Johnson WC, Thomas L, Adkisson CS. Dietary circumvention of acorn tannins by blue jays - Implications for oak demography. Oecologia, 1993, 94(2): 159-164. DOI:10.1007/BF00341312 |
| [38] | Shimada T, Nishii E, Saitoh T. Interspecific differences in tannin intakes of forest-dwelling rodents in the wild revealed by a new method using fecal proline content. Journal of Chemical Ecology, 2011, 37(12): 1277-1284. DOI:10.1007/s10886-011-0045-y |
| [39] | Onodera R, Akimoto Y, Shimada T, Saitoh T. Different population responses of three sympatric rodent species to acorn masting—the role of tannin tolerance. Population Ecology, 2017, 59(1): 29-43. DOI:10.1007/s10144-017-0570-9 |
| [40] | Zhang ZB, Wang ZY, Chang G, Yi XF, Lu JQ, Xiao ZS, Zhang HM, Cao L, Wang FS, Li HJ, Yan C. Trade-off between seed defensive traits and impacts on interaction patterns between seeds and rodents in forest ecosystems. Plant Ecology, 2016, 217(3): 253-265. DOI:10.1007/s11258-016-0566-0 |
