李静1, 徐美娟1,2

 , 舒群峰1, 赵雅雯1, 唐蜜1, 张显1, 杨套伟1, 许正宏1, 饶志明1
, 舒群峰1, 赵雅雯1, 唐蜜1, 张显1, 杨套伟1, 许正宏1, 饶志明1

1. 江南大学生物工程学院, 工业生物技术教育部重点实验室, 江苏 无锡 214122;
2. 江南大学(如皋)食品生物技术研究所, 江苏 如皋 226500
收稿日期:2019-01-12;修回日期:2019-02-26;网络出版日期:2019-03-07
基金项目:国家自然科学基金(31770058,31570085);江苏省自然科学基金(BK20181205);教育部重点研究项目(113033A);中央高校基本科研业务费专项资金资助(JUSRP51708A);国家双一流轻工业技术与工程一级学科计划(LITE2018-06)
*通信作者:徐美娟, E-mail:xumeijuan@jiangnan.edu.cn;
饶志明, Tel/Fax:+86-510-85916881, E-mail:raozhm@jiangnan.edu.cn.
摘要:[目的] 通过改造谷氨酸棒杆菌JNR中双功能尿苷酰转移/去除酶GlnD,减弱尿苷酰去除酶的活性,增强NH4+的转运和利用,提高L-精氨酸的合成。[方法] 本文对来源于谷氨酸棒杆菌的突变菌株JNR中的双功能尿苷酰转移/去除酶GlnD进行整合突变,采用同源重组的方法将H414和D415位点突变为两个丙氨酸AA,在此菌株的基础上过量表达PII蛋白GlnK,并对其进行尿苷酰化研究,离子色谱检测摇瓶发酵过程中NH4+的浓度,并对最终的改造菌株进行连续流加发酵分析。[结果] 该双功能尿苷酰转移/去除酶在谷氨酸棒杆菌中成功进行整合突变,有效减弱了尿苷酰去除酶的活性;同时过表达PII蛋白GlnK,其酰基化程度明显增强。摇瓶发酵结果表明菌株L4消耗NH4+增加,L-精氨酸产量为36.2±1.2 g/L,比对照菌株L3高出22.7%。5-L发酵罐实验结果显示改造菌株L4的L-精氨酸的产量为52.2 g/L,较野生型菌株L0提高了25.3%。[结论] 谷氨酸棒杆菌合成L-精氨酸的过程中氮源是必不可少的。减弱GlnD尿苷酰去除酶的活性后,胞内尿苷酰化的GlnK-UMP增加,GlnK-UMP与氮转录调控因子AmtR结合,转运至胞内的NH4+浓度提高,促使L-精氨酸产量显著提高。
关键词:谷氨酸棒杆菌JNR双功能尿苷酰转移/去除酶GlnDNH4+L-精氨酸整合突变
Function of uridylyltransferase and uridylyl-removing enzyme GlnD in Corynebacterium glutamicum JNR
Jing Li1, Meijuan Xu1,2

 , Qunfeng Shu1, Yawen Zhao1, Mi Tang1, Xian Zhang1, Taowei Yang1, Zhenghong Xu1, Zhiming Rao1
, Qunfeng Shu1, Yawen Zhao1, Mi Tang1, Xian Zhang1, Taowei Yang1, Zhenghong Xu1, Zhiming Rao1

1. The Key Laboratory of Industrial Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi 214122, Jiangsu Province, China;
2. Institute of Food Biotechnology, Jiangnan University(Rugao), Rugao 226500, Jiangsu Province, China
Received: 12 January 2019; Revised: 26 February 2019; Published online: 7 March 2019
*Corresponding author: Meijuan Xu, E-mail: xumeijuan@jiangnan.edu.cn;
Zhiming Rao, Tel/Fax:+86-510-85916881, E-mail: raozhm@jiangnan.edu.cn.
Foundation item: Supported by the National Natural Science Foundation of China (31770058, 31570085), by the Natural Science Foundation of Jiangsu Province (BK20181205), by the Research Project of the Chinese Ministry of Education (113033A), by the Fundamental Research Funds for the Central Universities (JUSRP51708A), and by the Thenational First-Class Discipline Program of Light Industry Technology and Engineering (LITE2018-06)
Abstract: [Objective] We modified the bifunctional uridylyltransferase and uridylyl-removing enzyme GlnD to reduce the activity of uridylyl-removing enzyme, thus, increasing the NH4+ transition and application, to improve L-arginine production. [Methods] PII protein GlnK was overexpressed and its uridylation was studied. A glnDAA(including H414A and D415A) mutant was generated from C. glutamicum JNR using the double-crossover chromosome replacement technique. The concentration of NH4+ in the fermentation medium was measured by ion chromatography. Then the resulting strain was cultivated in a 5-L stirring bioreactor to performed a fed-batch fermentation. [Results] The bifunctional uridylyltransferase and uridylyl-removing enzyme was successfully mutated in C. glutamicum JNR and the resulting strain L4 showed a weakened activity of uridylyl-removing enzyme. The shaking flask fermentation showed that the L4 strain consumed more NH4+, and the L-arginine yield was 36.2±1.2 g/L, 22.7% higher than the control strain. The production of L-arginine of L4 strain was 52.2 g/L, which was 25.3% higher than that of L0 strain in 5-L fermentation. [Conclusion] In C. glutamicum, nitrogen is necessary for the L-arginine biosynthesis. We conclude that reducing uridylyl-removing activity resulted in more intracellular GlnK-UMP. The GlnK-UMP interacts with the nitrogen regulator AmtR and enhances the intracellular consumption of NH4+. Subsequently, the increased uptake of NH4+ could promote the L-arginine production in C. glutamicum.
Keywords: Corynebacterium glutamicum JNRuridylyltransferase and uridylyl-removing enzyme GlnDNH4+L-arginineintegration mutation
氮元素是构成生物体大分子核酸、蛋白质及其他含氮化合物的材料,是各类微生物最基本的生长因子之一[1-2]。经过长期的进化,微生物形成了非常精细的全局氮调控网络,以应对不同环境下的生存压力[3-5]。NH4+是绝大多数微生物最优先利用的氮源。
谷氨酸棒杆菌作为革兰氏阳性菌,被广泛应用于氨基酸的生产。在谷氨酸棒杆菌中,氮代谢受到全局氮转录调控因子AmtR的调控,据报道谷氨酸棒杆菌AmtR可调控至少35个基因的转录,包括编码铵吸收相关的转运子和酶的基因(amtA,amtB,glnA,gltBD,dapA)、肌酐(codA,crnT)和尿素(urtABCDE,ureABCEFG)代谢相关基因;信号转导蛋白及其修饰酶(glnK,glnD)等氮代谢相关的基因[6]。如图 1-A所示,当胞内NH4+限量时,PII信号转导蛋白GlnK被双功能尿苷酰转移/去除酶GlnD (UTase/UR,glnD)尿苷酰化后形成GlnK-UMP[7],GlnK-UMP可与氮转录调控因子AmtR结合,AmtR对铵吸收利用途径的阻遏作用解除,相关基因的转录正常进行[8]。同时,尿苷酰化的GlnK-UMP不能与膜上的铵转运蛋白AmtB结合形成复合物,因此激活了铵转运通道,NH4+可通过AmtB铵转运通道进入细胞内。相反如图 1-B所示,当胞内NH4+充足时,GlnD蛋白将GlnK-UMP蛋白去尿苷酰化为GlnK,此时GlnK处于激活状态,被激活的GlnK会与细胞膜上的靶蛋白AmtB结合为复合物AmtB-GlnK,阻碍AmtB对铵的转运。激活后的GlnK不与氮转录调控因子AmtR结合,AmtR结合到相关基因的启动子区,阻遏了胞内氮代谢相关基因转录反应的发生,细胞内代谢活动减弱[9-10]。
 |
| 图 1 棒杆菌中GlnK,GlnD,AmtB和AmtR之间的关系 Figure 1 Relationship of GlnK, GlnD, AmtB and AmtRin Corynebacterium. A: Under conditions of intracellular ammonium starvation, GlnK is modified by GlnD, and the resulting adenylated-GlnK (GlnK-UMP) subsequently interacts with AmtR, which causes the transcriptional upregulation of nitrogen-regulated genes, thereby activating the intracellular transport of ammonium. B: under conditions of nitrogen surplus, de-adenylated GlnK interacts with the ammonium transporter AmtB, blocking the ammonium flux into the cell and therefore resulting in a GlnK-free state, allowing AmtR to be reactivated and bind to the promoter regions of nitrogen-controlled genes, repressing their transcription |
| 图选项 |
L-精氨酸(C6H12N4O2)作为半必需氨基酸,具有多种生理功能,是一种重要的工业氨基酸,广泛应用于医药、食品、化工等行业,是氨基酸工业研究与开发的热点之一[11]。在L-精氨酸形成过程中,需要大量的氮源供给[12]。高氮源的环境中,为了减少AmtR的抑制,增加胞内NH4+的浓度,提高L-精氨酸的产量,我们通过改造双功能尿苷酰转移/去除酶GlnD,减弱尿苷酰去除酶UR的活性。棒杆菌中,双功能尿苷酰转移/去除酶GlnD中的HD区域控制尿苷酰去除酶的活性[13]。通过同源比对谷氨酸棒杆菌与钝齿棒杆菌中的基因glnD的序列,选定HD区域的H414和D415位点突变为两个丙氨酸AA。在谷氨酸棒杆菌JNR中成功整合突变,并将PII信号转导蛋白GlnK在此基础之上进行过量表达,考察GlnK-UMP的活性。摇瓶发酵改造后的菌株L4,检测胞内NH4+的浓度和L-精氨酸的产量,L-精氨酸产量达到36.2±1.2g/L,比对照菌株L3提高了22.7%。5-L发酵罐实验结果表明改造菌株L4的L-精氨酸的产量为52.2 g/L,比野生型菌株L0提高了25.3%。
1 材料和方法 1.1 试剂 ClonExpress? Ultra One Step Cloning Kit购自南京诺维赞公司;质粒DNA小量试剂盒、DNA胶回收试剂盒、细菌DNA基因组提取试剂盒购自上海生物工程有限公司;Extaq DNA聚合酶、PrimerStar DNA聚合酶、T4 DNA连接酶、Xba I、EcoR I、Sal I、Hind III限制性内切酶购自宝公司;卡那霉素、氯霉素及L-精氨酸等购自阿拉丁生物试剂公司;其他试剂均购自国药集团化学试剂有限公司。
1.2 菌株和质粒 Escherichia coli BL21(DE3)购自上海生物工程有限公司;重组质粒pXMJ19-glnD、pXMJ19-glnK、pK18-glnDAA均由本实验室构建。本实验使用的菌株、质粒和引物见表 1和表 2。
表 1. 本实验所用的菌株及质粒 Table 1. Strains and plasmid used in this study
| Strain/Plasmid | Characteristic | Source |
| Strains | ||
| E. coli BL21(DE3) | F– ompT gal dcm lon hsdSB (rB– mB–)λ (DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Invitrogen |
| C. glutamicum JNR(L0) L1 | An arginine production strain, His?, SGr, D-Argr, ArgR– L1 with pXMJ19-glnD in the L0 | Our lab This study |
| L2 L3 L4 | L2 with glnDAA intergration on the L0 chromosome L3 with pXMJ19-glnK in the L0 L4with pXMJ19-glnK in the L2 | This study This study This study |
| Plasmids | ||
| pXMJ19 pK18mobsacB | A shutter expression vector, Ptac promoter Mobilizable vector allows for sections of double crossover I C. glutamicum, Km, sacB | This study This study |
| pXMJ19-glnD pK18-glnDAA pXMJ19-glnK | A derivative of pXMJ19, harboring glnD gene A derivative of pK18mobsacB, harboring glnDAA gene A derivative of pXMJ19, harboring glnK gene | This study This study This study |
表选项
表 2. 本研究所用到的引物 Table 2. Primers used in this study
| Name | DNA sequence (5?→3?) | Restriction site |
| P1 F | GCTCTAGAAAAGGAGGACAACCATGAATGACCCAGCCCAGC | Xba I |
| P1 R | CGGAATTCTTAGTGGTGGTGGTGGTGGTGTCAGCTTGCG GCTAAGACC | EcoR I |
| P2 F1 | CGGAATTCATGAATGACCCAGCCCAGC | EcoR I |
| P2 R1 | AATGTCCGGGTACAACGCGCCCAGCACCAATAGGTCAG | - |
| P2 F2 | CTGGGCGCGTTGTACGCCGCCATTGGTAAAGGAACT | - |
| P2 R2 | GCTCTAGAGGTCTTAGCCGCAAGCTGA | Xba I |
| P3 F | CCCAAGCTTAAAGGAGGGAAATCATGAAA CTCATCACCG CAATTG | Hind III |
| P3R | ACGCGTCGACTTAGTGGTGGTGGTGGTGGTGAAGGGCT GCTTCGCCG | Sal I |
表选项
1.3 培养基
1.3.1 LB培养基(g/L): 蛋白胨10,酵母提取物5,氯化钠10,LB平板,琼脂15。
1.3.2 LBG培养基(g/L): 蛋白胨10,酵母提取物5,氯化钠10,葡萄糖7。
1.3.3 LBGS培养基(g/L): 蛋白胨10,酵母提取物5,氯化钠10,葡萄糖10,蔗糖100。
1.3.4 种子培养基(g/L): 葡萄糖50,酵母抽提物20,(NH4)2SO4 20,MgSO4·7H2O 0.5,KH2PO4 1。
1.3.5 摇瓶发酵培养基(g/L): 葡萄糖120,酵母抽提物10,(NH4)2SO4 40,MgSO4·7H2O 0.02,KH2PO4 1,FeSO4·7H2O 0.02,MnSO4 0.02,CaCO3 20。
1.3.6 5-L发酵罐培养基(g/L): 葡萄糖100,酵母抽提物20,(NH4)2SO4 40,MgSO4·7H2O 0.02,KH2PO4 1.50,FeSO4·7H2O 0.02,MnSO4 0.02。
1.4 菌株培养 大肠杆菌BL21(DE3)采用LB液体培养基在37 ℃下进行培养,转速180 r/min;谷氨酸棒杆菌JNR在LBG培养基中培养,培养温度为30 ℃,转速180 r/min。
谷氨酸棒杆菌JNR摇瓶发酵的培养方法:保藏的甘油冻管菌种在LBG平板上划线活化,接一环活化后的菌种于种子培养基中,装液量50/250 mL,往复式摇床140 r/min,培养48 h;将种子培养液以10%的接种量转接到摇瓶发酵培养基中,规格为250 mL装液量25 mL,于30 ℃、220 r/min的往复式摇床培养72 h。5 L发酵方法参照文献[12],发酵过程中,每隔8 h取5 mL样品检测相关指标。
1.5 离子色谱检测NH4+的浓度 采用赛默飞ICS-5000+离子色谱检测NH4+的浓度,IonPac CS12A分离柱(4 mm×250 mm)与IonPac CG12A保护柱(4 mm×50 mm),流动相20 mmol/L甲基磺酸,流速1 mL/min,时间15 min,柱箱温度30 ℃,进样体积25 μL。样品处理如下:分别取1 mL摇瓶发酵12、24、36、48、60和72 h后的发酵液,12000 r/min离心10 min,收集上清液,采用0.22 μm滤膜过滤,数据分析参照Zhu的方法[14]。
1.6 生长量OD562的测定 采用分光光度计UNICOTM-UV2000测定细胞的生长量,1 OD562=0.375 g/L细胞干重。测定方法:收集发酵液,加入0.1 mol/L稀盐酸将发酵液稀释至适当倍数,同时也可以去除CaCO3,排除干扰,测定稀释后的发酵液在562 nm处的吸光值[15]。
1.7 L-精氨酸产量的测定 取1 mL摇瓶发酵72 h后的发酵液,12000 r/min离心10 min,收集上清液,1:1加入10%的三氯乙酸反应2 h,12000 r/min离心10 min,收集上清液备用。采用改良坂口反应测定L-精氨酸产量,反应液的配方为(g/L):NaOH 40,甲萘酚-正丙醇80,双乙酰溶液0.5 mL,将以上三种试剂各加1 mL至10 mL离心管中作为显色液,加入适当稀释的发酵液100 μL。30 ℃水浴加热20 min后测定OD521值。
2 结果和分析 2.1 谷氨酸棒杆菌JNR中过表达GlnD 以谷氨酸棒杆菌13032的基因组DNA为模板,引物P1 F/R进行PCR扩增,得到大小2079 bp基因片段。将glnD基因用Xba I和EcoR I双酶切,回收glnD片段后,与线性化质粒pXMJ19连接后转化,阳性转化子提取质粒经Xba I和EcoR I酶切验证,释放6601 bp和2127 bp大小的片段,分别对应pXMJ19和glnD的大小,结果表明质粒pXMJ19-glnD构建成功,如图 2所示。
 |
| 图 2 基因PCR结果及质粒pXMJ19-glnD酶切验证 Figure 2 PCR results of glnD genes and identification of pXMJ19-glnD plasmids by double enzyme digestion. A: M: DL10000 marker; lane 1, 2: glnD. B: M: DL10000 marker; lane 1: result of double enzyme digestion of empty pXMJ19-glnD |
| 图选项 |
将BL21中构建好的质粒pXMJ19-glnD电转到谷氨酸棒杆菌JNR(L0)中,挑选阳性转化子L1菌株进行诱导表达,菌液经过超声破碎后离心取上清,处理样品对其进行SDS-PAGE分析。如图 3所示,可以清晰地看见分子量约为76.23 kDa的特异性条带,说明尿苷酰转移/去除酶能够在谷氨酸棒杆菌JNR中表达。
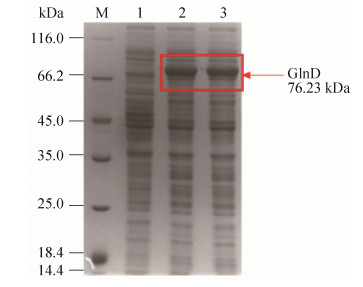 |
| 图 3 重组菌株L1的GlnD粗蛋白SDS-PAGE分析 Figure 3 SDS-PAGE analysis of GlnD crude proteins. M: Unstained protein Ladder; lane 1: crude enzyme of C. glutamicum JNR with plasmid pXMJ19; lane 2, 3: crude enzyme of L1 strain with plasmid pXMJ19-glnD |
| 图选项 |
2.2 同源比对分析GlnD的位点 据报道,在棒杆菌中,双功能尿苷酰转移/去除酶GlnD (UTase/UR)中HD区域控制尿苷酰去除酶UR的活性。我们通过DNAman软件对谷氨酸棒杆菌13032 (Cg)、钝齿棒杆菌(Cc)、分枝杆菌(Mt)和大肠杆菌(Ec)中的glnD基因进行比对分析,如图 4所示。通过同源比对,我们可以发现四株菌在414和415位点同时存在HD的位点。为了减弱尿苷酰去除酶的活性,增加GlnK-UMP,所以将414和415位点作为突变的位点,突变为两个丙氨酸AA,软件分析结果见图 4。
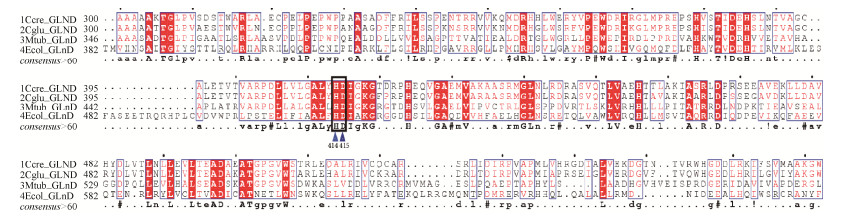 |
| 图 4 GlnD蛋白的同源序列比对 Figure 4 Homologous sequence alignment of GlnD protei |
| 图选项 |
2.3 整合突变 采用同源重组的方法进行整合突变,pK18mobsacB质粒作为载体[16]。首先通过融合PCR的方法,将编码双功能尿苷酰转移/去除酶的glnD基因中的H414与D415位点分别替换为编码丙氨酸(A)的序列。以pXMJ19-glnD质粒为模板,利用引物P2F1和P2R1、P2F2和P2R2进行单片段PCR扩增。PCR结束后将获得的两段同源臂arm1和arm2进行融合PCR[17],得到大小为2079 bp的glnDAA片段,如图 5所示。
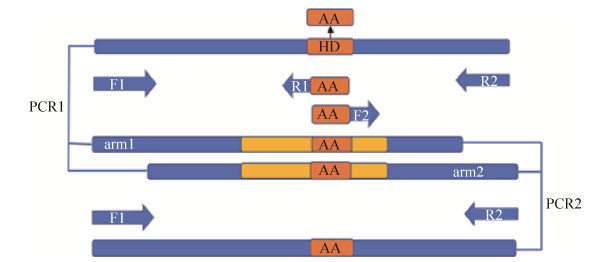 |
| 图 5 融合PCR示意图 Figure 5 Fusion PCR for the glnDAA gene |
| 图选项 |
采用EcoR I和Xba I两个酶同时酶切获得的glnDAA片段和pK18mobsacB载体,16 ℃过夜连接,将连接后的质粒转化到E. coli BL21(DE3)感受态中,挑取阳性转化子,如图 6所示。
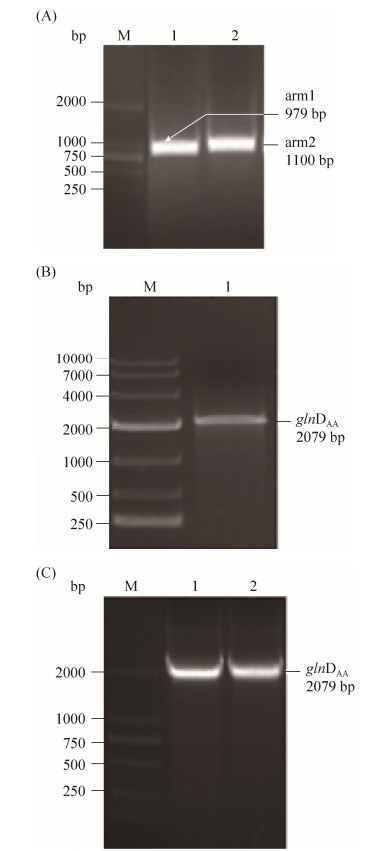 |
| 图 6 同源臂PCR,融合PCR和菌落PCR验证pK18-glnDAA质粒 Figure 6 Fragment and fusion PCR and identification of pK18-glnDHD plasmids double enzyme digestion. A: M: DL2000 Maker; lane 1: homologous arm1; lane 2: homologous arm2; B: M: DL10000 maker; lane 1:glnDAA; C: M: DL2000 maker; lane 1, 2: results of colony PCR of pK18-glnDAA in L3 strain |
| 图选项 |
将构建成功的质粒pK18-glnDAA电转到L0的感受态中,筛选第一次同源重组后的菌株转接至10%的LBGS培养基中进行第二次同源重组筛选,分别划线无抗性的LBG平板和含有卡那霉素抗性的平板,挑选在无抗性平板上生长但在卡那抗性平板上不生长的菌株进行测序。测序结果表明整合突变成功,glnDAA成功替换到基因组上,菌株命名为L1。
2.4 比较整合突变前后GlnD对GlnK的修饰作用 分别将PII信号转导蛋白GlnK在原始菌株L0和整合突变成功的菌株L2中进行过量表达,形成菌株L3和L4,如图 7所示。
 |
| 图 7 基因PCR结果及菌落PCR验证pXMJ19-glnK Figure 7 PCR results of glnK genes and identification of pXMJ19-glnKplasmids by colony PCR. A: M: DL2000 maker; lane 1, 2: glnK. B: M: DL2000 maker; lane 1, 2: result of pXMJ19-glnK plasmids by colony PCR |
| 图选项 |
为了验证整合突变的效果,我们采用验证GlnK蛋白酰基化的强弱来反映整合突变的效果。首先在大肠杆菌BL21(DE3)中构建质粒pXMJ19-glnK,构建方法参考2.1。将构建好的质粒pXMJ19-glnK分别电转到菌株L0和L2中,挑取转化子进行诱导表达。经超声波破碎,将获得的上清液于镍柱纯化并SDS-PAGE分析。如图 8所示,泳道2中GlnK蛋白呈现两条表达带,上面的条带为尿苷酰化条带,即GlnK-UMP;下面的条带为未修饰的GlnK蛋白。泳道1是没有进行整合突变的单纯表达GlnK的菌株,GlnK也是呈现出两条带,但是我们对比泳道1和泳道2、3可以明显发现,泳道2和3的上面一条带要比通道1中的条带粗,说明菌株L4中GlnK蛋白多数为GlnK-UMP状态,说明双功能尿苷酰化转移/去除酶的去尿苷酰化酶活被钝化而降低。
 |
| 图 8 重组谷氨酸棒杆菌L3及L4的GlnK蛋白SDS-PAGE分析 Figure 8 SDS-PAGE analysis of the overexpression of GlnK in recombinant L3 and L4 strain. M: unstained protein ladder; lane 1: L3; lane 2, 3: L4 |
| 图选项 |
2.5 重组菌发酵L-精氨酸性能测定 前面的实验证明,对双功能尿苷酰转移/去除酶GlnD进行整合突变有效地减弱了去尿苷酰化酶的活力,胞内的GlnK蛋白多呈尿苷酰化状态。通过检测胞外NH4+的残留量和L-精氨酸的产量验证我们的猜想,胞内的GlnK蛋白更多呈现尿苷酰化有助于铵通道的打开,将改造后的L3和L4菌株以及野生型L0菌株分别进行摇瓶发酵,收集发酵12、24、36、48、60和72 h的发酵液。比较各时间段3株菌的生长量、葡萄糖浓度和L-精氨酸的产量,如图 9-A所示,随着发酵时间的增加,菌株的生长量逐渐积累,基本上同步生长,说明分子改造没有对菌体造成伤害。将上述各时间段的样品进行L-精氨酸产量检测,结果表明L-精氨酸的产量逐渐增加,前36 h,L-精氨酸基本上没有产生,发酵36 h以后,L-精氨酸呈指数增长。比较L3和L4菌株,L4菌株最终产量高于L3菌株,达到(36.2±1.2) g/L,比L3菌株提高了22.7%,比野生型菌株L0高出31%。同时,L4菌株最终剩余葡萄糖明显低于L3菌株,浓度约为(10.2±0.6) g/L,由此可以证明整合突变的效果显著。
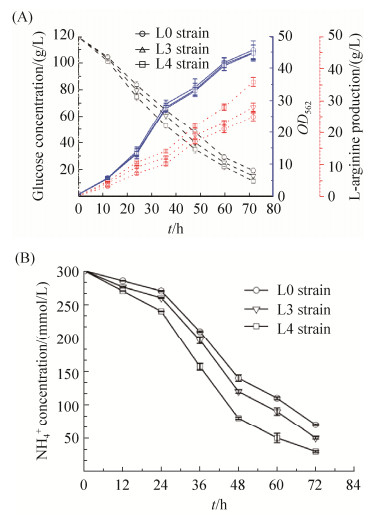 |
| 图 9 不同菌株在摇瓶发酵中的生长量、葡萄糖消耗、L-精氨酸的产量和NH4+浓度 Figure 9 The OD562, glucose concentration, L-arginine production and NH4+ concentration of strains. A: Comparison of OD562, glucose concentration and L-arginineproduction in the L3, L4 and L0 strains under shake flask fermentation. B: Detection of NH4+ concentration by ion chromatography, comparison of NH4+ concentrationin the L3, L4 and L0 strains under shake flask fermentation. The empty circle represents L0 strain, the empty triangle represents L3 strain, the empty square represents L4 strain. The data represent the mean values and standard deviations obtained from at least three independent experiments |
| 图选项 |
检测不同发酵时间段内NH4+的残留量,如图 9-B所示,随着发酵时间的增加,NH4+的浓度逐渐降低,对比L3和L4菌株,可以明显看出L4菌株对NH4+的消耗要高于L3菌株,发酵72 h后,NH4+残留量远远低于L3菌株。结果表明对GlnD蛋白进行的改造有效地加强了GlnK蛋白的尿苷酰化,使得铵转运蛋白处于活性状态,更多的NH4+转运到胞内,进一步证明了整合突变效果显著。
对菌株L4和野生型菌株L0进行5-L发酵罐研究,并考察发酵过程中的参数的变化。每隔8 h取一次样检测发酵液的OD562、残糖及L-精氨酸的产量。根据发酵结果可以分析出重组菌株L4产酸能力较出发菌株L0强,且消耗了更多的葡萄糖,共进行了3次葡萄糖的流加,每次流加15 g。如图 10-A所示,发酵前期,重组菌株的OD不及出发菌株,但是稳定期后两者相差甚微;40至72 h为菌体积累精氨酸的重要时期,此时重组菌株L4发酵产酸能力相对出发菌株L0来说优势较为明显,L-精氨酸的生成率明显提升。发酵72 h后,L4的L-精氨酸的产量为52.2 g/L,产率为0.73 g/(L·h),出发菌株L0的L-精氨酸的产量为39 g/L,产率为0.54 g/(L·h),L4比L0菌株提高了25.3%。另外,L4共消耗了137 g葡萄糖,L0消耗了120 g葡萄糖,说明更多的葡萄糖消耗用于L-精氨酸的产生。
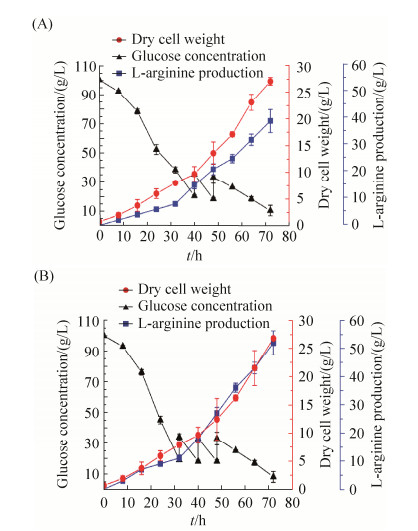 |
| 图 10 L0和L4菌株在5-L发酵罐中的生长量、葡萄糖消耗以及L-精氨酸的产量 Figure 10 The OD562, glucose concentration and L-arginine production of strains in 5-L fermentation. Comparison of OD562, glucose concentration and L-arginineproduction in the L4 and L0 strains under 5-L fermentation. A: L0 strain. B: L4 strain. The data represent the mean values and standard deviations obtained from at least three independent experiments |
| 图选项 |
3 讨论 L-精氨酸具有多种生理功能,是合成蛋白质和肌酸的重要原料,是人类和动物的半必需氨基酸,在工业上应用广泛[18-19]。关于L-精氨酸的研究,目前报道基本都以谷氨酸棒杆菌及其变种作为出发菌株,从L-精氨酸的合成途径入手。Ikeda等通过敲除argR和弱化N-乙酰谷氨酸激酶反馈抑制使得L-精氨酸产量达到52 g/L[20]。Park SH和Shin JH团队系统性地优化NADPH水平及发酵优化,L-精氨酸产量达到92.5 g/L,目前为L-精氨酸的最高水平[21-22]。总的来看,国内外关于L-精氨酸的研究基本上都是从碳源的角度[23-24]和L-精氨酸本身的合成途径入手[25-26],几乎没有涉及到氮代谢对L-精氨酸合成的影响。L-精氨酸的分子式中含有4个氮原子,氮占到分子量的32.1%,在其发酵过程中需要大量的氮源供给,因此提高胞内氮源的浓度,有助于L-精氨酸产量的增加[27]。
本研究的主旨主要是通过提高胞内氮源的浓度从而增加L-精氨酸的产量。结合前人关于棒杆菌中氮调节网络的研究,如图 1所示,氮转录调控因子AmtR在不同的氮源环境下对胞内的转录运行存在调控[28]。以产L-精氨酸的谷氨酸棒杆菌JNR为出发菌株,探究尿苷酰转移/去除酶GlnD(UTase, UR)与信号转导蛋白GlnK之间的修饰关系。尿苷酰转移/去除酶是一个双功能的酶,具有尿苷酰转移酶(UTase)和尿苷酰去除酶(UR)两种活性[29]。L-精氨酸的发酵过程是一个高氮源的环境,高氮源环境下尿苷酰去除酶的活力较强,未被修饰的GlnK蛋白处于激活状态,会结合膜上的铵转运的蛋白AmtB,胞外的NH4+不能被运输进胞内,激活状态下的GlnK蛋白不与AmtR结合,AmtR结合在启动子DNA上,抑制胞内的转录进行。
通过整合突变改造双功能尿苷酰转移/去除酶,减弱尿苷酰去除酶的活性,有助于胞内形成GlnK-UMP,更多的NH4+进入胞内,L-精氨酸的产量增加。如图 8所示,在完全培养基中,L4菌株GlnK-UMP明显高于L3菌株。最终,改造菌株L4在摇瓶发酵时L-精氨酸产量达到(36.2±1.2) g/L,比对照菌株L3提高了22.7%,同时,改造菌株消耗了更多的NH4+,发酵结束胞外NH4+浓度为25±1 mmol/L。实验结果说明研究的切入点是无误的,有效地提高了胞内的NH4+和L-精氨酸的产量,5-L发酵罐流加发酵,最终L-精氨酸的产量为52.2 g/L,比野生型菌株提高了25.3%。后期我们会从NH4+的吸收和利用以及氮源固定的角度去开展实验,进一步地将氮代谢与提高L-精氨酸的产量结合起来。本研究扩宽了对L-精氨酸研究的角度,从碳代谢转到氮代谢,为改造谷氨酸棒杆菌产L-精氨酸的深入研究以及应用开发提供指导。
References
| [1] | Rehm N, Burkovski A. Engineering of nitrogen metabolism and its regulation in Corynebacterium glutamicum: influence on amino acid pools and production. Applied Microbiology and Biotechnology, 2011, 89(2): 239-248. DOI:10.1007/s00253-010-2922-7 |
| [2] | Chen QH, Han YL, Ma Y, Yan YL, Ping SZ, Lu W. Research progress on structure and evolution of biological nitrogen-fixation gene cluster. Journal of Agricultural Science and Technology, 2013, 15(4): 129-138. (in Chinese) 陈清华, 韩云蕾, 马尧, 燕永亮, 平淑珍, 陆伟. 生物固氮基因簇结构与进化研究进展. 中国农业科技导报, 2013, 15(4): 129-138. DOI:10.3969/j.issn.1008-0864.2013.04.20 |
| [3] | Merrick MJ, Edwards RA. Nitrogen control in bacteria. Microbiological Reviews, 1995, 59(4): 604-622. |
| [4] | Hodgson DA. Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Advances in Microbial Physiology, 2000, 42: 47-238. DOI:10.1016/S0065-2911(00)42003-5 |
| [5] | Martín JF. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. Journal of Bacteriology, 2004, 186(16): 5197-5201. DOI:10.1128/JB.186.16.5197-5201.2004 |
| [6] | Jakoby M, Kr?mer R, Burkovski A. Nitrogen regulation in Corynebacterium glutamicum: isolation of genes involved and biochemical characterization of corresponding proteins. FEMS Microbiology Letters, 1999, 173(2): 303-310. DOI:10.1111/j.1574-6968.1999.tb13518.x |
| [7] | Nolden L, Ngouoto-Nkili CE, Bendt AK, Kr?mer R, Burkovski A. Sensing nitrogen limitation in Corynebacterium glutamicum: the role of glnK and glnD. Molecular Microbiology, 2001, 42(5): 1281-1295. |
| [8] | Meier-Wagner J, Nolden L, Jakoby M, Siewe R, Kr?mer R, Burkovski A. Multiplicity of ammonium uptake systems in Corynebacterium glutamicum: role of Amt and AmtB. Microbiology, 2001, 147(1): 135-143. |
| [9] | Beckers G, Str?sser J, Hildebrandt U, Kalinowski J, Farwick M, Kr?mer R, Burkovski A. Regulation of AmtR-controlled gene expression in Corynebacterium glutamicum: mechanism and characterization of the AmtR regulon. Molecular Microbiology, 2005, 58(2): 580-595. DOI:10.1111/j.1365-2958.2005.04855.x |
| [10] | Buchinger S, Str?sser J, Rehm N, H?n?ler E, Hans S, Bathe B, Schomburg D, Kr?mer R, Burkovski A. A combination of metabolome and transcriptome analyses reveals new targets of the Corynebacterium glutamicum nitrogen regulator AmtR. Journal of Biotechnology, 2009, 140(1/2): 68-74. |
| [11] | Mels CMC, Loots I, Schwedhelm E, Atzler D, B?ger RH, Schutte AE. Nitric oxide synthesis capacity, ambulatory blood pressure and end organ damage in a black and white population: the SABPA study. Amino Acids, 2016, 48(3): 801-810. |
| [12] | Xu ZH, Dou WF, Wang X, Tao WY. Effects of nitrogen source and its supply manner on production of L-arginine by Corynebacterium crenatum JDN28-75. Chinese Journal of Applied and Environmental Biology, 2006, 12(3): 381-385. (in Chinese) 许正宏, 窦文芳, 王霞, 陶文沂. 氮源及其添加模式对钝齿棒杆菌JDN28-75合成L-精氨酸的影响. 应用与环境生物学报, 2006, 12(3): 381-385. DOI:10.3321/j.issn:1006-687X.2006.03.021 |
| [13] | Zhang YP, Pohlmann EL, Serate J, Conrad MC, Roberts GP. Mutagenesis and functional characterization of the four domains of GlnD, a bifunctional nitrogen sensor protein. Journal of Bacteriology, 2010, 192(11): 2711-2721. DOI:10.1128/JB.01674-09 |
| [14] | Zhu Y, Guo YY, Ye ML, James FS. Separation and simultaneous determination of four artificial sweeteners in food and beverages by ion chromatography. Journal of Chromatography A, 2005, 1085(1): 143-146. |
| [15] | Xu MJ, Rao ZM, Dou WF, Yang J, Jin J, Xu ZH. Site-directed mutagenesis and feedback-resistant N-acetyl-L-glutamate kinase (NAGK) increase Corynebacterium crenatumL-arginine production. Amino Acids, 2012, 43(1): 255-266. |
| [16] | Sch?fer A, Tauch A, J?ger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletionsin the chromosome of Corynebacterium glutamicum. Gene, 1994, 145(1): 69-73. |
| [17] | Wang HL, Postier BL, Burnap RL. Optimization of fusion PCR for in vitro construction of gene knockout fragments. Biotechniques, 2002, 33(1): 26-32. DOI:10.2144/02331bm02 |
| [18] | Lu ZQ, Gong JH, Ding JY, Chen Q. Studies on the breeding of L-arginine-producing strain. Acta Microbiologica Sinica, 1988, 28(2): 131-135. (in Chinese) 路志强, 龚建华, 丁久元, 陈琦. L-精氨酸产生菌诱变育种的研究. 微生物学报, 1988, 28(2): 131-135. |
| [19] | Chen XL, Tang L, Jiao HT, Xu F, Xiong YH. Construction of Corynebacterium crenatum AS 1.542 △argR and analysis of transcriptional levels of the related genes of arginine biosynthetic pathway. Acta Microbiologica Sinica, 2013, 53(1): 92-98. (in Chinese) 陈雪岚, 汤立, 焦海涛, 徐峰, 熊勇华. 钝齿棒杆菌argR基因缺失株构建及其缺失对精氨酸生物合成途径相关基因转录水平的影响. 微生物学报, 2013, 53(1): 92-98. |
| [20] | Ikeda M, Mitsuhashi S, Tanaka K, Hayashi M. Reengineering of a Corynebacterium glutamicum L-arginine and L-citrulline producer. Applied and Environmental Microbiology, 2009, 75(6): 1635-1641. DOI:10.1128/AEM.02027-08 |
| [21] | Park SH, Kim HU, Kim TY, Park JS, Kim SS, Lee SY. Metabolic engineering of Corynebacterium glutamicum for L-arginine production. Nature Communications, 2014, 5: 4618. DOI:10.1038/ncomms5618 |
| [22] | Shin JH, Lee SY. Metabolic engineering of microorganisms for the production of L-arginine and its derivatives. Microbial Cell Factories, 2014, 13(1): 166. DOI:10.1186/s12934-014-0166-4 |
| [23] | Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels - Corynebacterium glutamicum as versatile cell factory. Current Opinion in Biotechnology, 2012, 23(4): 631-640. DOI:10.1016/j.copbio.2011.11.012 |
| [24] | Jensen JVK, Eberhardt D, Wendisch VF. Modular pathway engineering of Corynebacterium glutamicum for production of the glutamate-derived compounds ornithine, proline, putrescine, citrulline, and arginine. Journal of Biotechnology, 2015, 214: 85-94. DOI:10.1016/j.jbiotec.2015.09.017 |
| [25] | Zhao QQ, Luo YC, Dou WF, Zhang X, Zhang XM, Zhang WW, Xu MJ, Geng Y, Rao ZM, Xu ZH. Controlling the transcription levels of argGH redistributed L-arginine metabolic flux in N-acetylglutamate kinase and ArgR-deregulated Corynebacterium crenatum. Journal of Industrial Microbiology andBiotechnology, 2016, 43(1): 55-66. DOI:10.1007/s10295-015-1692-8 |
| [26] | Zhang JJ, Xu MJ, Ge XX, Zhang X, Yang TW, Xu ZH, Rao ZM. Reengineering of the feedback-inhibition enzyme N-acetyl-L-glutamate kinase to enhance L-arginine production in Corynebacterium crenatum. Journal of Industrial Microbiology and Biotechnology, 2017, 44(2): 271-283. DOI:10.1007/s10295-016-1885-9 |
| [27] | Guo J, Man ZW, Rao ZM, Xu MJ, Yang TW, Zhang X, Xu ZH. Improvement of the ammonia assimilation for enhancing L-arginine production of Corynebacterium crenatum. Journal of Industrial Microbiology and Biotechnology, 2017, 44(3): 443-451. DOI:10.1007/s10295-017-1900-9 |
| [28] | Jakoby M, Nolden L, Meier-Wagner J, Kr?mer R, Burkovski A. AmtR, a global repressor in the nitrogen regulation system of Corynebacterium glutamicum. Molecular Microbiology, 2000, 37(4): 964-977. DOI:10.1046/j.1365-2958.2000.02073.x |
| [29] | Zhang YP, Pohlmann EL, Roberts GP. GlnD is essential for NifA activation, NtrB/NtrC-regulated gene expression, and posttranslational regulation of nitrogenase activity in the photosynthetic, nitrogen-fixing bacterium Rhodospirillum rubrum. Journal of Bacteriology, 2005, 187(3): 342-348. |
