刘阳1,2, 姜丽晶2, 邵宗泽1,2


1.哈尔滨工业大学市政环境工程学院, 黑龙江 哈尔滨 150090;
2.国家海洋局第三海洋研究所, 海洋生物遗传资源重点实验室, 福建 厦门 361005
收稿日期:2017-03-28;修回日期:2017-05-12;网络出版日期:2017-05-25
基金项目:国家自然科学基金(41672333);中国大洋专项(DY135);国家微生物资源平台项目(NIMR-2017-9)
*通信作者:邵宗泽, Tel:+86-592-2195321;Fax:+86-592-2085376;E-mail:shaozz@163.com
摘要:硫,作为生物必需的大量营养元素之一,参与了细胞的能量代谢与蛋白质、维生素和抗生素等物质代谢。自然界中,硫以多种化学形态存在,包括单质硫、还原性硫化物、硫酸盐和含硫有机物。硫氧化是硫元素生物地球化学循环的重要组成部分,通常是指单质硫或还原性硫化物被微生物氧化的过程。硫氧化细菌种类繁多,其硫氧化相关基因、酶和途径也多种多样。近几年,相关方面的研究已取得很多进展,但在不同层面仍存在一些尚未解决的科学问题。本文主要围绕硫氧化细菌的种类及硫氧化途径的研究进展进行了综述。
关键词: 硫氧化菌 类群 硫氧化途径
Advances in sulfur-oxidizing bacterial taxa and their sulfur oxidation pathways
Yang Liu1,2, Lijing Jiang2, Zongze Shao1,2


1.School of Municipal and Environmental Engineering, Harbin Institute of Technology, Harbin 150090, Heilongjiang Province, China;
2.Key Laboratory of Marine Biogenetic Resources, Third Institute of Oceanography, State Oceanic Administration, Xiamen 361005, Fujian Province, China
Received 28 March 2017; Revised 12 May 2017; Published online 25 May 2017
*Corresponding author: Zongze Shao, Tel:+86-592-2195321;Fax:+86-592-2085376;E-mail:shaozz@163.com
Supported by the National Natural Science Foundation of China (41672333), by the COMRA Program (DY135) and by the Fund of National Infrastructure of Microbial Resources (NIMR-2017-9)
Abstract: Sulfur, as an essential element for organisms, is involved in cell energy and substance metabolism, i.e. protein, vitamins, antibiotics. In nature, sulfur exists in multiple valence states, represented respectively by element sulfur, reduced inorganic sulfur compounds (RISC) and sulfate as well as sulfur-containing organics. Sulfur oxidation is of significance in the biogeochemical sulfur cycle, which refers to the oxidation process of element sulfur and RISC by various microorganisms. Among sulfur-oxidizing microorganisms, sulfur-oxidizing bacteria are highly diverse, and their genes, enzymes and pathways of sulfur oxidation are varied. In recent years, significant progresses have been gained in many aspects, but some important issues at different levels still need to be solved. In this paper, we summarized the progresses of sulfur-oxidizing bacterial taxa and their sulfur oxidation pathways.
Key words: sulfur-oxidizing bacteria taxa sulfur oxidation pathway
硫,在自然界中广泛存在,是构成生物有机体所必需的大量元素之一。硫原子最外层有6个电子(e-),它们能以多种方式成键,进而形成单质或多种化合价,比如-2、0、+2、+4和+6价。常见的还原性硫化物包括H2S、硫化物、硫代硫酸盐(S2O32-)和亚硫酸盐(SO32-)等。微生物参与的生物地球化学硫循环包括硫氧化和硫还原两个过程[1],本文将阐述细菌参与的硫氧化过程。硫氧化菌(Sulfur-oxidizing bacteria,SOB)通过氧化单质硫或还原性硫化物产生能量或者参与光合作用[1]。目前,多个类群的SOB已被分离和鉴定。这些SOB能够在不同温度、pH和氧气浓度等条件下进行硫氧化,暗示SOB可能存在多种酶和硫氧化途径[2]。为了获得更加清晰的认识和理解,本文对硫氧化菌的种类及其硫氧化途径的研究进行了综述。
1 硫氧化细菌 SOB是指将低价态的还原性硫化物或单质硫完全氧化为硫酸盐(SO42-)或部分氧化为更高价态的硫化物的类群。目前研究表明:SOB不但种类多样,而且分布广泛,在海洋、热液口、冷泉、土壤、河流和湖泊等多种环境都有发现[2]。本部分将重点介绍4类研究较为深入的SOB,依次为绿硫细菌、紫硫细菌、紫色非硫细菌和无色硫细菌(图 1和表 1)。其他类群SOB不再详述,比如Firmicutes中的Alicyclobacillus spp.、绿色非硫细菌中的Chloroflexus aurantiacus和Aquificae中的Sulfurihydrogenibium spp.等。
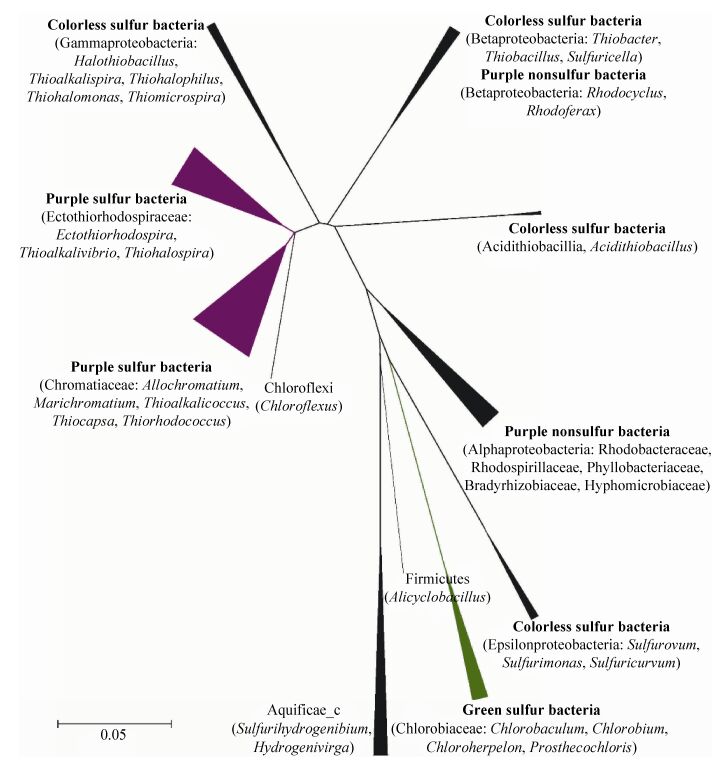 |
| 图 1 本研究基于硫氧化菌代表菌株16S rRNA基因序列构建的邻位相接系统发育树 Figure 1 The neighbor-joining phylogenetic tree based on 16S rRNA gene sequences of the representative members of sulfur-oxidizing bacteria in this study. Bar, 0.05 nucleotide substitution rate, (Knuc) units. |
| 图选项 |
表 1. 硫氧化菌主要类群代表菌株的代谢特征[3] Table 1. Metabolic features of representative SOB from four main taxa[3]
| Taxonomic affiliation | Metabolic features | Representative species | Molecular system(s) |
| GSB Chlorobi | Obligate phototrophy; S2-, S0 or S2O32- as e- donors for reduction of CO2; extracellular S0 globules; potential mixotrophy | Chlorobaculum tepidum, Chlorobaculum thiosulfatiphilum | SoxXAYZB, APS reductase, Qmo complex and Fcc. |
| PSB Chromatiaceae | Photoautotrophy except for Rheinheimera spp.; S2- and S0 as e- donors of photosynthesis; intracellular S0 globules | Allochromatium warmingi, Isochromatium buderi | - |
| Ectothiorhodospiraceae | Oxidation of S2- for all the members; extracellular S0 globules; polysulfides under alkaline conditions; Some can oxidize S2O32- to SO42- | Allochromatium vinosum, Ectothiorhodospira vacuolata | SoxXAYZB, Sqr, DsrABEFHCMKLJOPNRS, APS reductase and Fcc. |
| PNSB Alphaproteobacteria | The preferred photoheterotrophy under anaerobic conditions; photolithoautotrophy with S2-/S2O32- | Rhodopseudomonas palustris | SoxXAYZBCD, SoxEF and Sqr. |
| Betaproteobacteria | Chemoorganotrophy/chemolithoautot-rophy under aerobic or microaerobic conditions | Rhodocyclus purpureus | - |
| CSB Alphaproteobacteria | Facultative chemolithoautotrophy; Oxidation of S2-, S0, S2O32- or SO32- to SO42- | Paracoccus spp. | SoxXAYZBCD and SoxEF. |
| Acidithiobacillia | Obligate chemolithoautotrophy; Oxidation of S0, S2O32- or S4O62- by the incomplete Sox system; S0 globules as intermediates | Acidithiobacillus ferrooxidans | SoxXAYZB, DoxDA, GSSH, and Sqr. |
| Gammaproteobacteria | Obligate chemolithoautotrophy; extracellular S0 globules under low oxygen/pH; Transient accumulation of SO32- or polythionate during S0 globules or S2O32- oxidation | Thiomicrospira crunogena | SoxXAYZBCD and Sqr. |
| Gammaproteobacteria | Chemolithoheterotrophy/mixotrophy; intracellular S0 globules | Beggiatoa spp. | Dsr, Sqr and APS reductase |
表选项
绿硫细菌(Green sulfur bacteria,GSB)主要是指隶属于Chlorobi门Chlorobia纲Chlorobiales目Chlorobiaceae科的Chlorobaculum、Chlorobium、Chloroherpeton和Prosthecochloris等属的菌株[3]。而最近确定的Chlorobi门Ignavibacteriales目Ignavibacteriaceae科的Ignavibacterium album和Melioribacter roseus两个新类群虽然也属于GSB,但它们均没有硫氧化能力。GSB属于厌氧菌,能够厌氧氧化单质硫和H2S,少数菌株可以厌氧氧化S2O32-[2]。GSB硫化物氧化生成胞外单质硫。Gregersen等发现GSB中许多硫氧化基因是通过水平转移从其他类群中获得的[4]。
紫硫细菌(Purple sulfur bacteria,PSB)主要是指隶属于Gammaproteobacteria纲Chromatiales目中的Chromatiaceae科和Ectothiorhodospiraceae科的菌株[2]。这两个科的代谢中间物单质硫的贮存部位不同,前者在周质空间,后者在细胞外[2];而且,周质空间的单质硫存在蛋白包囊,但细胞外单质硫没有。这两个科中硫氧化研究最为详细的代表分别是Allochromatium vinosum DSM 180T[5]和Thioalkalivibrio属中的菌株[6]。
紫色非硫细菌(Purple nonsulfur bacteria,PNSB)主要包括Alphaproteobacteria纲中的Rhodospirillaceae、Acetobacteraceae、Rhodobacteraceae、Bradyrhizobiaceae和Hyphomicrobiaceae等科的菌株和Betaproteobacteria纲(比如Rhodocyclus和Rhodoferax)的菌株[2]。大多数PNSB硫化物氧化的终产物是SO42-,但是少数菌是单质硫。虽然PNSB耐受硫化物能力有差异,但总体而言,PNSB耐受硫化物的浓度比GSB和PSB更低。同时,与GSB和PSB相比,PNSB能够在好氧和微氧条件下进行硫氧化。因此,PNSB的分布更广泛。
无色硫细菌(Colorless sulfur bacteria,CSB)主要是指Betaproteobacteria、Gammaproteobacteria、Epsilonproteobacteria和少数Alphaproteobacteria纲的菌株[7]。比较常见的属是Paracoccus、Thiobacillus、Acidithiobacillus、Thiomicrospira和Thioalkalimicrobium等。CSB是因其缺少光合色素而得名。最近研究表明,CSB的硫氧化能力和途径与其分类地位没有相关性。与上述三类SOB相比,大多数CSB是在好氧条件下进行硫氧化,少数可以在微氧或厌氧条件下进行硫氧化。
2 硫氧化途径 硫元素具有多个化合价,不同的还原性硫化物和单质硫存在不同的氧化途径[8]。下文将重点介绍硫化物、单质硫、硫代硫酸盐和亚硫酸盐等氧化途径。
2.1 硫化物氧化 硫化物氧化是指把HS-/S2-氧化成聚硫化物,而后形成单质硫的过程,通常发生在几乎所有GSB、PSB和大多数PNSB、CSB中。如图 2所示,这个过程通常涉及两种酶,Sulfide:quinone reductase (Sqr)和Flavocytochrome c sulfide dehydrogenase (Fcc)[8]。目前鉴定的Sqr均是膜结合单亚基黄素蛋白,根据其结构差异可划分为6种类型[4]。同一菌株中可以存在多种类型的Sqr。比如,Chlorobaculum tepidum TLS存在3种功能相异的Sqr[9]。而Fcc通常以可溶周质蛋白或者膜结合蛋白两种形式存在。Fcc由大亚基黄素蛋白FccB和小亚基血红素蛋白FccA组成[8]。在硫化物氧化过程中,Sqr将产生的e-通过它的辅因子FDA传递至醌池;Fcc则把e-传递给细胞色素c。在C. tepidum TLS中,Sqr和Fcc分别对高浓度和低浓度的硫化物具有高亲和性,因此它们在这两种情况下分别发挥作用[9]。但是,目前关于聚硫化物是如何形成单质硫;不同类型的单质硫是如何形成的和细胞内或周质空间产生的单质硫是如何被分泌到细胞外等问题有待于进一步研究。
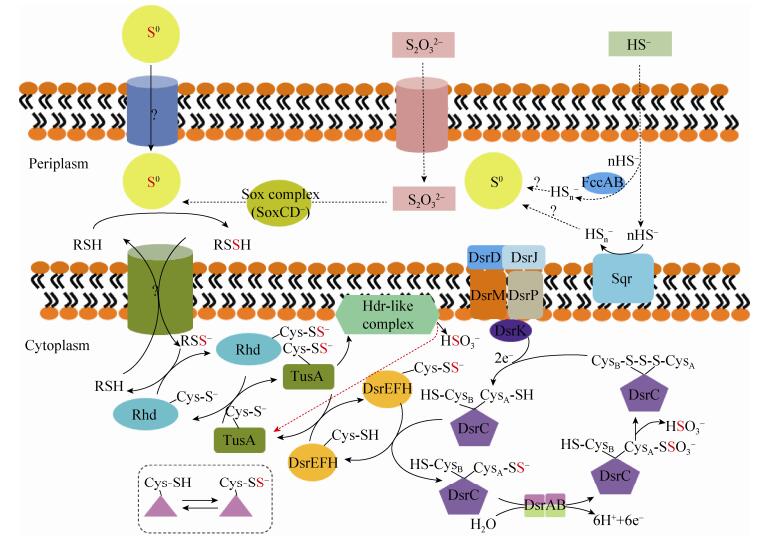 |
| 图 2 细菌硫化物和单质硫氧化途径[10] Figure 2 The pathways of sulfide and elemental sulfur oxidation in bacteria[10]. |
| 图选项 |
2.2 单质硫氧化 单质硫氧化是指把单质硫氧化为SO32-的过程,通常发生在几乎所有GSB、PSB和一些PNSB、CSB中。单质硫通常来自于环境中存在或者人工添加的外源单质硫和硫化物氧化产生的内源单质硫。目前,它包括Dsr (Reverse dissimilatory sulfite reduction,Dsr)和Hdr (Heterodisulide reductases-like,Hdr)两种途径(图 2)。
Dsr途径涉及到许多反向异化亚硫酸盐还原酶。Dahl课题组对A. vinosum DSM 180T的Dsr途径进行了详细的研究[11]。A. vinosum DSM 180T存在一个基于Cys-SSH的硫传递系统,它将单质硫中的硫原子依次通过蛋白Rhd、TusA、DsrEFH和DsrC转移至亚硫酸盐还原酶的活性位点,进而被氧化为SO32-[10]。在这个过程中,低分子量的有机过硫化物(比如谷氨酰胺过硫化物)被认为是将周质空间的硫转移至细胞质的载体[12],但是这些载体是如何产生的和是否有酶参与载体产生的过程等问题还不清楚。蛋白Rhd催化谷胱甘肽过硫化物中的硫转移到Cys残基上。蛋白TusA接受来自于Rhd蛋白的硫原子。在A. vinosum DSM 180T中,与蛋白TusA相互作用的复合体是DsrEFH,它是1个α2β2γ2结构的六聚体[13]。随后,硫通过DsrEFH复合体转移给蛋白DsrC。蛋白DsrC属于DsrC/TusE/RpsA超家族,包含2个保守的氧化还原活性Cys:CysA和CysB[14]。蛋白DsrC的2个活性Cys与接受到的硫原子在膜结合蛋白复合体DsrMKJOP的催化下形成1个三硫过氧化物,最后在DsrAB蛋白的催化下产生SO32-[15]。
Hdr途径存在于Aquifex aeolicus VF5[16]和Ectothiorhodospiraceae科[8]等的菌株。在Aquifex aeolicus VF5中,硫原子在蛋白Rhd和TusA间的转移与Dsr途径一致。随后,其在Hdr复合体的作用下,生成SO32-。Hdr复合体是1个膜结合蛋白,至少包含5个亚基[16]。最新研究表明,编码包含硫辛酰基结构域的蛋白LbpA在Hdr途径中也可能发挥重要功能[17]。但是,目前建立的Hdr途径仍需更多的研究来完善。另外,在Dsr和Hdr的途径中,SOB是如何结合、活化和吸收单质硫等问题也有待于研究。
2.3 硫代硫酸盐氧化 硫代硫酸盐氧化是指把S2O32-氧化为S4O62-或SO42-的过程,通常发生在大多数PNSB、CSB和少数GSB、PSB中。SOB存在2种S2O32-氧化途径(图 3):(1)酸性条件下,2S2O32-被氧化为S4O62-;(2)碱性和中性条件下,S2O32-被氧化为SO42-[8]。在A. vinosum DSM 180T[18]和Thiomicrospira thermophila EPR85[19]中,2种途径共存。
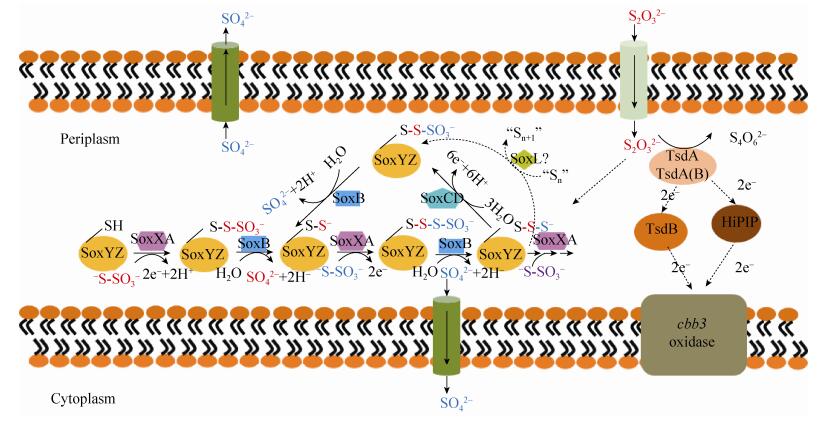 |
| 图 3 细菌硫代硫酸盐氧化途径[20-21] Figure 3 The pathways of thiosulfate oxidation in bacteria[20-21]. |
| 图选项 |
第1种途径是由蛋白TsdA (Thiosulfate dehydrogenase,Tsd)氧化2S2O32-生成S4O62-和2e-(图 3)。在A. vinosum DSM 180T中,蛋白TsdA是一个27.2 kDa含2个血红素的细胞色素c型的硫代硫酸盐脱氢酶[22]。在不同SOB中,2e-的受体不尽相同。在Thiomonas intermedia K12和Pseudomonas stutzeri A1501,蛋白TsdB被认为是e-受体[18]。在PSB中,铁硫蛋白HiPIP具有+350 mV氧化还原电位,据此推测它可能是TsdA的e-受体[8]。因此,这个途径中e-传递途径有待确认。
第2种氧化途径发生在周质空间中,S2O32-在完整Sox多酶复合体的作用下被彻底氧化生成SO42- (比如Paracoccus pantotrophus GB17) (图 3);或者在缺少SoxCD的多酶复合体的作用下被不完全氧化生成单质硫(比如A. vinosum DSM 180T) (图 3),然后如3.2所述,单质硫被进一步氧化为SO32- (图 2)。
Friedrich课题组的研究表明,P. pantotrophus GB17包含完整的Sox多酶复合体编码基因(soxRSVWXYZABCDEFGH),其中蛋白SoxXA、SoxYZ、SoxCD和SoxB构成了Sox多酶复合体的核心[1]。SoxXA复合体是由单血红素c型细胞色素SoxX和双血红素c型细胞色素SoxA组成,功能是催化S2O32-结合至SoxY蛋白的Cys138的巯基上。SoxYZ复合体是载体,SoxY蛋白的Cys138结合的硫依次与SoxXA、SoxCD和SoxB相互作用,最终被氧化为SO42-。SoxCD是包含钼辅因子的细胞色素复合体,催化6e-转移,氧化Cys-S-S-至硫砜氧化态,进而产生-Cys-S-SO32-,因此,SoxCD行使硫脱氢酶功能。SoxB蛋白是5′段包含锌核苷酸酶的同系物,催化-Cys-S-SO32-水解产生SO42-,同时释放SoxYZ,行使硫酸盐硫酯酶功能。SoxR是ArsR家族结合DNA的阻遏蛋白[23]。硫氧化蛋白SoxS是膜蛋白SoxV氧化还原伴侣[24]。膜蛋白SoxV包括六通道形成跨膜螺旋和两个在内侧相对的Cys,进而形成蛋白质二硫键来运输还原性物质。SoxW是成熟后包含166个氨基酸的周质硫氧还蛋白。P. pantotrophus GB17的多酶复合体需要SoxV发挥功能。核黄素蛋白SoxF可能激活SoxYZ复合体[25]。
Sox多酶复合体介导的S2O32-氧化过程如下:S2O32-中的硫原子首先与SoxYZ中SoxY亚基位于C端的Cys共价结合;然后,SoxXA催化SoxY中的Cys与S2O32-中的硫原子形成二硫化物,进而在SoxB的催化下释放SO42-;随后,SoxY结合的硫烷在SoxCD复合体的催化下形成硫砜;最后再在SoxB的催化下释放SO42-。因此,这个过程共释放2SO42-,产生8e-。如上所述,在Sox多酶复合体缺少SoxCD的情况时,S2O32-氧化过程会产生中间代谢物单质硫。目前推测:SoxY结合的硫烷在周质中以一种未知的机制形成单质硫。在此过程中,SoxL蛋白可能参与了SoxCD的循环[26]。
2.4 亚硫酸盐氧化 亚硫酸盐氧化是指SO32-在相应酶的作用下被氧化成SO42-。SO32-氧化包括由含钼因子亚硫酸盐氧化蛋白介导的直接途径和依赖腺苷酰硫酸(Adenosine-5-phosphosulfate,APS)的间接途径(图 4)。许多β-和γ-变形菌、绿硫细菌和G+菌中同时存在这两种途径。
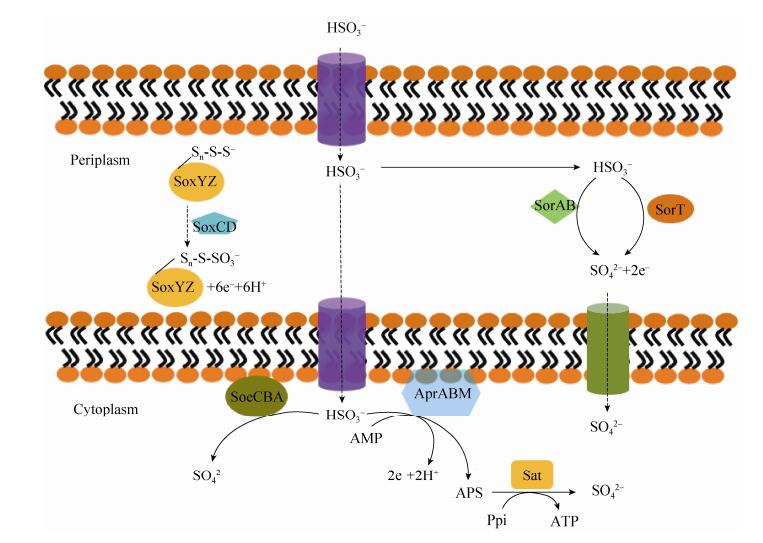 |
| 图 4 细菌亚硫酸盐氧化途径[27-29] Figure 4 The pathways of sulfite oxidation in bacteria[27-29]. |
| 图选项 |
直接途径中催化SO32-氧化的酶或复合体目前包括三种,即位于周质空间的SorAB (SorT)、SoxCD和位于细胞质中的SoeABC (图 4)。在Starkeya novella DSMZ 506T的SorAB复合体中,SorA亚基存在一个钼辅因子;SorB小亚基存在一个c522类型的血红素结合结构域[30]。在Campylobacter jejuni NCTC 11168中,类似的SorAB复合体也被鉴定[31]。而在Thermus thermophilus AT62,单独存在SorA被称为SorT[32]。包含钼辅因子和血红素的SoxCD复合体类似于SorAB,但SoxCD复合体介导的是6e-的反应,而SorAB复合体介导的是2e-的反应[29](图 4)。有研究表明,SoxCD复合体并不是SO32-氧化所必需的[33-34]。因此,SoxCD复合体介导SO32-氧化的途径目前存在争议,需要进一步研究。SoeABC复合体氧化SO32-的途径首先是在Ruegeria pomeroyi DSS-3中确定。这个复合体属于FeS钼家族,包含一个NrfD/PsrC类型的膜结合亚基SoeC[35]。A. vinosum DSM 180T的SoeABC复合体也包括3个亚基:包含钼和N端Fe4S4簇的SoeA亚基,结合4个Fe4S4簇的SoeB亚基,以及包括8个跨膜螺旋的膜蛋白SoeC亚基[27]。由于SoeAB蛋白没有TAT信号肽,因此,细胞质中的SoeAB被认为是与膜结合蛋白SoeC组成复合体,然后催化氧化SO32-(图 4)。另外,在A. vinosum DSM 180T利用SoeABC复合体氧化SO32-的过程中,SoxYZ复合体是必需的[27],但它们的作用细节有待于进一步确认。
许多GSB和PSB使用APS间接途径在细胞质氧化SO32-。在这个途径中,SO32-和腺苷单磷酸(Adenosine 5'-monophosphate,AMP)在AprABM复合体的作用下生成APS和2e-;随后,APS在腺苷磷酸硫酰酶(Sulfate adenylate transferase,Sat)的作用下生成SO42- (图 4),而产生的e-通过包含5个跨膜螺旋膜结合蛋白AprM[36]或者QmoABC复合体进入电子传递链[37]。
3 展望 综述SOB种类及其硫氧化途径研究不但有助于全面认识和理解硫元素生物地球化学循环,而且为SOB在农业、环境工程和冶金工业等领域的应用提供坚实的理论基础[38]。尽管如此,后续的研究仍需要重点关注3个方面。(1)硫氧化菌株资源需深入挖掘。自然界存在大量的SOB,但是,目前鉴定的SOB集中在上述四个类群。因此,我们需要利用各种方法来丰富SOB资源。目前,本课题组已从多种海洋生境(近海、深远海和热液区等)和多种样品(海水、沉积物、热液羽流和烟囱等)分离鉴定了众多SOB[39],包括许多新种[40]。同时,我们和Meier等采用免培养方法分别发现Sulfurimonas、Sulfurovum、Desulfobulbus、Arcobacter和Thiomicrospira等是热液区SOB的优势类群[41-42]。这表明海洋环境存在着大量新颖的SOB。因此,我们需要加大海洋环境(尤其是热液区)SOB资源的挖掘。同时,更多SOB的获得也将有助于发现新的硫氧化基因、酶类或途径等。(2)已建立的硫氧化途径需要重新认识。前些年,由于实验方法限制和认识的局限,对硫氧化途径理解可能存在偏差,随着各种组学方法的出现为重新认识硫氧化的途径提供了好的机遇。比如,本课题组对热液区分离的Thiomicrospira sp. S5的硫氧化途径的研究表明,该菌在利用Sox多酶复合体系的同时,可能还存在新的细胞内硫氧化途径。同时,Grabarczyk等通过一系列生化试验确认,在Sox多酶复合体参与的硫氧化过程中,SoxYZ-S才是S2O32-的载体,而不是SoxYZ[20]。(3)硫氧化调控机制研究需要加强。目前,细菌硫氧化途径的框架已基本建立,但是硫氧化的调控机制研究却很少。林建群课题组最近发现,在Acidithiobacillus caldus MTH-04中,σ45依赖性的双组分系统调控Sox系统[43];而双组分系统RsrS-RsrR调控S2O32-生成S4O62-的氧化途径[44];在Rhodobacter capsulatus BP503中,调控蛋白SqrR调控Sqr表达[45]。这些研究为后续探索硫氧化调控机制提供了很好的参考。本课题组也将重点关注热液环境来源SOB的硫氧化调控机制。我们发现Thiomicrosipa sp. S5在不同氧气浓度和pH等条件下,氧化S2O32-产生胞外单质硫的特性不同。但是,这些差异具体是由哪些基因调控则有待进一步研究。
References
| [1] | Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J. Prokaryotic sulfur oxidation. Current Opinion in Microbiology, 2005, 8(3): 253-259. DOI:10.1016/j.mib.2005.04.005 |
| [2] | Ghosh W, Dam B. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiology Reviews, 2009, 33(6): 999-1043. DOI:10.1111/j.1574-6976.2009.00187.x |
| [3] | Imhoff JF, Thiel V. Phylogeny and taxonomy of Chlorobiaceae. Photosynthesis Research, 2010, 104(2/3): 123-136. |
| [4] | Gregersen LH, Bryant DA, Frigaard NU. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Frontiers in Microbiology, 2011, 2: 116. |
| [5] | Weissgerber T, Dobler N, Polen T, Latus J, Stockdreher Y, Dahl C. Genome-wide transcriptional profiling of the purple sulfur bacterium Allochromatium vinosum DSM 180T during growth on different reduced sulfur compounds. Journal of Bacteriology, 2013, 195(18): 4231-4245. DOI:10.1128/JB.00154-13 |
| [6] | Sorokin DY, Muntyan MS, Panteleeva AN, Muyzer G. Thioalkalivibrio sulfidiphilus sp. nov., a haloalkaliphilic, sulfur-oxidizing gammaproteobacterium from alkaline habitats. International Journal of Systematic and Evolutionary Microbiology, 2012, 62(8): 1884-1889. |
| [7] | Muyzer G, Kuenen JG, Robertson LA. Colorless sulfur bacteria//Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F. The prokaryotes. Berlin Heidelberg:Springer, 2013:555-588. |
| [8] | Dahl C. Sulfur metabolism in phototrophic bacteria//Hallenbeck PC. Modern topics in the phototrophic prokaryotes:metabolism, bioenergetics, and omics. Cham:Springer International Publishing, 2017:27-66. |
| [9] | Chan LK, Morgan-Kiss RM, Hanson TE. Functional analysis of three sulfide:quinone oxidoreductase homologs in Chlorobaculum tepidum. Journal of Bacteriology, 2009, 191(3): 1026-1034. DOI:10.1128/JB.01154-08 |
| [10] | Dahl C. Cytoplasmic sulfur trafficking in sulfur-oxidizing prokaryotes. IUBMB Life, 2015, 67(4): 268-274. DOI:10.1002/iub.1371 |
| [11] | Dahl C, Engels S, Pott-Sperling AS, Schulte A, Sander J, Lübbe Y, Deuster O, Brune DC. Novel genes of the dsr gene cluster and evidence for close interaction of dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. Journal of Bacteriology, 2005, 187(4): 1392-1404. DOI:10.1128/JB.187.4.1392-1404.2005 |
| [12] | Frigaard NU, Dahl C. Sulfur metabolism in phototrophic sulfur bacteria. Advances in Microbial Physiology, 2009, 54: 103-200. |
| [13] | Dahl C, Schulte A, Stockdreher Y, Hong C, Grimm F, Sander J, Kim R, Kim SH, Shin DH. Structural and molecular genetic insight into a widespread sulfur oxidation pathway. Journal of Molecular Biology, 2008, 384(5): 1287-1300. DOI:10.1016/j.jmb.2008.10.016 |
| [14] | Stockdreher Y, Venceslau SS, Josten M, Sahl HG, Pereira IAC, Dahl C. Cytoplasmic sulfurtransferases in the purple sulfur bacterium Allochromatium vinosum: evidence for sulfur transfer from DsrEFH to DsrC. PLoS One, 2012, 7(7): e40785. DOI:10.1371/journal.pone.0040785 |
| [15] | Santos AA, Venceslau SS, Grein F, Leavitt WD, Dahl C, Johnston DT, Pereira IAC. A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science, 2015, 350(6267): 1541-1545. DOI:10.1126/science.aad3558 |
| [16] | Boughanemi S, Lyonnet J, Infossi P, Bauzan M, Kosta A, Lignon S, Giudici-Orticoni MT, Guiral M. Microbial oxidative sulfur metabolism:biochemical evidence of the membrane-bound heterodisulfide reductase-like complex of the bacterium Aquifex aeolicus. FEMS Microbiology Letters, 2016, 363(15): fnw156. DOI:10.1093/femsle/fnw156 |
| [17] | Ehrenfeld N, Levicán G, Parada P. Heterodisulfide reductase from Acidithiobacilli is a key component involved in metabolism of reduced inorganic sulfur compounds. Advanced Materials Research, 2013, 825: 194-197. DOI:10.4028/www.scientific.net/AMR.825 |
| [18] | Denkmann K, Grein F, Zigann R, Siemen A, Bergmann J, van Helmont S, Nicolai A, Pereira IAC, Dahl C. Thiosulfate dehydrogenase:a widespread unusual acidophilic c-type cytochrome. Environmental Microbiology, 2012, 14(10): 2673-2688. DOI:10.1111/emi.2012.14.issue-10 |
| [19] | Houghton JL, Foustoukos DI, Flynn TM, Vetriani C, Bradley AS, Fike DA. Thiosulfate oxidation by Thiomicrospira thermophila:metabolic flexibility in response to ambient geochemistry. Environmental Microbiology, 2016, 18(9): 3057-3072. DOI:10.1111/1462-2920.13232 |
| [20] | Grabarczyk DB, Berks BC. Intermediates in the Sox sulfur oxidation pathway are bound to a sulfane conjugate of the carrier protein SoxYZ. PLoS One, 2017, 12(3): e0173395. DOI:10.1371/journal.pone.0173395 |
| [21] | Kurth JM, Brito JA, Reuter J, Flegler A, Koch T, Franke T, Klein EM, Rowe SF, Butt JN, Denkmann K, Pereira IAC, Archer M, Dahl C. Electron accepting units of the diheme cytochrome c TsdA, a bifunctional thiosulfate dehydrogenase/tetrathionate reductase. The Journal of Biological Chemistry, 2016, 291(48): 24804-24818. DOI:10.1074/jbc.M116.753863 |
| [22] | Brito JA, Denkmann K, Pereira IAC, Archer M, Dahl C. Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum:structural and functional insights into thiosulfate oxidation. The Journal of Biological Chemistry, 2015, 290(14): 9222-9238. DOI:10.1074/jbc.M114.623397 |
| [23] | Rother D, Orawski G, Bardischewsky F, Friedrich CG. SoxRS-mediated regulation of chemotrophic sulfur oxidation in Paracoccus pantotrophus. Microbiology, 2005, 151(5): 1707-1716. DOI:10.1099/mic.0.27724-0 |
| [24] | Orawski G, Bardischewsky F, Quentmeier A, Rother D, Friedrich CG. The periplasmic thioredoxin SoxS plays a key role in activation in vivo of chemotrophic sulfur oxidation of Paracoccus pantotrophus. Microbiology, 2007, 153(4): 1081-1086. DOI:10.1099/mic.0.2006/004143-0 |
| [25] | Bardischewsky F, Quentmeier A, Friedrich CG. The flavoprotein SoxF functions in chemotrophic thiosulfate oxidation of Paracoccus pantotrophus in vivo and in vitro. FEMS Microbiology Letters, 2006, 258(1): 121-126. DOI:10.1111/fml.2006.258.issue-1 |
| [26] | Welte C, Hafner S, Kr?tzer C, Quentmeier A, Friedrich CG, Dahl C. Interaction between Sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Letters, 2009, 583(8): 1281-1286. DOI:10.1016/j.febslet.2009.03.020 |
| [27] | Dahl C, Franz B, Hensen D, Kesselheim A, Zigann R. Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum:identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology, 2013, 159(12): 2626-2638. |
| [28] | Kappler U. Bacterial sulfite-oxidizing enzymes. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 2011, 1807(1): 1-10. DOI:10.1016/j.bbabio.2010.09.004 |
| [29] | Quentmeier A, Kraft R, Kostka S, Klockenk?mper R, Friedrich CG. Characterization of a new type of sulfite dehydrogenase from Paracoccus pantotrophus GB17. Archives of Microbiology, 2000, 173(2): 117-125. DOI:10.1007/s002039900118 |
| [30] | Kappler U, Bailey S. Molecular basis of intramolecular electron transfer in sulfite-oxidizing enzymes is revealed by high resolution structure of a heterodimeric complex of the catalytic molybdopterin subunit and a c-type cytochrome subunit. The Journal of Biological Chemistry, 2005, 280(26): 24999-25007. DOI:10.1074/jbc.M503237200 |
| [31] | Myers JD, Kelly DJ. A sulphite respiration system in the chemoheterotrophic human pathogen Campylobacter jejuni. Microbiology, 2005, 151(1): 233-242. DOI:10.1099/mic.0.27573-0 |
| [32] | Di Salle A, D'Errico G, La Cara F, Cannio R, Rossi M. A novel thermostable sulfite oxidase from Thermus thermophilus:characterization of the enzyme, gene cloning and expression in Escherichia coli. Extremophiles, 2006, 10(6): 587-598. DOI:10.1007/s00792-006-0534-z |
| [33] | Rother D, Henrich HJ, Quentmeier A, Bardischewsky F, Friedrich CG. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. Journal of Bacteriology, 2001, 183(15): 4499-4508. DOI:10.1128/JB.183.15.4499-4508.2001 |
| [34] | Zander U, Faust A, Klink BU, de Sanctis D, Panjikar S, Quentmeier A, Bardischewsky F, Friedrich CG, Scheidig AJ. Structural basis for the oxidation of protein-bound sulfur by the sulfur cycle molybdohemo-enzyme sulfane dehydrogenase SoxCD. The Journal of Biological Chemistry, 2011, 286(10): 8349-8360. DOI:10.1074/jbc.M110.193631 |
| [35] | Lehmann S. Sulfite dehydrogenases in organotrophic bacteria:enzymes, genes and regulation. Doctoral Dissertation of Universit?t Konstanz, 2013. |
| [36] | Meyer B, Kuever J. Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5'-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology, 2007, 153(10): 3478-3498. DOI:10.1099/mic.0.2007/008250-0 |
| [37] | Ramos AR, Keller KL, Wall JD, Pereira IAC. The membrane QmoABC complex interacts directly with the dissimilatory adenosine 5'-phosphosulfate reductase in sulfate reducing bacteria. Frontiers in Microbiology, 2012, 3: 137. |
| [38] | Pokorna D, Zabranska J. Sulfur-oxidizing bacteria in environmental technology. Biotechnology Advances, 2015, 33(6): 1246-1259. DOI:10.1016/j.biotechadv.2015.02.007 |
| [39] | Xu HX, Jiang LJ, Li SN, Zhong TH, Lai QL, Shao ZZ. Diversity of culturable sulfur-oxidizing bacteria in deep-sea hydrothermal vent environments of the South Atlantic. Acta Microbiologica Sinica, 2016, 56(1): 88-100. (in Chinese) 徐鈜绣, 姜丽晶, 李少能, 钟添华, 赖其良, 邵宗泽. 南大西洋深海热液区可培养硫氧化微生物多样性及其硫氧化特性. 微生物学报, 2016, 56(1): 88-100. |
| [40] | Liu Y, Lai QL, Du J, Xu HX, Jiang LJ, Shao ZZ. Thioclava indica sp. nov., isolated from surface seawater of the Indian Ocean. Antonie van Leeuwenhoek, 2015, 107(1): 297-304. DOI:10.1007/s10482-014-0320-3 |
| [41] | Cao HL, Wang Y, Lee OO, Zeng X, Shao ZZ, Qian PY. Microbial sulfur cycle in two hydrothermal chimneys on the Southwest Indian Ridge. mBio, 2014, 5(1): e00980-13. |
| [42] | Meier DV, Pjevac P, Bach W, Hourdez S, Girguis PR, Vidoudez C, Amann R, Meyerdierks A. Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. The ISME Journal, 2017, doi:10.1038/ismej.2017.37. (in Press) |
| [43] | Li LF, Fu LJ, Lin JQ, Pang X, Liu XM, Wang R, Wang ZB, Lin JQ, Chen LX. The σ54-dependent two-component system regulating sulfur oxidization (Sox) system in Acidithiobacillus caldus and some chemolithotrophic bacteria. Applied Microbiology and Biotechnology, 2017, 101(5): 2079-2092. DOI:10.1007/s00253-016-8026-2 |
| [44] | Wang ZB, Li YQ, Lin JQ, Pang X, Liu XM, Liu BQ, Wang R, Zhang CJ, Wu Y, Lin JQ, Chen LX. The two-component system RsrS-RsrR regulates the tetrathionate intermediate pathway for thiosulfate oxidation in Acidithiobacillus caldus. Frontiers in Microbiology, 2016, 7: 1755. |
| [45] | Shimizu T, Shen JC, Fang MX, Zhang YX, Hori K, Trinidad JC, Bauer CE, Giedroc DP, Masuda S. Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(9): 2355-2360. DOI:10.1073/pnas.1614133114 |
