万琪慧1, 王书玲1, 赵伟烨1, 马黎华1, 贾仲君2, 蒋先军1


1.西南大学资源环境学院, 重庆 400715;
2.中国科学院南京土壤研究所, 江苏 南京 210008
收稿日期:2018-03-31;修回日期:2018-05-31;网络出版日期:2018-08-09
基金项目:国家自然科学基金项目(41671232);国家重点研发计划项目(2016TFD0300901)
*通信作者:蒋先军, Tel:+86-10-68251249;E-mail:jiangxj@swu.edu.cn
摘要:[目的] 系统评估全程氨氧化细菌(complete ammonia oxidizing bacteria,Comammox bacteria)、半程氨氧化细菌(AOB)和古菌(AOA)在典型水稻土剖面的垂直分异规律。2015年发现的"全程"氨氧化细菌(Comammox Nitrospira)可将氨分子一步氧化为硝酸盐,实现硝化作用。而经典的"半程"氨氧化细菌(AOB)或古菌(AOA)将氨分子氧化为亚硝酸盐后,再由系统发育完全不同的硝化细菌将其氧化为硝酸盐。全程氨氧化细菌实现了一步硝化全过程,根本改变了学术界对2类微生物分步硝化的经典认知,但相关研究仍处于初步阶段。[方法] 选择重庆北碚地区2017年典型水稻土并采集5、10、20和40 cm不同深度土壤(剖面采样点的上下误差不超过1 cm),提取水稻土总DNA后,利用标靶功能基因amoA,通过实时荧光定量PCR技术分析全程氨氧化细菌(Comammox)、半程氨氧化细菌(AOB)和古菌(AOA)在水稻土不同深度的数量变异规律。[结果] 半程氨氧化细菌AOB和古菌AOA均随土壤深度增加呈显著下降趋势。然而,全程氨氧化细菌的两大类微生物则表现出相反的规律,Comammox Clade A的丰度随着土壤剖面的加深而显著增加(P < 0.05),但Clade B并未有类似规律。Clade A在水稻土不同层次的土层中均比Clade B高出1个数量级,在5 cm和40 cm处的最低和最高值分别为3.42×107、8.46×107 copies/g。AOA与AOB的丰度大致相当,5 cm剖面处数量最高分别为1.23×107、1.83×105 copies/g,但其平均丰度远低于全程氨氧化细菌,Comammox与AOA、AOB amoA功能基因拷贝数之比为10-2000。[结论] 全程氨氧化细菌(Comammox bacteria)广泛分布于水稻土不同土层中,且数量远高于"半程"氨氧化细菌和古菌,意味着Comammox可能在水稻土硝化作用中起重要作用。
关键词:氮循环氨氧化微生物硝化作用亚硝酸细菌
Vertical abundance variations of "incomplete ammonia oxidizers" and "comammox" in purple paddy soil in Chongqing
Qihui Wan1, Shuling Wang1, Weiye Zhao1, Lihua Ma1, Zhongjun Jia2, Xianjun Jiang1


1.College of Resources and Environment, Southwest University, Chongqing 400715, China;
2.Institute of Soil Science, Chinese Academy of Sciences, Nanjing 210008, Jiangsu Province, China
Received 31 March 2018; Revised 31 May 2018; Published online 9 August 2018
*Corresponding author: Xianjun Jiang, Tel:+86-10-68251249;E-mail:jiangxj@swu.edu.cn
Supported by the National Natural Science Foundation of China (41671232) and by the National High Technology Research and Development Program of China (2016TFD0300901)
Abstract: [Objective] Nitrification has been thought to be as a two-step process, where ammonia-oxidizing bacteria and archaea (AOB and AOA) first oxidize ammonia to nitrite, which nitrite-oxidizing bacteria (NOB) subsequently transfer to nitrate. Recently, the ability to oxidize ammonia has also been discovered in members of the genus Nitrospira, which were formerly supposed to only be capable of nitrite oxidation. The discovery of these bacteria that oxidize ammonia to nitrate (complete ammonia oxidizing bacteria, Comammox bacteria), refuted the dogma that the oxidation of ammonia and nitrite requires two distinct groups of microorganisms. The discovery of one-step nitrification and Comammox Nitrospira triggered many important scientific issues of the global nitrogen cycle, but relevant studies were still in the preliminary stage. [Methods] Using real-time quantitative PCR (Q-PCR), we characterized the distribution patterns of functional amoA gene abundances of Comammox (Clade A and Clade B), ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) at 5, 10, 20, and 40 cm soil depths (up-down error within 1 cm) of flooded paddy field (FPF) in Beibei, Chongqing in 2017. [Results] Quantitative PCR (Q-PCR) revealed that soil depth had a significant effect on different nitrifiers. Comammox Nitrospira were ecologically widespread and numerically abundant in all depths of the standard profile. The Comammox Clade A amoA gene copies were higher at deeper layers, while Comammox Clade B didn't show the same trends and its abundances varied between 1.85×106 copies/g and 3.26×106 copies/g in different depths of the standard profile. However, the abundance of Comammox Clade A was about to an order of magnitude more abundant than Comammox Clade B in paddy soils. On the contrary, the amoA gene abundances of AOA and AOB significantly decreased with increasing depth (P < 0.05), both AOA and AOB amoA gene abundances were highest in the top layer (5 cm, 1.23×107 copies/g and 1.83×105 copies/g, respectively). The ratio of abundances of Comammox amoA genes to those of AOA and AOB increased significantly with the increase of soil depth, ranged from 10 to 2000. [Conclusion] The complete ammonia oxidizing bacteria (Comammox bacteria) are widely distributed in all soil depths of paddy soil and their abundances were significantly higher than "incomplete ammonia oxidizers (AOA, AOB)", which implicated that Comammox cannot be neglected in rice soil ecosystem.
Keywords: N cycleammonia-oxidizing microorganismsnitrificationnitrite-oxidizing bacteria
一个世纪以来,俄罗斯科学家Sergei Winogradsky提出的“分步硝化作用”理论奠定了硝化作用的基础:由2组不同的微生物分2步完成——首先通过氨氧化微生物AOM(氨氧化细菌AOB,氨氧化古菌AOA)将氨氧化为亚硝酸盐(NO2-),再通过亚硝酸氧化细菌(NOB)将亚硝酸盐氧化为硝酸盐(NO3-)。根据最理想动力学理论[1-2],Costa[3]等认为单步硝化比分步硝化(氨氧化和亚硝酸盐氧化)代谢途径更短,ATP合成效率更高,在热力学上由1种微生物完成将氨2步氧化为硝酸盐是可行的。直到2015年,研究者们分别在不同环境中发现了3种可纯培养的细菌(Candidatus Nitrospira nitrosa、Candidatus Nitrospira nitrificans)和(Candidatus Nitrospira inopinata)和1种未纯培养的细菌(类Nitrospira)[4-6],这些微生物都具备单独将氨氧化为硝酸盐的能力,均属于亚硝酸盐氧化细菌(NOB)中硝化螺菌属(Nitrospira),被定义为全程氨氧化微生物(complete ammonia oxidizers, comammox)。只能够将氨氧化为亚硝酸盐的AOA、AOB和只能氧化亚硝酸盐的NOB则可被称为“半程”氨氧化微生物(incomplete ammonia oxidizers)[3, 5]。****们根据已发表和更新的宏基因组数据库,进行基因序列比对后发现,全程氨氧化细菌广泛分布于农业土壤(稻田水域)[5],淡水环境、工程系统如污水处理厂(WWTPs)[4],饮用水处理厂(DWTPs)[6],水产养殖系统(RAS)[7],甚至大气颗粒物中[8]。此外,全程氨氧化微生物(Comammox)的发现,引发了许多与硝化作用相关的科学问题。例如不同生态系统中,全程氨氧化微生物(Comammox)、AOA和AOB的分布及其对硝化作用的相对贡献。
最新研究表明,相比氨氧化微生物(AOA和AOB),全程氨氧化细菌Nitrospira inopinata对氨具有更高的亲和力[9],且具有2种能在低氧浓度下高效表达的氨氧化单拷贝基因[10],在低氨和微氧环境中比AOA和AOB更具竞争优势[11-12]。由于不同深度土壤剖面层次全氮等养分、pH等指标表现出强烈的环境梯度,氧化还原电位作为水稻土剖面层次重要变化因素,亦影响硝化作用[13-15]。Wang等[16]研究了中国由北至南(海伦、常熟、鹰潭)的3种水稻土0-1 m不同土壤剖面层次微生物群落的变化,结果表明AOA和AOB amoA功能基因均随土壤层次加深显著降低。全程氨氧化细菌的发现引起硝化作用过程中,“半程”氨氧化微生物和全程氨氧化微生物在不同环境中的生态位分化,将成为最近研究的热点问题。本文选取重庆长期耕作的冬水田水稻土为研究对象,采用微生物分子生物学研究方法,旨在了解水稻土不同层次全程氨氧化微生物以及“半程”氨氧化微生物的丰度变化,重新评估Comammox(Clade A和Clade B)、AOA和AOB amoA功能基因的生态位分化及其对硝化作用的相对贡献,为更深一步理解农业生产土壤的氮循环提供前期基础。
1 材料和方法 1.1 试验地概况 试验地位于重庆市北碚区西南大学试验农场(106°26′E,30°26′N),海拔230 m,属亚热带湿润季风气候,年平均气温18.3 ℃,年平均降雨量1105 mm。土壤为中生代侏罗系沙溪庙组灰棕紫色沙泥岩母质上发育的中性紫色水稻土。该试验点自1990年开始实施长期定位试验,每个处理4个重复,小区面积为20 m2。本研究选择的试验处理取自冬水田(flooded paddy field,FPF),稻田终年处于淹水状态,保持水层深度为3 cm左右;水稻收获后将稻茬[50-60 cm,3850-4100 kg/(hm2·a)]和杂草[2750-3150 kg/(hm2·a)]翻入土中,春季翻耕种植中稻,耕作深度为25-30 cm,按传统方法每年三犁三耙翻耕种植水稻,水稻收获后灌冬水。
各小区施肥量均相同,施肥量均为:尿素273.1 kg/hm2;过磷酸钙500.3 kg/hm2;氯化钾150.1 kg/hm2。每年水稻都是过磷酸钙作底肥一次施用;尿素用量的2/3作底肥,1/3作追肥;氯化钾底肥和追肥各半。播种、施肥、水分管理一致。
1.2 土壤样品制备与培养 土壤样品采集于2017年11月,在试验农场水稻田4个小区分别取样,每个小区按五点取样法,分别用取土钻采取水稻田4个小区中5、10、20、40 cm (剖面采样点的上下误差不超过1 cm)土壤剖面层次的土样,带回实验室后去除枯枝落叶等非土壤成分,分两部分储存:一部分鲜土风干至含水量15%-20%,过2 mm筛,采取四分法均匀采集部分样品,立即进行土壤硝化势的测定和土壤DNA的提取,另一部分土样风干磨细后过1 mm及0.25 mm筛,用于测定土壤pH、有机质、全氮等基本理化性质。土壤性质见图 1。
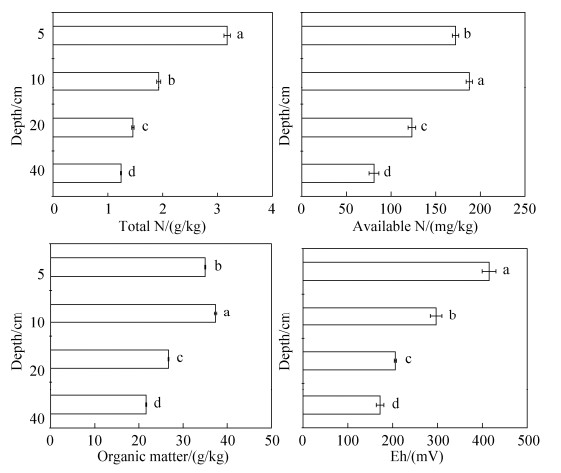 |
| 图 1 冬水田不同剖面层次土壤的基本理化性质 Figure 1 Basic physical and chemical properties at different soil layers in flooded paddy field. Values followed by different small letters within a column are significantly different at the 0.05 probability level. |
| 图选项 |
1.3 土壤基本理化性质的测定 土壤的含水量(H2O)采用烘干法进行测定;土壤pH值于1:2.5土水比条件下,采用梅特勒LE 438复合电极(梅特勒-托利多仪器有限公司,上海,中国)进行测定;有机质用K2Cr2O7-H2SO4容量法测定,全氮采用改进的凯氏法测定[17];速效磷采用碳酸氢钠-分光光度计法测定;速效钾采用醋酸铵-火焰光度计法测定[18]。所有样品均设置3次重复。土壤硝态氮和铵态氮均采用流动分析仪测定(AA3全自动连续流动分析仪,SEAL Analitical,德国)。所有样品均设置3次重复。
硝化势(potential nitrifcation rate,PNR)的测定采用好气培养法[19],稍作修改,具体如下:称取15 g鲜土置于三角瓶中,加入(NH4)2SO4调节NH4+--N的浓度到40 mg/100 g,按干土的质量计算,田间持水量调节到60%。将三角瓶放到摇床,在室温25 ℃下振荡24 h。在第2、4、22、24小时分别采取10 mL的振荡悬浊液,以1 mol/LKCl溶液振荡浸提,过滤,用AA3全自动流动分析仪(SEAL Analytical,德国)测定悬浊液中NO3--N的含量。另按上述土:水比测定原始土壤中NO3--N含量,均2次重复。以培养时间为横坐标,NO3--N含量为纵坐标,求出斜率,即土壤硝化势[mg N/(kg·d)]。
1.4 DNA的提取 采用FastDNA? SPIN Kit for Soil(MP Biomedicals)试剂盒进行提取土壤基因组总DNA,称取0.5 g于-80 ℃保存的土壤样品,按试剂盒提供的操作步骤进行。FastPrep?FP120细胞破碎仪中进行细胞破碎处理,速度6 m/s,时间40 s。提取的DNA在1%的琼脂糖凝胶中进行电泳检测,并将提取的DNA保存于-20 ℃用于后续分析。
1.5 定量PCR 实时荧光定量PCR法在QuantStudioTM6 Flex定量PCR仪(Thermo Fisher Scientific, Singapore)上测定氨氧化古菌(AOA)、氨氧化细菌(AOB)、全程氨氧化菌Comammox Clade A和Clade B amoA基因的拷贝数。AOB、AOA、Comammox Clade A和Clade B的amoA基因定量PCR所用扩增引物见表 1。AOB、AOA、Comammox Clade A和Clade B的amoA基因定量PCR所用反应体系均为20 μL,主要包含:待扩增的模板DNA 1 μL、上下引物及ROX染料各0.4 μL,Taq DNA聚合酶10 μL、灭菌水7.8 μL。AOA amoA基因定量PCR扩增条件为:95 ℃,1.5 min;40×(95 ℃,30 s;55 ℃,45 s;72 ℃,45 s)。AOB amoA基因定量PCR扩增条件为:95 ℃,3.0 min;38×(95 ℃,30 s;60 ℃,1.5 min)。Comammox Clade A、Comammox Clade B的amoA基因定量PCR扩增条件均为:95 ℃,3.0 min;45×(95 ℃,30 s;52 ℃,45 s;72 ℃,1 min)。
表 1. AOB、AOA、Comammox Clade A、Comammox Clade B amoA基因的引物序列 Table 1. Primers for AOB, AOA, Comammox Clade A, Comammox Clade B amoA gene
| Target gene | Primer sequence (5?→3?) | Reference |
| AOB amoA gene | amoA-1F (GGGGTTTCTACTGGTGGT) | [20] |
| amoA-2R(CCCCTCKGSAAAGCCTTCTTC) | ||
| AOA amoA gene | Arch-amoAF(STAATGGTCTGGCTTAGACG | [21] |
| Arch-amoAR(GCGGCCATCCATCTGTATGT) | ||
| Comammox Clade A amoA gene | comaA-244f_a TACAACTGGGTGAACTA | [22] |
| comaA-659r_a AGATCATGGTGCTATG | ||
| comaA-244f_b TATAACTGGGTGAACTA | ||
| comaA-659r_b AAATCATGGTGCTATG | ||
| comaA-244f_c TACAATTGGGTGAACTA | ||
| comaA-659r_c AGATCATGGTGCTGTG | ||
| comaA-244f_d TACAACTGGGTCAACTA | ||
| comaA-659r_d AAATCATGGTGCTGTG | ||
| comaA-244f_e TACAACTGGGTCAATTA | ||
| comaA-659r_e AGATCATCGTGCTGTG | ||
| comaA-244f_f TATAACTGGGTCAATTA | ||
| comaA-659r_f AAATCATCGTGCTGTG | ||
| Comammox Clade B amoA gene | comaB-244f_a TAYTTCTGGACGTTCTA | [22] |
| comaB-659r_a ARATCCAGACGGTGTG | ||
| comaB-244f_b TAYTTCTGGACATTCTA | ||
| comaB-659r_b ARATCCAAACGGTGTG | ||
| comaB-244f_c TACTTCTGGACTTTCTA | ||
| comaB-659r_c ARATCCAGACAGTGTG | ||
| comaB-244f_d TAYTTCTGGACGTTTTA | ||
| comaB-659r_d ARATCCAAACAGTGTG | ||
| comaB-244f_e TAYTTCTGGACATTTTA | ||
| comaB-659r_e AGATCCAGACTGTGTG | ||
| comaB-244f_f TACTTCTGGACCTTCTA | ||
| comaB-659r_f AGATCCAAACAGTGTG |
表选项
通过液体LB培养基培养的含有目的基因的克隆子,按试剂盒MiniBEST Plasmid Purification Kit说明书提取、纯化质粒后,首先对质粒进行测序验证,然后在NanoDrop?ND-1000 UV-Vis分光光度计上测定质粒的浓度,并计算出目的基因的拷贝数。最后用TE缓冲溶液将质粒连续稀释6-8个梯度以制作定量PCR的标准曲线。
1.6 数据处理与分析 数据采用Microsoft Excel 2010和SPSS 17.0进行统计分析。采用单因素(one-way ANOVA)、Duncan法进行方差分析及多重比较,显著性水平设定为α=0.05。利用Origin 8.6软件作图。图表中数据为平均值±标准差。
2 结果和分析 2.1 冬水田水稻土不同剖面层次土壤基本性质 水稻土不同剖面层次土壤的基本性质如图 1所示。由图 1可见,在水稻土不同剖面层次土壤的基本性质存在差异,所列指标表现为表层土壤指标含量高于下层土壤。土壤有机质的分解能够为硝化作用提供能量,但也可以抑制硝化作用。在本研究中,有机质、全氮、碱解氮、Eh随土壤剖面加深均呈显著下降趋势,全磷含量和有效磷含量在5 cm处,显著高于下层土壤(分别为1.22-0.43 g/kg和31.57-6.96 mg/kg)。不同剖面层次的全钾和有效钾含量具有相似规律,分别显著降低(23.5-14.0 g/kg和93.76-74.48 mg/kg)。土壤不同剖面层次的pH变化不显著,在7.7-7.9之间,土壤含水量从上至下依次为76.38%、61.67%、42.10%和62.98%。
2.2 全程氨氧化细菌(Comammox)在水稻土不同层次的分布 全程氨氧化细菌分支A(Comammox Clade A)和分支B(Comammox Clade B)丰度随土壤剖面不同层次的变化情况如图 2所示。由图 2-A可得,随土壤剖面层次加深,5、10、20及40 cm处Comammox Clade A amoA功能基因拷贝数依次显著性增加(P < 0.05),10 cm、20 cm、40 cm分别为5 cm (3.42×107 copies/g)基因拷贝数(以干土计)的1.6、1.7和2.5倍。而冬水田土壤不同剖面层次Comammox Clade B amoA功能基因拷贝数(以干土计),并没有类似规律(图 2-B),整体比Comammox Clade A低1个数量级,表现为10 cm、40 cm处(3.26×106 copies/g、2.97×106 copies/g)基因拷贝数显著高于5 cm、20 cm(1.99×106 copies/g、1.85×106 copies/g)。
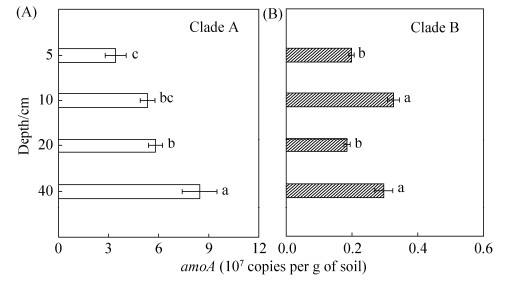 |
| 图 2 冬水田不同剖面层次的Comammox aomA功能基因拷贝数 Figure 2 The abundances of Comammox Clade A (a) and Clade B (b) amoA genes at different depths in flooded paddy field (FPF). Values followed by different small letters within a column are significantly different at the 0.05 probability level. |
| 图选项 |
2.3 氨氧化古菌(AOA)、氨氧化细菌(AOB)在水稻土不同层次的分布 由图 3-A可见,冬水田5 cm、10 cm和20 cm中AOA amoA功能基因拷贝数(以干土计)依次显著降低,分别为1.23×107 copies/g、1.19×107 copies/g和6.30×106 copies/g (P < 0.05)。40 cm处基因拷贝数又显著增加到1.16×107 copies/g。水稻土的土壤不同层次的AOB amoA功能基因拷贝数(图 3-B)显示出与AOA的变化规律类似,5 cm (1.83×105 copies/g)与10 cm (1.37×105 copies/g)无显著差异,分别为20 cm处基因拷贝数的5.6和4.2倍,40 cm处基因拷贝数又显著增加到20 cm处的3.3倍,但整体较AOA amoA功能基因拷贝数降低了2个数量级。
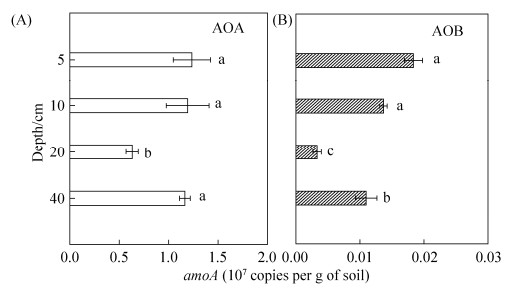 |
| 图 3 冬水田不同剖面层次AOA、AOB aomA功能基因拷贝数 Figure 3 The abundances of ammonia-oxidizing archaea AOA (A) and ammonia-oxidizing bacteria AOB (B) amoA genes at different depths in flooded paddy field (FPF). Values followed by different small letters within a column are significantly different at the 0.05 probability level. |
| 图选项 |
2.4 冬水田水稻土不同剖面层次硝化过程中硝化速率 从图 4可见,水稻土5cm的土壤剖面层次中硝化速率最高[40.70 mg N /(kg·d)],分别是水稻土10、20、40 cm处硝化速率的1.52倍、7.45倍和8.69倍,进一步表明在水稻土土壤上层剖面层次中硝化作用更强烈。而20 cm与40 cm的潜在硝化速率无显著差异,分别是5.46和4.69 mg N /(kg·d),且稻田土壤剖面层次加深后,硝化作用显著降低(P < 0.05)。这与前人报道相似[15]。
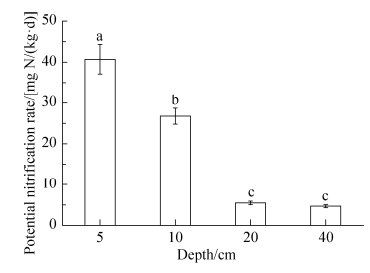 |
| 图 4 水稻土不同深度土壤潜在硝化速率的变化 Figure 4 Potential nitrification rate (PNR) at different depths in flooded paddy field (FPF). Error bars represent standard deviations, n=3. The different letters above the columns indicates a significant differences at P < 0.05. |
| 图选项 |
2.5 冬水田水稻土不同层次中土壤化学性质与硝化微生物丰度的关系 对水稻土不同土壤层次5、10、20、40 cm的Eh、有机质、全氮、碱解氮与硝化微生物丰度相关性分析结果如表 2所示。从表中可见,水稻土剖面不同层次的Eh、有机质、全氮、碱解氮与Comammox Clade A amoA基因拷贝数具有显著负相关关系(P < 0.05);与Comammox Clade B amoA基因拷贝数也呈负相关关系,但不显著。说明随着土壤剖面加深Eh显著降低后,Comammox(分支A与分支B之和)的丰度呈上升趋势,推测Comammox更适应于土壤深层低氧及贫营养条件。水稻土中,不同土壤剖面层次的Eh与AOB amoA基因拷贝数具有极显著正相关关系(P < 0.01),与AOA amoA基因拷贝数也呈正相关关系,但不显著。说明AOB相较于AOA,对氧气的响应更敏感。
表 2. 水稻土不同剖面深度土壤化学性质与硝化微生物丰度相关性 Table 2. Pearson's correlation coefficients among abundance of nitrification microorganism and soil properties in different soil depths of paddy field
| Eh | OM | TN | AN | Comammox Clade A | Comammox Clade B | AOA | AOB | |
| Eh | 1.000 | |||||||
| OM | 0.804* | 1.000 | ||||||
| TN | 0.981** | 0.714* | 1.000 | |||||
| AN | 0.788* | 0.994** | 0.699 | 1.000 | ||||
| Comammox Clade A | -0.842** | -0.769* | -0.895** | -0.779* | 1.000 | |||
| Comammox Clade B | -0.125 | 0.133 | -0.262 | 0.080 | 0.402 | 1.000 | ||
| AOA | 0.166 | 0.370 | 0.050 | 0.323 | 0.102 | 0.793** | 1.000 | |
| AOB | 0.751** | 0.595 | 0.706 | 0.523 | -0.379 | 0.371 | 0.565 | 1.000 |
| *Significant at P < 0.05, ** significant at P < 0.01. | ||||||||
表选项
3 讨论 据已发表的宏基因数据库中DNA序列比对后,研究者推测全程氨氧化微生物可能广泛分布于土壤、淡水等生态环境和工程系统中[4-6, 10]。Bartelme等[8]在再循环水产养殖系统(RAS)的生物过滤器中,检测到Comammox丰度达到108 copies/g,高于AOA丰度。本研究稻田土壤不同剖面点位5、10、20和40 cm中,全程氨氧化细菌(Comammox bacteria) amoA基因拷贝数,数量级均较高,分支A (Comammox Clade A)与分支B(Comammox Clade B) amoA基因拷贝数分别为3.42×107-8.46×107和1.85×106-3.26×106 copies/g,为全程氨氧化细菌广泛分布于农田土壤提供了可参考的数据。其次土壤剖面4个点位的Comammox amoA基因拷贝数(Clade A和Clade B之和)与AOA、AOB的比值及AOA与AOB的比值均大于3 (表 3),表明冬季休闲水稻土不同层次中的全程氨氧细菌amoA基因拷贝数最高,其次是氨氧化古菌,而氨氧化细菌最低。表明在冬季休闲水稻土的硝化微生物中,全程氨氧化细菌比AOA、AOB占有更大的比例。
表 3. 氨氧化微生物丰度比值 Table 3. The ratios of nitrifying microorganisms at different depths in flooded paddy field (FPF)
| Depth | AOA /AOB | Comammox /AOA | Comammox /AOB | Clade A /Clade B | Clade A /AOA | Clade A /AOB | Clade B /AOA | Clade B /AOB |
| 5 cm | 57.0±8.80c | 3.40±0.40c | 192±2.30c | 22.3±2.40b | 3.28±0.34c | 183±6.20d | 0.15±0.01c | 8.58±0.29c |
| 10 cm | 118±15.4b | 3.50±0.20c | 415±8.63bc | 16.3±0.50c | 3.34±0.22c | 391±17.7c | 0.21±0.02b | 24.0±0.54b |
| 20 cm | 199±18.4a | 10.2±0.30a | 2044±23.1a | 33.6±1.30a | 9.93±0.32a | 1984±7.32a | 0.30±0.01a | 59.5±3.21a |
| 40 cm | 110±12.9b | 7.50±0.70b | 816±8.34b | 28.5±2.40a | 7.23±0.67b | 788±25.2b | 0.25±0.01ab | 27.7±1.66b |
| Values followed by different small letters within a column are significantly different at the 0.05 probability level. | ||||||||
表选项
已有研究表明,AOA主要在较苛刻的环境包括低氮、强酸性和高温的环境中发挥功能活性[23-24],AOB在高氮投入的中性和碱性土壤中是硝化作用的主要驱动者。前人传统观念普遍认为相较于AOB,AOA对氨有更强的亲和力。Kit等最新研究结果[9]显示根据氨氧化动力学分析,Comammox (Nitrosopumilus inopinata)的Km(μmol/L NH3)值为0.063,显著低于AOA(Nitrososphaera, N. maritimus SCM1,Ca. N. uzonensis)的Km(0.41- 0.96 μmol/L NH3),且比AOB低了4-2500倍。这就表明氨寡营养条件下Comammox可与AOA共存,并直接竞争可用的氨,如饮用水处理系统、地下水井[5-8, 25]。此外,****们还发现相比于AOA、AOB的耐氧卡尔文循环(Calvin-Benson-Bassham)和羟基丙酸-羟基丁酸循环,Comammox Nitrospira具有微量需氧的rTCA途径[10],基因序列中有与氧化应激抵抗和氧清除相关的2/2血红蛋白Ⅱ(TrHb2),这些特征表明在微量氧气环境中,Comammox可能更具有竞争优势。Lawson等发现Comammox Nitrospira的化学计量耗氧量([mol O2: NH3] 2.0)比AOB(1.5)大,但Nitrospira仍会形成较小的典型细胞簇[26],因而推测菌落大小对KO(app)影响最强[27],也会使Comammox在氧含量不足条件下的活性污泥中具有竞争优势。研究显示滨海新区水稻土中仅表土0-4 cm可以检测到溶解氧的含量[28],深层土壤溶解氧极低,淹水水稻土中3 cm处氧浓度已降为0[29]。本研究结果显示Comammox分支A的丰度与水稻土不同层次的Eh、全氮具有极显著负相关关系(表 2),Comammox(分支A和分支B之和)与AOA、AOB的amoA gene丰度比值,分别在3.4-10.2和192-2044之间(表 3),比AOA、AOB分别高了1和3个数量级。由此推断,可能在深剖面层氮磷含量不足、微量氧浓度的营养环境中,Comammox Nitrospira单步硝化作用相对于两步进行的硝化作用将更具有竞争优势[30-31]。
此外,Pjevac[22]等在德国沃尔芬比特尔的水厂地下水井(GWW)的悬浮泥浆中检测到,Comammox Clade A和Comammox Clade B的丰度均高于104 gene copies/ng DNA,远远高于AOB amoA的基因拷贝数,这与Daims[5]的研究结果一致。而水稻土中Comammox Clade A amoA基因拷贝数高于Comammox Clade B,范围在102-103 gene copies/ng DNA,表明Comammox Clade A对于地下水井等工程系统及水稻土的硝化作用可能有重要贡献。本研究结果表明全程氨氧化细菌Clade A amoA基因丰度达107 copies/g,也就是103 gene copies/ng DNA,Clade B amoA基因丰度在102 gene copies/ng DNA,且全程氨氧化细菌分支A的amoA基因丰度随土层加深显著增加,说明中性冬季休闲水稻土中Comammox Clade A是优势种群。
水稻土壤剖面不同层次的硝化作用受氨(NH3)浓度、Eh、pH等多种因素影响。Zheng等[32]采用定量PCR技术分析白洋淀湖泊沉积物(距离水平面6 m) 0-100 cm分布的土样中,发现在溶解氧含量较低的土壤层次中,AOA丰度比AOB高,意味着环境中除氨浓度之外,溶解氧含量也是影响硝化作用[33]以及AOA、AOB丰度[34]的关键因素。Das等[35]对印度孟加拉湾北海岸的红树林森林土0-60 cm土壤剖面的微生物进行分析,也发现总微生物量、硝化细菌、固氮细菌均随土层加深而降低。本研究结果显示水稻土5 cm、10 cm、20 cm和40 cm剖面点位的土壤有机质、全氮等养分、Eh以及硝化势,均呈显著降低趋势(图 1和图 4),AOA与AOB amoA基因丰度均呈显著降低趋势,全程氨氧化细菌Clade A与AOA、AOB的amoA基因丰度比值却显著增加(3.28-9.93和183-1984),值得一提的是微生物的丰度并不代表其硝化活性,因此推测水稻土深层贫瘠营养条件下Comammox Clade A可能起作用。
Comammox的发现让人们走出传统两步硝化的“盒子思维”,但是基因丰度数值并不意味着基因全部进行了表达,RNA反转录、蛋白质等代谢水平的实验有可能更适应于评估全程氨氧化细菌的相对贡献度[36]。此外,以PCR为基础的实验,标靶功能基因amoA的选择对实验结果的影响尤为关键[37]。Orellana等[38]从美国中西部2个农业区(乌尔班纳和哈瓦那),分别在周年内(4、6、9、11月)采集了长期玉米和大豆轮作的沙土和壤土0-5 cm以及20-30 cm的土样,结果表明添加N肥后,奇古门菌Thaumarchaeota的6个深分支和3种全程氨氧化细菌(Comammox)Nitrospira的数量均增加了5倍。也就是共营养条件下,氨氧化古菌对氨浓度的响应可能更敏感。因此,未来的研究方向集中在检测Comammox Nitrospira的原位活性,能更好地确定它们在自然和工程系统中对硝化作用和其他营养循环的相对贡献。Nitrospira Ⅱ是唯一能够进行单步硝化的系统发育分支,还是具有这种能力的微生物在Nitrospira及其以外的种属中也有分布,目前仍是个谜。由于功能基因分析仅限于已知的基因,不能提供这个问题的答案,因此有必要采取新策略来评估Comammox Nitrospira对全球氮循环的实际贡献。
References
| [1] | Pfeiffer T, Bonhoeffer S. Evolution of cross-feeding in microbial populations. The American Naturalist, 2004, 163(6): E126-E135. DOI:10.1086/383593 |
| [2] | Heinrich R, Schuster S. The regulation of cellular systems. Berlin, MA: Springer, 1996. |
| [3] | Costa E, Pérez J, Kreft JU. Why is metabolic labour divided in nitrification?. Trends in Microbiology, 2006, 14(5): 213-219. DOI:10.1016/j.tim.2006.03.006 |
| [4] | van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op Den Camp HJM, Kartal B, Jetten MSM, Lücker S. Complete nitrification by a single microorganism. Nature, 2015, 528(7583): 555-559. DOI:10.1038/nature16459 |
| [5] | Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. Complete nitrification by Nitrospira bacteria. Nature, 2015, 528(7583): 504-509. DOI:10.1038/nature16461 |
| [6] | Pinto AJ, Marcus DN, Ijaz UZ, Bautista-De Lose Santos QM, Dick GJ, Raskin L. Metagenomic evidence for the presence of comammox Nitrospira-like bacteria in a drinking water system. mSphere, 2016, 1(1): e00054-15. |
| [7] | Gao JF, Fan XY, Pan KL, Li HY, Sun LX. Diversity, abundance and activity of ammonia-oxidizing microorganisms in fine particulate matter. Scientific Reports, 2016, 6: 38785. DOI:10.1038/srep38785 |
| [8] | Bartelme RP, McLellan SL, Newton RJ. Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing Archaea and comammox Nitrospira. Frontiers in Microbiology, 2017, 8: 101. |
| [9] | Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, Daebeler A, Romano S, Albertsen M, Stein LY, Daims H, Wagner M. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature, 2017, 549(7671): 269-272. DOI:10.1038/nature23679 |
| [10] | Palomo A, Pedersen AG, Fowler SJ, Dechesne A, Sicheritz-Pontén T, Smets BF. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. The ISME Journal, 2018, 12(7): 1779-1793. DOI:10.1038/s41396-018-0083-3 |
| [11] | Ouellet H, Ranguelova K, LaBarre M, Wittenberg JB, Wittenberg BA, Magliozzo RS, Guertin M. Reaction of Mycobacterium tuberculosis truncated hemoglobin O with hydrogen peroxide: evidence for peroxidatic activity and formation of protein-based radicals. Journal of Biological Chemistry, 2007, 282(10): 7491-7503. DOI:10.1074/jbc.M609155200 |
| [12] | Torge R, Comandini A, Catacchio B, Bonamore A, Botta B, Boffi A. Peroxidase-like activity of Thermobifida fusca hemoglobin: the oxidation of dibenzylbutanolide. Journal of Molecular Catalysis B: Enzymatic, 2009, 61(3/4): 303-308. |
| [13] | H?fferle ?, Nicol GW, Pal L, Hacin J, Prosser JI, Mandi?-Mulec I. Ammonium supply rate influences archaeal and bacterial ammonia oxidizers in a wetland soil vertical profile. FEMS Microbiology Ecology, 2010, 74(2): 302-315. DOI:10.1111/fem.2010.74.issue-2 |
| [14] | Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature, 2006, 442(7104): 806-809. DOI:10.1038/nature04983 |
| [15] | Qin HL, Zhang ZX, Lu J, Zhu YJ, Webster R, Liu XL, Yuan HZ, Hou HJ, Chen CL, Wei WX. Change from paddy rice to vegetable growing changes nitrogen-cycling microbial communities and their variation with depth in the soil. European Journal of Soil Science, 2016, 67(5): 650-658. DOI:10.1111/ejss.2016.67.issue-5 |
| [16] | Wang HH, Li X, Li X, Li XY, Wang J, Zhang HW. Changes of microbial population and N-cycling function genes with depth in three Chinese paddy soils. PLoS One, 2017, 12(12): e0189506. DOI:10.1371/journal.pone.0189506 |
| [17] | 杨剑虹, 王成林, 代亨林. 土壤农化分析与环境监测. 北京: 中国大地出版社, 2008. |
| [18] | 鲁如坤. 土壤农业化学分析方法. 北京: 中国农业科技出版社, 2000. |
| [19] | Chu HY, Fujii T, Morimoto S, Lin XG, Yagi K, Hu JL, Zhang JB. Community structure of ammonia-oxidizing bacteria under long-term application of mineral fertilizer and organic manure in a sandy loam soil. Applied and Environmental Microbiology, 2007, 73(2): 485-491. DOI:10.1128/AEM.01536-06 |
| [20] | Beman JM, Francis CA. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahía del Tóbari, Mexico. Applied and Environmental Microbiology, 2006, 72(12): 7767-7777. DOI:10.1128/AEM.00946-06 |
| [21] | Rotthauwe JH, Witzel KP, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Applied and Environmental Microbiology, 1997, 63(12): 4704-4712. |
| [22] | Pjevac P, Schauberger C, Poghosyan L, Herbold CW, van Kessel MAHJ, Daebeler A, Steinberger M, Jetten MSM, Lücker S, Wagner M, Daims H. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. Frontiers in Microbiology, 2017, 8: 1508. DOI:10.3389/fmicb.2017.01508 |
| [23] | Zhang LM, Hu HW, Shen JP, He JZ. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. The ISME Journal: Multidisciplinary Journal of Microbial Ecology, 2012, 6(5): 1032-1045. |
| [24] | Xia WW, Zhang CX, Zeng XW, Feng YZ, Weng JH, Lin XG, Zhu JG, Xiong ZQ, Xu J, Cai ZC, Jia ZJ. Autotrophic growth of nitrifying community in an agricultural soil. The ISME Journal, 2011, 5(7): 1226-1236. DOI:10.1038/ismej.2011.5 |
| [25] | Gonzalez-Martinez A, Rodriguez-Sanchez A, van Loosdrecht MCM, Gonzalez-Lopez J, Vahala R. Detection of comammox bacteria in full-scale wastewater treatment bioreactors using tag-454-pyrosequencing. Environmental Science and Pollution Research, 2016, 23(24): 25501-25511. DOI:10.1007/s11356-016-7914-4 |
| [26] | Lawson CE, Lücker S. Complete ammonia oxidation: an important control on nitrification in engineered ecosystems?. Current Opinion in Biotechnology, 2018, 50: 158-165. DOI:10.1016/j.copbio.2018.01.015 |
| [27] | Picioreanu C, Pérez J, van Loosdrecht MCM. Impact of cell cluster size on apparent half-saturation coefficients for oxygen in nitrifying sludge and biofilms. Water Research, 2016, 106: 371-382. DOI:10.1016/j.watres.2016.10.017 |
| [28] | Bai R, Xi D, He JZ, Hu HW, Fang YT, Zhang LM. Activity, abundance and community structure of anammox bacteria along depth profiles in three different paddy soils. Soil Biology and Biochemistry, 2015, 91: 212-221. DOI:10.1016/j.soilbio.2015.08.040 |
| [29] | Lüdemann H, Arth I, Liesack W. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Applied and Environmental Microbiology, 2000, 66(2): 754-762. DOI:10.1128/AEM.66.2.754-762.2000 |
| [30] | Kageyama H, Tripathi K, Rai AK, Cha-um S, Waditee-Sirisattha R, Takabe T. An alkaline phosphatase/phosphodiesterase, PhoD, induced by salt stress and secreted out of the cells of Aphanothece halophytica, a halotolerant cyanobacterium. Applied and Environmental Microbiology, 2011, 77(15): 5178-5183. DOI:10.1128/AEM.00667-11 |
| [31] | Shen YC, Hu YN, Shaw GC. Expressions of alkaline phosphatase genes during phosphate starvation are under positive influences of multiple cell wall hydrolase genes in Bacillus subtilis. The Journal of General and Applied Microbiology, 2016, 62(2): 106-109. DOI:10.2323/jgam.62.106 |
| [32] | Zheng L, Zhao X, Zhu GB, Yang W, Xia C, Xu T. Occurrence and abundance of ammonia-oxidizing archaea and bacteria from the surface to below the water table, in deep soil, and their contributions to nitrification. Microbiology Open, 2017, 6(4): e00488. DOI:10.1002/mbo3.2017.6.issue-4 |
| [33] | Dong YM, Zhang ZJ, Jin YW, Li ZR, Lu J. Nitrification performance of nitrifying bacteria immobilized in waterborne polyurethane at low ammonia nitrogen concentrations. Journal of Environmental Sciences, 2011, 23(3): 366-371. DOI:10.1016/S1001-0742(10)60418-4 |
| [34] | Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiology Reviews, 2009, 33(5): 855-869. DOI:10.1111/j.1574-6976.2009.00179.x |
| [35] | Das S, Ganguly D, Maiti TK, Mukherjee A, Jana TK, De TK. A depth wise diversity of free living N2 fixing and nitrifying bacteria and its seasonal variation with nitrogen containing nutrients in the mangrove sediments of Sundarban, WB, India. Open Journal of Marine Science, 2013, 3(2): 112-119. DOI:10.4236/ojms.2013.32012 |
| [36] | Liu S, Shen LD, Lou LP, Tian GM, Zheng P, Hu BL. Spatial distribution and factors shaping the niche segregation of ammonia-oxidizing microorganisms in the Qiantang River, China. Applied and Environmental Microbiology, 2013, 79(13): 4065-4071. DOI:10.1128/AEM.00543-13 |
| [37] | Jin T, Zhang T, Ye L, Lee OO, Wong YH, Qian PY. Diversity and quantity of ammonia-oxidizing Archaea and Bacteria in sediment of the Pearl River Estuary, China. Applied Microbiology and Biotechnology, 2011, 90(3): 1137-1145. DOI:10.1007/s00253-011-3107-8 |
| [38] | Orellana LH, Chee-Sanford JC, Sanford RA, L?ffler FE, Konstantinidis KT. Year-round shotgun metagenomes reveal stable microbial communities in agricultural soils and novel ammonia oxidizers responding to fertilization. Applied and Environmental Microbiology, 2018, 84(2): e01646-17. |
