田克俭1, 孟繁星1, 霍洪亮1,2


1.东北师范大学环境学院, 吉林 长春 130117;
2.吉林省水污染控制与资源化工程实验室, 吉林 长春 130117
收稿日期:2018-05-14;修回日期:2018-08-26;网络出版日期:2018-11-28
基金项目:国家自然科学基金(51478096);吉林省自然科学基金(20180101083JC)
*通信作者:霍洪亮,Tel:+86-431-89165602;E-mail:huohl@nenu.edu.cn
摘要:雌激素作为环境内分泌干扰物的一类重要物质,生物降解法是最为经济、最为绿色和最为适用的去除方法。通过分析雌激素的主要来源和危害、国内外雌激素降解菌的筛选与鉴定、类固醇雌激素降解酶的表达与检测、雌激素降解菌的基因组学特征以及雌激素的降解途径和机制,进一步阐述了雌激素生物降解的研究现状与进展,对未来的研究工作作出了展望。
关键词:雌激素降解菌降解酶生物降解机制
Microbial degradation of environmental estrogens
Kejian Tian1, Fanxing Meng1, Hongliang Huo1,2


1.School of Environment, Northeast Normal University, Changchun 130117, Jilin Province, China;
2.Engineering Laboratory for Water Pollution Control and Resources Recovery, Changchun 130117, Jilin Province, China
Received 14 May 2018; Revised 26 August 2018; Published online 28 November 2018
*Corresponding author: Hongliang Huo, Tel:+86-431-89165602; E-mail: huohl@nenu.edu.cn
Supported by the National Natural Science Foundation of China (51478096) and by the Natural Science Foundation of Jilin Province of China (20180101083JC)
Abstract: Estrogens are major substances of environmental endocrine disruptors, and biodegradation is the most economical, greenest and most suitable method to remove them. We elaborated the research status and progress of estrogen biodegradation by analyzing the major sources and hazards of estrogen, the isolation and identification of estrogen-degrading bacteria, the expression and detection of steroidal estrogen-degrading enzymes, the genomics study of estrogen-degrading bacteria and the estrogen degradation pathways. We also address future works of biodegradation of environmental estrogens.
Keywords: estrogendegrading bacteriadegrading enzymedegradation mechanism
环境雌激素污染已经成为全球性问题,严重影响人类健康和生态平衡,治理环境雌激素污染成为人们关注热点。环境雌激素广泛存在于人类和动物排泄物、植物性化合物以及塑料制品和残留农药中,其中人类和各种大型家畜排泄物中类固醇雌激素是城镇和农村地表水重要污染源。主要的环境雌激素有雌酮(Estrone,E1)、17β-雌二醇(17β-estradiol,E2)、雌三醇(Estriol,E3)和孕酮等。另外,人工合成雌激素17α-乙炔基雌二醇(17α-ethynyl estradiol,EE2)也是主要环境雌激素污染物之一。它们均具有脂溶性和很强的生物活性,与其他内分泌干扰素(EDCs)相比较有更强的内分泌干扰作用,对人类健康和生态环境的影响最为显著,在极低的浓度下(1.0 ng/L)就会对生物体产生明显的影响,有严重的致畸致癌作用,能诱发雄性个体雌性化,引发女性肿瘤、癌症、男性不育和儿童性早熟等。因此,世界卫生组织已将雌激素列为一类致癌物和重要环境污染物。目前,去除污水中环境雌激素的方法主要采用微生物降解法。筛选强化高效雌激素降解菌、建立活性污泥降解菌菌群优势生态位和调控菌体雌激素降解酶表达是污水处理场降解环境雌激素的关键性工作。本文将主要总结近年来开展环境雌激素主要降解菌、类固醇雌激素降解酶以及雌激素微生物代谢途径等相关研究工作,为今后在污水处理中构建和利用工程微生物以及开展相关研究提供理论依据。
1 雌激素降解菌 雌激素的微生物降解主要是通过细菌以及少部分真菌和藻类植物完成,降解菌大多是在污水处理厂活性污泥中分离或是根据雌激素来源在农田土壤或堆肥中分离。目前,被报道的雌激素降解菌已经有80余株,它们在一定时间范围内对雌激素的降解效率都能达到90%以上。在这其中,变形菌门最多,有44株,其次是放线菌门有16株,厚壁菌门12株。其他菌门菌株数量较少,如拟杆菌门只有3株。
在变形菌门,对高浓度雌激素降解率大多高于95%。α-变形菌纲有6株,主要以鞘氨醇单胞菌属为主。如鞘氨醇单胞菌(Sphingomonas sp.)ED8菌株,对E2降解率达到100%[1];γ-变形菌纲有4株,主要以假单胞菌属为主,如铜绿假单胞菌(Pseudomonas aeruginosa.)BP3菌株,对浓度为3.6 mg/L的E3在24 h降解率达到99%[2];β-变形菌纲有3株,其中2株是皮氏罗尔斯顿菌属,如皮氏罗尔斯顿菌(Ralstonia pickettii) BP2雌激素降解率达到100%。在放线菌门,菌株对高浓度雌激素具有更好的底物适应性,菌体生长无延迟。同时,雌激素降解效率和降解率相对较高。目前在已经筛选鉴定的16株放线菌中,高效降解菌有5株,均属于红球菌属(Rhodococcus sp.),并且对雌激素E1、E2、E3和EE2的降解率全部都大于95%,例如马红球菌(Rhodococcus equi) Y50156对浓度为100 mg/L的上述4种主要雌激素降解率在24 h内均大于95%[3]。因此,红球菌可能是构建雌激素降解工程菌的优势备选菌株之一。
除以上菌株外,表 1列出23种根据雌激素的初始浓度、培养时间以及降解率等因素筛选出的雌激素高效降解菌,另有60余种未列出。
表 1. 已分离的雌激素降解菌 Table 1. Isolated estrogen-degrading bacteria
| Phylogenetic affiliation | Name | Degradation ability and mechanism | Concentration | Degradation rate/% | Time/h | References |
| Alpha-proteobacteria | Sphingomonas sp. ED8 | E1, E2 | E2: 0.8 mg/4 mL | 100.0 | 120 | [1] |
| E1: 0.8 mg/4 mL | 90.0 | 120 | ||||
| Sphingomonas sp. KC8 | E1, E2 | E1: product | 100.0 | 72 | [10] | |
| E2: 3 mg/L | 100.0 | 24 | ||||
| Sphingomonas sp. CYH | E1, E2 | E2: 500 μg/L | 100.0 | 48 | [11] | |
| E1: 500 μg/L | 100.0 | 48 | ||||
| Novosphingobium tardaugens ARI-1 | E1, E2, E3 | E3: 10 mg/30 mL | 100.0 | 240 | [12] | |
| Phyllobacterium myrsinacearum BP1 | E1, E2, E3, co-metabolism EE2 | E1: 3.2 mg/L | 99.0 | 24 | [2] | |
| Brevundimonas diminuta I | E1, E2, EE2 | E2: 3.5 mg/L | 100.0 | 24 | [13] | |
| E1: 2 mg/L | 95.0 | 96 | ||||
| EE2: 3 mg/L | 99.0 | 360 | ||||
| Beta-proteobacteria | Ralstonia pickettii BP2 | E1, E2, E3, co-metabolism EE2 | E2: 2.3 mg/L | 100.0 | 48 | [2] |
| Ralstonia sp. picketii | E1, E2 | 100.0 | 72 | [14] | ||
| Achromobacter xylosoxidans | E1, E2 | E2: 1.5 mg/L | 100.0 | 3 | ||
| Gamma-proteobacteria | Pantoea agglomerans ES1 | EE2 | EE2: 10 mg/L | 97.0 | 288 | [15] |
| Pseudomonas citronellolis SS-2 | E1, E2, EE2 | E1: 2 mg/L | 99.0 | 36 | [16] | |
| E2: 2 mg/L | 99.0 | 36 | ||||
| EE2: 4 mg/L | 93.6 | 168 | ||||
| Phyllobacterium myrsinacearum BP3 | E1, E2, E3, co-metabolism EE2 | E1: 3.6 mg/L | 99.0 | 24 | [2] | |
| Pseudomonas aeruginosa TJ1 | E2 | E2: 5-15 μg/L | 100.0 | 2 | [17] | |
| Firmicutes | Virgibacillus halotolerans LF1 | E1, E2 | E2: 5 mg/L | 100.0 | 504 | [18] |
| E1: product | 100.0 | 768 | ||||
| Bacillus flexus LF3 | E1, E2 | E2: 5 mg/L | 98.0 | 768 | ||
| Bacillus sp. E2Y1 | E1, E2 | E2: 1 mg/L | 100.0 | 144 | [19] | |
| Bacillus sp. E2Y2 | E2 | E2: 1 mg/L | 100.0 | 96 | ||
| Bacillus sp. E2Y4 | E1, E2 | E2: 1 mg/L | 100.0 | 144 | ||
| Actinobacteria | Rhodococcus equi Y50155 | E1, E2, E3, EE2 | E2: 100 mg/L | 99.0 | 24 | [3] |
| E1: 100 mg/L | 99.0 | 24 | ||||
| E3: 100 mg/L | 72.0 | 24 | ||||
| EE2: 100 mg/L | 80.0 | 24 | ||||
| Rhodococcus equi Y50156 | E1, E2, E3, EE2 | E1: 100 mg/L | 99.0 | 24 | ||
| E2: 100 mg/L | 99.0 | 24 | ||||
| E3: 100 mg/L | 95.0 | 24 | ||||
| EE2: 100 mg/L | 96.0 | 24 | ||||
| Rhodococcus zopfii Y50158 | E1, E2, E3, EE2 | E1: 100 mg/L | 100.0 | 24 | ||
| E2: 100 mg/L | 100.0 | 24 | ||||
| E3: 100 mg/L | 100.0 | 24 | ||||
| EE2: 100 mg/L | 100.0 | 24 | ||||
| Rhodococcus rubber KC4 | E2 | E2: 3 mg/L | 99.0 | 24 | [4] | |
| Rhodococcus equi DSSKP-R-001 | E1, E2, E3, EE2 | E1: 30 mg/L | 100.0 | 96 | This study | |
| E2: 30 mg/L | 100.0 | 96 | ||||
| E2: 30 mg/L | 90.0 | 96 |
表选项
2 雌激素降解酶 微生物降解雌激素主要是通过酶促反应来完成。Chen等[4]对鞘氨醇单胞菌KC8降解酶研究表明,由OecA编码的3β, 17β-羟基类固醇脱氢酶、OecB编码的雌酮-4-羟化酶、OecC编码的4-羟基雌酮4, 5-双加氧酶是菌株雌激素降解的关键酶。Khunjar等[5]研究表明,欧洲亚硝化单胞菌(Nitrosomonas europaea)在降解EE2时,氨单加氧酶发挥重要作用,并且可以与异养菌产生协同作用。通过对不动杆菌(Acinetobacter sp.) Sphe3和AGAT-W研究发现,双加氧酶和邻苯二酚双加氧酶也是降解类固醇雌激素的重要酶系,它们分别负责苯环氧分子的催化反应以及芳香环的彻底开环裂解,是多环芳烃化合物降解起始和彻底代谢的关键酶[6-7]。王平等[8]在以雌二醇为唯一碳源的不动杆菌DSSKY-A-001培养中,双加氧酶和邻苯二酚1, 2双加氧酶得到表达,并推测可能参与雌二醇的生物降解过程。
****对放线菌门雌激素降解酶的研究更为详细。以红球菌属为例,已知参与类固醇雌激素分解代谢的酶共有60余种,主要分为脱氢酶、羟化酶/单加氧酶、双加氧酶、水解酶、水合酶以及其他辅酶等。前三种酶在类固醇雌激素分解代谢过程中起主要作用。脱氢酶以3-类固醇-Δ1-脱氢酶为代表的类固醇脱氢酶(KSTD)研究最为深入。Geize等[9]通过活性染色的方法还发现,在红平红球菌(Rhodococcus erythropolis) SQ1中,存在2种KSTD,分别是KSTD1和KSTD2。两种酶均在4-雄甾烯-3, 17-二酮(AD)和9α-羟基-4-雄甾烯-3, 17-二酮(9OHAD)的降解过程中起作用。通过基因敲除进一步证明,其中KSTD1失活,该菌株仍能使类固醇底物A环发生C1、2位脱氢反应[20],说明KSTD1同工酶在反应过程中还起到一定作用。羟化酶中3-甾酮-9α-羟基化酶(KSH)也是广泛存在于甾醇化合物降解菌中的一种双组分铁含硫单加氧酶,由终端氧化酶(kshA)和还原酶两部分(kshB)组成[21],能在多元环的9位加入一个羟基(9α-OH),并且与KSTD一同参与类固醇母核B环的碳骨架断裂[22-23]。2002年,Geize等[24]首先对红平红球菌SQ1的3-甾酮-9α-羟基化酶编码基因kshA和kshB进行了分子鉴定。2009年,Petrusma等[25]在大肠杆菌中表达了来自紫红红球菌(Rhodococcus rhodochrous) DSM 43269的kshA和kshB基因,并证明kshB将来自还原型辅酶Ⅰ (NADH)的还原力传递给kshA,后者催化底物的羟基化反应。2011年,Petrusma等[26]分析了存在于紫红红球菌DSM43269基因组中的5个kshA基因产物的酶学特性,发现kshA1到kshA5分属于4个不同的基因簇,对于类固醇底物选择有很大的重叠性,其中kshA1仅对胆酸具有一定的分解代谢能力,而KshA5的多功能性与KshA1形成鲜明对比,底物特异性较为广泛没有明显的底物偏好。红球菌属其他菌株的典型类固醇雌激素降解的主要酶见表 2。
表 2. 红球菌属雌激素降解相关酶 Table 2. Estrogen degradation related enzymes of Rhodococcus sp.
| Categories | Abbreviation | Encoding gene | Name | References |
| Dehydrogenase | HSD | hsd | 3-hydroxysteroid dehydrogenase/5-4 isomerase | [27] |
| hsd4A | 17-hydroxysteroid dehydrogenase | [28] | ||
| 3α-hydroxysteroid dehydrogenase | [29] | |||
| KSTD | ksdD | 3-indolone Δ1-dehydrogenase | [28] | |
| kstD1 | 3-indolone Δ1-dehydrogenase | |||
| kstD2 | 3-indolone Δ1-dehydrogenase | [30] | ||
| kstD3 | 3-keto-5-steroidal Δ1-dehydrogenase | |||
| Hydroxylase/Monooxygenase | KSH | kshA | 3-ketosteroid 9α-hydroxylase oxygenase component | |
| kshB | 3-iketosteroid 9α-hydroxylase | [31] | ||
| Hsa | hsaA | Flavin-dependent monooxygenase | [31-32] | |
| hsaB | Flavin-dependent monooxygenase | [28, 31] | ||
| Cyp | Cyp 450 | Cytochrome P450 | [33] | |
| cyp125 | Member of the cholesterol catabolic gene cluster | [34] | ||
| Dioxygenase | hsaC | 2, 3-dihydroxybiphenyl 1, 2-dioxygenase | [28] |
表选项
3 雌激素降解菌基因组 基因组学在污染物降解菌的研究中的作用日渐重要,对菌株降解能力以及底物偏好作出了更合理的判断和预测。雌激素降解菌的基因组学研究尚处于起步阶段,目前已有部分类固醇降解菌进行了全基因组测序。例如,α-变形菌纲的鞘氨醇单胞菌KC8、新鞘氨醇杆菌Chol 11以和交替赤杆菌MH-B5,β-变形菌纲的睾丸酮丛毛单胞菌ATCC11996和伯克氏菌CQ001,γ-变形菌纲的恶臭假单胞菌SJTE-1、香茅醇假单胞菌SJTE-3和不动杆菌DSSKY-A-001,放线菌门的红球菌DSSKP-R-001、红球菌P14和红球菌RHA1等。其中红球菌RHA1基因组最大,为9.7 Mb;伯克氏菌CQ001次之,为7.27 Mb;不动杆菌DSSKY-A-001最小,仅为3.13 Mb。大多数菌株基因组大小在5 Mb左右。红球菌RHA1和新鞘氨醇杆菌Chol 11除染色体外还包含有3个质粒,其他菌株质粒数量较少或没有。除不动杆菌DSSKY-A-001之外,其他菌株均有较高的GC含量,最高为红球菌P14达到70.42%。部分类固醇激素降解菌的基因组特征见表 3。
表 3. 雌激素降解菌基因组特征 Table 3. Genomic characteristics of estrogen-degrading bacteria
| Name | Total sequence length/bp | Contig N50/bp | GC content/% | Total number of Chr and Pla | CDS | Coding gene in COG | Coding gene in KEGG | References |
| Pseudomonas putida SJTE-1 | 5551505 | 79005 | 62.25 | 1 | 4915 | 4408 | - | [35] |
| Sphingomonas sp. KC8 | 4074265 | 142404 | 63.70 | 1 | 3950 | 3531 | - | [36] |
| Altererythrobacter MH-B5 | 3668441 | 339753 | 60.00 | 1 | 3475 | - | - | [37] |
| Pseudomonas citronellolis SJTE-3 | 7309421Chr | - | 67.04Chr | 2 | 6756 | - | - | [38] |
| 370338Pla | - | 56.57Pla | - | - | ||||
| Acinetobacter DSSKY-A-001 | 3132860 | 18130 | 41.64 | 1 | 2963 | 2174 | 1770 | [18] |
| Rhodococcus DSSKP-R-001 | 5438826 | 5252360 | 68.72 | 3 | 5180 | 3736 | 2590 | This study |
| Rhodococcus sp. P14 | 5669990 | 23966 | 70.42 | 1 | 5501 | - | - | [39] |
| Rhodococcus sp. RHA1 | 9702737 | - | 67.00 | 4 | 9145 | - | - | [40] |
| Comamonas testosteroni ATCC 11996 | 5415699 | 250929 | 61.48 | 1 | 4985 | - | - | [41] |
| Novosphingobium sp. Chol11 | 3660000 | - | 62.44 | 4 | 3532 | - | - | [42] |
| Burkholderia sp. CQ001 | 7570308 | 82290 | 66.90 | 1 | 8632 | 6487 | 6931 | [43] |
| Chr: Chromosome; Pla: Plasmid. | ||||||||
表选项
红球菌属对类固醇化合物表现出很强的降解能力因而研究较为深入。例如,红球菌DSSKP-R-001核苷酸序列全长为5438826 bp,基因全长为4917591 bp,占总核苷酸全长的90.42%。通过数据库注释,得到编码蛋白基因3736个,参与代谢通路基因2590个,描述功能基因3472个以及物种分类基因4292个。虽然该菌株基因组不大,但包含的功能基因却非常丰富。又如伯克氏菌CQ001也具有较强的类固醇降解功能[43],其基因组测序结果表明,基因全长为7.57 Mb。70.1%的基因可以在COG数据库注释到。其中,氨基酸转运和代谢的基因741个,脂质运输和新陈代谢的基因308个,碳水化合物的运输和新陈代谢的基因395个,次生代谢生物合成、转运和分解代谢的基因260个。在KEGG数据库注释发现80.3%的基因参与了8种类固醇化合物代谢途径。其中,碳水化合物代谢基因268个,氨基酸代谢基因253个,外源性化学物质生物降解和新陈代谢基因165个,脂质代谢基因90个等。
对于筛选的大量污染物降解菌,全基因组测序工作尚有很大的开展空间。基因组学、转录组学和蛋白质组学三者的相互融合也是未来污染物降解菌研究的重要方向。
4 雌激素降解途径 类固醇雌激素是一类环戊烷多氢菲衍生物,具有相同的母核结构,一般能降解环戊烷多氢菲母核的菌株对E1、E2和E3均有降解能力,其降解方式也很相似。EE2是含有双苯环芳香族化合物,是一类难降解物质,能够单独降解EE2的微生物只有少量报道。但是,当以E1、E2或者E3为碳源与EE2进行共代谢时,EE2则能够被较高程度地降解。另外,E2与EE2的比值对EE2本身的降解起重要作用,E2浓度越高EE2的降解效率也越高[2]。E2和E3一般在脱氢酶的作用下首先代谢为E1,之后再进一步降解。E3也可在脱氢酶作用下转化为E2之后再进行降解。
由于E2是活性最高的天然雌激素,对E2生物降解的研究相对广泛而深入,目前已发现E2有多个不同的降解途径,根据E2降解菌种属不同,本文归纳为变形菌门途径、放线菌门途径和其他菌门途径。
变形菌门的E2降解菌主要有鞘氨醇单胞菌、欧洲亚硝化单胞菌、不动杆菌、木糖氧化无色杆菌和罗尔斯通氏菌,各菌降解E2主要通过A-D四种路径完成,如图 1所示。鞘氨醇单胞菌ED8降解E2路径用A代表。主要通过加氧酶以及元劈裂作用从B环开始逐步裂解并进行进一步代谢[1],该途径的中间及末端产物尚未完全研究清楚。此外,菌株ED8的双加氧酶还能通过途径D1将E2降解为4-OH-E1,但进一步降解产物未见报道。B路径是欧洲亚硝化单胞菌将E2的C-17位置脱水生成雌甾四烯(Estratetraenol,E0)[44],并沿着这一路径继续降解,这一过程主要是脱氢酶的作用。C路径是木糖氧化无色杆菌(Achromobacter xylosoxidans)和罗尔斯通氏菌的混合菌液通过单加氧酶把E2降解为16α-OH-E1[8],之后进入未知降解途径。D路径是不动杆菌(Acinetobacter sp.) DSSKY-A-001通过脱氢酶作用把E2降解为E1,之后在加氧酶作用下在C4位置首先裂解形成代谢下游产物,之后经过一系列的未知代谢途径[18]。
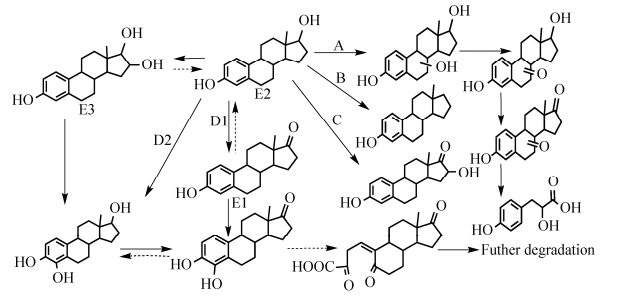 |
| 图 1 变形菌门5种代表菌株对E2的生物降解途径 Figure 1 Biodegradation pathway of E2 by five representative strains of Proteobacteria. |
| 图选项 |
放线菌门降解E2途径主要是红球菌属研究的比较详细,均由菌株将E2首先降解成E1,之后再降解为4-羟基雌酮,然后主要通过E-G三种路径完成最终降解,如图 2所示。E路径是马红球菌DSSKP-R-001在单加氧酶作用下通过在C-16位置上经单加氧酶羟基化生成4, 16-二羟基-雌酮,再经过一系列降解过程,最终进入三羧酸循环。此途径是目前已知雌激素降解菌降解E1最为高效的途径。F路径是在加氧酶作用下通过在C-4和C-5之间加入1分子氧,之后C-5位置被氧化成羧基,通过一系列氧化还原反应以及裂解作用最终进入三羧酸循环。G路径是红球菌DS201在C-4和C-5之间裂解,使4-羟基雌酮形成一种中间产物,并经过进一步裂解形成2-乙基-3-羟基-6-甲基环己烷-1-羧酸,之后再进入未知的裂解途径[45],在这一过程中双加氧酶可能发挥重要作用。E2其他降解途径如图 3所示,主要是在活性污泥菌群的作用下先将E2降解为E1,之后E1的D环再通过加氧酶作用裂解形成一种不稳定的内脂化合物,最终进入三羧酸循环[46]。
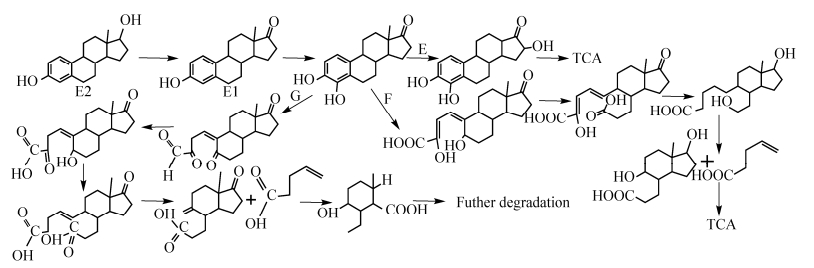 |
| 图 2 放线菌门2种代表菌株对E2的生物降解途径 Figure 2 Biodegradation pathway of E2 by two representative strains of actinomycetes. |
| 图选项 |
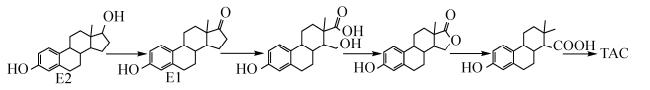 |
| 图 3 活性污泥代表性菌群对E2的生物降解途径 Figure 3 Biodegradation pathway of E2 by the representative flora of activated sludge. |
| 图选项 |
人工合成雌激素EE2具有很高的雌激素活性,在环境中不易降解。根据目前研究结果,EE2的生物降解可分为微藻途径和变形菌门途径。微藻途经主要有四尾栅藻、纤维藻和羊角月牙藻3个路径,用H-J表示,如图 4所示。H路径是EE2在四尾栅藻加氧酶作用下,通过羟基化和糖基化将3-羟基转化为3-酮,从而将EE2转化为新的代谢产物。I路径是EE2在纤维藻作用下引起C-6羟基化进行降解,之后进入未知代谢途径,该路径过程中EE2生物降解产物最为复杂。J路径是羊角月牙藻在EE2的C-2和C-6位置进行羟基化,在C-3位置糖基化形成共轭EE2[47]。变形菌门途径主要由亚消化单胞菌、亚硝化球菌、亚硝化螺菌组成的菌群和鞘氨醇杆菌参与的2个路径组成,分别用K和L表示,如图 5所示。K路径是在菌群反应器中通过双加氧酶首先在A环C-2位置发生羟化形成2-OH-EE2,之后使A环裂解形成产物ETDC[48]。L路径是鞘氨醇杆菌JCR5将EE2降解为E1之后在双加氧酶作用下使E1的B环发生羟化和酮化,将B环裂解后在A环羟化形成3, 4-邻苯二酚,之后经过水解最终形成水和二氧化碳[49]。
 |
| 图 4 3种典型微藻对EE2的生物降解途径 Figure 4 Biodegradation pathway of EE2 by three typical microalgae. |
| 图选项 |
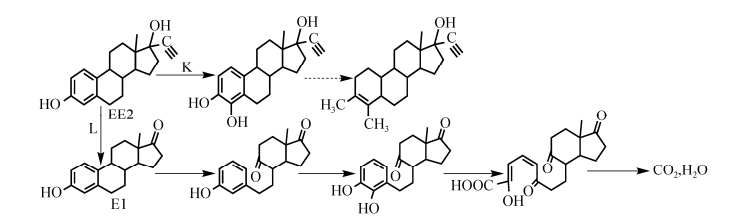 |
| 图 5 变形菌门4种代表性菌株对EE2的生物降解途径 Figure 5 Biodegradation pathway of EE2 by four representative strains of Proteobacter. |
| 图选项 |
总之,固醇类雌激素主要在微生物加氧酶和脱氢酶作用下进行生物降解,不同种类微生物降解作用所产生的中间产物也大不相同,中间产物还可以相互转化,但大多数以E1为第一级中间产物,之后按不同路径再进一步代谢。所以,雌激素的降解机制还需要深入研究,降解途径还需要进一步完善。
5 结论和展望 雌激素污染问题已经成为全球性环境问题,主要治理手段是利用微生物对其进行代谢。已经筛取的雌激素高效降解菌主要以变形菌门、厚壁菌门和放线菌门为主。在已知的雌激素降解菌中,放线菌门红球菌属对高浓度雌激素具有更好的底物适应性,具有相对更高的降解效率和降解率。同时对于其他污染物,如苯酚、萘、联苯和多氯联苯等都具有很好的去除效果[50],可以成为污水处理过程中工程菌构建的主要候选菌种。由于酶促反应是微生物实现雌激素降解的主要方式,雌激素降解酶研究也成****关注的热点问题。放线菌门雌激素降解酶研究是一个重要领域。在已经发现的雌激素降解酶中,羟化酶和脱氢酶在类固醇雌激素的开环裂解过程中发挥至关重要的作用。在酶促反应中,雌激素降解的中间产物因菌株不同而有很大差异,但大多是以E1为一级产物再进一步代谢。能够最终降解为CO2和H2O或者进入TCA的报道并不多,并且降解产物的雌激素生物活性也尚不明确。
随着人口不断增长以及养殖产业的迅猛发展,固醇类雌激素的排放量持续增加,城市生活污水和农村纳污水体雌激素含量日趋升高,对人类健康以及生态环境造成的影响也日趋加大。然而,我国对雌激素污染的研究工作开展较晚,污水处理排放雌激素指标体系还没有建立,对雌激素污染物处理还缺乏相应的制度约束。治理雌激素环境污染还需要从以下几方面做好工作。
(1) 在国家政策方面要对雌激素污染问题给予更高保障。将环境雌激素污染列为须着力解决的突出环境污染问题,制定明确的污水处理厂雌激素排放标准,提高污染排放标准,坚持源头防治,强化排污者责任,健全环保信用评价、信息强制性披露、严惩重罚等制度,以满足人民群众对水质安全和优美生态环境的需要。
(2) 提升雌激素降解菌株的应用技术水平。加强高效雌激素降解工程菌的构建,完善菌种固定化技术和投放方式使其可以在多菌种、多污染物的实际水体中长期存活、保持对雌激素的降解功能。同时不会对原有生态环境造成破坏,减小生态威胁以达到生态位平衡。针对实际水体中雌激素浓度较低,应对现有降解菌进行驯化,使其在实际污染水体中发挥最大的降解效用。
(3) 加强雌激素降解菌种和污水处理工艺的研发投入。从基因组、转录组和蛋白组水平对雌激素降解菌及其降解机制进行更为深入的研究,确定不同的降解酶或酶系的诱导条件以及在各个降解途径的功能和作用,以便于在基因水平优化和促进降解酶的表达水平,提高雌激素的降解效率。对污水处理厂现有工艺进行优化,采用深度处理技术等。
References
| [1] | Kurisu F, Ogura M, Saitoh S, Yamazoe A, Yagi O. Degradation of natural estrogen and identification of the metabolites produced by soil isolates of Rhodococcus sp. and Sphingomonas sp. Journal of Bioscience and Bioengineering, 2010, 109(6): 576-582. DOI:10.1016/j.jbiosc.2009.11.006 |
| [2] | Pauwels B, Wille K, Noppe H, de Brabander H, van de Wiele T, Verstraete W, Boon N. 17α-Ethinylestradiol cometabolism by bacteria degrading estrone, 17β-estradiol and estriol. Biodegradation, 2008, 19(5): 683-693. DOI:10.1007/s10532-007-9173-z |
| [3] | Yoshimoto T, Nagai F, Fujimoto J, Watanabe K, Mizukoshi H, Makino T, Kimura K, Saino H, Sawada H, Omura H. Degradation of estrogens by Rhodococcus zopfii and Rhodococcus equi isolates from activated sludge in wastewater treatment plants. Applied and Environmental Microbiology, 2004, 70(9): 5283-5289. DOI:10.1128/AEM.70.9.5283-5289.2004 |
| [4] | Chen YL, Yu CP, Lee TH, Goh KS, Chu KH, Wang PH, Ismail W, Shih CJ, Chiang YR. Biochemical mechanisms and catabolic enzymes involved in bacterial estrogen degradation pathways. Cell Chemical Biology, 2017, 24(6): 712-724. DOI:10.1016/j.chembiol.2017.05.012 |
| [5] | Khunjar WO, Mackintosh SA, Skotnicka-Pitak J, Baik S, Aga DS, Love NG. Elucidating the relative roles of ammonia oxidizing and heterotrophic bacteria during the biotransformation of 17α-ethinylestradiol and trimethoprim. Environmental Science & Technology, 2011, 45(8): 3605-3612. |
| [6] | Koukkou AI, Drainas C. Addressing PAH biodegradation in Greece: biochemical and molecular approaches. IUBMB Life, 2008, 60(5): 275-280. DOI:10.1002/(ISSN)1521-6551 |
| [7] | Ghosal D, Dutta A, Chakraborty J, Basu S, Dutta TK. Characterization of the metabolic pathway involved in assimilation of acenaphthene in Acinetobacter sp. strain AGAT-W. Research in Microbiology, 2013, 164(2): 155-163. DOI:10.1016/j.resmic.2012.11.003 |
| [8] | 王平.不动杆菌DSSKY-A-001雌激素降解酶生物信息学分析.东北师范大学学位论文, 2017. https://www.ixueshu.com/document/3a44b64ec38b15c08323d9024fb876cc.html |
| [9] | van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Δ1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiology Letters, 2001, 205(2): 197-202. |
| [10] | Yu CP, Roh H, Chu KH. 17β-Estradiol-degrading bacteria isolated from activated sludge. Environmental Science & Technology, 2007, 41(2): 486-492. |
| [11] | Ke JX, Zhuang WQ, Gin KYH, Reinhard M, Hoon LT, Tay JH. Characterization of estrogen-degrading bacteria isolated from an artificial sandy aquifer with ultrafiltered secondary effluent as the medium. Applied Microbiology and Biotechnology, 2007, 75(5): 1163-1171. DOI:10.1007/s00253-007-0923-y |
| [12] | Fujii K, Kikuchi S, Satomi M, Ushio-Sata N, Morita N. Degradation of 17β-estradiol by a Gram-negative bacterium isolated from activated sludge in a sewage treatment plant in Tokyo, Japan. Applied and Environmental Microbiology, 2002, 68(4): 2057-2060. DOI:10.1128/AEM.68.4.2057-2060.2002 |
| [13] | Muller M, Patureau D, Godon JJ, Delgenès JP, Hernandez-Raquet G. Molecular and kinetic characterization of mixed cultures degrading natural and synthetic estrogens. Applied Microbiology and Biotechnology, 2010, 85(3): 691-701. |
| [14] | Weber S, Leuschner P, K?mpfer P, Dott W, Hollender J. Degradation of estradiol and ethinyl estradiol by activated sludge and by a defined mixed culture. Applied Microbiology and Biotechnology, 2005, 67(1): 106-112. |
| [15] | 曾庆玲.活性污泥法去除城市污水中雌激素E2与EE2的机理研究.同济大学博士学位论文, 2007. http://www.wanfangdata.com.cn/details/detail.do?_type=degree&id=Y1450948 |
| [16] | Shi JH, Han R, Su LY, Cao JL, Hu LJ. Isolation and identification of a 17α-ethynylestradiol-degrading strain from a wastewater treatment plant and its degradation characteristics. Acta Scientiae Circumstantiae, 2010, 30(12): 2414-2419. (in Chinese) 史江红, 韩蕊, 宿凌燕, 曹金玲, 呼丽娟. 某污水处理厂中17α-乙炔基雌二醇降解菌的分离鉴定及其降解特性. 环境科学学报, 2010, 30(12): 2414-2419. |
| [17] | Zeng QL, Li YM, Gu GW, Zhao JM, Zhang CJ, Luan JF. Sorption and biodegradation of 17β-estradiol by acclimated aerobic activated sludge and isolation of the bacterial strain. Environmental Engineering Science, 2009, 26(4): 783-790. DOI:10.1089/ees.2008.0116 |
| [18] | Fernández L, Louvado A, Esteves Ⅵ, Gomes NCM, Almeida A, Cunha ?. Biodegradation of 17β-estradiol by bacteria isolated from deep sea sediments in aerobic and anaerobic media. Journal of Hazardous Materials, 2017, 323: 359-366. DOI:10.1016/j.jhazmat.2016.05.029 |
| [19] | Jiang LY, Yang J, Chen JM. Isolation and characteristics of 17β-estradiol-degrading Bacillus spp. strains from activated sludge. Biodegradation, 2010, 21(5): 729-736. DOI:10.1007/s10532-010-9338-z |
| [20] | van der Geize R, Hessels GI, van Gerwen R, Vrijbloed JW, van der Meijden P, Dijkhuizen L. Targeted disruption of the kstD gene encoding a 3-ketosteroid Δ1-dehydrogenase isoenzyme of Rhodococcus erythropolis strain SQ1. Applied and Environmental Microbiology, 2000, 66(5): 2029-2036. DOI:10.1128/AEM.66.5.2029-2036.2000 |
| [21] | Knol J, Bodewits K, Hessels GI, Dijkhuizen L, van der Geize R. 3-Keto-5α-steroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochemical Journal, 2008, 410(2): 339-346. |
| [22] | Rohman A, van Oosterwijk N, Thunnissen AM, Dijkstra BW. Crystal structure and site-directed mutagenesis of 3-ketosteroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 explain its catalytic mechanism. Journal of Biological Chemistry, 2013, 288(49): 35559-35568. DOI:10.1074/jbc.M113.522771 |
| [23] | van der Geize R, van der Meijden P, Hessels GI, Dijkhuizen L. Identification of 3-ketosteroid 9-alfa-hydroxylase genes and microorganisms blocked in 3-ketosteroid 9-alfa-hydroxylase activity. US: 7, 514, 236 B2. 2009-04-07. http://europepmc.org/patents/PAT/US7514236 |
| [24] | van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9α-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Molecular Microbiology, 2002, 45(4): 1007-1018. DOI:10.1046/j.1365-2958.2002.03069.x |
| [25] | Petrusma M, Dijkhuizen L, van der Geize R. Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9α-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity. Applied and Environmental Microbiology, 2009, 75(16): 5300-5307. DOI:10.1128/AEM.00066-09 |
| [26] | Petrusma M, Hessels G, Dijkhuizen L, van der Geize R. Multiplicity of 3-Ketosteroid-9α-Hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for specific degradation of different classes of steroids. Journal of Bacteriology, 2011, 193(15): 3931-3940. DOI:10.1128/JB.00274-11 |
| [27] | Yang XX, Dubnau E, Smith I, Sampson NS. Rv1106c from Mycobacterium tuberculosis is a 3β-hydroxysteroid dehydrogenase. Biochemistry, 2007, 46(31): 9058-9067. DOI:10.1021/bi700688x |
| [28] | van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW, Eltis LD. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(6): 1947-1952. DOI:10.1073/pnas.0605728104 |
| [29] | Marcus PI, Talalay P. Induction and purification of α- and β-hydroxysteroid dehydrogenases. Journal of Biological Chemistry, 1956, 218(2): 661-674. |
| [30] | Fernández de las Heras L, van der Geize R, Drzyzga O, Perera J, María Navarro Llorens J. Molecular characterization of three 3-ketosteroid-Δ1-dehydrogenase isoenzymes of Rhodococcus ruber strain Chol-4. Journal of Steroid Biochemistry and Molecular Biology, 2012, 132(3/5): 271-281. |
| [31] | Mohn WW, Wilbrink MH, Casabon I, Stewart GR, Liu J, van der Geize R, Eltis LD. Gene cluster encoding cholate catabolism in Rhodococcus spp.. Journal of Bacteriolog, 2012, 194(24): 6712-6719. DOI:10.1128/JB.01169-12 |
| [32] | Donova MV. Transformation of steroids by actinobacteria: a review. Prikladnaia Biokhimiia Ⅰ Mikrobiologiia, 2007, 43(1): 5-18. |
| [33] | Guevara G, Fernández de las Heras L, Perera J, María Navarro Llorens J. Functional differentiation of 3-ketosteroid Δ1-dehydrogenase isozymes in Rhodococcus ruber strain Chol-4. Microbial Cell Factories, 2017, 16(1): 42. DOI:10.1186/s12934-017-0657-1 |
| [34] | Wilbrink MH. Microbial sterol side chain degradation in Actinobacteria. Groningen: Doctor Dissertation of University of Groningen, 2011. |
| [35] | Sang YY, Xiong GM, Masera E. Identification of a new steroid degrading bacterial strain H5 from the Baltic Sea and isolation of two estradiol inducible genes. The Journal of Steroid Biochemistry and Molecular Biology, 2012, 129(1/2): 22-30. |
| [36] | Hu AY, He JB, Chu KH, Yu CP. Genome sequence of the 17β-estradiol-utilizing bacterium Sphingomonas strain KC8. Journal of Bacteriology, 2011, 193(16): 4266-4267. DOI:10.1128/JB.05356-11 |
| [37] | Qin D, Ma C, Hu AY, Zhang FF, Hu HB, Yu CP. Altererythrobacter estronivorus sp. nov., an estrogen- degrading strain isolated from Yundang lagoon of Xiamen city in China. Current Microbiology, 2016, 72(5): 634-640. DOI:10.1007/s00284-016-0995-y |
| [38] | Zheng DN, Wang XL, Wang PP, Peng WL, Ji NN, Liang RB. Genome sequence of Pseudomonas citronellolis SJTE-3, an estrogen- and polycyclic aromatic hydrocarbon-degrading bacterium. Genome Announcements, 2016, 4(6): e01373-16. DOI:10.1128/genomeA.01373-16 |
| [39] | Zhang Y, Qin FJ, Qiao J, Li GM, Shen CH, Huang TW, Hu Z. Draft genome sequence of Rhodococcus sp. strain p14, a biodegrader of high-molecular-weight polycyclic aromatic hydrocarbons. Journal of Bacteriology, 2012, 194(13): 3546. DOI:10.1128/JB.00555-12 |
| [40] | McLeod MP, Warren RL, Hsiao WWL, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, Dosanjh M, Hara H, Petrescu A, Morin RD, Yang G, Stott JM, Schein JE, Shin H, Smailus D, Siddiqui AS, Marra MA, Jones SJM, Holt R, Brinkman FSL, Miyauchi K, Fukuda M, Davies JE, Mohn WW, Eltis LD. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(42): 15582-15587. DOI:10.1073/pnas.0607048103 |
| [41] | Gong WJ, Kisiela M, Schilhabel MB, Xiong GM, Maser E. Genome sequence of Comamonas testosteroni ATCC 11996, a representative strain involved in steroid degradation. Journal of Bacteriology, 2012, 194(6): 1633-1634. DOI:10.1128/JB.06795-11 |
| [42] | Yücel O, Wibberg D, Philipp B, Kalinowski J. Genome sequence of the bile salt-degrading bacterium Novosphingobium sp. strain Chol11, a model organism for bacterial steroid catabolism. Genome Announcements, 2018, 6(1): e01372-17. DOI:10.1128/genomeA.01372-17 |
| [43] | Si D, Xiong YX, Li XY, Ma LJ, Deng XC, Wang Y, Yang ZB. Isolation and genome sequence analysis of a bacterium degrading dexamethasone. Biomedical Research, 2017, 28(11): 4825-4831. |
| [44] | Nakai S, Yamamura A, Tanaka S, Shi JH, Nishikawa M, Nakashimada Y, Hosomi M. Pathway of 17β-estradiol degradation by Nitrosomonas europaea and reduction in 17β-estradiol-derived estrogenic activity. Environmental Chemistry Letters, 2011, 9(1): 1-6. |
| [45] | Yu QM, Wang P, Liu DB, Gao RX, Shao HH, Zhao HY, Ma Z, Wang D, Huo HL. Degradation characteristics and metabolic pathway of 17β-estradiol (E2) by Rhodococcus sp. DS201. Biotechnology and Bioprocess Engineering, 2016, 21(6): 804-813. DOI:10.1007/s12257-016-0283-5 |
| [46] | Lee HB, Liu D. Degradation of 17β-estradiol and its metabolites by sewage bacteria. Water, Air, and Soil Pollution, 2002, 134(1/4): 351-366. DOI:10.1023/A:1014117329403 |
| [47] | Della Greca M, Pinto G, Pistillo P, Pollio A, Previtera L, Temussi F. Biotransformation of ethinylestradiol by microalgae. Chemosphere, 2008, 70(11): 2047-2053. DOI:10.1016/j.chemosphere.2007.09.011 |
| [48] | Yi T, Harper WF Jr. The link between nitrification and biotransformation of 17α-ethinylestradiol. Environmental Science & Technology, 2007, 41(12): 4311-4316. |
| [49] | Ren HY, Ji SL, ud din Ahmad N, Wang D, Cui CW. Degradation characteristics and metabolic pathway of 17α-ethynylestradiol by Sphingobacterium sp. JCR5. Chemosphere, 2007, 66(2): 340-346. DOI:10.1016/j.chemosphere.2006.04.064 |
| [50] | Sun Y, Chao YP, Qian SJ. Study on the degradation pathway of biphenyl by Rhodococcus pyridinovorans R04. Acta Microbiologica Sinica, 2003, 43(5): 653-658. (in Chinese) 孙艳, 钞亚鹏, 钱世钧. 嗜吡啶红球菌R04的联苯降解途径的研究. 微生物学报, 2003, 43(5): 653-658. DOI:10.3321/j.issn:0001-6209.2003.05.017 |
