Yu Sun1,2, Zhihong Xie1

 , Wei Liu1, Hongen Guo3
, Wei Liu1, Hongen Guo3 1. Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai 264003, Shandong Province, China;
2. University of Chinese Academy of Sciences, Beijing 100049, China;
3. The Sericultural Research Institute of Shandong Province, Yantai 264002, Shandong Province, China
Received: 4 December 2018; Revised: 17 January 2019; Published online: 24 January 2019
Foundation item: Supported by the National Natural Science Foundation of China (31570063, 31870020), by the Shandong Key Research and Development Program (2017GSF17129), by the Shandong Key Scientific and Technological Innovation Program (2017CXGC0303), by the Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences (+CXGC2016B10)
Corresponding author: Zhihong Xie, Tel:+86-535-2109183, Fax:+86-535-2109000, E-mail:zhxie@yic.ac.cn.
Abstract: [Objective] c-di-GMP, an important second messenger regulating multiple functions of bacteria, is generally synthesized and hydrolysed by proteins containing GGDEF or EAL domain. In this study, we analyzed the genome-wide GGDEF/EAL domain-containing proteins of Azorhizobium caulinodans ORS571, and selected three GGDEF-EAL composite proteins (AZC_3085, AZC_3226 and AZC_4658) for functional analysis.[Methods] SMART and CLUSTALW were used for prediction and multi-alignment of GGDEF/EAL domain-containing proteins. Mutants were constructed by homologous recombination. Phenotypes including cell motility, exopolysaccharide (EPS) production, biofilm formation and nodulation with legume host were investigated.[Results] There were 37 GGDEF/EAL domain-containing proteins in A. caulinodans ORS571. Mutant △4658 showed deficiency in cell motility, while its EPS production and biofilm formation were higher than that of wild type. Mutant △4658 showed stronger competitiveness than wild type in competitive nodulation assay. The loss of AZC_4658 led to the increase of intracellular c-di-GMP level. Mutants △3085 and △3226 did not show obvious difference in comparison with wild type.[Conclusion] The vast number of GGDEF/EAL domain-containing proteins suggested that c-di-GMP may play an important role in signal transduction of ORS571. The GGDEF-EAL composite protein AZC_4658 was involved in cell motility, EPS production, biofilm formation and nodulation of A. caulinodans ORS571.
Keywords: Azorhizobium caulinodans ORS571c-di-GMPGGDEF/EAL domain
茎瘤固氮根瘤菌ORS571 GGDEF和EAL结构域蛋白的预测及功能研究
孙雨1,2, 解志红1

 , 刘卫1, 郭洪恩3
, 刘卫1, 郭洪恩3 1. 中国科学院烟台海岸带研究所, 山东 烟台 264003;
2. 中国科学院大学, 北京 100049;
3. 山东省蚕业研究所, 山东 烟台 264002
收稿日期:2018-12-04;修改日期:2019-01-17;网络出版日期:2019-01-24
基金项目:国家自然科学基金(31570063,31870020);山东省重点研发计划(2017GSF17129);山东省重大科技创新工程(2017CXGC0303);山东省农业科学院农业科技创新工程(+CXGC2016B10)
通信作者:解志红, Tel:+86-535-2109183, Fax:+86-535-2109000, E-mail:zhxie@yic.ac.cn.
摘要:[目的] 环二鸟苷酸c-di-GMP是细菌中广泛存在的第二信使,能够调控多种细胞功能。c-di-GMP的合成与水解分别由含有GGDEF结构域和EAL结构域的蛋白催化。本研究针对茎瘤固氮根瘤菌ORS571的GGDEF和EAL结构域相关蛋白进行基因组学分析,并对三个同时含有GGDEF和EAL结构域的蛋白(AZC_3085、AZC_3226和AZC_4658)进行功能研究。[方法] 利用SMART数据库对含有GGDEF和EAL结构域的蛋白进行结构域预测。利用CLUSTALW程序对蛋白序列进行比较分析。通过同源重组的方法构建突变株,并对突变株的细胞运动能力、胞外多糖合成、生物膜形成及与豆科宿主的结瘤等表型进行测定。[结果] 茎瘤固氮根瘤菌ORS571中一共存在37个GGDEF和EAL结构域蛋白。突变株△4658的运动能力较野生型有下降,但是其胞外多糖合成能力、生物膜形成能力和竞争性结瘤能力较野生型有提高。此外,实验结果表明突变株△4658的胞内c-di-GMP水平高于野生型。突变株△3085和△3226的各种表型与野生型相比没有明显差异。[结论] 茎瘤固氮根瘤菌ORS571编码如此大数量的GGDEF和EAL结构域蛋白,表明c-di-GMP可能在其信号转导过程中起到非常重要的作用。同时具有GGDEF和EAL结构域的蛋白AZC_4658对茎瘤固氮根瘤菌ORS571的运动能力、胞外多糖合成、生物膜形成及与宿主的结瘤起到一定的调节作用。
关键词:茎瘤固氮根瘤菌ORS571环二鸟苷酸GGDEF/EAL结构域
Azorhizobium caulinodans ORS571 is a symbiont of legume Sesbania rostrata and a versatile nitrogen fixer that could fix nitrogen not only in symbiotic state but also in free-living state[1]. The symbiotic relationship between A. caulinodans and S. rostrata leads to formation of nitrogen-fixing nodules[2]. Within these nodules, internalized A. caulinodans have suitable microaerobic conditions to fix atmospheric nitrogen used for S. rostrata nutrition.
c-di-GMP (bis-(3′-5′)-cyclic dimeric guanosine monophosphate) is an important bacterial secondary messenger that regulates bacterial motility, exopolysaccharide (EPS) production, biofilm formation, and virulence[3]. Generally, c-di-GMP is synthesized from GTP by diguanylate cyclase (DGC) containing GGDEF domain, and hydrolyzed into pGpG by phosphodiesterase (PDE) containing EAL domain[4-5]. GGDEF and EAL domains are often coupled to diverse sensory domains such as PAS, GAF, and REC, which could regulate the enzymatic activities by sensing of different extracellular or intracellular signals[6-7]. Many GGDEF/EAL domain-containing proteins have been identified to play roles in catabolism of c-di-GMP and bacterial physiology. In Vibrio cholerae, an EAL domain-containing protein VieA was identified to regulate biofilm formation and virulence to its host[8]. In Salmonella typhimurium, all 12 GGDEF domain-containing proteins were shown to be involved in cell motility, biofilm formation, and virulence[9]. In Pseudomonas aeruginosa, all GGDEF/EAL domain-containing proteins encoded in its whole genome have been analyzed and the results revealed a role for these proteins in biofilm formation and virulence[10].
Recently, 14 GGDEF/EAL domain-containing proteins of Sinorhizobium meliloti were shown to play different roles in motility, EPS production, and competitive nodulation on host legume[11]. And functions of two GGDEF-EAL composite proteins of Rhizobium etli were also analyzed[12]. To date, little is known about GGDEF/EAL domain-containing proteins of A. caulinodans ORS571. In this study, genomic analysis of all putative GGDEF/EAL domain-containing proteins is performed and 3 GGDEF-EAL composite proteins are selected for functional analysis. The results may benefit further investigations on roles of these proteins in the rhizobia-legume interaction.
1 Materials and Methods 1.1 Bioinformatic analysis The GGDEF/EAL domain-containing proteins of A. caulinodans ORS571 were identified by searching the SMART database (http://smart.embl.de/). The domain structures of proteins were predicted by the SMART program. The sequences of GGDEF/EAL domain-containing proteins were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/nuccore/158328513) (A. caulinodans ORS571, Accession number in GenBank: AP009384.1). Multi-alignment of sequences was performed by using CLUSTALW (https://www.genome.jp/tools-bin/clustalw).
1.2 Strains, plasmids, and culture conditions All strains and plasmids used in this study are listed in Table 1. A. caulinodans ORS571 and mutants were grown at 37 ℃ in TY medium or L3 minimal medium (–N, without NH4Cl; +N, with 10 mmol/L NH4Cl). The E. coli strains used for mutant construction were routinely grown at 37 ℃ in LB medium. Different antibiotics (nalidixic acid 25 μg/mL, ampicillin 100 μg/mL, kanamycin 25 μg/mL, gentamycin 50 μg/mL, and tetracycline 10 μg/mL) were added to select for appropriate bacterial strains.
Table 1. Strains and plasmids used in this study
| Strains or plasmids | Relevant characteristics | Source or reference |
| Strains | ||
| ??E. coli | ||
| ??DH5α | F- supE44 AlacU169 (?80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Transgen |
| ??Azorhizobium caulinodans | ||
| ??ORS571 | Wild type strain; AmpR, NalR | [13] |
| ???3085 | ORS571 derivative; AZC_3085 was substituted with a gentamycin cassette, AmpR, NalR, GenR | This study |
| ???3226 | ORS571 derivative; AZC_3226 was substituted with a gentamycin cassette, AmpR, NalR, GenR | This study |
| ???4658 | ORS571 derivative; AZC_4658 was substituted with a gentamycin cassette, AmpR, NalR, GenR | This study |
| Plasmids | ||
| ??pCM351 | Recombination vector for gentamycin substitution, cre/loxp, GenR, TcR | [14] |
| ??pCM351-3085UD | pCM351 with 714-bp upstream fragment and 709-bp downstream fragment of AZC_3085 flanking the gentamycin cassette | This study |
| ??pCM351-3226UD | pCM351 with 685-bp upstream fragment and 700-bp downstream fragment of AZC_3226 flanking the gentamycin cassette | This study |
| ??pCM351-4658UD | pCM351 with 718-bp upstream fragment and 728-bp downstream fragment of AZC_4658 flanking the gentamycin cassette | This study |
| ??pRK2013 | Helper plasmid, ColE1 replicon; Tra+, KmR | [15] |
表选项
1.3 Construction of mutants The mutants were constructed using the allelic exchange vectors pCM351, a vector carrying the cre/lox system and gentamycin cassette[14]. DNA fragments flanking upstream and downstream of each gene were amplified from ORS571 genomic DNA and cloned into pCM351. The recombinant vectors were introduced into A. caulinodans ORS571 by tri-parental conjugation with the helper plasmid pRK2013. Subsequent allelic exchange between the ORS571 chromosome and the recombinant plasmid led to the substitution of target genes with gentamycin. The resulting gentamycin resistant mutants were further confirmed by PCR with corresponding primer pairs. All the primers used in mutant construction are listed in Table 2.
Table 2. Primers used in this study
| Primers | Sequences (5′→3′) |
| 3085 Up-Xba I-F | TCTAGAGCGCACATAGGTCTGG |
| 3085 Up-Nde I-R | CATATGCGCAGGGATTCCTCTT |
| 3085 Down-Age I-F | ACCGGTAGATGGAAGGTGTCAG |
| 3085 Down-Sac I-R | GAGCTCCAAGCAGGTGGACTCT |
| 3226 Up-Xba I-F | TCTAGACGCTGCAGGATCCAGC |
| 3226 Up-Nde I-R | CATATGGAGAGGTCCGCAACAG |
| 3226 Down-Age I-F | ACCGGTAGAACGCTCCTACCTC |
| 3226 Down-Sac I-R | GAGCTCATCTCGCGGGATCTTG |
| 4658 Up-Xba I-F | TCTAGAGAAGGACTTCCCGCAC |
| 4658 Up-Nde I-R | CATATGAAGACATGCGTGCCCG |
| 4658 Down-Age I-F | ACCGGTTGCAGGGCTTCCACTT |
| 4658 Down-Sac I-R | GAGCTCTTCGAGAATGCGACGC |
| Restriction sites are underlined. | |
表选项
1.4 Growth curve assay The growth curves of ORS571 and mutants were measured based on the method descried before[16]. ORS571 and mutants were grown overnight in TY liquid medium at 37 ℃. The overnight-grown culture was then diluted to 20 mL TY liquid medium at an initial OD600 of 0.02 and incubated at 37 ℃. The cell density was monitored by spectrophotometer Nanodrop 2000c (Thermo Scientific) every 2 h (0–12 h), 3 h (12–24 h), and 6 h (24–48 h).
1.5 Motility assay The motility assay was performed according to the protocol reported previously[17], with some modifications. Overnight grown culture of strains was normalized to an OD600 of 1.0. Aliquots of 5 μL of normalized cell suspensions were inoculated on 0.3% soft agar TY plates. After 48 h of incubation at 37 ℃, the swimming diameters of strains on soft agar plates were measured.
1.6 Extraction and quantification of intracellular c-di-GMP The extraction of intracellular c-di-GMP was performed as described previously[18]. The concentration of extracted c-di-GMP was analysed by HPLC-MS and normalized to the amount of whole cell protein. The HPLC column was reverse phase C18 column (Thermo Scientific Hypersil GOLD, 3 μm particle size), column temperature was 25 ℃, and the mobile phases were solvent A: 15 mmol/L acetic acid in 97% water and 3% methanol, and solvent B: 100% methanol. The solvents were used to create a 50 min gradient: 0 min, 100% A : 0 B; 5 min, 100% A : 0 B; 10 min, 80% A : 20% B; 15 min, 80% A : 20% B; 30 min, 35% A : 65% B; 33 min, 5% A : 95% B; 37 min, 5% A : 95% B; 38 min, 100% A : 0 B; 50 min, 100% A : 0 B. The flow rate was 200 μL/min and the UV absorbance was measured at 254 nm. The eluent was then introduced into a tandem quadrupole mass spectrometer to detect the ions.
1.7 Quantification of EPS production Overnight grown cell cultures of strains were washed with L3+N liquid medium and normalized to an OD600 of 1.0. 20 μL aliquots of cell suspensions were inoculated on L3+N agar plates (0.8% agar) with or without 40 μg/mL of Congo red. Plates were incubated at 37 ℃ for 72 h. The plates containing Congo red were used for qualitative assay and the plates without Congo red were used for quantitative anthrone-sulfuric assay[13]. Cells harvested from plates without Congo red were resuspended in 500 μL of λ buffer (10 mmol/L Tris, 10 mmol/L MgSO4, pH 7.0). After centrifugation, 100 μL of the supernatants were mixed with 1 mL 0.2% anthrone dissolved in concentrated sulfuric acid. The mixtures were then incubated at 100 ℃ for 10 min and chilled on ice immediately. The harvested cells were resuspended with another 500 μL of λ buffer. The OD600 of the cell suspensions were measured and the OD620 of chilled mixtures were also measured with D-glucose as standard. EPSs production of each cell colony was then calculated by normalizing to the amount of cells.
1.8 Quantification of biofilm formation The biofilm formation was assayed by using Crystal Violet staining method as described[19], with some modifications. Overnight grown cell cultures of strains were washed with L3+N liquid medium and normalized to an OD600 of 0.2. 2 mL of normalized cell culture was added into each sterile glass tube. After 5 days of incubation at 37 ℃, cell cultures were removed and the tubes were washed with sterile water. For biofilm stain, 3 mL of 1% Crystal Violet was added into each tube. Crystal Violet was removed after 20 min of staining, and 1 mL of 30% acetic acid was added to dissolve the Crystal Violet-stained biofilms. The OD570 of each tube was then measured to determine the amount of biofilms.
1.9 Competitive nodulation assay The competitive nodulation assay was carried out according to Jiang et al[20]. S. rostrata seeds were treated with concentrated sulfuric acid for 30 min and washed thoroughly with sterile water. After that, the seeds were soaked in sterile water for germination in the dark at 37 ℃ for 48 h. Overnight grown cell cultures were washed with sterile L3-N liquid medium and normalized to an OD600 of 1.0. Normalized cell suspensions of ORS571 and mutants were then mixed with a 1:1 ratio, respectively. The seedlings were then inoculated with cell culture mixtures and planted in Leonard jars filled with vermiculite. All plants were grown in a greenhouse at 26 ℃ with a daylight illumination period of 12 h. The nodules were harvested after four weeks. The nodules were crushed and the strains were re-isolated by serial dilutions plated on TY plates. The re-isolated strains (at least 200 colonies) were then detected by PCR with corresponding primer pairs, and the relative nodulation ratios were calculated.
2 Results and Discussion 2.1 Identification of 37 putative GGDEF/EAL domain-containing proteins in A.caulinodans ORS571 The whole genome sequence of A. caulinodans ORS571 has been determined previously[21]. According to the prediction of SMART database, there are a total of 37 proteins containing GGDEF and/or EAL domains in A. caulinodans ORS571, including 23 proteins containing GGDEF domain, 4 proteins containing EAL domains, and 10 proteins containing both domains. The predicted domain structures of all proteins are shown in Figure 1. It has been reported that about 1/3 of known GGDEF/EAL domain-containing proteins contain both domains (known as GGDEF-EAL composite protein)[22]. Some composite proteins are shown to be single-function proteins, which are either PDEs or DGCs[5, 23-24], while only few bi-function composite proteins have been reported[25-26].
 |
| Figure 1 All GGDEF and EAL domains present in the genome of A. caulinodans ORS571. A: There were a total of 37 proteins containing GGDEF and/or EAL domains in ORS571, including 23 proteins containing GGDEF domain; B: 10 proteins containing both GGDEF and EAL domain; C: 4 proteins containing EAL domains. Domain structures of all these proteins were predicted by using the SMART program. Blue rectangles stand for transmembrane regions. |
| 图选项 |
There are 14 proteins containing different sensory domains, which are HAMP, REC, GAF, PAS and PAC respectively. HAMP domain generally functions as an intramolecular relay module transmitting conformational change and REC domain functions as receiver of the phosphorylation relay system[27-28]. GAF and PAS domains are capable of binding diverse small molecules, and PAC domains are associated with PAS domains and contribute to the PAS domain fold[29-30]. The present of diverse accompanying sensory domains suggest that the functions of GGDEF/EAL domain- containing proteins may be regulated by sensing of different signals. About 65% (24/37) of proteins have transmembrane regions, indicating that these proteins are targeted to the inner membrane of A. caulinodans ORS571. The association with inner membrane has been shown to facilitate co-localization of signals with target proteins and enhance specificity of signal transduction, thus allowing different signaling pathways to work independently[31]. In A. caulinodans ORS571, the inner membrane associated GGDEF/EAL domain- containing proteins may function in parallel without potential crosstalk.
It has been reported that rhizobia generally encode a high number of GGDEF/EAL domain-containing proteins[12]. The vast number of proteins in A. caulinodans ORS571 are comparable to other well-studied rhizobial strains: Bradyrhizobium japonicum USDA110 encodes 39 proteins, Rhizobium leguminosarum bv. viciae 3841 encodes 37 proteins, Rhizobium etli CFN42 encodes 35 proteins, and Mesorhizobium loti MAFF303099 encodes 34 proteins. The numbers indicate that c-di-GMP may play an important role in signal transduction of rhizobia.
2.2 Sequence analysis of GGDEF and EAL domains The multi-alignment was then performed to analyze the sequences of these domains. As shown in Figure 2, all GGDEF domains share about 45% sequence similarity. GGDEF domains are characterized by the conserved GG[D/E]EF motif, which forms catalytically active site (A-site) of DGCs[32]. Among 33 GGDEF domains, there are only 4 GGDEF domains from GGDEF/EAL composite proteins have degenerate motifs. This is consistent with previous report that about 62% of GGDEF-EAL composite proteins have the conserved GG[D/E]EF motifs[33]. It has been reported that GGDEF domain with a degenerate A-site (GEDEF motif) could allosterically activate the PDE activity of its neighbor EAL domain[23]. The inhibition site (I-site) RxxD motif is located five residues upstream of the conserved GG[D/E]EF motif, and c-di-GMP binding of I-site leads to allosteric inhibition of DGC activity[34]. Only 57% of 33 GGDEF domains contain intact inhibitory site, indicating that allosteric inhibition mechanism is not applied by all GGDEF domains. And it is interesting that GGDEF domains of all GGDEF-EAL composite proteins have degenerate inhibitory sites.
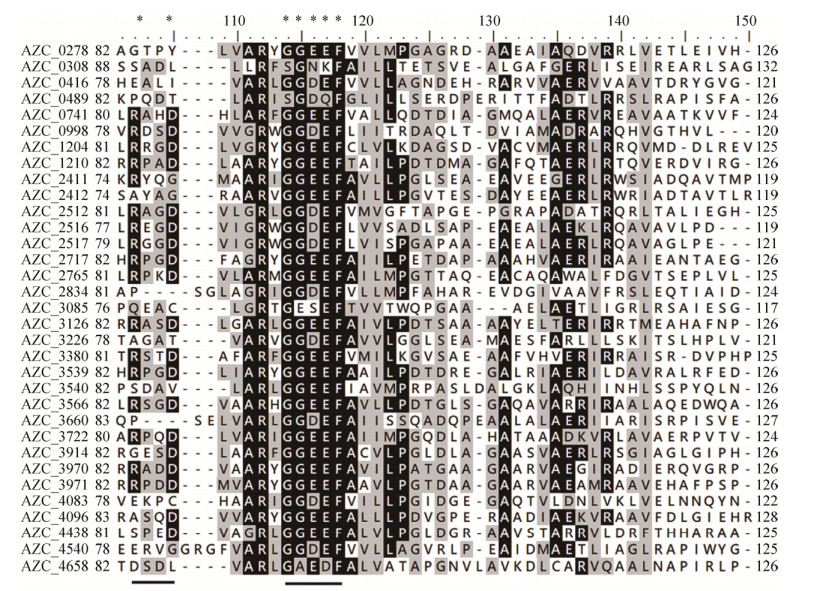 |
| Figure 2 Multi-alignment of selected sequences of all GGDEF domains. The backgrounds of conserved residues are labeled black and grey according to their conservation degree. The asterisks and underlines indicate the conserved RxxD and GGDEF motifs. |
| 图选项 |
As shown in Figure 3, all EAL domains share about 60% sequence similarity. 12 of 14 aligned EAL domains have the conserved EAL motif, while only one EAL domain has an EVL motif, and another EAL domain has an EAF motif. The most conservative residue of the EAL motif is the first Glu (E) residue, which is responsible for metal coordination in catalyzing[35]. The other two residues are considered as non-conservative residues. Although with an altered second residue (A→V), the EVL motif is a functional variant of EAL motif in documented PDEs. Previously, the EVL motif-containing proteins YahA from Escherichia coli and RocR from Pseudomonas aeruginosa have been proved to be functional PDEs[24, 36]. However, it needs to be investigated that whether EAF motif-containing EAL domain is truly functional. In addition to the EAL motif, there are other crucial motifs present in EAL domains: N, ExxE, DD and KxD, which are essential for metal coordination and catalysis[22]. And these motifs are mostly conservative for all aligned EAL domains.
 |
| Figure 3 Multi-alignment of selected sequences of all EAL domains. The backgrounds of conserved residues are labeled black and grey according to their conservation degree. The asterisks and underlines indicate the conserved EAL, DD and KxD motifs. |
| 图选项 |
2.3 Motility assay of GGDEF-EAL composite protein mutants In this study, three GGDEF-EAL composite proteins, AZC_3085, AZC_3226 and AZC_4658, were selected based on their motifs and sensory domains. To determine the roles of these composite proteins in physiology of A. caulinodans ORS571, single-gene mutants, ?3085, ?3226, and ?4658, were constructed and characterized. In S. meliloti Rm1021, several GGDEF/EAL proteins have been shown to be involved in motility[11]. Swimming motility of wild type and mutants was then examined on 0.3% soft agar TY plates. To exclude any effect on motility resulted from potential growth deficiency, the growth rates of strains were first determined in TY liquid medium. And all mutants had no obvious difference from wild type during 48 h of incubation (data not shown). The result also suggested that loss of these GGDEF-EAL composite proteins did not affect the growth of A. caulinodans ORS571. In Figure 4, the soft agar plates showed that the swimming diameters of mutants ?3085 and ?3226 were about the same with wild type. Different from these two mutants, mutant ?4658 showed deficiency in swimming motility (about 75% of wild type). It has been reported that c-di-GMP could repress swimming motility at high levels[37]. In this study, there is a possibility that the loss of protein AZC_4658 led to the increase of intracellular c-di-GMP level, and the motility of mutant ?4658 was then repressed. It could be speculated that the composite protein AZC_4658 functions as a PDE.
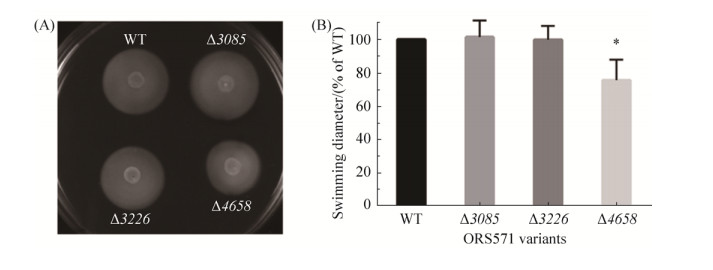 |
| Figure 4 The motility assay of wild-type ORS571 and mutants ?3085, ?3226 and ?4658. A: The swimming circles of wild-type ORS571 and mutants ?3085, ?3226 and ?4658 on TY plates. B: The quantitative determination of swimming diameters of wild-type ORS571 and mutants ?3085, ?3226, and ?4658. Error bars indicate standard deviations from three parallel replicates, and asterisks represent significant differences. *, P < 0.01. |
| 图选项 |
2.4 Intracellular c-di-GMP level of GGDEF- EAL composite protein mutants To determine whether the loss of composite protein AZC_4658 led to the increase of intracellular c-di-GMP level, extraction and quantification of intracellular c-di-GMP of wild type and mutants were performed. As shown in Figure 5, the intracellular c-di-GMP level of mutant ?4658 was about 2-fold higher than that of wild type, and the result favor the hypothesis that the composite protein AZC_4658 was a functional PDE. Different from mutant ?4658, the intracellular c-di-GMP level of mutants ?3085 and ?3226 had no obvious difference compared with wild type.
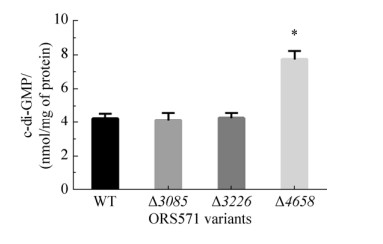 |
| Figure 5 The intracellular c-di-GMP level of wild-type ORS571 and mutants ?3085, ?3226, and ?4658. Error bars indicate standard deviations from three parallel replicates, and asterisks represent significant differences. *, P < 0.01. |
| 图选项 |
2.5 EPS production of GGDEF-EAL composite protein mutants To investigate the roles of GGDEF-EAL composite proteins in EPS production of ORS571, we determined the amount of EPS produced by wild type and mutants. Congo red is known to bind to polysaccharide and a qualitative assay based on the Congo red-binding was first applied to compare the EPS production. As shown in Figure 6, the appearance of colony formed by mutant ?4658 was stained deeper compared with wild type. The result of quantitative anthrone-sulfuric assay further confirmed that the EPS produced by mutant ?4658 was about 2-fold higher than that of wild type. However, the amount of EPS produced by mutants ?3085 and ?3226 was about the same with wild type. Generally, high level of intracellular c-di-GMP level stimulates production of EPS[38]. And the regulation of EPS production by c-di-GMP could function at different levels including transcriptional level or allosteric regulation of EPS synthase[39-40]. In this study, the regulation of AZC_4658 on EPS production may need further investigation.
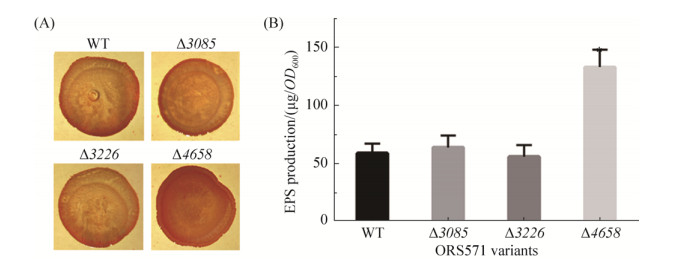 |
| Figure 6 The EPS production assay of wild-type ORS571 and mutants ?3085, ?3226, and ?4658. A: Colony phenotypes of wild-type ORS571 and mutants ?3085, ?3226, and ?4658 on L3+N Congo red plates. B: Quantification of EPS production of wild-type ORS571 and mutants ?3085, ?3226, and ?4658. Error bars indicate standard deviations from three parallel replicates, and asterisks represent significant differences. *, P < 0.01. |
| 图选项 |
2.6 Biofilm formation of GGDEF-EAL composite protein mutants In addition to EPS production, GGDEF/EAL domain-containing proteins have also been shown to be associated with biofilm formation in bacteria[9]. The biofilm formation of wild type and mutants was then assessed by using Crystal violet staining. As shown in Figure 7, the biofilm of mutant ?4658 formed at the air-liquid interface was much more than that of wild type. EPS is a major component of biofilm. The increased biofilm formation of mutant ?4658 was in consistence with increased EPS production. However, mutants ?3085 and ?3226 formed similar amount of biofilm with wild type. It has been reported that many single gene mutations of GGDEF/EAL proteins did not show obvious changes in phenotypes typically associated with c-di-GMP, such as motility, EPS production, and biofilm formation [41]. For the composite protein AZC_3085 and AZC_3226, it remains to be further investigated that whether they are functional or degenerate.
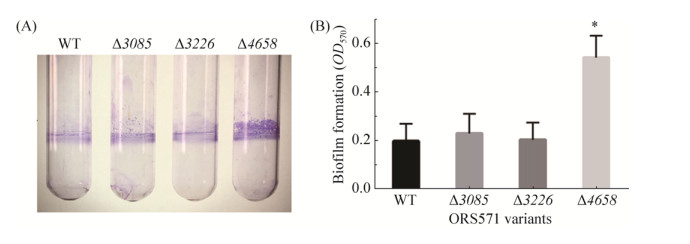 |
| Figure 7 The biofilm formation assay of wild-type ORS571 and mutants ?3085, ?3226, and ?4658. A: Crystal violet staining of biofilm formed by wild-type ORS571 and mutants ?3085, ?3226, and ?4658. B: Quantification of biofilm formation of wild-type ORS571 and mutants ?3085, ?3226, and ?4658. Error bars indicate standard deviations from three parallel replicates, and asterisks represent significant differences. *, P < 0.01. |
| 图选项 |
2.7 Nodulation of GGDEF-EAL composite protein mutants A competitive nodulation assay was performed to analyze the nodulation efficiency of these mutants. Wild type was inoculated on roots of S. rostrata in combination with each mutant in a 1:1 ratio, respectively, and the relative nodulation occupation ratios were determined when root nodules were harvested. As shown in Figure 8, the nodules induced by mutant ?3085 were about the same with wild type when inoculated together with a 1:1 ratio. And the mutant ?3226 also had similar nodulation efficiency compared with wild type in competitive nodulation assay. The nodulation occupation ratio of mutant ?4658 was about 1.5-fold higher than wild type, indicating that mutant ?4658 was more competitive than wild type when inoculated together in 1:1 ratio. The biofilm formation has been suggested to be involved in nodulation process[16]. The increased biofilm formation of mutant ?4658 could-at least partially-have contributed to its stronger competitiveness in nodulation. Together, the in vivo functional analysis revealed that the protein AZC_4658 may function as a PDE, and the in vitro analysis of protein function needs to be conducted to further confirm the speculation.
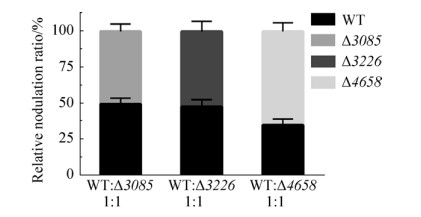 |
| Figure 8 The competitive nodulation assay of wild-type ORS571 and mutants ?3085, ?3226, and ?4658. Wild type and each mutant were mixed in 1:1 ratio, respectively. Error bars indicate standard deviations from three parallel replicates. |
| 图选项 |
In conclusion, the genomic analysis of all putative GGDEF/EAL domain-containing proteins of A. caulinodans ORS571 has provided new insights into the c-di-GMP signal transduction of A. caulinodans ORS571. And functional analysis of three GGDEF-EAL composite proteins showed that these proteins may play different roles in regulating physiological functions of A. caulinodans ORS571. Given the large number of GGDEF/EAL domain-containing proteins present in genomes of rhizobia, the study on c-di-GMP signal transduction may benefit our further understanding of rhizobia-legume symbiosis.
References
| [1] | Dreyfus B, Garcia JL, Gillis M. Characterization of Azorhizobium caulinodans gen. nov., sp. nov., a stem-nodulating nitrogen-fixing bacterium isolated from Sesbania rostrata. International Journal of Systematic Bacteriology, 1988, 38(1): 89-98. DOI:10.1099/00207713-38-1-89 |
| [2] | Goormachtig S, Capoen W, James EK, Holsters M. Switch from intracellular to intercellular invasion during water stress-tolerant legume nodulation. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(16): 6303-6308. DOI:10.1073/pnas.0401540101 |
| [3] | Hengge R. Principles of c-di-GMP signalling in bacteria. Nature Reviews Microbiology, 2009, 7(4): 263-273. DOI:10.1038/nrmicro2109 |
| [4] | Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(49): 17084-17089. DOI:10.1073/pnas.0406134101 |
| [5] | Rao F, Yang Y, Qi Y, Liang ZX. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. Journal of Bacteriology, 2008, 190(10): 3622-3631. DOI:10.1128/JB.00165-08 |
| [6] | Kazmierczak BI, Lebron MB, Murray TS. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Molecular Microbiology, 2006, 60(4): 1026-1043. DOI:10.1111/j.1365-2958.2006.05156.x |
| [7] | Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Molecular Microbiology, 2007, 65(6): 1474-1484. DOI:10.1111/j.1365-2958.2007.05879.x |
| [8] | Tamayo R, Tischler AD, Camilli A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. Journal of Biological Chemistry, 2005, 280(39): 33324-33330. DOI:10.1074/jbc.M506500200 |
| [9] | Solano C, García B, Latasa C, Toledo-Arana A, Zorraquino V, Valle J, Casals J, Pedroso E, Lasa I. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(19): 7997-8002. DOI:10.1073/pnas.0812573106 |
| [10] | Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3'-5')-cyclic-GMP in virulence. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(8): 2839-2844. DOI:10.1073/pnas.0511090103 |
| [11] | Wang YW, Xu J, Chen AM, Wang YZ, Zhu JB, Yu GQ, Xu L, Luo L. GGDEF and EAL proteins play different roles in the control of Sinorhizobium meliloti growth, motility, exopolysaccharide production, and competitive nodulation on host alfalfa. Acta Biochimica et Biophysica Sinica, 2010, 42(6): 410-417. DOI:10.1093/abbs/gmq034 |
| [12] | Gao SJ, Romdhane SB, Beullens S, Kaever V, Lambrichts I, Fauvart M, Michiels J. Genomic analysis of cyclic-di-GMP-related genes in rhizobial type strains and functional analysis in Rhizobium etli. Applied Microbiology and Biotechnology, 2014, 98(10): 4589-4602. DOI:10.1007/s00253-014-5722-7 |
| [13] | Nakajima A, Aono T, Tsukada S, Siarot L, Ogawa T, Oyaizu H. Lon protease of Azorhizobium caulinodans ORS571 is required for suppression of reb gene expression. Applied and Environmental Microbiology, 2012, 78(17): 6251-6261. DOI:10.1128/AEM.01039-12 |
| [14] | Marx CJ, Lidstrom ME. Broad-host-range cre-lox system for antibiotic marker recycling in Gram-negative bacteria. BioTechniques, 2002, 33(5): 1062-1067. DOI:10.2144/02335rr01 |
| [15] | Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proceedings of the National Academy of Sciences of the United States of America, 1979, 76(4): 1648-1652. DOI:10.1073/pnas.76.4.1648 |
| [16] | Liu W, Yang JB, Sun Y, Liu XL, Li Y, Zhang ZP, Xie ZH. Azorhizobium caulinodans transmembrane chemoreceptor TlpA1 involved in host colonization and nodulation on roots and stems. Frontiers in Microbiology, 2017, 8: 1327. DOI:10.3389/fmicb.2017.01327 |
| [17] | Wei C, Jiang WD, Zhao MR, Ling JJ, Zeng X, Deng J, Jin DL, Dow JM, Sun WX. A systematic analysis of the role of GGDEF-EAL domain proteins in virulence and motility in Xanthomonas oryzae pv. oryzicola. Scientific Reports, 2016, 6: 23769. DOI:10.1038/srep23769 |
| [18] | Russell MH, Bible AN, Fang X, Gooding JR, Campagna SR, Gomelsky M, Alexandre G. Integration of the second messenger c-di-GMP into the chemotactic signaling pathway. mBio, 2013, 4(2): e00001-13. |
| [19] | Liu XL, Liu W, Sun Y, Xia CL, Elmerich C, Xie ZH. A cheZ-like gene in Azorhizobium caulinodans is a key gene in the control of chemotaxis and colonization of the host plant. Applied and Environmental Microbiology, 2017, 84(3): e01827-17. |
| [20] | Jiang N, Liu W, Li Y, Wu HL, Zhang ZH, Alexandre G, Elmerich C, Xie ZH, Voordouw G. A chemotaxis receptor modulates nodulation during the Azorhizobium caulinodans-Sesbania rostrata symbiosis. Applied and Environmental Microbiology, 2016, 82(11): 3174-3184. DOI:10.1128/AEM.00230-16 |
| [21] | Lee KB, de Backer P, Aono T, Liu CT, Suzuki S, Suzuki T, Kaneko T, Yamada M, Tabata S, Kupfer DM, Najar FZ, Wiley GB, Roe B, Binnewies TT, Ussery DW, D'Haeze W, den Herder J, Gevers D, Vereecke D, Holsters M, Oyaizu H. The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics, 2008, 9: 271. DOI:10.1186/1471-2164-9-271 |
| [22] | Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nature Reviews Microbiology, 2009, 7(10): 724-735. DOI:10.1038/nrmicro2203 |
| [23] | Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. Journal of Biological Chemistry, 2005, 280(35): 30829-30837. DOI:10.1074/jbc.M504429200 |
| [24] | Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. Journal of Bacteriology, 2005, 187(14): 4774-4781. DOI:10.1128/JB.187.14.4774-4781.2005 |
| [25] | Tarutina M, Ryjenkov DA, Gomelsky M. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. Journal of Biological Chemistry, 2006, 281(46): 34751-34758. DOI:10.1074/jbc.M604819200 |
| [26] | Bharati BK, Sharma IM, Kasetty S, Kumar M, Mukherjee R, Chatterji D. A full-length bifunctional protein involved in c-di-GMP turnover is required for long-term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology, 2012, 158: 1415-1427. DOI:10.1099/mic.0.053892-0 |
| [27] | Taylor BL. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Molecular Microbiology, 2007, 65(6): 1415-1424. DOI:10.1111/j.1365-2958.2007.05889.x |
| [28] | Bourret RB. Receiver domain structure and function in response regulator proteins. Current Opinion in Microbiology, 2010, 13(2): 142-149. DOI:10.1016/j.mib.2010.01.015 |
| [29] | Heikaus CC, Pandit J, Klevit RE. Cyclic nucleotide binding GAF domains from phosphodiesterases: structural and mechanistic insights. Structure, 2009, 17(12): 1551-1557. DOI:10.1016/j.str.2009.07.019 |
| [30] | Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annual Review of Microbiology, 2011, 65(1): 261-286. DOI:10.1146/annurev-micro-121809-151631 |
| [31] | Romling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiology and Molecular Biology Reviews, 2013, 77(1): 1-52. |
| [32] | Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annual Review of Genetics, 2006, 40(1): 385-407. DOI:10.1146/annurev.genet.40.110405.090423 |
| [33] | Galperin M, Nikolskaya A, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiology Letter, 2001, 203(1): 11-21. DOI:10.1111/j.1574-6968.2001.tb10814.x |
| [34] | Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. Allosteric control of cyclic di-GMP signaling. Journal of Biological Chemistry, 2006, 281(42): 32015-32024. DOI:10.1074/jbc.M603589200 |
| [35] | Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature, 2009, 459(7249): 1015-1018. DOI:10.1038/nature07966 |
| [36] | Chen MW, Kotaka M, Vonrhein C, Bricogne G, Rao F, Chuah MLC, Svergun D, Schneider G, Liang ZX, Lescar J. Structural insights into the regulatory mechanism of the response regulator RocR from Pseudomonas aeruginosa in cyclic di-GMP signaling. Journal of Bacteriology, 2012, 194(18): 4837-4846. DOI:10.1128/JB.00560-12 |
| [37] | Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. Journal of Bacteriology, 2012, 194(13): 3307-3316. DOI:10.1128/JB.00100-12 |
| [38] | Pérez-Mendoza D, Sanjuán J. Exploiting the commons: cyclic diguanylate regulation of bacterial exopolysaccharide production. Current Opinion in Microbiology, 2016, 30: 36-43. DOI:10.1016/j.mib.2015.12.004 |
| [39] | Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Molecular Microbiology, 2008, 69(2): 376-389. DOI:10.1111/j.1365-2958.2008.06281.x |
| [40] | Pérez-Mendoza D, Bertinetti D, Lorenz R, Gallegos MT, Herberg FW, Sanjuán J. A novel c-di-GMP binding domain in glycosyltransferase BgsA is responsible for the synthesis of a mixed-linkage β-glucan. Scientific Reports, 2017, 7: 8997. DOI:10.1038/s41598-017-09290-2 |
| [41] | Sch?per S, Krol E, Skotnicka D, Kaever V, Hilker R, S?gaard-Andersen L, Becker A, Stock AM. Cyclic di-GMP regulates multiple cellular functions in the symbiotic alphaproteobacterium Sinorhizobium meliloti. Journal of Bacteriology, 2016, 198(3): 521-535. DOI:10.1128/JB.00795-15 |
