 ,1, 焦嘉杰1, 董宁宁1, 潘圆月2, 崔孟梅1, 潘玉善
,1, 焦嘉杰1, 董宁宁1, 潘圆月2, 崔孟梅1, 潘玉善 ,1
,1Analysis of Plasmid-Mediated AmpC β-lactamases Gene and Plasmid in Poultry Proteus mirabilis Strains
ZHAO ShiYu ,1, JIAO JiaJie1, DONG NingNing1, PAN YuanYue2, CUI MengMei1, PAN YuShan
,1, JIAO JiaJie1, DONG NingNing1, PAN YuanYue2, CUI MengMei1, PAN YuShan ,1
,1通讯作者:
责任编辑: 林鉴非
收稿日期:2020-08-28接受日期:2021-01-25
| 基金资助: |
Received:2020-08-28Accepted:2021-01-25
作者简介 About authors
赵世玉,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1604KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
赵世玉, 焦嘉杰, 董宁宁, 潘圆月, 崔孟梅, 潘玉善. 禽源奇异变形杆菌质粒介导AmpC酶基因型检测与质粒分析. 中国农业科学, 2021, 54(17): 3780-3788 doi:10.3864/j.issn.0578-1752.2021.17.018
ZHAO ShiYu, JIAO JiaJie, DONG NingNing, PAN YuanYue, CUI MengMei, PAN YuShan.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】奇异变形杆菌(Proteus mirabilis)是一种条件性致病菌,是引起人尿路、伤口、血液等感染的重要病原之一[1,2]。兽医临床中,奇异变形杆菌可以感染鸡、猕猴、狐狸、水貂、大熊猫、断奶仔猪等多种动物[3]。近几年,动物感染奇异变形杆菌的病例屡见报道,但有关奇异变形杆菌的流行调查比较少。近几年,杨睿等[4]对腹泻仔猪、王道宁等[5]对腹泻犬、路佳琦等[6]对某鸽场、袁东芳[7]对健康肉鸡进行了奇异变形杆菌的流行性调查,分离率在10%—30%。β-内酰胺酶是肠杆菌科细菌对β-内酰胺类抗生素耐药的主要机制,AmpC酶属于β-内酰胺酶的一个重要分支。目前,人源奇异变形杆菌对β-内酰胺类抗生素耐药率明显升高[8,9],但有关产AmpC酶动物源奇异变形杆菌的研究比较少。因此,本试验对禽源奇异变形杆菌进行AmpC酶检测与质粒分析,对掌握AmpC酶基因在该菌中的传播规律具有重要意义。【前人研究进展】根据Bush-Jacoby-Medeiros的分类方法,AmpC β-内酰胺酶属第一组,是一类不被克拉维酸和乙二胺四乙酸抑制的头孢菌素酶,按分子结构类型属于C类,也称为AmpC酶[10]。按ampC的来源分为染色体编码的AmpC酶和质粒介导的AmpC酶。对于染色体介导AmpC酶来说,ampC是该酶的结构基因,另外是几个不连锁的调节基因ampR、ampD、ampG、ampE等,可被β-内酰胺类抗生素诱导,主要见于阴沟肠杆菌、弗劳地枸橼酸杆菌、鲍曼不动杆菌、粘质沙雷菌及铜绿假单胞菌[11,12]。奇异变形杆菌染色体虽也能编码AmpC酶,但ampC主要是位于整合接合元件(integrating conjugative elements,ICEs)上,能进一步在大肠杆菌、肺炎克雷伯菌、沙门菌中转移[2,13]。质粒介导的AmpC酶主要出现在肺炎克雷伯菌、大肠杆菌、沙门菌及奇异变形杆菌,这些菌株无论有无β-内酰胺类抗生素存在均能持续高水平产AmpC酶。携带ampC的质粒可以通过接合方式在同种属和不同种属菌之间传播,这是ampC快速传播的一个重要途径[11]。质粒介导的AmpC酶已在世界范围内流行,CMY-2是其最流行的酶型[12]。【本研究切入点】奇异变形杆菌是一种常见的条件性致病菌。目前,有关禽源奇异变形杆菌产AmpC酶的研究比较少。前期研究中,对临床分离的21株禽源奇异变形杆菌已完成了超广谱β-内酰胺酶的检测和blaCTX-M基因的上下游环境研究[3],但有关AmpC酶的检测、基因亚型、质粒特征还未深入研究。【拟解决的关键问题】对临床分离的21株禽源奇异变形杆菌进行AmpC β-内酰胺酶和耐药基因检测,利用接合试验和脉冲场凝胶电泳(pulsed-field gel electrophoresis,PFGE)技术研究blaCMY-2在奇异变形杆菌中垂直和水平传播规律,利用高通量测序技术获得接合质粒的全序列,解析携带blaCMY-2质粒的分子特征,为防控多重耐药禽源奇异变形杆菌的传播提供理论基础。1 材料与方法
本研究于2017至2020年在河南农业大学牧医工程学院完成。1.1 菌株材料
1.1.1 菌株 2009年5—10月,从河北、山东、河南、四川、江西、浙江等8省12个地区收集的43例病死肉鸡、蛋鸡、鹌鹑肝脏中分离,并经实验室VITEK-32全自动微生物鉴定系统鉴定的21株奇异变形杆菌,其中10株产超广谱β-内酰胺酶(extended- spectrum β-lactamases,ESBLs),检出率为47.6%[3]。沙门菌H9812用于PFGE的DNA分子量标准,为实验室保存菌株。耐利福平大肠杆菌C600作为接合试验的受体菌,由河南省人民医院惠赠。大肠埃希菌(ATCC 25922),购自中国普通微生物菌种保存中心。1.1.2 主要试剂 麦康凯琼脂、MH琼脂、MH肉汤、营养肉汤均购自北京陆桥技术有限责任公司;质粒中量提取试剂盒购自德国QIAGEN公司;Marker 2000、Premix EX Taq购自宝生物工程(大连)有限公司。
1.1.3 药品 氨苄西林、头孢他啶、头孢噻肟、头孢西丁、环丙沙星、恩诺沙星、卡那霉素、阿米卡星、庆大霉素、氟苯尼考、多西环素、黏菌素,均为国产原料药,使用时均在有效期内。
1.2 试验方法
1.2.1 头孢西丁三维试验 通过三维试验检测21株奇异变形杆菌分离菌是否产生AmpC酶[14]。具体步骤如下:将0.5麦氏单位浊度的大肠杆菌ATCC 25922 按临床实验室标准化协会(CLSI)的K-B 法均匀涂布在M-H 平板上,在平板的中央贴一张头孢西丁(30 mg/片)纸片,从距离纸片5 mm 处用无菌刀片在平板琼脂上向外缘方向切一裂隙,在裂隙中加入25 μL 的一种待检菌株的β-内酰胺酶粗提物,35℃培养24 h,观察裂隙的内侧端(头孢西丁纸片的抑菌环内)周围有无细菌生长。结果判断:头孢西丁纸片的抑菌环有缺失者为阳性,提示待检菌株产AmpC酶。1.2.2 MIC值的测定 采用肉汤微量稀释法,测定受试菌株的最小抑菌浓度(MIC),相同过程重复3次,其结果根据临床实验室标准化协会(CLSI)的标准进行耐药和敏感的判断[15]。
1.2.3 耐药基因检测 依据文献[16],由上海生工生物工程公司合成6对通用引物。利用水煮法制备细菌DNA模板,对ampC基因进行PCR扩增。PCR扩增产物经1.0%琼脂糖凝胶电泳,最后由凝胶成像系统摄像。将PCR扩增阳性产物送上海生工生物工程公司测序,测序结果用NCBI数据库BLAST(
Table 1
表1
表1扩增AmpC酶基因的引物
Table 1
| 引物名称 Primer name | 引物方向(5′→3′) Primer sequence (5′→3′) | 产物长度 Size (bp) | 参考文献 Reference |
|---|---|---|---|
| MOX-F | GCTGCTCAAGGAGCACAGGAT | 520 | [16] |
| MOX-R | CACATTGACATAGGTGTGGTGC | [16] | |
| CIT-F | TGGCCAGAACTGACAGGCAAA | 462 | [16] |
| CIT-R | TTTCTCCTGAACGTGGCTGGC | [16] | |
| DHA-F | AACTTTCACAGGTGTGCTGGGT | 405 | [16] |
| DHA-R | CCGTACGCATACTGGCTTTGC | [16] | |
| ACC-F | AACAGCCTCAGCAGCCGGTTA | 346 | [16] |
| ACC-R | TTCGCCGCAATCATCCCTAGC | [16] | |
| EBC-F | TCGGTAAAGCCGATGTTGCGG | 302 | [16] |
| EBC-R | CTTCCACTGCGGCTGCCAGTT | [16] | |
| FOX-F | AACATGGGGTATCAGGGAGATG | 190 | [16] |
| FOX-R | CAAAGCGCGTAACCGGATTGG | [16] | |
| blaCMY-2-F | ATGATGAAAAAATCGTTATGC | 1146 | 本研究 This study |
| blaCMY-2-R | TTATTGCAGCTTTTCAAGAATG | 本研究 This study |
新窗口打开|下载CSV
1.2.4 接合试验 以耐头孢西丁奇异变形杆菌为供体菌,耐利福平大肠杆菌C600为受体菌进行接合试验。先把供体菌、受体菌分别接种到5 mL的LB肉汤中,于37℃振荡培养过夜。然后取供体菌0.1 mL、受体菌1 mL接种到5 mL LB肉汤中混匀,37℃低速振荡培养4 h。将混合培养后的菌液涂布在含药的麦康凯琼脂平板(利福平500 µg·mL-1,头孢西丁20 µg·mL-1),于37℃培养18 h,筛选阳性接合子。通过PCR和药物敏感性试验测定接合子的耐药基因和最小抑菌浓度(MIC)。
1.2.5 PFGE分型 利用PFGE技术对耐头孢西丁的奇异变形杆菌进行分型[17]。先将耐头孢西丁奇异变形杆菌和沙门菌H9812复苏、单菌落37℃过夜培养,然后制备含菌胶块、利用蛋白酶K进行菌体裂解、胶块洗涤、再利用Sfi I酶切奇异变形杆菌、用Xba I酶切沙门菌H9812,最后凝胶电泳、成像。CHEF Mapper脉冲场凝胶电泳系统的电泳参数设置为:片段大小为10—1 000 kb,电压6 V,角度120°,脉冲时间2.2—64 s,电泳时间18 h。
1.2.6 质粒测序与序列分析 利用Qiagen质粒中提试剂盒对获得的阳性接合子进行DNA提取,然后送天津生物芯片技术有限公司进行高通量测序。用NovaSeq 6000系统进行二代测序,测序结果用SOAP denovo v2.04 软件组装,然后用二代组装结果校正PacBio RSII三代测序结果,矫正后的三代测序结果用SMRT v2.3.0 软件进行组装,最终获得质粒的完整序列[18]。质粒的全序列先用RAST v2.0服务器(
2 结果
2.1 AmpC酶与基因亚型检测
三维试验结果显示,6株奇异变形杆菌的头孢西丁纸片的抑菌环有缺失,即这些菌株产AmpC酶。PCR扩增和测序表明,6株耐头孢西丁的奇异变形杆菌均携带blaCMY-2耐药基因,阳性率为28.6%(6/21)。2.2 接合试验
6株blaCMY-2阳性奇异变形杆菌中,只有菌株C12携带的blaCMY-2发生了接合转移,获得的相应接合子命名为TC12,其余5株接合不成功。药敏试验和PCR扩增结果表明,接合子TC12携带blaCMY-2,具有头孢西丁耐药表型(MIC=32 μg·mL-1),说明blaCMY-2位于一个可接合型质粒上,该质粒可从奇异变形杆菌转移到大肠杆菌。2.3 药敏试验
6株blaCMY-2阳性禽源奇异变形杆菌及接合子TC12对12种受试药物敏感性检测结果见表2。由表2可以看出,6株奇异变形杆菌对氨苄西林、头孢西丁、多西环素、氟苯尼考、粘菌素全部耐药;对头孢他啶、阿米卡星全部敏感;但有两株对头孢噻肟耐药,3株中介,1株敏感;有5株对环丙沙星、卡那霉素耐药,1株敏感。接合子TC12与原菌C12一样呈现出对氨苄西林、头孢西丁、多西环素、氟苯尼考、卡那霉素、庆大霉素耐药。Table 2
表2
表212种抗菌药物对6株blaCMY-2阳性禽源奇异变形杆菌及接合子TC12的MIC值
Table 2
| 抗菌药物 Antimicrobial agents MICs (µg·mL-1) | 菌株 Strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CY12 | CY32 | WS2 | S31 | S52 | C12 | TC12 | E. coli C600 | E. coli ATCC 25922 | |
| 氨苄西林Ampicillin | >256 | >256 | >256 | >256 | >256 | >256 | 256 | 4 | 2 |
| 头孢他啶Ceftazidime | 8 | 8 | 4 | 4 | 4 | 2 | 2 | <0.125 | <0.125 |
| 头孢噻肟Cefotaxime | 4 | 1 | 2 | 2 | 2 | 4 | 4 | <0.125 | <0.125 |
| 头孢西丁Cefoxitin | 32 | 32 | 64 | 32 | 32 | 32 | 32 | 2 | 2 |
| 环丙沙星Ciprofloxacin | 4 | 4 | 1 | 8 | 8 | 4 | <0.125 | <0.125 | <0.125 |
| 恩诺沙星Enrofloxacin | 16 | 16 | 4 | 32 | 32 | 32 | <0.125 | <0.125 | <0.125 |
| 卡那霉素Kanamycin | 256 | 256 | 2 | >256 | 128 | 128 | 64 | 4 | 4 |
| 阿米卡星Amikacin | 4 | 2 | 0.5 | 8 | 4 | 1 | 1 | <0.125 | <0.125 |
| 庆大霉素Gentamycin | 8 | 16 | 0.25 | 2 | 2 | 64 | 32 | <0.125 | <0.125 |
| 氟苯尼考Florfenicol | >256 | 256 | 8 | 128 | 128 | 64 | 128 | 2 | 0.5 |
| 多西环素Doxycycline | 128 | 64 | 128 | 64 | 64 | 64 | 64 | 0.25 | <0.125 |
| 粘菌素Colistin | >256 | >256 | >256 | >256 | >256 | >256 | <0.125 | <0.125 | <0.125 |
新窗口打开|下载CSV
2.4 PFGE分型
对上述检测到的6株blaCMY-2阳性奇异变形杆菌进行PFGE分型,根据菌株同源性判别标准,可将其分为3种PFGE型别,记为A—C,其中CY12、CY32和C12属于A型,S31和S52属于B型,WS2属于C型。CY12和CY32,S31和S52分别是分离自同一个养殖场的奇异变形杆菌菌株,这说明在一个养殖场里存在同一克隆菌株,耐药基因blaCMY-2存在垂直传播,如图1。图1
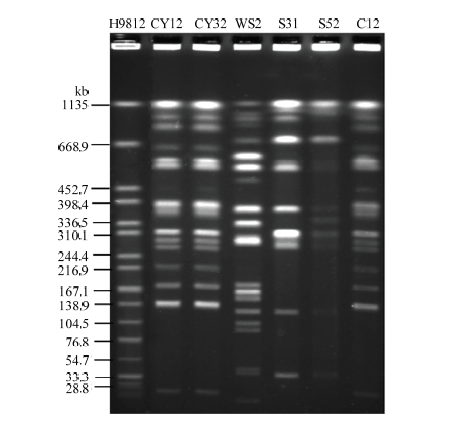 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图16株blaCMY-2阳性奇异变形杆菌的PFGE分型
Fig. 1Pulsed-Field Gel Electrophoresis of six Proteus mirabilis strains carrying blaCMY-2
2.5 质粒的分子特征
为进一步研究接合菌TC21携带的blaCMY-2阳性质粒的分子特征,利用全基因组测序技术对提取的质粒进行全序列测定。序列分析表明该接合菌含有一个广宿主的1b型IncC质粒,命名为pC12,其全长161 319 bp,GC含量为52.45%,预测有161个开放阅读框,提交NCBI并获得序列号MT320534。进一步序列分析表明,该质粒包含了基本的骨架区和3个耐药区。骨架区包含负责质粒复制、接合和稳定的功能基因。3个耐药区分散在质粒骨架区中:第一个耐药区ARI-B(antibiotic resistance islands,ARI-B)携带floR、tet(A)、strA、strB、sul2;第二个耐药区是一个典型的ISEcp1-blaCMY-2-blc-sugE结构,其中的ISEcp1被插入的IS10R截断;第三个耐药区ARI-A包含一个1类整合子基因盒(aac(6')-Ib-cr|arr3|dfrA27|aadA16)和汞抗性基因簇merRTPABDE,其插入到质粒骨架区产生两个重复序列TTGTA(图2)。ARI-A是一个Tn1696tnp-pDUmer杂合型转座子,tnpAR与Tn1696的tnpAR(序列号,U12338)序列相似性达100%,汞抗性基因簇merRTPABDE与质粒pDU1358的汞抗性基因簇相似度达99.6%。ARI-A两端的38 bp末端反向重复序列(IRtnp 和IRmer)分别由IS4321 和 IS5075插入而被截断,如图2。图2
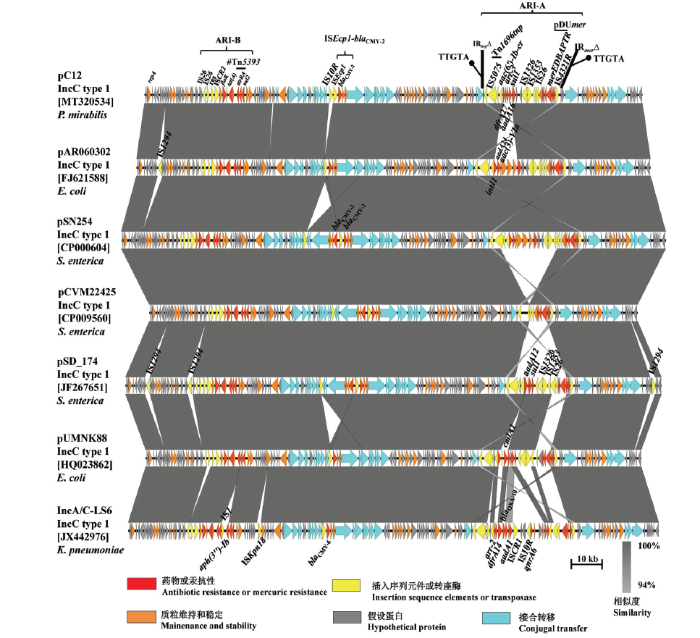 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2质粒pC12与其他blaCMY阳性IncC质粒的序列比较
奇异变形杆菌质粒pC12 (MT320534),大肠杆菌质粒pAR060302 (FJ621588)和pUMNK88 (HQ023862),沙门氏菌质粒pSN254 (CP000604),pCVM22425 (CP009560)和pSD_174 (JF267651),肺炎克雷伯氏菌质粒IncA/C-LS6(JX442976)
红色箭头代表药物或汞抗性基因;黄色箭头代表插入序列或转座酶基因;蓝绿色代表与接合转移相关的基因;橙色箭头代表与质粒维持和稳定相关的基因;灰色箭头代表编码假设蛋白的基因。灰色背景代表≥94%的相似度序列
Fig. 2Sequence comparisons of pC12 and other blaCMY-positive IncC plasmids
pAR060302 (FJ621588), pSN254 (CP000604), pCVM22425 (CP009560), pSD_174 (JF267651), pUMNK88 (HQ023862), IncA/C-LS6 (JX442976) from P. mirabilis, E. coli, S. enterica, and K. pneumoniae
Red arrows represent genes coding for antibiotic resistance or mercury resistance; yellow arrows represent genes coding for insertion sequence elements or transposase; cyan arrows represent genes coding for conjugal transfer; orange arrows represent genes coding for maintenance and stability; gray arrows represent genes coding for hypothetical proteins. Homologous segments generated by a BLASTn comparison (≥94% identity of nucleotide sequence) are shown as grey boxes
3 讨论
3.1 奇异变形杆菌的流行特征
奇异变形杆菌是动物源常见细菌之一。2019年杨睿等[4]从107份腹泻仔猪肛门拭子样本中分离出15株奇异变形杆菌,分离率为14.02%。2020年王道宁等[5]从62份犬腹泻粪便中分离得到12株奇异变形杆菌,分离率为19.35%。同年,路佳琦等[6]从某鸽场采集72份样品,分离获得22株奇异变形杆菌,分离率为30.6%;袁东芳[7]从肉鸡场采集健康鸡泄殖腔肛拭子516份,鉴定出61株奇异变形杆菌,分离率为11.8%。本研究从临床43份病死鸡中分离获得21株禽源奇异变形杆菌,分离率达48.84%。这些研究表明动物源奇异变形杆菌已成为养殖中常见的条件性致病菌,特别是在疾病状态下分离率更高。目前,人源奇异变形杆菌对β-内酰胺类抗生素耐药率明显升高[8,9]。在一项连续10年(2001—2010年)监测人源奇异变形杆菌耐药性变迁的研究中,2008、2009、2010年产超广谱β-内酰胺酶菌株分别占45.63%、56.98%、37.2%[9]。另一项人源奇异变形杆菌β-内酰胺酶检测及耐药性分析研究显示,302株分离菌中有60株只产ESBLs,检出率为19.87%;40株只产AmpC,检出率为13.25%;36株同时产ESBLs及AmpC,检出率为11.92%[8]。2015年冯福英等[19]从20株人源奇异变形杆菌中检测到10株携带blaCMY-2,阳性率达50%。2016年中国****LEI等[13]在125株动物源奇异变形杆菌中检测到16株携带blaCMY-2,阳性率为12.8%。2019年杨睿等[4]在分离的15株猪源奇异变形杆菌中检测到12株携带blaCMY-2,阳性率高达80%。在本课题组的前期研究[3]中,从21株禽源奇异变形杆菌中检测到10株携带blaCTX-M,检出率为47.6%,其中6株携带blaCTX-M-14,4株携带blaCTX-M-65。本试验进一步从21株菌中检测到6株单产AmpC酶,均携带blaCMY-2,阳性率为28.6%(6/21)。虽然本试验分离的禽源奇异变形杆菌数量有限,但一定程度上也反映出该菌对β-内酰胺类抗生素耐药率比较高。接合试验显示,6株耐头孢西丁奇异变形杆菌中只有1株接合成功,并获得了一株携带blaCMY-2阳性IncC型质粒的接合子,另外5株接合失败。blaCMY-2阳性菌株的酶切图谱分为3个PFGE型别。本次试验结果表明,blaCMY-2在禽源奇异变形杆菌中检出率比较高,blaCMY-2在该菌中存在垂直和水平传播。3.2 blaCMY-2在奇异变形杆菌的传播方式
AmpC β-内酰胺酶可以由染色体介导,也可以由质粒介导。一般认为介导AmpC酶的ampC基因起源于弗劳地枸橼酸杆菌的染色体,从染色体转座到质粒,再接合转移到肺炎克雷伯菌和大肠杆菌[20,21,22]。携带ampC基因的质粒通过接合方式可以在不同细菌之间传递,这种方式是基因快速传播的一个重要途径[11]。blaCMY-2是ampC基因的一种,与另外的ampC基因blaBIL-1和blaLAT-2实际是同一个基因,与大多数的blaCMY和blaLAT一样都来自于弗劳地枸橼酸杆菌的ampC [20]。blaCMY-2是肠杆菌科细菌携带ampC中最流行的一种[23,24]。有研究显示,117株人源AmpC阳性的肠杆菌科细菌中有78株携带blaCMY-2(66.7%),包括19株blaCMY-2阳性奇异变形杆菌,其中7株奇异变形杆菌基因组中含有携带blaCMY-2的ICEs(3株可发生接合转移,4株不接合),8株奇异变形杆菌携带blaCMY-2阳性IncC质粒[2,24]。ABERKANE等[25]报道显示,分离自法国的64株禽源和7株人源AmpC阳性奇异变形杆菌携带的ampC全部是blaCMY-2,其中67株奇异变形杆菌中的blaCMY-2位于ICEs上,另外4株的blaCMY-2位于接合性IncC质粒上。在中国,16株携带blaCMY-2的动物源奇异变形杆菌中检测到8株携带blaCMY-2的ICEs[13]。这些研究表明,SXT/R391型ICEs是blaCMY-2基因在奇异变形杆菌中传播的一种重要方式。本试验中5株blaCMY-2奇异变形杆菌接合失败,这些blaCMY-2的定位有待进一步研究。3.3 blaCMY-2阳性IncC质粒的结构特征
早期一些研究常把质粒IncA和IncC命名为IncA/C,但实际上IncA和IncC质粒的骨架区有着显著的序列差异[26,27]。IncC型质粒属广宿主不相容群,在大肠杆菌、沙门菌、奇异变形杆菌等肠杆菌科细菌中分布广泛[23,24]。近年来,作为多重耐药质粒类型之一的IncC型质粒,由于常携带AmpC酶基因blaCMY和金属β-内酰胺酶基因blaNDM而备受关注[28,29,30]。IncC型质粒可分为1型和2型,目前也有1/2杂合型IncC质粒的报道[26,31]。根据1型IncC质粒repA与ant-tox基因之间序列的单核苷酸多态性(SNP),该型质粒又进一步划分为1a和1b亚型[32]。ARI-B耐药区在1型和2型IncC质粒均有出现,但ARI-A和ISEcp1- blaCMY-2仅出现在1型IncC质粒中。1型IncC质粒是最流行的一种亚型,包含质粒骨架区和耐药区,其中质粒骨架区负责复制、接合、稳定等相关功能,3个耐药区分散在质粒骨架中,包括ARI-B、ISEcp1-blaCMY 和ARI-A[26,30]。第一个耐药区ARI-B由一个常见的ISCR2-floR-tet(A)-tetR-strBA-sul2结构组成。ARI-B被认为来源于一个15 kb的基因岛(genomic island GIsul2)。整合入IncC骨架区的GIsul2的右端被保留,中间片段被floR-tet(A)-tetR-strAB耐药区取代,左端丢失,由于IS26介导的插入或缺失导致产生不同大小的ARI-B[26]。第二个耐药区ISEcp1-blaCMY-2-blc-sugE是一个典型的结构,ISEcp1对blaCMY-2从弗劳地枸橼酸杆菌染色体转移到质粒和ampC表达发挥了重要作用。ISEcp1可以是完整的,也可以被其他插入序列元件IS1294,IS10,IS4等截短[12,33],部分IncC质粒在这个区域有两个拷贝的blaCMY-2,如pSN254、pCVM22425、pSD_174(图2)。第三个耐药区ARI-A由一个复合转座子构成。在pC12和其他IncC质粒pAR060302、pSN254、pCVM22425、pSD_174、pUMNK88和IncA/C-LS6中,ARI-A均由一个杂合的Tn1696tnp-pDUmer转座子组成,并插入到质粒骨架区产生两个重复序列TTGTA(图2)。ARI-A是多重耐药IncC质粒进化的主要耐药区,主要包含I型整合子盒,汞抗性基因簇以及与IS26相关的耐药模块。整合子是耐药基因的主要载体之一,可以携带多个不同耐药基因,进一步整合到IncC质粒的ARI-A耐药区,能够使质粒成为一个多重耐药基因散播的更大载体,加速耐药基因的扩散。冯福英等[19]从20株人源奇异变形杆菌中检测到10株携带blaCMY-2,阳性率达50%,19株携带I型整合子,阳性率达95%。杨睿等[4]对15株猪源奇异变形杆菌的分离鉴定与耐药基因分析中发现,奇异变形杆菌对β-内酰胺类抗生素、磺胺类抗生素、四环素类抗生素、氨基糖苷类抗生素和氯霉素类抗生素的耐药性主要是由blaCMY-2、sul2、tet(A)、aadA和floR耐药基因所导致,特别是blaCMY-2阳性率高达80%。本试验对一株禽源奇异变形杆菌携带的1型IncC质粒的全序列分析结果表明,IncC质粒是blaCMY-2、tet(A)、floR等多个耐药基因及整合子的重要载体之一,该质粒在动物源奇异变形杆菌的传播扩散进一步增加了治疗该菌感染的难度,应引起足够的重视。4 结论
本研究中,21株禽源奇异变形杆菌中有6株产AmpC β-内酰胺酶且均携带blaCMY-2,检出率为28.6%。6株blaCMY-2阳性菌株中有5株菌的blaCMY-2不发生接合转移,1株菌的blaCMY-2可以通过接合而发生水平传递。PFGE分型显示有3种型,blaCMY-2在同一场区中有垂直传播现象。序列分析表明,接合菌包含一个1b型IncC多重耐药质粒。该质粒携带了floR、tet(A)、strA、strB、sul2、blaCMY-2和一个杂合的Tn1696tnp-pDUmer转座子,其中转座子又携带了一个包含aac(6')-Ib-cr、arr3、drfA27、aadA16、sul1的1型整合子盒。应该注意的是,虽然头孢西丁在兽医临床应用很少,但在其他药物的选择压力下,动物源奇异变形杆菌中携带blaCMY-2的多重耐药IncC质粒会通过和其他基因的共转移而得到传播和扩散。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
DOI:10.1093/jac/dkr286URL [本文引用: 3]
[本文引用: 4]
[本文引用: 4]
[本文引用: 4]
[本文引用: 4]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 3]
[本文引用: 3]
[本文引用: 3]
[本文引用: 3]
DOI:10.1128/AAC.39.6.1211URL [本文引用: 1]
DOI:10.1128/AAC.46.1.1-11.2002URL [本文引用: 3]
DOI:10.1128/CMR.00036-08URL [本文引用: 3]
DOI:10.1128/AAC.02852-15URL [本文引用: 3]
DOI:10.1128/JCM.38.5.1791-1796.2000URL [本文引用: 1]
[本文引用: 1]
DOI:10.1128/JCM.40.6.2153-2162.2002URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/nbt.2280URL [本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.1128/AAC.46.5.1190-1198.2002URL [本文引用: 2]
DOI:10.1128/AAC.40.1.221URL [本文引用: 1]
DOI:10.1128/AAC.38.5.1182URL [本文引用: 1]
DOI:10.1371/journal.pone.0096738URL [本文引用: 2]
DOI:10.1093/jac/dkr412URL [本文引用: 3]
DOI:10.1128/AAC.01654-15URL [本文引用: 1]
DOI:10.1016/j.plasmid.2015.04.003URL [本文引用: 4]
DOI:10.1016/j.plasmid.2018.02.002URL [本文引用: 1]
DOI:10.1089/mdr.2014.0012URL [本文引用: 1]
DOI:10.1128/AAC.00944-15URL [本文引用: 1]
DOI:10.1016/j.plasmid.2018.08.001URL [本文引用: 2]
DOI:10.3389/fmicb.2019.02508URL [本文引用: 1]
DOI:10.1016/j.plasmid.2017.10.001URL [本文引用: 1]
DOI:10.1128/AAC.00753-08URL [本文引用: 1]
