 ,1, 王霞1, 马友记
,1, 王霞1, 马友记 ,1,2, 尹德恩1, 张勇3, 赵兴绪3
,1,2, 尹德恩1, 张勇3, 赵兴绪3Molecular Characterization of Tibetan Sheep BOLL and Its Expression Regulation and Functional Analysis in Testis
LI TaoTao ,1, WANG Xia1, MA YouJi
,1, WANG Xia1, MA YouJi ,1,2, YIN DeEn1, ZHANG Yong3, ZHAO XingXu3
,1,2, YIN DeEn1, ZHANG Yong3, ZHAO XingXu3通讯作者:
责任编辑: 林鉴非
收稿日期:2020-06-1接受日期:2020-09-3网络出版日期:2020-10-16
| 基金资助: |
Received:2020-06-1Accepted:2020-09-3Online:2020-10-16
作者简介 About authors
李讨讨,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (13900KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李讨讨, 王霞, 马友记, 尹德恩, 张勇, 赵兴绪. 藏绵羊BOLL的分子特征及其在睾丸中的表达调控与功能分析[J]. 中国农业科学, 2020, 53(20): 4297-4312 doi:10.3864/j.issn.0578-1752.2020.20.017
LI TaoTao, WANG Xia, MA YouJi, YIN DeEn, ZHANG Yong, ZHAO XingXu.
0 引言
【研究意义】藏绵羊作为我国三大原始绵羊品种之一,是分布于海拔3 000 m以上的青藏高原及其毗邻地区种群数量最多的家畜[1],为当地农牧民提供了极其重要的生活物资和经济收入,并在高寒草甸草地生物多样性及生态系统功能中发挥重要作用。但由于藏绵羊所具有的发育周期长、繁殖力低下、性成熟晚等繁殖生理特点,严重制约了其种群的数量。目前有关雄性藏绵羊睾丸发育及精子发生的分子生物学认识仍十分匮乏。睾丸作为维持雄性动物生殖能力的最关键器官,主要生物学功能是产生雄性配子(精子),而精子发生是精子产生所必需的发育过程,其有序运行是雄性生殖系统建立和维持的前提保障[2]。精子发生过程受到许多基因在多个层面(表观遗传、转录、转录后、翻译、翻译后等)的精密调控[3,4]。研究精子发生相关基因在藏绵羊睾丸发育及精子发生中的表达特征及调控作用,对于理解绵羊及其他高原动物睾丸功能维持的分子调控机理具有重要科学意义。【前人研究进展】BOLL(也称BOULE)作为无精症缺失(Deleted in Azoospermia,DAZ)基因家族中最古老的成员,目前已被广泛报道特异性存在于从昆虫至哺乳动物的性腺中,并且主要在雄性动物睾丸中表达,如锯蝇[5]、果蝇[6]、虹鳟[7]、鸡[8]、小鼠[9]、地鼠[10]、山羊[11]、牛(黄牛、牦牛、犏牛)[12]、猕猴[13]、人[14,15,16]等。在睾丸发育期间,BOLL在精子发生(尤其减数分裂阶段)中发挥着重要的调控作用,当其表达缺失或功能异常时,可导致精子发生过程停滞,从而因精子生成障碍而造成精液品质不良甚至雄性不育。在BOLL缺失或突变的人和果蝇睾丸中均表现出减数分裂紊乱和停滞,进而导致无精症,并且人的BOLL可以挽救果蝇睾丸中BOLL缺失导致的减数分裂缺陷[14, 17],提示BOLL在昆虫和人上具有类似的减数分裂调控作用。反之,在无精症患者睾丸中,也发现BOLL的表达显著降低,并且随着睾丸功能衰竭程度的加深其降低程度愈发明显[18]。在小鼠中,BOLL缺失的睾丸体积明显缩小,虽然精母细胞的减数分裂过程可以正常进行,但减数分裂产生的圆形精细胞丧失进一步发育为成熟精子的能力[9],表明BOLL在精子发生阶段的调控作用因物种而异。此外,在人原始生殖细胞中BOLL的过表达可诱导联会复合体的形成从而进入减数分裂阶段,形成在细胞形态上类似于圆形精细胞的单倍体生殖细胞[15]。与之相类似,在奶山羊雄性生殖干细胞中过表达BOLL也可启动并促进其减数分裂进程[11]。【本研究切入点】当前研究表明,BOLL尽管在雄性动物的精子发生过程中扮演不可或缺的角色,且在很多物种精子发生的减数分裂阶段起关键调控作用,但不难发现,其所执行的生物学功能因物种的不同存在差异,并且目前在藏绵羊睾丸中有关BOLL的序列特征、表达模式及调控作用尚不清楚。【拟解决的关键问题】本研究以藏绵羊作为试验动物,旨在解读BOLL的分子特征及在绵羊睾丸发育过程中的动态表达与分布规律,进而探究其表达调控作用及所赋予的生物学功能,为进一步探索BOLL在绵羊乃至其他哺乳动物睾丸中的时空表达转录调控机制及在精子发生中的作用机理提供依据。1 材料与方法
试验于2019年3月至2020年8月在甘肃农业大学进行。1.1 试验动物与样品采集
以甘肃省夏河甘加藏羊养殖合作社提供的24只健康雄性藏绵羊(同父异母)作为试验动物。试验羊来自相同父本,不同母本,分为3个发育阶段:性成熟前(3月龄,n=8)、性成熟(1岁龄,n=8)和成年(3岁龄,n=8)。所有羊只在海拔3 000—3 500 m的天然草场围栏放牧,昼夜自由采食,无补饲。屠宰后收集每只绵羊的右侧睾丸组织,一部分液氮速冻后存放于-80 ℃,另一部分置于4%多聚甲醛溶液中固定用于制作石蜡切片。1.2 总RNA提取及质检
冷冻的各睾丸组织经液氮快速研磨后,使用Trizol试剂盒(全式金生物公司, 北京)进行总RNA提取;使用NanoDrop ND-2000分光光度计(德国)对提取的RNA浓度和纯度进行检测。选取OD260/ OD280介于1.8—2.1,OD260/OD230大于2.0的RNA样品,并经1.5%的琼脂糖凝胶电泳对其完整性进行检测。1.3 第一链cDNA的合成
以质量和浓度检测合格的RNA样品作为模板,根据反转录试剂盒(Evo M-MLV RT Kit with gDNA Clean for qPCR,艾克瑞,湖南)的操作说明合成第一链cDNA。1.4 引物设计与合成
从NCBI数据库中获取绵羊BOLL(XM_ 004004798.4)和β-actin(NM_001009784.2)的核苷酸序列,随后利用Primer Premier 6.0软件设计引物并送至擎科生物公司(西安)进行合成。1.5 BOLL的克隆
以合成的cDNA为模板,利用RT-PCR技术进行BOLL CDS区的扩增。扩增总反应体系为50 μL:2×TransStart FastPfu Fly PCR SuperMix(全式金生物公司,北京)25 μL,PCR Stimulant 10 μL(全式金生物公司,北京),模板1 μL,上下游引物(BOLL-F:5′-ATG GAGACCGAGTCCGGG-3′,BOLL-R:5′-TTAATAAT GAATGCTCCACAC-3′)各2 μL, ddH2O 10 μL。PCR扩增程序:95 °C 5 min;95 °C 30 s,55 °C 30 s,72 °C 1 min,30个循环;72 °C 5 min。对PCR产物进行1%琼脂糖凝胶电泳分离后,用胶回收试剂盒对目的扩增产物进行纯化并回收,随后与pEASY-Blunt载体相连并转化Trans1-T1化学感受态细胞。随机挑取8个独立的阳性克隆通过菌落PCR法鉴定后,由擎科生物公司(西安)进行测序。1.6 生物信息学分析
利用BLAST工具将获得的藏绵羊BOLL序列与NCBI中现有的核苷酸序列数据库进行比对,以确定最近的亲缘关系和序列相似度,并用DNAMAN 8.0软件对不同物种BOLL的氨基酸序列同源性进行比较。使用NCBI中的ORFfinder在线工具(https://www.ncbi.nlm.nih.gov/orffinder/)对藏绵羊CDS区所有开放阅读框(ORF)进行查找分析。分别利用ProtParam(https://web.expasy.org/protparam/)、InterPro(http://www.ebi.ac.uk/interpro/)、SOPMA(https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html)和Phyre 2.0(http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index)在线软件对藏绵羊BOLL蛋白的理化性质、功能结构域、二级结构和三级结构进行分析。用WebLogo 在线工具(http://weblogo.berkeley.edu/logo.cgi)对结构域区段的核苷酸和氨基酸序列保守性进行分析。该蛋白的跨膜区和信号肽区分别通过TMHMM 2.0(http://www.cbs.dtu.dk/services/TMHMM-2.0)和SignalP 4.0(http://www.cbs.dtu.dk/services/SignalP-4.0/)软件进行预测。采用邻接算法通过MEGA 7.0软件构建BOLL蛋白的系统发育树。采用STRING 11.0数据库(https://string-db.org)对与绵羊BOLL蛋白相互作用的潜在蛋白质进行分析,并使用Cytoscape 3.8.0软件进行可视化。1.7 qRT-PCR分析
采用qRT-PCR技术在罗氏LightCycler 96荧光定量PCR仪(瑞士)上对各年龄组睾丸中BOLL mRNA的表达量进行实时定量检测。以β-actin作为内参。采用两步法反应程序:95 ℃ 30 s;95 ℃ 5 s,60 ℃ 30 s,共40个循环。反应体系如下:1 μL cDNA, 每0.4 μL上下游引物(表1),10 μL 2×SYBR Green Pro Taq HS Premix(艾克瑞,湖南),补充ddH2O至20 μL。每个样本设计3个重复。Table 1
表1
表1qRT-PCR引物信息
Table 1
| 引物名称 Primer name | 上游引物 Forward primer (5′-3′) | 下游引物 Reverse primer (5′-3′) | 产物长度 Product length(bp) |
|---|---|---|---|
| BOLL | GGCGCAAACATCAAATCAGAC | GGGCACTCGTTGGGTTATTC | 92 |
| β-actin | CTTCCAGCCTTCCTTCCTGG | GCCAGGGCAGTGATCTCTTT | 180 |
| LOC105602204 | ATATGACACGACGGGACAGC | CACAACCCGGTGCGTATCTA | 242 |
| LOC105603195 | GGGATTTGTCACTGGGCTCT | TGTGTTCTTCCCATTCGCCT | 258 |
| LOC105616228 | GTCTGGTCGGGAAATGCTGG | GGGCTCTCGTAAAACCTCCC | 156 |
| Circ-ECT2L | AGAGACTCATCTGGGGGTGC | TCTGGTTCGCTTTTCGGCT | 154 |
| Circ-SPHKAP | TTATTTCCGAATGCAGCCCC | TCCTCCTTAGTGGTTTCCTTTT | 182 |
| oar-miR-760-3p | CGGCTCTGGGTCTGTGGG | Universal reverse* | - |
| oar-miR-382-5p | GAAGTTGTTCGTGGTGGATTC | Universal reverse* | - |
| oar-miR-127-5p | CTGAAGCTCAGAGGGCTCTG | Universal reverse* | - |
| U6 | GGAACGATACAGAGAAGATTAGC | TGGAACGCTTCACGAATTTGCG | - |
新窗口打开|下载CSV
1.8 Western blot检测
使用含PMSF的RIPA裂解液(索莱宝生物公司, 北京)提取各睾丸组织总蛋白,BCA试剂盒(碧云天生物公司,上海)对其浓度进行测定,并经5×SDS上样缓冲液(索莱宝生物公司,北京)变性后,用12% SDS-PAGE进行电泳分离。电泳结束后,转移蛋白至PVDF膜上,并经5%脱脂奶粉室温封闭2.5 h后,用一抗BOLL(稀释比1:500;博奥森,北京)或β-actin(稀释比1:1 500;博奥森,北京)4 ℃孵育过夜。0.1% PBST洗膜后,添加山羊抗兔二抗(稀释比1:5 000;博奥森,北京)孵育2 h。使用ECL发光液进行化学发光、曝光和显影。用AlphaEaseFC软件对条带灰度值进行量化,并对BOLL蛋白的相对表达丰度进行计算。1.9 免疫荧光染色
使用常规方法对石蜡切片进行脱蜡、梯度乙醇脱水后,用微波炉加热法加入EDTA抗原修复缓冲液对组织切片进行抗原修复。5%BSA室温封闭30 min后,添加一抗BOLL(1:200;博奥森,北京)4 ℃孵育过夜。同时用5%BSA代替一抗BOLL作为阴性对照。PBS清洗后,切片中滴加带有FITC绿色荧光标签的山羊抗兔二抗(稀释比1:200;赛维尔,武汉),室温避光孵育1 h。PBS冲洗后,DAPI核染10 min,用含抗荧光猝灭剂的溶液封片,荧光显微镜(尼康,日本)观察并拍照。1.10 绵羊BOLL的功能注释与ceRNA调控网络分析
利用非冗余蛋白质(NR)数据库和基因本体论(GO)数据库,对绵羊BOLL所参与的生物学过程或功能进行注释。基于课题组前期通过RNA测序(RNA-seq)技术获得的藏绵羊睾丸组织全转录组测序数据,利用RNAhybrid 2.1.2(https://bibiserv.cebitec.uni-bielefeld)+SVM_light 6.01(http://svmlight.joachims.org/),Miranda 3.3a(http://www.microrna.org/microrna/home)和TargetScan 7.0(http://www.targetscan.org/)数据库对潜在的调控BOLL的长链非编码RNA(long noncoding RNA,lncRNA)/环状RNA(circular RNA,circRNA)-微小RNA(microRNA,miRNA)-BOLL作用网络进行预测分析,并对上述数据库的预测结果取交集作为ceRNA整合分析结果,使用Cytoscape 3.8.0软件对网络进行可视化。1.11 潜在靶向调控BOLL的miRNAs及ceRNAs表达量验证
对通过上述ceRNA网络分析获得的潜在调控BOLL的lncRNAs、circRNAs和miRNAs表达量进行qRT-PCR验证,并与RNA-seq结果进行比较。以1.2中提取的总RNA为模板,根据Evo M-MLV RT Kit with gDNA Clean for qPCR(艾克瑞,湖南)和Mir-XTM miRNA First-Strand Synthesis Kit(Takara,日本)反转录试剂盒操作说明分别合成用于lncRNA/circRNA和miRNA qRT-PCR扩增的第一链cDNA。qRT-PCR操作方法同1.7所述。运用Primer Premier 6.0软件设计lncRNA引物和circRNA发散引物,并经Blast对其特异性检测;采用加尾法设计miRNA的上游引物。引物(表1)均由擎科生物公司(西安)进行合成。每个样本设计3个重复。分别以β-actin和U6作为内参对lncRNA/circRNA和miRNA的表达量进行校正。1.12 荧光素酶报告基因载体构建及双荧光素酶活性检测
将分别含有oar-miR-760-3p、oar-miR-127-5p和oar-miR-382-5p假定靶结合位点的绵羊源BOLL 3′UTR区序列(XM_004004798.4),即BOLL-m760- 3′UTR WT(核苷酸位点1539-1843)、BOLL-m127- 3′UTR WT(核苷酸位点1599-1918)和BOLL-m382- 3′UTR WT(核苷酸位点2430-2769);含有oar-miR- 760-3p假定靶结合位点的circ-SPHKAP、Circ-ECT2L、LOC105602204(XR_001020768,核苷酸位点16-325)、LOC105603195(XR_001022217,核苷酸位点50-369)和LOC105616228(XR_001043211,核苷酸位点190-494)野生型序列;以及它们分别对应的突变型(MUT)序列克隆至pmirGLO双荧光素酶miRNA靶向表达载体(Promega, 美国)的Xho I和Sal I位点。上述序列均由金唯智生物科技有限公司(苏州)合成。miR-760-3p、miR-127-5p和miR-382-5p的模拟物(mimic)及其阴性对照物(mimic NC)由吉玛制药技术有限公司(上海)合成。复苏购自北京北纳生物的HEK-293T细胞,以1.0×105/孔的密度接种于24孔板中,使用含10% FBS的DMEM高糖培养基,在5% CO2、37℃恒温箱静置培养24 h后(汇合率为70%—80%),采用LipofectamineTM 2000转染试剂盒(Invitrogen, 美国),按照操作说明将构建的野生型及突变型重组双荧光素酶报告载体分别与对应的mimics或mimic NC共转染293T细胞。48 h后收集细胞,采用E1910双荧光素酶报告基因检测试剂盒(Promega, 美国)分析相对荧光素酶活性,结果以萤火虫荧光素酶活性与海肾素荧光素酶活性的比值来表示。每个试验设置3个重复。1.13 数据统计分析
采用2-ΔΔCt法(ΔCt=CtmRNAcircRNA/lncRNA/miRNA-?Ctβ-actin/U6)对qRT-PCR结果进行计算。用SPSS21.0统计软件进行单因素方差分析,结果用均值±标准差(Mean±SD)表示。采用Duncan多重检验比较各组间的差异,P<0.05和P<0.01分别表示组间差异显著和极显著。2 结果
2.1 藏绵羊BOLL完整CDS区序列特征
绵羊BOLL位于2号常染色体上,总长为63 155 bp,由11个外显子和10个内含子组成(图1-A)。1%琼脂糖凝胶电泳显示,PCR扩增的cDNA片段大小位于预期位置(图1-B)。对该cDNA片段进行克隆并测序,获得了长度为888 bp的核苷酸序列,对应的序列信息已提交至GenBank(Accession No. MT424763)。经与NCBI中的核苷酸序列数据库进行Blast检索比对发现,与设计克隆引物所用绵羊BOLL CDS区序列(XM_004004798.4)完全一致。该克隆序列存在14个ORF,其中全长开放阅读框ORF1(即完整CDS区)为888 bp,可编码295个氨基酸(图1-C、D)。藏绵羊BOLL编码蛋白的分子量、等电点(pI)和分子式分别为32.43 kD、6.07和C1459H2217N379O440S10。该蛋白序列不含信号肽和跨膜区域。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1藏绵羊BOLL CDS区克隆及序列分析
A:绵羊BOLL结构图(序列数据来源于GenBank:accession No. NC_040253.1)。B:藏绵羊BOLL cDNA的PCR扩增。M,DL2000分子标记。C:BOLL cDNA扩增产物的开放阅读框(ORF)分析。D:克隆的BOLL CDS区核苷酸序列及推导的氨基酸序列。ATG和TAA分别为起始密码子和终止密码子
Fig. 1Cloning and sequence analysis of Tibetan sheep BOLL CDS region
A: Structural diagram of the ovine BOLL (The sequence data is available from GenBank: accession No. NC_040253.1). B: PCR amplification derived from cDNA of Tibetan sheep BOLL gene. M, DL2000 marker. C: Open reading frame (ORF) analysis of Tibetan sheep BOLL cDNA amplification product. D: The nucleotide and deduced amino acid sequences for the cloned BOLL CDS region. ATG and TAA are the start codon and the stop codon, respectively
2.2 藏绵羊与其他哺乳动物BOLL的同源性比较和结构域分析
不同哺乳动物BOLL的氨基酸序列多重比较结果发现:藏绵羊与山羊的序列相似度最高(100%),其次为川南山地黄牛(99.66%)、牦牛(99.66%)、水牛(98.98%)、威德海豹(98.31%)、白尾鹿(97.97%)、非洲象(97.97%)、家猫(97.63%)、单峰驼(97.29%)、瘤牛(97.29%)、双峰驼(97.29%)、灰熊(96.95%)、北极熊(96.95%)、家马(96.95%)、野猪(96.95%)、北海狮(96.61%)、家犬(96.27%)和抹香鲸(95.93%),与兔的序列相似度最低(94.92%)(图2-A)。结构域分析发现,在藏绵羊BOLL氨基酸序列靠近N端43—123位点处(核苷酸序列位点127—369)和172—196处(核苷酸序列位点514—588)分别存在RNA识别基序(RNA recognition motif,RRM)结构域和DAZ重复基序,并且对应的核苷酸和氨基酸序列在不同哺乳动物间具有高度保守性(图2-B—E)。用系统进化树对来自20种哺乳动物的BOLL氨基酸序列进行分析发现,藏绵羊与山羊聚为一支,在进化上与山羊、水牛、牦牛和川南山地黄牛的亲缘关系较近,这与氨基酸同源性比较结果基本一致(图3)。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2不同哺乳动物BOLL CDS区序列特征比较分析
A:20种不同哺乳动物BOLL氨基酸序列的多重比对。红色方框表示RNA识别基序(RRM)结构域;黄色方框表示DAZ重复基序。B:RRM结构域对应的核苷酸序列保守性分析。C:RRM结构域对应的氨基酸序列保守性分析。D:DAZ重复基序对应的核苷酸序列保守性分析。E:DAZ重复基序对应的氨基酸序列保守性分析
Fig. 2Comparative analysis of BOLL CDS sequence features among different mammals
图3
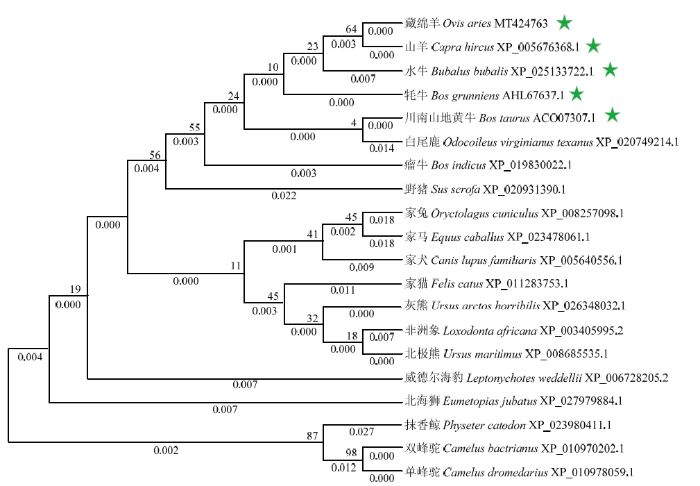 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3基于邻接算法的BOLL蛋白氨基酸序列系统进化树
每个分支上方和下方的数值分别表示引导值和分支长度。星号表示与藏绵羊BOLL有较近的亲缘关系
Fig. 3Phylogenetic tree for amino acid sequences of BOLL protein based on Neighbor-joining algorithm
The bootstrap values and branch lengths were showed above and below each branch, respectively. A closer phylogenetic relationship with Tibetan sheep BOLL is indicated by the asterisk
2.3 藏绵羊BOLL蛋白空间结构分析
藏绵羊BOLL蛋白的二级结构由无规则卷曲、延伸链、α螺旋和β转角4种结构组成,分别占64.07%、18.64%、13.56%和3.73%(图4-A)。BOLL蛋白的三级分子结构预测结果与其二级结构组成相似,主要含无规则卷曲、延伸链、α螺旋和β转角4种结构类型(图4-B)。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4藏绵羊BOLL蛋白的空间结构
A:二级结构;B:三级结构
Fig. 4Spatial structure of Tibetan sheep BOLL protein
A: Secondary structure; B: Tertiary structure
2.4 与绵羊BOLL蛋白互作蛋白分析
由蛋白相互作用网络分析结果(图5)可知,绵羊BOLL编码蛋白与10个蛋白可能存在相互作用,包括细胞质多腺苷酸化元件结合蛋白1(CPEB1)、细胞分裂周期蛋白25A(CDC25A)、细胞分裂周期蛋白20B(CDC20B)、DEAD家族的RNA解螺旋酶4(DDX4,也称VASA)、生长抑制蛋白家族成员2(ING2)、RNA结合基序蛋白25(RBM25)、核转运蛋白2(KPNA2)、核转运蛋白7(KPNA7)、鸟氨酸脱羧酶抗酶2(OAZ2)、泛素样修饰活化酶5(UBA5)。BOLL蛋白处于网络核心位置,且与CPEB1、UBA5和CDC20B蛋白间的相互作用具有更高的可信度。图5
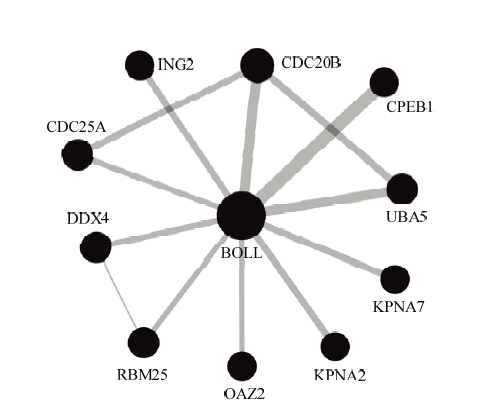 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5与绵羊BOLL蛋白互作蛋白网络分析
圆圈代表基因所编码蛋白,直线代表不同蛋白间的相互作用。圆圈的大小对应的是与该蛋白相互作用的蛋白数,圆圈越大表明相互作用的蛋白数越多。线的厚度表示蛋白间相互作用的置信度(置信度阈值为0.4),线越厚表明蛋白与蛋白之间相互作用的置信度越高
Fig. 5Network analysis of the proteins interacting with ovine BOLL protein
The circles represent proteins, and the straight lines represent the interactions between different proteins. Circle size corresponds to the protein numbers of interacting with this protein, with larger circles indicating the greater the numbers of interacting-proteins. The line thickness denotes the confidence of interactions (confidence threshold 0.4), with thicker lines indicating higher confidence in the protein-protein interaction
2.5 BOLL的表达模式
由图6可知:在mRNA和蛋白质水平,3月龄藏绵羊睾丸中BOLL表达量均较低;与3月龄相比,1岁龄和3岁龄睾丸中的BOLL mRNA和蛋白表达量极显著上调(P<0.01);相比1岁龄,3岁龄睾丸中BOLL mRNA表达呈上调趋势,而蛋白呈现一定的下调趋势。图6
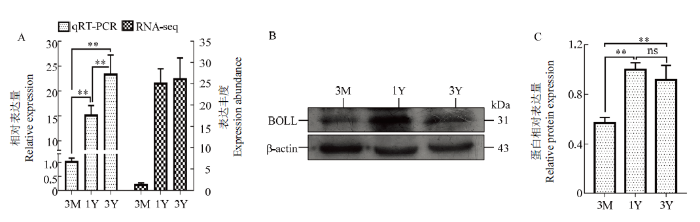 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6发育的藏绵羊睾丸中BOLL mRNA和蛋白的表达规律
A:RNA-Seq和qRT-PCR验证获得的BOLL mRNA表达变化规律的比较。所有RNA-seq试验由4个生物学重复组成;所有qRT-PCR试验由8个生物学重复组成,每个生物学重复由3技术重复组成。RNA-seq获得的BOLL mRNA表达丰度用FPKM值(每千个碱基的转录每百万映射读取的片段数)表示。B:BOLL 蛋白的Western blot分析;C:BOLL蛋白相对表达量。3M:3月龄;1Y:1岁龄;3Y:3岁龄。**:差异极显著(P<0.01);ns:差异不显著(P>0.05)
Fig. 6Expression patterns of BOLL mRNA and protein in developmental Tibetan sheep testes
A: Comparison of the changes in mRNA expression of BOLL acquired by RNA-seq and by qRT-PCR validation. All RNA-seq experiments consisted of four biological replicates; all qRT-PCR experiments consisted of eight biological replicates each consisting of three technical replications. The mRNA expression abundance of BOLL obtained from RNA-seq was expressed by the FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) value. B: Western blot analysis for BOLL protein; C: The relative level of BOLL protein expression. 3M: 3-month-old; 1Y: 1-year-old; 3Y: 3-year-old. **: Extremely significant difference (P<0.01); ns: Non-significant difference (P>0.05)
2.6 BOLL蛋白的免疫荧光染色
由BOLL蛋白的阳性免疫染色结果(图7)可知:在3月龄藏绵羊睾丸中,较弱的BOLL蛋白阳性信号定位于部分精原细胞;在1岁龄和3岁龄睾丸中,强的BOLL蛋白阳性信号主要定位于长形精细胞中,其次为圆形精细胞,也有较弱的阳性信号被发现存在于精原细胞和精母细胞中,并且随着年龄的增加,在精原细胞和精母细胞中的阳性信号强度减弱。图7
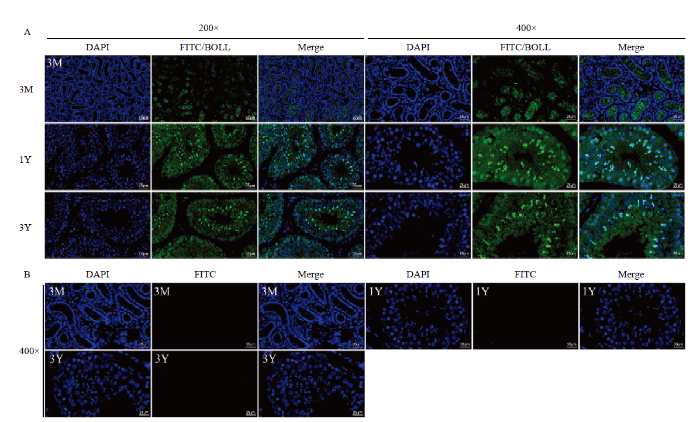 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7藏绵羊睾丸中BOLL蛋白的免疫荧光染色
A:BOLL蛋白的免疫定位;B:阴性对照。3M:3月龄;1Y:1岁龄;3Y:3岁龄。DAPI(蓝色):细胞核染色;FITC(绿色):荧光染色;Merge:共定位
Fig. 7Immunofluorescence staining of BOLL protein in Tibetan sheep testis
A: Immunolocalization of BOLL protein; B: Negative controls. 3M: 3-month-old; 1Y: 1-year-old; 3Y: 3-year-old. DAPI (blue): Nuclear staining; FITC (green): Fluorescent staining; Merge: Colocalization
2.7 绵羊BOLL潜在的ceRNA表达调控网络及功能注释
通过对BOLL的功能注释结果发现:在分子功能方面,其主要涉及结合、翻译调节活性等;在生物学过程方面,其主要参与组织细胞发育、生殖、配子生成、基因转录后及翻译起始调控等过程(图8)。ceRNA网络分析发现:绵羊BOLL受到3个潜在的miRNAs(oar-miR-127-5p、oar-miR-760-3p和oar-miR-382-5p)靶向调控,并且3个lncRNAs(LOC105602204、LOC105603195和LOC105616228)和2个circRNAs(circ-ECT2L和circ-SPHKAP)分子可与BOLL共同竞争性结合oar-miR-760-3p(图8)。图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8绵羊BOLL的功能注释及潜在的ceRNA调控网络
Fig. 8The functional annotation and potential ceRNA regulatory network for ovine BOLL
2.8 潜在调控BOLL的非编码RNAs在睾丸中的表达特征
实时荧光定量PCR结果(图9-A)显示:随着年龄的增加,oar-miR-127-5p、oar-miR-382-5p和oar-miR- 760-3p的表达量逐渐下降;LOC105616228和circ-ECT2L的表达量逐渐上调;LOC105602204、LOC105603195和circ-SPHKAP的表达量先上调而后呈现不同程度的下调趋势。由图9-A可知,qRT-PCR与RNA-seq获得的各非编码RNAs在睾丸中的表达模式基本一致:相比3月龄,在1岁龄和3岁龄睾丸中各miRNAs的表达量极显著下调(P<0.01),而lncRNAs和circRNAs的表达量极显著上调(P<0.01)。图9
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9潜在调控BOLL的非编码RNAs的时间表达模式及靶向关系验证
A:RNA-Seq(4个生物学重复)和qRT-PCR验证(8个生物学重复组成,每个由3个技术重复组成)获得的非编码RNA表达模式的比较。RNA-seq获得的miRNAs、circRNAs和lncRNAs的表达丰度分别用TPM(每千个碱基的转录每百万映射读取的转录本数)、RPM(每百万映射读取的reads数)和FPKM(每千个碱基的转录每百万映射读取的片段数)衡量。B:双荧光素酶报告法验证miRNAs与BOLL或其ceRNAs的靶向关系。3M:3月龄;1Y:1岁龄;3Y:3岁龄。**:差异极显著(P<0.01);*:差异显著(P<0.05);ns:差异不显著(P>0.05)
Fig. 9Temporal expression patterns of potential regulatory non-coding RNAs for BOLL gene and verification of the target relationship
A: Comparison of the changes in expression of non-coding RNAs obtained by RNA-seq (four biological replicates) and by qRT-PCR validation (eight biological replicates each consisting of three technical replications). The expression abundance of miRNAs, circRNAs, and lncRNAs obtained from RNA-seq was measured by the TPM (Transcripts Per Kilobase of exon model per Million mapped reads), RPM (Reads of exon model per Million mapped reads), and FPKM (Fragments Per Kilobase of exon model per Million mapped fragments), respectively. B: A dual luciferase reporter assay was used to validate the targeting relationships between miRNAs and BOLL or their ceRNAs. 3M: 3-month-old; 1Y: 1-year-old; 3Y: 3-year-old. **: Extremely significant difference (P<0.01); *: Significant difference (P<0.05); ns: Non-significant difference (P>0.05)
2.9 BOLL-miRNAs及miRNAs-circRNAs/lncRNAs靶向关系验证
由双荧光素酶报告基因检测结果(图9-B)可知,相比对照组,oar-miR-127-5p和oar-miR-760-3p模拟物均显著降低了293T细胞中的BOLL 3′UTR野生型报告者的荧光素酶活性(P<0.01),并且oar-miR-760- 3p模拟物显著降低了野生型Circ-ECT2L(P<0.05)以及LOC105616228(P<0.01)报告者的荧光素酶活性,但对相应的突变型报告者均无明显影响。oar-miR- 382-5p模拟物对BOLL 3′UTR野生型报告基因的荧光素酶活性无明显影响(P>0.05),oar-miR-760-3p模拟物对野生型circ-SPHKAP、LOC105602204和LOC105603195报告基因的荧光素酶活性也无明显影响(P>0.05;结果未显示)。这些结果表明,绵羊BOLL是oar-miR-127-5p和oar-miR-760-3p共有的直接作用靶标,并且circRNA Circ-ECT2L和lncRNA LOC105616228作为ceRNAs, 可与BOLL共同竞争性靶向结合oar-miR-760-3p。3 讨论
首先通过克隆并测序获得了藏绵羊BOLL的完整CDS区,由888 bp的核苷酸组成,可编码295个氨基酸,这与NCBI中提供的绵羊预测序列(XM_004004798.4)完全一致,并且与在山羊[19]、川南山地黄牛[12]和牦牛[20]上报道的该基因CDS区长度一致。通过氨基酸序列比对发现,藏绵羊BOLL编码区对应的氨基酸序列与山羊、川南山地黄牛和牦牛的相似度均在99%以上,与其他哺乳动物的序列相似度也高于95%(兔除外)。在进化上,其与山羊、牦牛、川南山地黄牛也具有比较近的亲缘关系。这些结果表明,藏绵羊BOLL同其他哺乳动物具有高度的序列同源性和进化保守性。DAZ家族成员(包括BOLL)所编码蛋白中均含有一个RRM结构域和至少一段DAZ重复基序[6]。RRM结构域作为真核生物细胞中最丰富的RNA结合元件,可通过与其他核酸或蛋白质相互作用,以调节mRNA的转录、翻译和衰变等环节而参与基因表达的精细调控(尤其转录后调控),进而发挥相应的生物学功能[21]。但对于DAZ重复基序,其在DAZ家族基因编码蛋白中所承担的具体功能目前仍不清楚。YEN等[22]依据自己的研究结果推测,DAZ重复基序可能参与RNA结合,也可能参与与其他蛋白质的相互作用。EWIS等[23]研究发现,DAZ重复基序的缺失可造成男性生精能力下降。综上所述,DAZ重复基序可能具有与RRM结构域相似的功能,即通过与其他蛋白质分子结合从而贡献于DAZ家族各成员自身的功能维持。与其他哺乳动物一样,本研究通过氨基酸序列分析表明,藏绵羊BOLL蛋白也具有一个RRM结构域(靠近N端)以及一段由25个氨基酸残基组成的DAZ重复基序,并且二者对应的氨基酸序列在不同物种中高度相似。由此推测,藏绵羊BOLL蛋白所含的RRM结构域和DAZ重复基序在进化和功能上是非常保守的,并且可能是其执行并维持正常的生物学功能所必需的。
鉴于BOLL蛋白可通过结合相应的蛋白质而发挥作用,本研究进一步借助STRING数据库对潜在的与绵羊BOLL蛋白互作的基因编码蛋白进行分析发现,CDC25A、CDC20B 等10个蛋白质分子与BOLL蛋白可能存在相互作用。其中,CDC25A和CDC20B作为细胞周期相关蛋白,被报道在哺乳动物精子发生中广泛参与生殖细胞的有丝分裂和减数分裂等过程[24,25]。在成年雄性小鼠睾丸中发现,BOLL可与CDC25A mRNA靶向结合以调节其翻译过程,从而对精母细胞的减数分裂进行调控[9]。LI等[11]在山羊上研究发现,过表达BOLL可通过上调减数分裂相关基因CDC25A的表达,进而启动山羊生殖干细胞的减数分裂过程。与细胞周期蛋白所发挥的功能类似,OAZ2和DDX4也被报道在雄性哺乳动物生殖细胞中表达并参与调控细胞的增殖与分化[26,27]。在女性原始生殖细胞中过表达BOLL可增加DDX4的表达,进而促进生殖细胞的减数分裂进程及单倍体卵子的形成[15]。同DAZ家族成员一样,CPEB1也是一种RNA结合蛋白,已被报道在小鼠卵母细胞成熟过程中,可靶向调控DAZ家族中另一成员DAZL的mRNA翻译和蛋白积累,并且CPEB1和DAZL可通过调节共同靶标mRNA的翻译表达从而对卵母细胞的减数分裂进行正向调控[28]。CPEB1的缺失不仅造成雌性小鼠不育,也可导致雄鼠精子发生停滞在减数分裂阶段而严重损害其生育力[29]。类似于CPEB1在睾丸中的调节功能,ING2可通过与相应的靶标结合参与精子发生过程,其缺失也可导致精子生成异常甚至雄性不育[30]。UBA5是泛素激活酶E1的主要成员之一,而KPNA2和KPNA7均属于核转运蛋白。泛素化作为广泛存在于真核生物细胞内的一种蛋白质翻译后修饰,在精子发生过程中可将泛素分子结合到靶蛋白上,以参与调节蛋白质的稳定性和功能[31,32]。同样,在雄性生殖细胞发育的特定阶段,核转运蛋白通过与不同的转录因子或蛋白质结合以转运其穿过核孔参与调控生殖细胞分化和精子发生[33]。RBM25作为RNA结合基序蛋白家族中的一员,被报道在小鼠睾丸精子细胞中特异表达[34]。综上,这些潜在与绵羊BOLL互作的蛋白分子均在雄性动物精子发生过程中表达并扮演不同的角色,且大多已被报道具有结合并调节其他蛋白质分子的能力。这为深入理解BOLL在哺乳动物,尤其绵羊睾丸发育期间的分子作用机理提供了新的见解和思路,但这些蛋白是否与BOLL存在相互作用并且通过何种机制贡献于生精细胞的表型维持及精子生成,仍有待进一步探索。
本研究发现,在性成熟后藏绵羊睾丸中BOLL在转录和蛋白质水平的表达均显著上调,这与以前研究报道其在具有正常生精能力(性成熟后)的哺乳动物睾丸中高表达[35,36]的结果相符,表明BOLL可能主要在性成熟后藏绵羊睾丸中发挥作用。尽管BOLL是一个减数分裂相关基因,但在睾丸中的表达不仅仅局限于减数分裂阶段的精母细胞中。如在成年小鼠睾丸中,BOLL蛋白不仅存在于精母细胞中,也在圆形和长形精细胞、精子及部分精原细胞中分布[37]。在地鼠睾丸中,BOLL蛋白除了在精原细胞、精母细胞和圆形精细胞中表达外,在睾丸间质细胞中也有分布[10]。为了明确BOLL蛋白在发育的藏绵羊睾丸细胞中所呈现的表达特征及发挥的生物学功能,本研究进一步通过免疫荧光染色发现,BOLL蛋白在性成熟前睾丸中存在于部分精原细胞中,而在性成熟后主要分布于精细胞中,在精母细胞和精原细胞中也有较弱的阳性信号且随着年龄的增加信号强度更加弱化。这与在小鼠睾丸中[9],研究发现该基因在圆形精细胞中高表达并发挥作用的结果类似,提示BOLL可能主要在藏绵羊精子发生中参与调控减数分裂后的单倍体精子细胞进一步向成熟精子发育。这也与在小尾寒羊(低海拔、性成熟早、繁殖力高)[36]和地鼠[10]上报道BOLL蛋白在睾丸生殖细胞中的分布特征基本一致,但与在小尾寒羊上研究结果不同的是,BOLL蛋白在性成熟后小尾寒羊睾丸中主要表达于精母细胞和圆形精细胞中。此外,有趣的是,在藏绵羊睾丸中,随着年龄的增加,BOLL在转录水平的表达逐渐上调,而在翻译水平的表达先(性成熟)上调而后(成年)出现下调,这与在转录和翻译水平都发现BOLL在成年小尾寒羊睾丸中表达上调(相比性成熟阶段)[36]的结果不一致。这些差异可能是由于基因的表达调控机制和表观遗传机制因品种的繁殖力和生存环境不同而异所造成的。已有研究报道,在具有不同繁殖力的绵羊品种卵巢中许多基因的表达及调控作用存在差异[38]。正如在雄性不育的人[18]和犏牛[20]睾丸中发现的BOLL表达很微弱甚至不表达,在发育的绵羊睾丸中,相比同处于成年期(精子产生旺盛期)的具有高繁殖力的小尾寒羊[36],藏绵羊睾丸内BOLL蛋白的表达呈现一定的下降趋势,这可能与其繁殖力较低有关,但其具体作用机制的差异性还需进一步研究与证实。这为从细胞和分子水平探究不同繁殖力的种公羊品种在睾丸发育分子遗传机理方面的差异提供了思路。
在藏绵羊睾丸中BOLL mRNA和蛋白的表达趋势及表达改变倍数存在差异,这可能是由于基因的表达受到转录、转录后等水平的严格调控所造成的。非编码RNAs(lncRNAs、miRNAs、circRNAs等)作为转录调节因子,在转录、转录后、表观遗传等层面参与调控基因的表达[39]。同样,哺乳动物精子发生过程也受到许多非编码RNAs的调控[40,41]。有研究发现,在斑马鱼原始生殖细胞中miR-430可靶向抑制DAZL(与BOLL属同一家族)的mRNA表达[42],但有关靶向调控BOLL的miRNAs研究仍处于空白阶段。本研究基于前期有关藏绵羊睾丸组织的全转录组测序数据并借助相关数据库对潜在调控BOLL的非编码RNAs,即ceRNAs作用网络进行了鉴定与分析,并通过qRT-PCR和双荧光素酶报告实验对其表达特征和靶向关系进行了验证。发现,oar-miR-127-5p和oar-miR-760-3p可直接靶向调控藏绵羊BOLL的表达并且circRNA Circ-ECT2L和lncRNA LOC105616228可通过竞争性结合oar-miR- 760-3p而调控BOLL的表达。为了进一步理解ceRNA介导调控绵羊BOLL所涉及的生物学功能,对BOLL参与的生物学过程和功能进行了详细注释,发现其主要参与配子生成、细胞分裂、基因转录后及翻译起始调控等生物学过程。以上结果表明,在藏绵羊睾丸中,BOLL的表达受miRNAs及ceRNAs调控,进而可能与可激活下游信号或事件的蛋白分子相互作用,以响应生精细胞的发育及精子生成。该研究填补了在绵羊睾丸中有关BOLL表达与调控作用认识的空白,丰富了对其在绵羊甚至其他哺乳动物精子发生中潜在的分子作用机制的理解,并为进一步的功能探究提供了新的理论视角。
4 结论
本研究成功克隆获得藏绵羊BOLL完整CDS区序列,其在哺乳动物间具有高度进化保守性,并发现该基因主要表达于性成熟后藏绵羊睾丸精子细胞中,且在精母细胞和整个发育阶段的精原细胞中也有表达。oar-miR-127-5p和oar-miR-760-3p直接靶向抑制绵羊BOLL的表达;circRNA Circ-ECT2L和lncRNA LOC105616228作为ceRNAs,可与BOLL共同竞争性吸附oar-miR-760-3p而正向调控绵羊BOLL的表达,进而通过与下游分子相互作用以激活细胞增殖或分化相关信号事件来调控精子发生,尤其减数分裂后的单倍体精子细胞的进一步发育。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/j.livsci.2010.08.007URL [本文引用: 1]

We investigated the macro-mineral status in free-grazing Tibetan sheep (Ovis aries) and their forage diets. The study was conducted in 4 counties in the northeast of the Qinghai-Tibetan Plateau. Forage samples and blood serum from 20 sheep in each county were collected in July (summer) and December (winter), and analyzed to determine the concentration of the main macro-minerals (Ca, K. Mg, Na and P). Macro-mineral concentrations varied with season, but values for forage were below the recommended levels for P and K in one or more counties in winter and for Na was low during both summer and winter for all counties. Similarly, serum samples were found to be within marginal range of P. with the exception of Tianzhu County (TC) in winter, and serum Na were all below the marginal level in both summer and winter in all counties. The results indicate that the winter forage in particular is insufficient in macro-minerals and that this may lead to the observed mineral deficiencies and marginal deficiencies in Tibetan sheep in these areas. Supplementary minerals may therefore play a role in balancing the minerals available from the forage in winter and lead to increases in animal production on the Tibetan Plateau. Crown Copyright (C) 2010 Published by Elsevier B.V.
URLPMID:30980107 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:26416010 [本文引用: 1]
DOI:10.1016/j.ydbio.2014.12.027URLPMID:25592223 [本文引用: 1]

boule (bol), a member of the Deleted in Azoospermia (DAZ) gene family plays an important role in meiosis (reductional maturation divisions) in a spermatogenesis-specific manner in animals by regulating translation of the downstream cell division cycle 25 (cdc25) phosphatase mRNA. Orthologues of bol are conserved among animals and found in the genomes of hymenopteran insects, in which the general mode of reproduction is haplodiploidy: female is diploid and male is haploid. In this mode of reproduction, haploid males produce haploid sperm through non-reductional maturation divisions. The question thus arises of whether the bol gene actually functions during spermatogenesis in these haploid males. In this study, we identified two transcriptional isoforms of bol orthologue (Ar bol and Ar bol-2), and one cdc25 orthologue (Ar cdc25) in the hymenopteran sawfly, Athalia rosae. Ar bol was expressed exclusively in the testis when maturation divisions occurred, while Ar bol-2 was expressed ubiquitously. Knockdown of all bol transcripts (both Ar bol and Ar bol-2) resulted in a lack of mature sperm, whereas males with sole knockdown of Ar bol-2 were able to produce a small number of mature sperm. The cell cycle was arrested before maturation divisions in the testis in which all bol transcripts were knocked down, as revealed by flow cytometry. Although no mature sperm was produced, sperm elongation was partially observed when Ar cdc25 alone was knocked down. These results indicate that Ar bol is essential for the entry and progression of maturation divisions and sperm differentiation in haploid males.
URLPMID:26327816 [本文引用: 2]
DOI:10.1016/j.ygcen.2015.05.011URLPMID:26027538 [本文引用: 1]

Boule, the ancestor of the DAZ (Deleted in AZoospermia) gene family, in most organisms is mainly involved in male meiosis. The present study investigates the effects of the plasticizer DEHP (50mg/kg body weight) and herbicide butachlor (0.39mg/L) on male rainbow trout (Oncorhynchus mykiss) for a 10-day period in two independent experiments. The results showed that plasma testosterone (T) concentrations were significantly lower in fish exposed to either DEHP or butachlor compared to the control fish (P<0.05). Fish showed a significantly elevated hepatosomatic index (HSI) in the butachlor treatment (P<0.05). However, no significant difference was observed in HSI values in the DEHP treatment (P>0.05). In addition, no significant differences were found in the gonadosomatic index (GSI) in both DEHP and butachlor treatments (P>0.05). Histologically, testes of male trout in the control groups were well differentiated and filled with large numbers of cystic structures containing spermatozoa. In contrast, the testes of male trout contained mostly spermatocytes with few spermatozoa in both treated group, suggesting that DEHP and butachlor may inhibit the progression of meiosis. Also, boule gene expression was significantly lower in the testes of male trout affected by DEHP and butachlor in comparison with their control groups (P<0.05), which confirmed the meiotic arrest in affected trout. Based on the results, the present study demonstrated that DEHP and butachlor can inhibit the progression of spermatogenesis in male trout, potentially by causing an arrest of meiosis, maybe due to down-regulation of boule gene expression through T and/or IGF1 via ERK1/2 signaling in T-independent pathways. In addition, these results confirmed that boule can be considered as a predictive marker to assess meiotic efficiency.
DOI:10.3892/ijmm.2018.3602URLPMID:29620249 [本文引用: 1]

In vitro production of functional spermatids has special significance in the research of spermatogenesis and the treatment of male infertility. Primordial germ cells (PGCs) are the precursors of oocyte and sperm, which generate the totipotent cells. Studies have shown that PGCs have the potential ability to develop meiotic spermatids in vitro. Here we have shown that retinoic acid (RA) leads to PGC differentiation, and SCF can improve the efficiency of induction. We indicate an efficient approach to produce haploid spermatids from chicken PGCs in the presence of RA and stem cell factor (SCF). Real-time RT-PCR assays showed that RA and SCF induced a remarkable increase in expression of SYCP1, ACR, BOULE and DCM1 of meiotic germ cells and haploid germ cells, respectively. DNA content assays revealed that RA and SCF induced a remarkable increase of haploid cells. This study provides a theoretical basis and a great animal model for spermatogenesis study.
DOI:10.1093/hmg/ddq109URLPMID:20335278 [本文引用: 4]

Reproduction is required for the survival of all animals, yet few reproductive genes have been shown to have a conserved requirement for fertility across the animal kingdom. Remarkably, the RNA binding protein BOULE, the oldest member of the DAZ (Deleted in AZoospermia) family of genes, appears to have maintained its conserved functional motif and spermatogenic expression from insects to humans. Boule mutations lead to a pachytene meiotic arrest before metaphase in Drosophila males and C. elegans females, and human BOULE can restore meiosis in the fly testis, suggesting a conserved meiotic function of human BOULE. However, the physiological function of BOULE in mammals is not yet known. We generated Boule knockout mice and found it to be required only for spermatogenesis, as in Drosophila. Interestingly, meiosis completed normally in the absence of Boule, and haploid round spermatids were readily detected. However, round spermatids did not progress beyond step 6, revealing a novel role for Boule in spermiogenesis, the differentiation of round spermatids into mature spermatozoa. Expression of key regulators of spermiogenesis was unaffected in Boule(-/-) mice, suggesting that Boule regulates germ-cell differentiation through a novel pathway.
DOI:10.1017/S0967199418000023URLPMID:29573758 [本文引用: 3]

SummaryThe Deleted in AZoospermia (DAZ) gene family regulates the development, maturation and maintenance of germ cells and spermatogenesis in mammals. The DAZ family consists of two autosomal genes, Boule and Dazl (Daz-like), and the Daz gene on chromosome Y. The aim of this study was to analyze the localization of DAZL and BOULE during testicular ontogeny of the seasonal-breeding Syrian hamster, Mesocricetus auratus. We also evaluated the testicular expression of DAZ family genes under short- or long-photoperiod conditions. In the pre-pubertal and adult testis, DAZL protein was found mainly in spermatogonia. BOULE was found in the spermatogonia from 20 days of age and during the pre-pubertal and adult period it was also detected in spermatocytes and round spermatids. DAZL and BOULE expression in spermatogonia was strictly nuclear only in 20-day-old hamsters. We also detected the novel mRNA and protein expression of BOULE in Leydig cells. In adult hamsters, Dazl expression was increased in regressed testis compared with non-regressed testis and DAZL protein expression was restricted to primary spermatocytes in regressed testis. These results show that DAZL and BOULE are expressed in spermatogonia at early stages in the Syrian hamster, then both proteins translocate to the cytoplasm when meiosis starts. In the adult regressed testis, the absence of DAZL in spermatogonia might be related to the decrease in germ cell number, suggesting that DAZ gene family expression is involved in changes in seminiferous epithelium during photoregression.
DOI:10.1002/jcb.24368URL [本文引用: 3]

Boule is a conserved gene in meiosis, which encodes RNA binding protein required for spermatocyte meiosis. Deletion of Boule was found to block meiosis in spermatogenesis, which contributes to infertility. Up to date, the expression and function of Boule in the goat testis are not known. The objectives of this study were to investigate the expression pattern of Boule in dairy goat testis and their function in male germline stem cells (mGSCs). The results first revealed that the expression level of Boule in adult testes was significantly higher than younger and immature goats, and azoospermia and male intersex testis. Over-expression of Boule promoted the expression of meiosis-related genes in dairy goat mGSCs. The expression of Stra8 was up-regulated by over-expression of Boule analyzed by Western blotting and Luciferase reporter assay. While, Cdc25a, the downstream regulator of Boule, was found not to affect the expression of Stra8, and our data illustrated that Cdc25a did not regulate meiosis via Stra8. The expression of Stra8 and Boule was up-regulated by RA induction. Taken together, results suggest the Boule plays an important role in dairy goat spermatogenesis and that over-expression of Boule may promote spermatogenesis and meiosis in dairy goat. J. Cell. Biochem. 114: 294302, 2013. (c) 2012 Wiley Periodicals, Inc.
DOI:10.1007/s00438-008-0394-6URLPMID:18987886 [本文引用: 2]

The Deleted in Azoospermia (DAZ) genes encode potential RNA-binding proteins that are expressed exclusively in the germ-line. The bovine Deleted in Azoospermia-like gene is a strong candidate for male cattle-yak infertility. In this work, with the goe goal to further reveal the genetic cause of male cattle-yak sterility, another bovine DAZ family gene, b-boule, was isolated and characterized. The b-boule gene is predicted to encode a polypeptide of 295 amino acids with an RNP-type RNA recognition domain. Tertiary structure analysis shows that b-boule binds specifically to polypyrimidine RNAs and might act as a nuclear ribonucleoprotein particle auxiliary factor during germ cell formation and morphological changes of germ cells. RT-PCR assays revealed that b-boule was expressed specifically in the adult testis. However, an extremely low level of expression was detected in the testis of sterile male cattle-yaks. Microstructure of the testes from sterile males showed that type A spermatogonia were the only germ cells present and that few germ cells developed further than the stage of pachytene spermatocytes. These results suggest that b-boule may function in bovine spermatogenesis, and that low levels of b-boule expression might lead to male sterility in cattle-yaks.
DOI:10.1371/journal.pone.0218194URLPMID:31226129 [本文引用: 1]

BACKGROUND: From a biological and clinical perspective, it is imperative to establish primate spermatogonial cultures. Due to limited availability of human testicular tissues, the macaque (Macaca fascicularis) was employed as non-human primate model. The aim of this study was to characterize the expression of somatic as well as germ cell markers in testicular tissues and to establish macaque testicular primary cell cultures. MATERIALS AND METHODS: Characterization of macaque testicular cell population was performed by immunohistochemical analyses for somatic cell markers (SOX9, VIM, SMA) as well as for germ cell markers (UTF1, MAGEA4, VASA). Testicular cells from adult macaque testes (n = 4) were isolated and cultured for 21 days using three stem cell culture media (SSC, PS and SM). An extended marker gene panel (SOX9, VIM, ACTA2; UTF1, FGFR3, MAGEA4, BOLL, DDX4) was then employed to assess the changes in gene expression levels and throughout the in vitro culture period. Dynamics of the spermatogonial population was further investigated by quantitative analysis of immunofluorescence-labeled MAGEA4-positive cells (n = 3). RESULTS: RNA expression analyses of cell cultures revealed that parallel to decreasing SOX9-expressing Sertoli cells, maintenance of VIM and ACTA2-expressing somatic cells was observed. Expression levels of germ cell marker genes UTF1, FGFR3 and MAGEA4 were maintained until day 14 in SSC and SM media. Findings from MAGEA4 immunofluorescence staining corroborate mRNA expression profiling and substantiate the overall maintenance of MAGEA4-positive pre- and early meiotic germ cells until day 14. CONCLUSIONS: Our findings demonstrate maintenance of macaque germ cell subpopulations in vitro. This study provides novel perspective and proof that macaques could be used as a research model for establishing in vitro germ cell-somatic cell cultures, to identify ideal culture conditions for long-term maintenance of primate germ cell subpopulation in vitro.
DOI:10.1093/molehr/gal101URLPMID:17114206 [本文引用: 2]

The complex process of spermatogenesis requires the expression and precise coordination of a multitude of genes. Abnormal function of such genes is frequently associated with male infertility. Among these candidates is the human BOULE gene that is a possible fundamental mediator of meiotic transition. In this study, we describe for the first time the existence of three BOULE transcript variants (B1, B2 and B3). We investigated their tissue specificity and mRNA transcript levels in 23 testis biopsies from infertile men. B1, B2 and B3 differed solely in their N-terminal sequences, which are encoded by three alternatively spliced exons 1. In humans, all three isoforms are exclusively expressed in the testes in a relative proportion of 80:220:1 for B1, B2 and B3, respectively. RT-PCR quantification revealed significantly reduced mRNA expression of all three variants in testicular biopsies with meiotic arrest (MA) compared with those with qualitatively complete spermatogenesis. Alteration of the B1/B2 and B1/B3 transcript ratios was correlated with reduced meiotic capacity of spermatocytes to produce round spermatids as assessed by flow cytometry. Furthermore, BOULE mRNA reduction in biopsies with MA paralleled the absence of BOULE protein as analysed by immunohistochemistry. In conclusion, the relative proportions of B1, B2 and B3 may serve as predictive markers for meiotic efficiency and thus the probability of finding haploid cells in the human testis. Among the three isoforms, B2 might have the major role for meiotic completion.
DOI:10.1038/nature08562URLPMID:19865085 [本文引用: 3]

The leading cause of infertility in men and women is quantitative and qualitative defects in human germ-cell (oocyte and sperm) development. Yet, it has not been possible to examine the unique developmental genetics of human germ-cell formation and differentiation owing to inaccessibility of germ cells during fetal development. Although several studies have shown that germ cells can be differentiated from mouse and human embryonic stem cells, human germ cells differentiated in these studies generally did not develop beyond the earliest stages. Here we used a germ-cell reporter to quantify and isolate primordial germ cells derived from both male and female human embryonic stem cells. By silencing and overexpressing genes that encode germ-cell-specific cytoplasmic RNA-binding proteins (not transcription factors), we modulated human germ-cell formation and developmental progression. We observed that human DAZL (deleted in azoospermia-like) functions in primordial germ-cell formation, whereas closely related genes DAZ and BOULE (also called BOLL) promote later stages of meiosis and development of haploid gametes. These results are significant to the generation of gametes for future basic science and potential clinical applications.
[本文引用: 1]
URLPMID:12499397 [本文引用: 1]
DOI:10.1093/humrep/deh647URL [本文引用: 2]
DOI:10.1016/j.theriogenology.2018.05.002URLPMID:29778921 [本文引用: 1]

A conserved gene in meiosis, the Boule gene is involved in meiosis and spermatogenesis. The deletion of this gene in males blocks meiosis and results in infertility. Alternative splicing variants of the Boule gene have been identified in humans, bovines, and bats, but in dairy goats remains unknown. This study was therefore to detect splicing variants of the goat Boule gene and explore their potential roles in meiosis. Three isoforms, denoted as Boule-a, Boule-b, and Boule-c, were identified in the testes of goats using real-time PCR (RT-PCR) and cloning sequencing. Compared to the normal Boule gene, Boule-a was found to lack exons 7 and 8, which corresponds to a predicted variant, X4, on the NCBI database. Boule-b lacked exon 8, and Boule-c only retained exons 1 and 2. Of these three variants, two were novel isoforms of the Boule gene. Quantitative RT-PCR (qRT-PCR) showed that the Boule-a and Boule-b expression patterns were significantly different between the adult goat testes and the postnatal testes of 42 and 56 days. Overexpression of Boule-a and Boule-c in GC-1 spg cells of model mice significantly repressed CDC2 expression. Bisulfite sequencing PCR (BSP) results showed that the promoter region of the Boule gene was hypermethylated in goat testes. A negative correlation between the methylation levels of the Boule gene promoter and total mRNA expression of its transcripts was found. Our data showed alternative splicing and promoter methylation in the goat Boule gene, suggesting that this gene may play an important role in the regulation of Boule expression and in meiosis processing.
URLPMID:25149018 [本文引用: 2]
DOI:10.4161/rna.7.3.12087URLPMID:20458169 [本文引用: 1]

In eukaryotic cells, a regulated import and export of factors is required to fulfill the requirements of precise gene expression. Post-transcriptional regulation of gene expression has proven to provide ubiquitous control, as well as a quick response to environmental changes when required. RNA-binding proteins (RBP) are involved in the several steps at which mRNA biogenesis, stability, translation and decay is exerted. The most characterized RBPs contain single or multiple copies of an RNA Recognition Motif (RRM). Here, we concentrate on RRMs mediating protein nuclear import by virtue of its ability to interact with proteins, besides interacting with nucleic acids. The consensus on how RRM-protein interactions take place is non-existent, and so is the involvement of the RRM as a nuclear localization signal (NLS). Within the cases examined, the single RRM from a trypanosome RBP behaves as a structural NLS, alternating nuclear import and RNA-binding.
DOI:10.1007/s003359900560URLPMID:9321470 [本文引用: 1]

The DAZ genes on the human Y Chromosome (Chr) are strong candidates for the azoospermia factor AZF. They are frequently deleted in azoospermic or severely oligospermic males and are expressed exclusively in germ cells. In addition, the DAZ genes share a high degree of similarity with a Drosophila male infertility gene, boule. The predicted DAZ proteins contain an RNA recognition motif (RRM), and multiple copies of a repeat (the DAZ repeat) in tandem array. To understand the DAZ gene family and its expression, the DAZ genomic structure and RNA transcripts in numerous males, as well as several DAZ cDNA clones were analyzed. The results of genomic Southern blot showed that each male contains multiple DAZ genes with varying numbers of DAZ repeats, and that the copy number of the DAZ repeats are polymorphic in the population. The presence of multiple species of DAZ transcripts with different copy number and arrangement of the DAZ repeats in an individual suggests that more than one DAZ gene are transcribed. The existence of multiple functional DAZ genes complicates the analysis of genotype/phenotype correlations among males with varying sperm counts.
DOI:10.1007/s100380200078URLPMID:12376741 [本文引用: 1]

A recently developed microsatellite marker on the Y chromosome, Yfm1, which was originally cloned from a cosmid clone mapped near the DAZ (Deleted in AZoospermia) genes, was used to classify Y chromosomes using an automatic sequencer. Yfm1 could detect multicopies on Y chromosomes in a single polymerase chain reaction, showing four main classes, A, A*, B, and C, according to the number of copies and peak patterns. Compound haplotype analysis of the Y chromosome using the Yfm1 marker with three other biallelic markers on the Y chromosome, SRY, DXYS5Y, and YAP, resulted in nine different haplotypes among different populations, including Japanese. Haplotype II (defined by YAP insertion) observed in the Japanese population was consistently associated with Yfm1 class A or A*, which showed the lowest number of copies of Yfm1. Haplotypes III and IV were consistently associated with Yfm1 class B. On the other hand, haplotype I showed a variety of Yfm1 patterns that were dubbed class C when not appropriately classified as A, A*, or B. These relationships among Yfm1 microsatellite and Y-specific biallelic markers could supply useful population genetic information. Moreover, because we have already shown that men with haplotype II have significantly lower spermatogenic ability than those with other haplotypes, Yfm1 class A or A* with the least number of copies may be related to the haplotype II-specific structure of the Y chromosome, such as deletion of DAZ or DAZ repeats, reflecting the lower spermatogenic abilities of Japanese haplotype II men. Thus, Yfm1 represents a very useful marker for analysis of genetic structure in different populations and studies on Y chromosome lineage-specific genotype-phenotype correlations.
DOI:10.1371/journal.pgen.1001147URLPMID:20941357 [本文引用: 1]

Chromosome missegregation in germ cells is an important cause of unexplained infertility, miscarriages, and congenital birth defects in humans. However, the molecular defects that lead to production of aneuploid gametes are largely unknown. Cdc20, the activating subunit of the anaphase-promoting complex/cyclosome (APC/C), initiates sister-chromatid separation by ordering the destruction of two key anaphase inhibitors, cyclin B1 and securin, at the transition from metaphase to anaphase. The physiological significance and full repertoire of functions of mammalian Cdc20 are unclear at present, mainly because of the essential nature of this protein in cell cycle progression. To bypass this problem we generated hypomorphic mice that express low amounts of Cdc20. These mice are healthy and have a normal lifespan, but females produce either no or very few offspring, despite normal folliculogenesis and fertilization rates. When mated with wild-type males, hypomorphic females yield nearly normal numbers of fertilized eggs, but as these embryos develop, they become malformed and rarely reach the blastocyst stage. In exploring the underlying mechanism, we uncover that the vast majority of these embryos have abnormal chromosome numbers, primarily due to chromosome lagging and chromosome misalignment during meiosis I in the oocyte. Furthermore, cyclin B1, cyclin A2, and securin are inefficiently degraded in metaphase I; and anaphase I onset is markedly delayed. These results demonstrate that the physiologically effective threshold level of Cdc20 is high for female meiosis I and identify Cdc20 hypomorphism as a mechanism for chromosome missegregation and formation of aneuploid gametes.
DOI:10.1093/biolre/ioz062URLPMID:30985893 [本文引用: 1]

MicroRNAs (miRNAs) have recently been shown to be important for spermatogenesis; both DROSHA and Dicer1 KO mice exhibit infertility due to abnormal miRNA expression. However, the roles of individual miRNAs in spermatogenesis remain elusive. Here we demonstrated that miR-15b, a member of the miR-15/16 family, is primarily expressed in testis. A miR-15b transgenic mouse model was constructed to investigate the role of miR-15b in spermatogenesis. Impaired spermatogenesis was observed in miR-15b transgenic mice, suggesting that appropriate expression of miR-15b is vital for spermatogenesis. Furthermore, we demonstrated that overexpression of miR-15b reduced CDC25A gene post-transcriptional activity by targeting the 3'-UTR region of CDC25A, thus regulating spermatogenesis. In vitro results further demonstrated that a mutation in CFTR could affect the interaction between Ago2 with Dicer1 and that Dicer1 activity regulates miR-15b expression. We extended our study to azoospermia patients and found that infertile patients have a significantly higher level of miR-15b in semen and plasma samples. Taken together, we propose that CFTR regulation of miR-15b could be involved in the post-transcriptional regulation of CDC25A in mammalian testis and that miR-15b is important for spermatogenesis.
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/s0378-1119(02)00978-2URLPMID:12426106 [本文引用: 1]

Ornithine decarboxylase antizyme 1 and 2 (OAZ1 and OAZ2) are expressed ubiquitously, and control the intracellular concentration of polyamines. Their testicular isoform, OAZt/Oaz3, is specifically expressed in differentiated haploid germ cells. We have identified and characterized the gene encoding OAZt in mice. The mouse OAZt gene contains, as does the human ortholog and paralogs, five exons and four introns. Comparison of the mouse OAZt with the human ortholog gene revealed that exon sizes are identical and nucleotide sequences in exons are highly homologous (83% identity). The major transcriptional start site was determined by primer extension assay. Promoter activity was confirmed by transgenic mouse assays, using the upstream region of the mouse OAZt gene fused to a EGFP reporter gene. The OAZt essential promoter located between -133 and +242, has two CREs and an Inr, and lacks a TATA box. These elements are conserved in the human ortholog but not in the paralogs, indicating that such a short upstream region including two CREs and Inr is sufficient to drive endogenous OAZt mRNA expression in the haploid testicular germ cells.
DOI:10.1242/jcs.179218URLPMID:26826184 [本文引用: 1]

Meiotic progression requires exquisitely coordinated translation of maternal messenger (m)RNA that has accumulated during oocyte growth. A major regulator of this program is the cytoplasmic polyadenylation element binding protein 1 (CPEB1). However, the temporal pattern of translation at different meiotic stages indicates the function of additional RNA binding proteins (RBPs). Here, we report that deleted in azoospermia-like (DAZL) cooperates with CPEB1 to regulate maternal mRNA translation. Using a strategy that monitors ribosome loading onto endogenous mRNAs and a prototypic translation target, we show that ribosome loading is induced in a DAZL- and CPEB1-dependent manner, as the oocyte reenters meiosis. Depletion of the two RBPs from oocytes and mutagenesis of the 3' untranslated regions (UTRs) demonstrate that both RBPs interact with the Tex19.1 3' UTR and cooperate in translation activation of this mRNA. We observed a synergism between DAZL and cytoplasmic polyadenylation elements (CPEs) in the translation pattern of maternal mRNAs when using a genome-wide analysis. Mechanistically, the number of DAZL proteins loaded onto the mRNA and the characteristics of the CPE might define the degree of cooperation between the two RBPs in activating translation and meiotic progression.
DOI:10.1016/s1534-5807(01)00025-9URLPMID:11702780 [本文引用: 1]

CPEB is a sequence-specific RNA binding protein that regulates translation during vertebrate oocyte maturation. Adult female CPEB knockout mice contained vestigial ovaries that were devoid of oocytes; ovaries from mid-gestation embryos contained oocytes that were arrested at the pachytene stage. Male CPEB null mice also contained germ cells arrested at pachytene. The germ cells from the knockout mice harbored fragmented chromatin, suggesting a possible defect in homologous chromosome adhesion or synapsis. Two CPE-containing synaptonemal complex protein mRNAs, which interact with CPEB in vitro and in vivo, contained shortened poly(A) tails and mostly failed to sediment with polysomes in the null mice. Synaptonemal complexes were not detected in these animals. CPEB therefore controls germ cell differentiation by regulating the formation of the synaptonemal complex.
DOI:10.1371/journal.pone.0015541URLPMID:21124965 [本文引用: 1]

ING2 (inhibitor of growth family, member 2) is a member of the plant homeodomain (PHD)-containing ING family of putative tumor suppressors. As part of mSin3A-HDAC corepressor complexes, ING2 binds to tri-methylated lysine 4 of histone H3 (H3K4me3) to regulate chromatin modification and gene expression. ING2 also functionally interacts with the tumor suppressor protein p53 to regulate cellular senescence, apoptosis and DNA damage response in vitro, and is thus expected to modulate carcinogenesis and aging. Here we investigate the developmental and physiological functions of Ing2 through targeted germline disruption. Consistent with its abundant expression in mouse and human testes, male mice deficient for Ing2 showed abnormal spermatogenesis and were infertile. Numbers of mature sperm and sperm motility were significantly reduced in Ing2(-/-) mice ( approximately 2% of wild type, P<0.0001 and approximately 10% of wild type, P<0.0001, respectively). Their testes showed degeneration of seminiferous tubules, meiotic arrest before pachytene stage with incomplete meiotic recombination, induction of p53, and enhanced apoptosis. This phenotype was only partially abrogated by concomitant loss of p53 in the germline. The arrested spermatocytes in Ing2(-/-) testes were characterized by lack of specific HDAC1 accumulation and deregulated chromatin acetylation. The role of Ing2 in germ cell maturation may extend to human ING2 as well. Using publicly available gene expression datasets, low expression of ING2 was found in teratozoospermic sperm (>3-fold reduction) and in testes from patients with defective spermatogenesis (>7-fold reduction in Sertoli-cell only Syndrome). This study establishes ING2 as a novel regulator of spermatogenesis functioning through both p53- and chromatin-mediated mechanisms, suggests that an HDAC1/ING2/H3K4me3-regulated, stage-specific coordination of chromatin modifications is essential to normal spermatogenesis, and provides an animal model to study idiopathic and iatrogenic infertility in men. In addition, a bona fide tumor suppressive role of Ing2 is demonstrated by increased incidence of soft-tissue sarcomas in Ing2(-/-) mice.
URLPMID:24632385 [本文引用: 1]
[本文引用: 1]
DOI:10.4103/1008-682X.154310URLPMID:25994647 [本文引用: 1]

Importin proteins were originally characterized for their central role in protein transport through the nuclear pores, the only intracellular entry to the nucleus. This vital function must be tightly regulated to control access by transcription factors and other nuclear proteins to genomic DNA, to achieve appropriate modulation of cellular behaviors affecting cell fate. Importin-mediated nucleocytoplasmic transport relies on their specific recognition of cargoes, with each importin binding to distinct and overlapping protein subsets. Knowledge of importin function has expanded substantially in regard to three key developmental systems: embryonic stem cells, muscle cells and the germ line. In the decade since the potential for regulated nucleocytoplasmic transport to contribute to spermatogenesis was proposed, we and others have shown that the importins that ferry transcription factors into the nucleus perform additional roles, which control cell fate. This review presents key findings from studies of mammalian spermatogenesis that reveal potential new pathways by which male fertility and infertility arise. These studies of germline genesis illuminate new ways in which importin proteins govern cellular differentiation, including via directing proteins to distinct intracellular compartments and by determining cellular stress responses.
DOI:10.1093/nar/gks342URLPMID:22570411 [本文引用: 1]

Members of the BET (bromodomain and extra terminal motif) family of proteins have been shown to be chromatin-interacting regulators of transcription. We previously generated a mutation in the testis-specific mammalian BET gene Brdt (bromodomain, testis-specific) that yields protein lacking the first bromodomain (BRDT(DeltaBD1)) and observed disrupted spermiogenesis and male sterility. To determine whether BRDT(DeltaBD1) protein results in altered transcription, we analyzed the transcriptomes of control versus Brdt(DeltaBD1/DeltaBD1) round spermatids. Over 400 genes showed statistically significant differential expression, and among the up-regulated genes, there was an enrichment of RNA splicing genes. Over 60% of these splicing genes had transcripts that lacked truncation of their 3'-untranslated region (UTR) typical of round spermatids. We selected four of these genes to characterize: Srsf2, Ddx5, Hnrnpk and Tardbp. The 3'-UTRs of Srsf2, Ddx5 and Hnrnpk mRNAs were longer in mutant round spermatids and resulted in reduced protein levels. Tardbp was transcriptionally up-regulated and a splicing shift toward the longer variant was observed. All four splicing proteins were found to complex with BRDT in control and mutant testes. We thus suggest that, along with modulating transcription, BRDT modulates gene expression as part of the splicing machinery. These modulations alter 3'-UTR processing in round spermatids; importantly, the BD1 is essential for these functions.
DOI:10.1002/cbf.2970URL [本文引用: 1]

Reproduction is required for the survival of all mammalian animals. Spermatogenesis is an essential and complex developmental process that ultimately results in production of haploid spermatozoa. Recent studies demonstrated that Boule and stimulated by retinoic acid 8 (Stra8) played important roles in initiation meiosis in male germ cells. miR-34c is indispensable in the late steps of spermatogenesis; remarkably, the main function of miR-34c is to reduce cell proliferation potentiality and promote cellular apoptosis. The objectives of this study were to investigate the expression patterns of Boule, Stra8, P53 and miR-34c in dairy goat testis and their relationship in male germ line stem cells (mGSCs). The results first revealed the expression patterns of Boule, Stra8, P53 and miR-34c in 30dpp, 90dpp and adult testes of dairy goats. The expression levels of Boule, Stra8, P53 and miR-34c in adult dairy goat testes were significantly higher than that of 30dpp. Overexpression of Boule and Stra8 promoted the expression of miR-34c in dairy goat mGSCs. In our previous study, we showed that miR-34c was P53 dependent in mGSCs. These results have shown that the up-regulation of miR-34c was not due to P53 protein activation but which might be caused by the up-regulation of Boule and Stra8 promoting the advance of meiosis. In addition, we found retinoic acid would decrease the expression of P53 and miR-34c, however, did not change the expression of c-Myc greatly. It suggested that the function of driving differentiation of dairy goat mGSCs by retinoic acid might not be caused by P53. Copyright (c) 2013 John Wiley & Sons, Ltd.
[本文引用: 4]
[本文引用: 1]
DOI:10.1038/srep35299URLPMID:27731399 [本文引用: 1]

miRNAs and lncRNAs, which represent one of the most highly expressed classes of ncRNAs in development, are attracting increasing interest. A variety of regulators is considered to be implicated in sheep species with different fecundity. However, interactions between miRNAs and lncRNAs and changes in the expression of regulatory lncRNAs in sheep fecundity have not yet been reported. To characterize the important roles of miRNAs and lncRNAs and elucidate their regulating networks in sheep prolificacy, a genome-wide analysis of miRNAs and lncRNAs from Small Tail Han sheep of genotypes FecB(B)FecB(B) (Han BB) and FecB(+) FecB(+) (Han++) and from Dorset sheep (Dorset) was performed. An integrated analysis of miRNAs and lncRNAs was performed to study the regulatory function of miRNAs and lncRNAs in fecundity, revealing significantly correlated patterns of expression. Dramatic changes of miRNAs and lncRNAs suggest their critical roles in sheep fecundity. In conclusion, this is the first study performing thorough investigations of regulatory relationships among lncRNAs, miRNA and mRNAs, which will provide a novel view of the regulatory mechanisms involved in sheep fecundity. These results may provide further insight into sheep fecundity and help us to improve sheep prolificacy.
DOI:10.3390/ijms21082774URL [本文引用: 1]
DOI:10.3864/j.issn.0578-1752.2017.02.016URL [本文引用: 1]

精子发生始于精原干细胞(spermatogonial stem cells, SSCs),SSCs一部分自我更新,另一部分首先分裂形成Asingle(As)型精原细胞,进而形成Aparied(Apr)型精原细胞和Aaligned(Aal)型精原细胞;随后,Aal型精原细胞再发育为A1-A4型精原细胞、中间型精原细胞以及B型精原细胞;B型精原细胞有丝分裂可形成初级精母细胞,经历前细线期、细线期、偶线期、粗线期,再经减数分裂形成次级精母细胞;当圆形精子细胞形成之后,则经细胞核浓缩等过程形成晚期的细长型成熟精子,随之最终变形成为精子。这一复杂的生理过程需要相关基因的适时表达,并受到转录和转录后水平的调控。研究表明,多种类型的非编码RNA (ncRNAs)在精子发生过程中发挥着重要作用。ncRNAs包括微小RNAs (miRNAs)、与Piwi蛋白相互作用的RNAs (piRNAs)、长链非编码RNAs (lncRNAs)、环状RNAs(circRNAs)以及内源性小干扰RNAs (endo-siRNAs)等。这些ncRNAs的表达具有细胞组织特异性和发育阶段特异性,可从时间和空间上精确调控精子发生的整个过程。miRNAs是一类长约21—25 nt的内源性非编码单链RNA分子,广泛存在于各种生物中,其形成至少需要Drosha和Dicer等两种RNA酶的参与,可降解靶mRNA或抑制靶mRNA翻译,对SSCs干性的维持、自我更新和分化的调控以及生殖细胞减数分裂和精子发生过程具有重要的调控作用。此外,精子发生过程中,在生殖细胞不同阶段所表达的基因也可调控miRNAs的生成加工过程。piRNAs是2006年发现的一种新的小RNA,长度约24—32nt,其作用与Dicer酶无关,能够与生殖细胞特异性蛋白Piwi蛋白家族成员结合,进而行使生物学功能,其主要表现为:在表观遗传水平和转录后水平沉默转座子、反转座子等基因组移动遗传元件,维持生殖细胞自身基因组稳定性和完整性,调控生殖细胞增殖、减数分裂及精子发生过程。LncRNAs是一类长度大于200 nt的ncRNAs,其生成加工过程与mRNA类似,并且与mRNA有着相似的结构。不同来源的lncRNAs可通过转录前与转录后多种机制进而调控SSCs的干性及分化,并且调控生殖细胞凋亡。有些lncRNAs还可调控miRNAs的表达,进而调控精子发生过程。circRNAs是区别于传统线性RNA的一类新型RNA,在不同物种中具有保守性,在组织及不同发育阶段呈特异性表达。其生成加工方式与其序列相关,同一基因位点可通过选择性环化产生多种circRNAs进而发挥功能。研究表明,circRNAs可结合miRNAs从而调控生精过程。相对于其他ncRNAs,endo-siRNAs的生成加工方式更为简单,并有着与miRNAs相同的作用方式,在精子发生和雄性生殖中扮演着重要角色。文章结合最新的研究进展,综述了几种ncRNAs的生成及其在精子发生过程中的调控作用,旨在为精子发生过程中ncRNAs的进一步研究提供参考。
DOI:10.3864/j.issn.0578-1752.2017.02.016URL [本文引用: 1]

精子发生始于精原干细胞(spermatogonial stem cells, SSCs),SSCs一部分自我更新,另一部分首先分裂形成Asingle(As)型精原细胞,进而形成Aparied(Apr)型精原细胞和Aaligned(Aal)型精原细胞;随后,Aal型精原细胞再发育为A1-A4型精原细胞、中间型精原细胞以及B型精原细胞;B型精原细胞有丝分裂可形成初级精母细胞,经历前细线期、细线期、偶线期、粗线期,再经减数分裂形成次级精母细胞;当圆形精子细胞形成之后,则经细胞核浓缩等过程形成晚期的细长型成熟精子,随之最终变形成为精子。这一复杂的生理过程需要相关基因的适时表达,并受到转录和转录后水平的调控。研究表明,多种类型的非编码RNA (ncRNAs)在精子发生过程中发挥着重要作用。ncRNAs包括微小RNAs (miRNAs)、与Piwi蛋白相互作用的RNAs (piRNAs)、长链非编码RNAs (lncRNAs)、环状RNAs(circRNAs)以及内源性小干扰RNAs (endo-siRNAs)等。这些ncRNAs的表达具有细胞组织特异性和发育阶段特异性,可从时间和空间上精确调控精子发生的整个过程。miRNAs是一类长约21—25 nt的内源性非编码单链RNA分子,广泛存在于各种生物中,其形成至少需要Drosha和Dicer等两种RNA酶的参与,可降解靶mRNA或抑制靶mRNA翻译,对SSCs干性的维持、自我更新和分化的调控以及生殖细胞减数分裂和精子发生过程具有重要的调控作用。此外,精子发生过程中,在生殖细胞不同阶段所表达的基因也可调控miRNAs的生成加工过程。piRNAs是2006年发现的一种新的小RNA,长度约24—32nt,其作用与Dicer酶无关,能够与生殖细胞特异性蛋白Piwi蛋白家族成员结合,进而行使生物学功能,其主要表现为:在表观遗传水平和转录后水平沉默转座子、反转座子等基因组移动遗传元件,维持生殖细胞自身基因组稳定性和完整性,调控生殖细胞增殖、减数分裂及精子发生过程。LncRNAs是一类长度大于200 nt的ncRNAs,其生成加工过程与mRNA类似,并且与mRNA有着相似的结构。不同来源的lncRNAs可通过转录前与转录后多种机制进而调控SSCs的干性及分化,并且调控生殖细胞凋亡。有些lncRNAs还可调控miRNAs的表达,进而调控精子发生过程。circRNAs是区别于传统线性RNA的一类新型RNA,在不同物种中具有保守性,在组织及不同发育阶段呈特异性表达。其生成加工方式与其序列相关,同一基因位点可通过选择性环化产生多种circRNAs进而发挥功能。研究表明,circRNAs可结合miRNAs从而调控生精过程。相对于其他ncRNAs,endo-siRNAs的生成加工方式更为简单,并有着与miRNAs相同的作用方式,在精子发生和雄性生殖中扮演着重要角色。文章结合最新的研究进展,综述了几种ncRNAs的生成及其在精子发生过程中的调控作用,旨在为精子发生过程中ncRNAs的进一步研究提供参考。
DOI:10.1016/j.acthis.2020.151506URLPMID:32008790 [本文引用: 1]

Circular RNA (CircRNA), a type of endogenous non-coding RNAs (ncRNAs), is generally generated from precursor mRNA (pre-mRNA) by canonical splicing and head-to-tail back splicing. The structure without a polyA tail renders circRNA highly insensitive to ribonuclease. Simultaneously, the distribution of circRNAs is tissue and developmental stage-specific. There are five potential biological functions of circRNAs: 1) promote transcription of their parental genes; 2) function as a miRNA sponge; 3) RNA binding protein (RBP) sponge; 4) encode protein; 5) act as an mRNA trap. Recently, circRNA has attracted attention because studies have shown that circRNAs are associated with follicular development, ovarian senescence, spermatogenesis, and germ cell development process, suggesting that circRNAs may function in germ cells regulation. The investigation of circRNAs in germ cells will provide an excellent opportunity to understand its potential molecular basis, and potentially improving reproduction status in human. In this article, the relationship between circRNA and germ cell development will be discussed.
DOI:10.1371/journal.pone.0007513URLPMID:19838299 [本文引用: 1]

BACKGROUND: During zebrafish embryogenesis, microRNA (miRNA) miR-430 contributes to restrict Nanos1 and TDRD7 to primordial germ cells (PGCs) by inducing mRNA deadenylation, mRNA degradation, and translational repression of nanos1 and tdrd7 mRNAs in somatic cells. The nanos1 and tdrd7 3'UTRs include cis-acting elements that allow activity in PGCs even in the presence of miRNA-mediated repression. METHODOLOGY/PRINCIPAL FINDINGS: Using a GFP reporter mRNA that was fused with tdrd7 3'UTR, we show that a germline-specific RNA-binding protein DAZ-like (DAZL) can relieve the miR-430-mediated repression of tdrd7 mRNA by inducing poly(A) tail elongation (polyadenylation) in zebrafish. We also show that DAZL enhances protein synthesis via the 3'UTR of dazl mRNA, another germline mRNA targeted by miR-430. CONCLUSIONS/SIGNIFICANCE: Our present study indicated that DAZL acts as an
