 ,, 吴然, 金晶, 丘萍, 张洁, 吴祖建
,, 吴然, 金晶, 丘萍, 张洁, 吴祖建 ,, 杜振国
,, 杜振国 ,福建农林大学植物病毒研究所/闽台作物有害生物生态防控国家重点实验室,福州 350002
,福建农林大学植物病毒研究所/闽台作物有害生物生态防控国家重点实验室,福州 350002An in vitro Cap-Snatching System of Rice Stripe Tenuivirus Based on Crude Virion Preparations
LIN WenZhong ,, WU Ran, JIN Jing, QIU Ping, ZHANG Jie, WU ZuJian
,, WU Ran, JIN Jing, QIU Ping, ZHANG Jie, WU ZuJian ,, DU ZhenGuo
,, DU ZhenGuo ,Plant Virus Research Institute, Fujian Agriculture and Forestry University/State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fuzhou 350002
,Plant Virus Research Institute, Fujian Agriculture and Forestry University/State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fuzhou 350002通讯作者:
责任编辑: 岳梅
收稿日期:2020-06-15接受日期:2020-07-13网络出版日期:2020-11-01
| 基金资助: |
Received:2020-06-15Accepted:2020-07-13Online:2020-11-01
作者简介 About authors
林文忠,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1219KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
林文忠, 吴然, 金晶, 丘萍, 张洁, 吴祖建, 杜振国. 基于粗提物的水稻条纹病毒体外“抓帽”体系[J]. 中国农业科学, 2020, 53(21): 4440-4448 doi:10.3864/j.issn.0578-1752.2020.21.012
LIN WenZhong, WU Ran, JIN Jing, QIU Ping, ZHANG Jie, WU ZuJian, DU ZhenGuo.
0 引言
【研究意义】水稻条纹病毒(rice stripe tenuivirus,RSV)是我国最重要的粮食作物病毒之一,深入认识RSV侵染机理,实现RSV的可持续防控,是关系到我国粮食安全的重大需求[1,2]。作为基因表达的第一步,“抓帽”(cap-snatching)是RSV侵染周期中最关键的环节之一[3]。建立一种便捷的体外体系,有助于深入研究RSV的“抓帽”机制。【前人研究进展】真核生物信使RNA(mRNA)5′端有一个帽子结构,能保护mRNA免受5′到3′外切酶降解的同时,在mRNA的翻译起始等过程中也起重要作用[4]。除非有一些替代机制,病毒也需要给自己的mRNA添加帽子结构。病毒以多种不同的机制实现这一目的,其中布尼亚病毒目(Bunyavirales)和正黏病毒科(Orthomyxoviridae)病毒普遍采用“抓帽”。在此过程中,它们从帽子结构下游10—20个碱基处切割寄主mRNA,然后利用5′端切割产物(帽子序列)作为病毒模板链的转录引物,使每条病毒mRNA都含一小段来源于寄主mRNA的帽子序列[5]。“抓帽”现象最早发现于流感病毒(正黏病毒科),迄今为止研究最多的也是该病毒[6,7]。相应地,“抓帽”已成为抗流感病毒药物开发的一个重要靶标[8]。RSV是布尼亚病毒目白纤病毒科(Phenuiviridae)纤细病毒属(Tenuivirus)的代表种,自然条件下由灰飞虱(Laodelphax striatellus)以持久增殖型方式传播,主要侵染水稻和小麦等禾本科作物[9,10],实验室条件下,也可以通过机械接种或昆虫传毒方式侵染模式植物本氏烟和拟南芥[11,12]。RSV基因组由4条单链RNA(ssRNA)组成,按分子量大小分别称为RNA1—RNA4。RNA1—RNA4在5′和3′末端有一段保守的反向互补序列,分别为5′-A1C2A3C4AAAGUC-和3′-U1G2U3G4UUUCAG-。RNA1采用负义编码,而RNA2—RNA4采用双义编码,即它们的病毒链(vRNA)和互补链(vcRNA)都可以作为转录的模板[9,10]。SHIMIZU等[13]最先克隆了RSV的mRNA,发现它们除拷贝自基因组的序列(下称拷贝序列),还含一段异源帽子序列,表明RSV使用“抓帽”机制;YAO等[14]以RSV与黄瓜花叶病毒(cucumber mosaic virus,CMV)共侵染本氏烟,发现RSV可从CMV抓取帽子序列。对RSV从CMV“抓帽”后合成的mRNA进行克隆和分析发现,大部分RSV mRNA在帽子序列和拷贝序列之间含一个或多个AC重复,表明RSV在利用从CMV抓取的帽子序列进行转录时,频繁使用引发与重配(prime- and-realign)机制[14,15]。RSV使用引发与重配机制的频率与帽子序列的长短有关,帽子序列越短,使用该机制的频率越高。本实验室首次对RSV mRNA进行了5′端的高通量测序,并根据所得数据,提出了RSV“抓帽”的基本模型[16]。在RSV侵染的本氏烟叶片浸润含pCHF3载体的农杆菌,RSV会从pCHF3表达的GFP mRNA“抓帽”。基于此,近期建立了一种向RSV呈递特定mRNA(供其“抓帽”)的实验体系,并应用该体系初步验证了RSV“抓帽”的基本模型[17]。【本研究切入点】目前,对RSV“抓帽”机制的很多细节还不清楚。体外“抓帽”体系在解析流感病毒“抓帽”机制的过程中发挥了重要作用,在一些布尼亚病毒上也有成功的应用[18,19,20,21]。利用这一体系,研究者可以定量地向病毒提供特定mRNA,甚至特定的帽子序列,以研究病毒如何选择和利用帽子序列[22,23,24,25,26]。显然,这样的体系对RSV“抓帽”和转录机制研究也具有重要意义。【拟解决的关键问题】确定RSV粗提物能否从外源mRNA“抓帽”,并研究RSV体外“抓帽”与体内“抓帽”在方式上是否存在差异。1 材料与方法
试验于2019年6月至2020年5月间在福建农林大学植物病毒研究所完成。1.1 试验材料
1.1.1 供试毒株与植物 携带RSV的灰飞虱由江苏省农业科学院季英华研究员提供,引入本实验室后,在水稻上继代培养。带毒灰飞虱取食过的水稻幼苗种植于日光温室,出现典型RSV感染症状后用于RSV的提取。1.1.2 主要试剂 主要试剂及其来源见表1,其他常规化学试剂均为国产分析纯。
Table 1
表1
表1本研究所用的主要试剂和试剂盒
Table 1
| 试剂名称 Reagents and kits | 来源 Manufacture | 用途 Usage |
|---|---|---|
| RCL(未经核酸酶处理 Not treated with nuclease) | Promega | 体外“抓帽”体系 In vitro cap-snatching |
| RNase inhibitor | Promega | |
| NTP (100 mmol·L-1 each) | TaKaRa | |
| m7G (5′) ppp (5′) G | NEB | |
| Prestained Protein Marker | GenStar | SDS-PAGE |
| RNA purification Kit (DP412) | TIANGEN | RSV mRNA的检测与克隆 Detection and cloning of RSV mRNAs |
| 1st Strand cDNA Synthesis Kit | Vazyme Biotech | |
| 2×ExTaq Mix | TaKaRa | |
| HiPure Gel Pure DNA Mini Kit | Magen | |
| pMD19-T | TaKaRa |
新窗口打开|下载CSV
1.2 试验方法
1.2.1 RSV的粗提和聚丙烯酰胺凝胶电泳检测 取感染RSV的新鲜水稻叶片约100 g,剪碎后加入研钵,以液氮浸没并迅速研磨至粉末状。随后参照WANG等[27]描述的步骤提取RSV,以0.01 mol·L-1 PBS缓冲液(0.27 g·L-1 KH2PO4,1.42 g·L-1 Na2HPO4,8 g·L-1 NaCl,0.2 g·L-1 KCl,pH 7.4)代替了文献[27]中的PB缓冲液。最终提取物以2 mL PBS溶解,分装后置于-80℃冰箱备用。为了解RSV粗提物的蛋白组分,取粗提液80 μL,加入5×蛋白上样缓冲液20 μL,混合均匀后,沸水浴5 min,于恒压120 V下进行聚丙烯酰胺凝胶电泳(SDS-PAGE,5%浓缩胶,12%分离胶)。每孔上样20 μL,以6 μL Direct-loadTM Color Prestained Protein Marker(GenStar)作为分子量标准。电泳完毕,凝胶经考马斯亮蓝染色和脱色液脱色后用相机进行拍照,并割取主要蛋白条带,送上海启研生物科技有限公司进行质谱鉴定。
1.2.2 体外“抓帽”反应体系 RSV体外“抓帽”体系参照VAN KNIPPENBERG等[28]的报道略有改动,含2 μL RSV粗提液,8 μL兔网织红细胞裂解液(rabbit reticulocyte lysate,RCL),4 mmol·L-1 MgCl2,2 mmol·L-1 NTP及0.8 U·μL-1 RNA酶抑制剂,总体积20 μL,30℃下恒温孵育1.5 h。
1.2.3 体外“抓帽”所产生的RSV mRNA的巢式RT-PCR检测和克隆测序 上述反应完成后,以RNA纯化试剂盒对总RNA进行纯化,具体步骤参照说明书进行,纯化产物以15 μL RNAase-free去离子水溶解。RCL中含大量珠蛋白-α和珠蛋白-β mRNA,本研究仅扩增了RSV从珠蛋白-α抓帽后的转录产物。所用巢式RT-PCR基本原理如图1所示,以primer 1合成第一链,然后以primer 2和3扩增cDNA。Primer 3的3′末端10个碱基与珠蛋白-α mRNA最5′端10个碱基相同,5′端15个碱基(下划线部分)主要作用是增加引物长度。RSV共转录7种不同的mRNA,本文仅扩增了其vRNA3的转录产物NP和vcRNA4的转录产物NCP[29,30]。扩增NP和NCP所用的primer 1和2如图1所示。反转录体系:RNA 2 μL(约0.6 μg),primer 1 0.25 μmol·L-1,HiScript? II Enzyme Mix 2 μL,10×RT Mix 2 μL,总体积20 μL。反转录程序参照试剂盒说明书进行。PCR体系:cDNA 2 μL,primer 2和3各0.5 μmol·L-1,2×ExTaq Mix 12.5 μL,总体积25 μL。PCR反应程序:94℃预变性3 min后,按照94℃ 30 s,55℃ 30 s,72℃ 30 s,进行31次循环,最后于72℃延伸10 min。PCR产物经1.5%琼脂糖凝胶电泳后,于凝胶成像仪下观察目的条带。需要时切割目的条带,以HiPure Gel Pure DNA Mini Kit进行DNA回收,回收产物连接pMD19-T后转化大肠杆菌,并挑取20—30个单菌落进行测序。作为对照,除以图1所示的巢式RT-PCR扩增RSV mRNA,还扩增了RSV的vcRNA3和vRNA4。其基本流程与巢式RT-PCR相同,但以病毒自身序列特异性引物取代了图1所示的primer 3 (vcRNA3:5′-GCTACAATGGGCACCAACAAGC-3′;vRNA4:5′-AGATGCAAGACGTACAAAGGAC-3′)。
图1
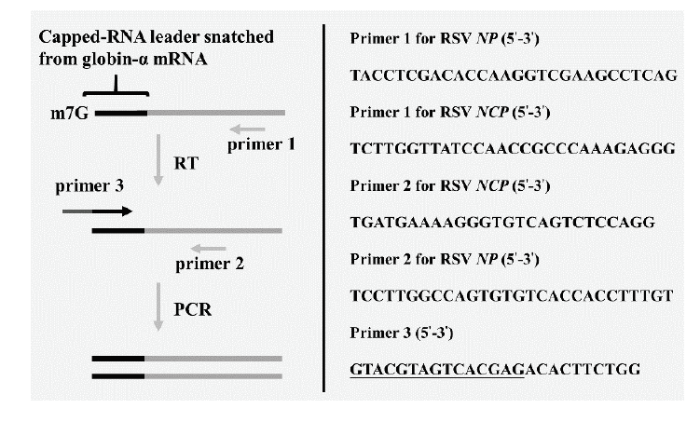 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1利用巢式RT-PCR扩增RSV从珠蛋白-α“抓帽”后合成的NP(转录自vRNA3)和NCP(转录自vcRNA4)mRNA
Fig. 1Schematic diagram showing the nested RT-PCR used to detect RSV NP (transcribed from vRNA3) and NCP (transcribed from vcRNA4) mRNAs with capped-RNA leaders obtained from globin-α mRNA
2 结果
2.1 RSV粗提物的获得
从100 g感染RSV的水稻叶片获得RSV粗提液2 mL。取少量粗提液进行SDS-PAGE,考马斯亮蓝染色后可观察到4条主要的蛋白条带,分子量依次约250、50、30和11 kD(图2)。质谱鉴定结果表明,条带1主要为光系统II相关蛋白,条带2主要为核酮糖1,5-二磷酸羧化酶(RuBisCO)大亚基和RuBisCO活化酶,条带3主要为RSV的NP,条带4主要为RuBisCO小亚基。由这些结果可知,RSV粗提物含大量杂质,而叶绿体是杂质成分的重要来源。RNA1编码的RdRp也是RSV的结构蛋白[31],但可能因为含量太低,未观察到该蛋白对应的条带。图2
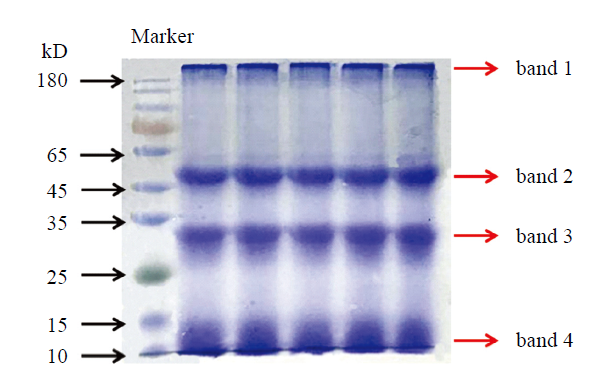 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2RSV粗提物的SDS-PAGE检测
Band 1—4:4个进行质谱鉴定的主要蛋白条带
Fig. 2SDS-PAGE detection of the RSV crude preparations
The 4 protein bands subjected to mass spectrometry
2.2 RSV粗提物“抓帽”产物的检测
取体外“抓帽”反应产物,用巢式RT-PCR扩增含珠蛋白-α mRNA帽子序列的RSV NP 和NCP mRNA,均能得到目的条带。与病毒粒子中含更多的vRNA相符,NP(vRNA转录产物)条带明显亮于NCP(vcRNA转录产物)条带(图3-A)。作为对照,以95℃热失活2 min的RSV粗提液配制反应体系,反应结束后进行同样的检测,未观察到目的条带。无论是否对RSV粗提液进行热失活,反应结束后均能检测到RSV基因组RNA(图3-A)。上述结果表明粗提液中的RSV能利用溶液中的mRNA作为帽子序列供体合成自身mRNA,且这种能力需要有活性的RSV蛋白来实现。图3
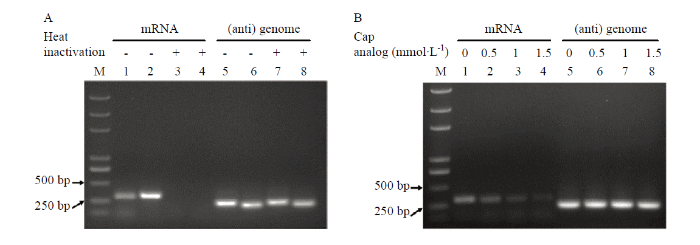 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3RSV粗提物从外源mRNA的“抓帽”
M:DNA marker。A:以正常或热失活RSV粗提物进行体外“抓帽”,然后利用巢式RT-PCR扩增含珠蛋白-α帽子序列的RSV NCP(孔1和3)或NP(孔2和4) mRNA,以RT-PCR扩增vcRNA4上NCP对应区域(孔6和8)或vRNA3上NP对应区域(孔5和7);B:在0—1.5 mmol·L-1帽子类似物m7G (5′) ppp (5′) G存在下进行体外“抓帽”,然后以巢式RT-PCR扩增含珠蛋白-α帽子序列的RSV NCP(孔1—4),以RT-PCR扩增vcRNA4上NCP对应区域(孔5—8)
Fig. 3The cap-snatching of crude RSV preparations from exogenously supplied mRNAs
M:DNA marker。A:Untreated or heat inactivated RSV crude preparations were used in the in vitro cap-snatching assay. Nested RT-PCR was used to detect RSV NCP (lanes 1 and 3) or NP (lanes 2 and 4) mRNAs with capped-RNA leaders derived from globin-α. RT-PCR was used to detect a NP-corresponding region on RSV vRNA3 (lanes 5 and 7) or a NCP-corresponding region on RSV vcRNA4 (lanes 6 and 8);B:The cap analog m7G (5′) ppp (5′) G was added at a final concentration of 0.5, 1 or 1.5 mmol·L-1. Nested RT-PCR was used to detect RSV NCP with capped-RNA leaders derived from globin-α (lanes 1 to 4) and RT-PCR was used to detect a NCP-corresponding region on RSV vcRNA4
为验证RSV需要识别帽子结构才能利用溶液中的mRNA作为帽子序列供体,在反应体系中加入了不同浓度的帽子结构类似物m7G (5′) ppp (5′) G。如图3-B所示,m7G (5′) ppp (5′) G明显减少含珠蛋白-α mRNA帽子序列的NCP的生成。当m7G (5′) ppp (5′) G浓度为1.5 mmol·L-1时,目的条带基本消失。相反,m7G (5′) ppp (5′) G对反应后的RSV基因组RNA的检出性没有影响,表明m7G (5′) ppp (5′) G不会造成反应体系中RNA的降解。
2.3 RSV粗提物“抓帽”产物分析
为进一步验证RSV粗提物能从外源mRNA“抓帽”,并分析这种情况下产生的RSV mRNA,对图3-A中Lane 1和2的目的条带进行了割胶回收。经连接、转化和挑取单克隆测序后,得到RSV NP mRNA序列19条,含非冗余单一序列7条(表2),NCP mRNA序列28条,含非冗余单一序列6条(表3)。Table 2
表2
表2RSV从珠蛋白-α mRNA“抓帽”后生成的NP mRNA
Table 2
| 行数 Row | 珠蛋白-α mRNA序列Globin-α mRNA sequence m7G-ACACUUCUGGUCCAGUCCGACUGAG- | 克隆数 Clone number | 引发与重配 Priming and realignment |
|---|---|---|---|
| 1 | m7G-ACACUUCUGGUC12------------ACACAAAGUC- | 1 | + |
| 2 | m7G-ACACUUCUGGUCCAGUCC18------ACACAAAGUC- | 7 | + |
| 3 | m7G-ACACUUCUGGUC12-----AC-----ACACAAAGUC- | 1 | + |
| 4 | m7G-ACACUUCUGGUC12----ACAC----ACACAAAGUC- | 1 | + |
| 5 | m7G-ACACUUCUGGUC12-ACACACACAC-ACACAAAGUC- | 3 | + |
| 6 | m7G-ACACUUCUGGUCCA14----------CACAAAGUC- | 5 | - |
| 7 | m7G-ACACUUCUGGUCCA14------------CAAAGUC- | 1 | - |
新窗口打开|下载CSV
Table 3
表3
表3RSV从珠蛋白-α mRNA“抓帽”后生成的NCP mRNA
Table 3
| 行数 Row | 珠蛋白-α mRNA序列Globin-α mRNA sequence m7G-ACACUUCUGGUCCAGUCCGACUGAG- | 克隆数 Clone number | 引发与重配 Priming and realignment |
|---|---|---|---|
| 1 | m7G-ACACUUCUGGUC12------------ACAAAGUC- | 12 | - |
| 2 | m7G-ACACUUCUGGUC12----------ACACAAAGUC- | 3 | + |
| 3 | m7G-ACACUUCUGGUC12--AC------ACACAAAGUC- | 3 | + |
| 4 | m7G-ACACUUCUGGUCCA14-----------CAAAGUC- | 7 | - |
| 5 | m7G-ACACUUCUGGUCCA14---------CACAAAGUC- | 1 | + |
| 6 | m7G-ACACUUCUGGUCCA14-C------ACACAAAGUC- | 2 | + |
新窗口打开|下载CSV
分析发现,这些序列都可用之前通过体内研究提出的模型解释[16,17]:(1)RSV从C12或C18切割珠蛋白-α mRNA,得到末端为C的帽子序列m7G---C。该帽子序列与RSV基因组3′端G2配对后从U3引发转录,生成m7G---CAC。m7G---CAC可以继续按照模板延伸,直至生成完整的RSV mRNA m7G---CACAAAGUC- (表3,第一行,下划线部分为能Map到病毒基因组的碱基),也可以与模板链脱离,以新延伸出的AC与RSV基因组3′端的U1G2配对,重新从U3引发转录,即发生引发与重配(图4-A)。引发与重配可发生多次,其结果是RSV mRNA在帽子序列与所谓的拷贝序列之间出现1或多个AC重复(表2第2、4和5行及表3的第3行);(2)除C12或C18外,RSV还可以在A14切割珠蛋白-α mRNA,得到末端为A的帽子序列m7G---C13A14。该帽子序列可以利用A14与RSV基因组3′端的U1配对,或利用C13A14与RSV基因组3′端的G2U3配对(图4-B、4-C)。同样,在转录起始之后,RSV可以使用0到多次引发与重配,生成多种不同形式的RSV mRNA。按照这种解释,RSV利用珠蛋白-α mRNA帽子序列生成NP(58%)时比生成NCP(33%)时更频繁使用引发与重配,这与之前的发现也完全相符[16]。
图4
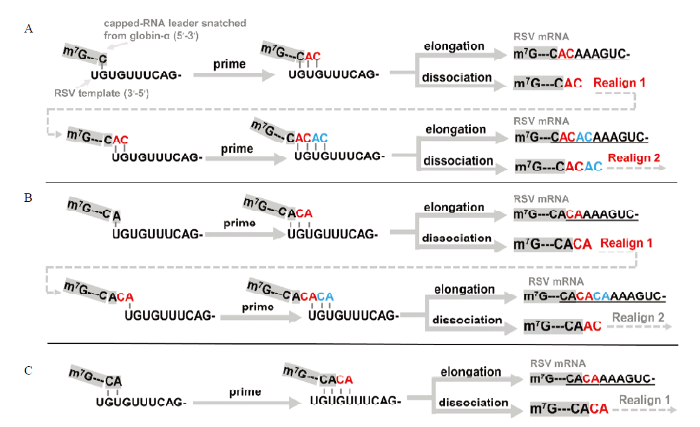 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4RSV利用珠蛋白-α mRNA帽子序列生成自身mRNA的可能机制
A:帽子序列与RSV基因组G2配对;B:帽子序列与RSV基因组U1配对;C:帽子序列与RSV基因组G2U3配对
Fig. 4Possible mechanisms by which RSV produces its own mRNAs using capped-RNA leaders snatched from globin-α mRNA
A:Capped-RNA leaders base pair with the G2 of RSV template RNA;B:Capped-RNA leaders base pair with the U1 of RSV template RNA;C:Capped-RNA leaders base pair with the G2U3 of RSV template RNA
3 讨论
体外体系已被广泛应用于研究流感和一些布尼亚病毒的“抓帽”及与“抓帽”紧密伴随的转录[18,19,20,21,22,23,24,25,26]。然而,这些研究都使用纯度较高的病毒粒子,需要繁杂的预备实验。为获得一种便捷的实验体系,本文研究了RSV粗提物的“抓帽”活性。叶绿体是RSV粗提物中杂质成分的重要来源(图2)。根据条带亮度判断,叶绿体蛋白在粗提物总蛋白中的占比可能超过2/3。推测其原因是叶绿体在离心过程中与RSV病毒粒子发生了共沉淀。虽然它们在后续过程中被破坏,但本研究未以密度梯度离心将它们释放出的蛋白与RSV进行分离。按照这种推测,RSV粗提物中应该还含有来源于线粒体及核糖体等细胞器或大分子的蛋白,只是相较于叶绿体蛋白来讲,这些蛋白的占比较低。
尽管含有大量杂质,本研究结果表明RSV粗提物具有体外“抓帽”活性,且在切割和利用帽子序列的方式上,其表现与细胞内的RSV没有明显差别。主要证据有以下两点:(1)以RSV粗提物与RCL配制反应体系,短时间的孵育后,可以检测到含珠蛋白-α mRNA帽子序列的RSV mRNA,而帽子类似物可以降低这类mRNA的合成(图3),表明RSV能利用溶液中的mRNA作为帽子序列供体来合成自身mRNA,且同在体内一样,这一过程需要帽子结构的识别;(2)对体外合成的mRNA进行分析发现,RSV粗提物在A或C处切割珠蛋白-α mRNA,得到能与自身模板链3′端U或G配对的帽子序列。在以这些帽子序列进行mRNA合成时,RSV频繁使用引发与重配(图4)。这些现象都完全符合之前通过体内研究提出的模型。值得指出的是,虽然纤细病毒的体外转录活性已有报道[31,32],但还未有研究者对体外转录产物进行测序验证。从这方面讲,本文是首次明确表明纤细病毒可在体外从外源mRNA“抓帽”。
布尼亚病毒和正黏病毒在“抓帽”后的转录过程中普遍使用引发与重配机制[3,33]。笔者前期发现,RSV也使用引发与重配,而且RSV在转录vRNA时比在转录vcRNA时更频繁使用该机制[16]。起初,笔者推测这可能是RSV与寄主互作的结果,或者反映了RSV vRNA和vcRNA在表达上的时空差异。但后来发现,这种现象在RSV建立侵染的极早期就已出现[34]。因而,推测RSV对引发与重配机制的差异化使用可能是其模板链的固有差异造成的,与其他因素无关。本研究发现RSV在体外合成NP(转录自vRNA)时也比合成NCP(转录自vcRNA)时更频繁使用引发与重配,这进一步支持了第二种推测。
RSV可与多种双生病毒共侵染本氏烟。在共侵染植株中,RSV大量从双生病毒抓取帽子序列。通过高通量测序技术获得RSV mRNA的帽子序列后,将它们Map到双生病毒基因组,可以得到一个精细的双生病毒转录起始位点(transcriptional start site,TSS)图谱。基于此,本实验室提出“抓帽”可被利用于病毒甚至寄主植物TSS的鉴定[35]。然而,该方法有其明显缺点,即需要病毒的共侵染。一方面,这在很多情况下是无法实现的;另一方面,RSV有可能会影响双生病毒TSS的选择。RSV体外“抓帽”体系的建立为解决这一缺点提供了可能。因而,除解析“抓帽”和转录机制外,本实验建立的体系还有潜在的应用价值。
4 结论
RSV粗提物能从外源mRNA“抓帽”,并利用抓取的帽子序列转录自身模板链。在切割外源mRNA和利用其帽子序列的方式上,粗提物的表现与细胞内的RSV无明显差别,因而,基于粗提物的体外体系可以用于解析RSV的“抓帽”机制,甚至研究该病毒“抓帽”之后的转录机制。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.tim.2019.12.006URLPMID:31948728 [本文引用: 2]

In common with all segmented negative-sense RNA viruses, bunyavirus transcripts contain heterologous sequences at their 5' termini originating from capped host cell RNAs. These heterologous sequences are acquired by a so-called cap-snatching mechanism. Whereas for nuclear replicating influenza virus the source of capped primers as well as the cap-binding and endonuclease activities of the viral polymerase needed for cap snatching have been functionally and structurally well characterized, our knowledge on the expected counterparts of cytoplasmic replicating bunyaviruses is still limited and controversial. This review focuses on the cap-snatching mechanism of bunyaviruses in the light of recent structural and functional data.
URLPMID:27317694 [本文引用: 1]
URLPMID:22138959 [本文引用: 1]
[本文引用: 1]
URLPMID:30642766 [本文引用: 1]
URLPMID:31150614 [本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
DOI:10.1128/JVI.69.9.5754-5762.1995URLPMID:7637020 [本文引用: 1]

We examined the 5' ends of Hantaan virus (HTN) genomes and mRNAs to gain insight into the manner in which these chains were initiated. Like those of all members of the family Bunyaviridae described so far, the HTN mRNAs contained 5' terminal extensions that were heterogeneous in both length and sequence, presumably because HTN also
DOI:10.1128/JVI.01414-17URLPMID:29046442 [本文引用: 4]

Most segmented negative-sense RNA viruses employ a process termed cap snatching, during which they snatch capped RNA leaders from host cellular mRNAs and use the snatched leaders as primers for transcription, leading to the synthesis of viral mRNAs with 5' heterogeneous sequences (HSs). With traditional methods, only a few HSs can be determined, and identification of their donors is difficult. Here, the mRNA 5' ends of Rice stripe tenuivirus (RSV) and Rice grassy stunt tenuivirus (RGSV) and those of their host rice were determined by high-throughput sequencing. Millions of tenuiviral HSs were obtained, and a large number of them mapped to the 5' ends of corresponding host cellular mRNAs. Repeats of the dinucleotide AC, which are complementary to the U1G2 of the tenuiviral template 3'-U1G2U3G4UUUCG, were found to be prevalent at the 3' termini of tenuiviral HSs. Most of these ACs did not match host cellular mRNAs, supporting the idea that tenuiviruses use the prime-and-realign mechanism during cap snatching. We previously reported a greater tendency of RSV than RGSV to use the prime-and-realign mechanism in transcription with leaders cap snatched from a coinfecting reovirus. Besides confirming this observation in natural tenuiviral infections, the data here additionally reveal that RSV has a greater tendency to use this mechanism in transcribing genomic than in transcribing antigenomic templates. The data also suggest that tenuiviruses cap snatch host cellular mRNAs from translation- and photosynthesis-related genes, and capped RNA leaders snatched by tenuiviruses base pair with U1/U3 or G2/G4 of viral templates. These results provide unprecedented insights into the cap-snatching process of tenuiviruses.IMPORTANCE Many segmented negative-sense RNA viruses (segmented NSVs) are medically or agriculturally important pathogens. The cap-snatching process is a promising target for the development of antiviral strategies against this group of viruses. However, many details of this process remain poorly characterized. Tenuiviruses constitute a genus of agriculturally important segmented NSVs, several members of which are major viral pathogens of rice. Here, we for the first time adopted a high-throughput sequencing strategy to determine the 5' heterogeneous sequences (HSs) of tenuiviruses and mapped them to host cellular mRNAs. Besides providing deep insights into the cap snatching of tenuiviruses, the data obtained provide clear evidence to support several previously proposed models regarding cap snatching. Curiously and importantly, the data here reveal that not only different tenuiviruses but also the same tenuivirus synthesizing different mRNAs use the prime-and-realign mechanism with different tendencies during their cap snatching.
URLPMID:31710910 [本文引用: 2]
URLPMID:12606583 [本文引用: 2]
DOI:10.1016/j.antiviral.2018.10.008URLPMID:30316915 [本文引用: 2]

Cap-dependent endonuclease (CEN) resides in the PA subunit of the influenza virus and mediates the critical
[本文引用: 2]
[本文引用: 2]
DOI:10.1016/j.virol.2005.01.041URLPMID:15823611 [本文引用: 2]

Transcription of segmented negative-strand RNA viruses is initiated by cap snatching: a host mRNA is cleaved generally at 10-20 nt from its 5' capped end and the resulting capped leader used to prime viral transcription. For Tomato spotted wilt virus (TSWV), type species of the plant-infecting Tospovirus genus within the Bunyaviridae, cap donors were previously shown to require a single base complementarity to the ultimate or penultimate viral template sequence. More recently, the occurrence in vitro of
DOI:10.1016/j.virol.2010.09.003URLPMID:21051068 [本文引用: 2]

The requirements for alignment of capped leader sequences along the viral genome during influenza transcription initiation (cap-snatching) have long been an enigma. In this study, competition experiments using an in vitro transcription assay revealed that influenza virus transcriptase prefers leader sequences with base complementarity to the 3'-ultimate residues of the viral template, 10 or 11 nt from the 5' cap. Internal priming at the 3'-penultimate residue, as well as prime-and-realign was observed. The nucleotide identity immediately 5' of the base-pairing residues also affected cap donor usage. Application to the in vitro system of RNA molecules with increased base complementarity to the viral RNA template showed stronger reduction of globin RNA leader initiated influenza transcription compared to those with a single base-pairing possibility. Altogether the results indicated an optimal cap donor consensus sequence of (7m)G-(N)(7-8)-(A/U/G)-(A/U)-AGC-3'.
DOI:10.1128/JVI.01775-17URLPMID:29142123 [本文引用: 2]

The RNA-dependent RNA polymerase (RdRp) of the influenza A virus replicates and transcribes the viral genome segments in the nucleus of the host cell. To transcribe these viral genome segments, the RdRp
DOI:10.1261/rna.436607URLPMID:17400818 [本文引用: 2]

Messenger RNA transcription by Bunyaviridae family members is unique within the group of negative-strand RNA viruses as it requires on-going protein synthesis. The long-standing model explaining this phenomenon proposes that the translational requirement is not for a protein product, but instead is for ribosomal translocation along nascent mRNAs. This movement is proposed to disrupt spurious transcription termination signals that otherwise cause premature mRNA truncation leading to a fatal loss of gene expression. This model was tested by introducing translational stop codons into model RNA genomes of Bunyamwera virus, the prototypic member of the Bunyaviridae family. This directly showed that translation of nascent mRNAs prevents transcription termination. While such coupling of transcription and translation is common in prokaryotic systems, these results represent the first report of such obligatory coupling in a eukaryotic cell environment. The results also provide insight into the bunyavirus termination mechanism and suggest it is mechanistically similar to prokaryotic intrinsic termination.
[本文引用: 2]
DOI:10.1038/ncomms5768URLPMID:25203424 [本文引用: 2]

Rice stripe virus (RSV) causes one of the most serious viral diseases of rice (Oryza sativa L.), but the molecular basis of RSV resistance has remained elusive. Here we show that the resistant allele of rice STV11 (STV11-R) encodes a sulfotransferase (OsSOT1) catalysing the conversion of salicylic acid (SA) into sulphonated SA (SSA), whereas the gene product encoded by the susceptible allele STV11-S loses this activity. Sequence analyses suggest that the STV11-R and STV11-S alleles were predifferentiated in different geographic populations of wild rice, Oryza rufipogon, and remained prevalent in cultivated indica and japonica rice varieties, respectively. Introgression of the STV11-R allele into susceptible cultivars or heterologous transfer of STV11-R into tobacco plants confers effective resistance against RSV. Our results shed new insights into plant viral defense mechanisms and suggest effective means of breeding RSV-resistant crops using molecular marker-assisted selection or genetic engineering.
DOI:10.1006/viro.2002.1632URLPMID:12490389 [本文引用: 1]

Purified Tomato spotted wilt virus particles were shown to support either genome replication or transcription in vitro, depending on the conditions chosen. Transcriptional activity was observed only upon addition of rabbit reticulocyte lysate, indicating a dependence on translation. Under these conditions RNA molecules of subgenomic length were synthesized that hybridized to strand-specific probes for the N and NSs genes. Cloning of these transcripts demonstrated the presence of nonviral leader sequences at their 5' ends, confirming the occurrence of genuine viral transcription initiation known as
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
DOI:10.1128/JVI.71.4.2621-2627.1997URLPMID:9060614 [本文引用: 1]

An RNA-dependent RNA polymerase (RdRp) activity associated with the ribonucleoproteins of rice hoja blanca tenuivirus (RHBV) was detected and analyzed. Conditions for in vitro RNA synthesis and for coupled RNA synthesis-translation of RHBV were established. In both cases, synthesis of the viral and viral complementary genomic and subgenomic RNA3 and RNA4 were observed, demonstrating that both transcription and replication occurred. Though coupling of RNA synthesis to translation allowed efficient translation of the newly synthesized subgenomic RNAs, studies of the effect of various inhibitors of protein synthesis revealed that RNA synthesis was independent of translation. Primer extension experiments demonstrated that in the presence of capped exogenous RNAs, a stretch of 10 to 16 nonviral nucleotides was added to the 5' end of a population of newly synthesized viral complementary RNA4. It appears that in addition to RdRp activity, RHBV-associated protein(s) also possessed cap-snatching capacity.
DOI:10.1093/nar/gkv333URLPMID:25901029 [本文引用: 1]

The influenza polymerase cleaves host RNAs approximately 10-13 nucleotides downstream of their 5' ends and uses this capped fragment to prime viral mRNA synthesis. To better understand this process of cap snatching, we used high-throughput sequencing to determine the 5' ends of A/WSN/33 (H1N1) influenza mRNAs. The sequences provided clear evidence for nascent-chain realignment during transcription initiation and revealed a strong influence of the viral template on the frequency of realignment. After accounting for the extra nucleotides inserted through realignment, analysis of the capped fragments indicated that the different viral mRNAs were each prepended with a common set of sequences and that the polymerase often cleaved host RNAs after a purine and often primed transcription on a single base pair to either the terminal or penultimate residue of the viral template. We also developed a bioinformatic approach to identify the targeted host transcripts despite limited information content within snatched fragments and found that small nuclear RNAs and small nucleolar RNAs contributed the most abundant capped leaders. These results provide insight into the mechanism of viral transcription initiation and reveal the diversity of the cap-snatched repertoire, showing that noncoding transcripts as well as mRNAs are used to make influenza mRNAs.
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.3389/fmicb.2017.02519URLPMID:29312219 [本文引用: 1]

Identification of the transcription start sites (TSSs) of a virus is of great importance to understand and dissect the mechanism of viral genome transcription but this often requires costly and laborious experiments. Many segmented negative-sense RNA viruses (sNSVs) cleave capped leader sequences from a large variety of mRNAs and use these cleaved leaders as primers for transcription in a conserved process called cap snatching. The recent developments in high-throughput sequencing have made it possible to determine most, if not all, of the capped RNAs snatched by a sNSV. Here, we show that rice stripe tenuivirus (RSV), a plant-infecting sNSV, co-infects Nicotiana benthamiana with two different begomoviruses and snatches capped leader sequences from their mRNAs. By determining the 5' termini of a single RSV mRNA with high-throughput sequencing, the 5' ends of almost all the mRNAs of the co-infecting begomoviruses could be identified and mapped on their genomes. The findings in this study provide support for the using of the cap snatching of sNSVs as a tool to map viral TSSs.
