 ,, 邹修平, 张靖芸, 张庆雯, 姚家欢, 何永睿, 李强, 陈善春
,, 邹修平, 张靖芸, 张庆雯, 姚家欢, 何永睿, 李强, 陈善春 ,西南大学/中国农业科学院柑桔研究所国家柑桔品种改良中心,重庆 400712
,西南大学/中国农业科学院柑桔研究所国家柑桔品种改良中心,重庆 400712Verification of SNPs Associated with Citrus Bacterial Canker Resistance and Induced Expression of SNP-Related Calcium-Dependent Protein Kinase Gene
PENG Yun, LEI TianGang ,, ZOU XiuPing, ZHANG JingYun, ZHANG QingWen, YAO JiaHuan, HE YongRui, LI Qiang, CHEN ShanChun
,, ZOU XiuPing, ZHANG JingYun, ZHANG QingWen, YAO JiaHuan, HE YongRui, LI Qiang, CHEN ShanChun ,National Center for Citrus Variety Improvement, Citrus Research Institute, Southwest University/Chinese Academy of Agricultural Sciences, Chongqing 400712
,National Center for Citrus Variety Improvement, Citrus Research Institute, Southwest University/Chinese Academy of Agricultural Sciences, Chongqing 400712通讯作者:
责任编辑: 岳梅
收稿日期:2019-12-9接受日期:2020-01-28网络出版日期:2020-05-16
| 基金资助: |
Received:2019-12-9Accepted:2020-01-28Online:2020-05-16
作者简介 About authors
彭蕴,E-mail:pengyun1995@icloud.com。

摘要
关键词:
Abstract
Keywords:
PDF (1163KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
彭蕴, 雷天刚, 邹修平, 张靖芸, 张庆雯, 姚家欢, 何永睿, 李强, 陈善春. 柑橘溃疡病抗性SNP验证及其相关钙依赖性蛋白激酶基因诱导表达[J]. 中国农业科学, 2020, 53(9): 1820-1829 doi:10.3864/j.issn.0578-1752.2020.09.010
PENG Yun, LEI TianGang, ZOU XiuPing, ZHANG JingYun, ZHANG QingWen, YAO JiaHuan, HE YongRui, LI Qiang, CHEN ShanChun.
0 引言
【研究意义】柑橘溃疡病(citrus bacterial canker,CBC)是世界柑橘产业的一种极具威胁性的检疫性病害,现已在30多个国家和地区发生[1]。目前,柑橘溃疡病没有特别有效的防治药剂,生产上主要采用喷施铜制剂、铲除病树等措施来防止病害的严重发生。铜制剂和农业抗生素的大量施用不仅增加生产成本,而且还会对环境和人造成一定影响[2]。开发与柑橘溃疡病耐/感性相关联的单核苷酸多态性(single nucleotide polymorphism,SNP)标记[3],分子标记辅助选择(marker-assisted selection,MAS),将大幅度提高柑橘溃疡病抗性育种的效率[4]。【前人研究进展】大量研究表明,培育抗性品种是防治病虫害最经济有效的方法之一[5],而杂交育种是柑橘新品种选育的主要途径之一[6]。生产实践证明,利用耐病品种作亲本,选育的新品种对溃疡病多具有较强的耐性[7,8]。传统的溃疡病抗性鉴定方法必须采用大量的离体叶片或田间活体接种溃疡病菌(Xanthomonas citri subsp. citri,Xcc)进行评价,这种方法不仅费力费时,且鉴定结果还易受环境温度和湿度等因素的影响[9]。目前,分子标记广泛用于优良性状基因型选择以改良作物品种[10,11,12]。KIM等[13]通过全基因组序列分析发现,番茄青枯病感病品种和抗病品种之间存在多态性SNP,并可利用高分辨率熔解曲线(high resolution melting,HRM)技术有效鉴别抗性基因型,实现抗青枯病番茄品种的辅助选择;在苹果中利用分子标记定位到苹果白粉病抗性基因Pl-1[14]以及苹果黑星病抗性基因Vb[15];刘升锐[16]构建了柑橘高密度遗传连锁图谱,利用分子标记分析了控制枳落叶性状的基因位点;BARBA等[17]通过分子标记鉴定出葡萄白粉病抗性和敏感性位点,确定了葡萄与白粉病抗性相关的SNP;据报道,柑橘中也开发出与柑橘杂色萎黄病[18]、柑橘麻风病病毒[19]等抗病性相关的分子标记。【本研究切入点】目前,柑橘溃疡病抗性相关SNP的研究尚未见报道,笔者研究室前期基于转录组数据挖掘SNP位点,并以柑橘自然群体为材料,采用关联分析法,筛选到14个与柑橘溃疡病抗性关联的SNP。本研究将进一步利用耐/感柑橘品种的杂交F1群体,采用离体叶片针刺接种法鉴定溃疡病抗性表型,利用HRM技术进行SNP分型,采用相关性分析,验证这14个柑橘溃疡病抗性关联SNP的可靠性。【拟解决的关键问题】筛选与溃疡病耐/感性相关的SNP,以期获得可用于柑橘溃疡病抗性分子标记辅助选择的SNP,并分析SNP相关基因柑橘钙依赖性蛋白激酶基因(CsCDPK)受溃疡病菌和激素诱导处理后的表达情况,为基因功能研究打下基础。1 材料与方法
试验于2018年4月至2019年6月在西南大学柑桔研究所国家柑桔品种改良中心完成。1.1 材料
以对溃疡病高抗的品种‘金弹’、中抗品种‘新生系3号椪柑’和敏感品种‘冰糖橙’,以及耐/感品种杂交F1代群体共143个材料为试材,其中‘沙田柚’♀ב梁平柚’♂杂交后代27个、‘沃柑’♀ב米柑’♂后代 9个、‘茂谷柑’♀ב椪柑’♂后代17个、‘清见’♀ב冰糖橙’♂后代21个、‘沃柑’♀ב南丰蜜橘’♂后代15个、‘清见’♀ב南丰蜜橘’♂后代 18个、‘清见’♀ב长叶香橙’♂后代33个。F1群体经SSR分子标记鉴定均为真实杂种。叶片均于2018年采自中国农业科学院柑桔研究所国家柑桔品种改良中心育种圃。1.2 方法
1.2.1 溃疡病抗性评价 采用离体叶片针刺接种法进行溃疡病抗性鉴定,溃疡病菌为亚洲种A株系(由柑桔研究所胡军华老师提供),该病原菌接种‘晚锦橙’发病率高达91%,病情指数为73.3,病斑面积为4.5 mm2,单个病斑病原菌含量为2.99×106 cfu/mm2。接种方法参照文献[20],选取成熟度一致且完全展开的春梢叶片,经75%的乙醇擦拭消毒后用灭菌水清洗干净,无菌脱脂棉擦干叶片,用接种针(直径为0.5 mm)在每片叶片的背面扎取4—6组小孔,每组6个;移液枪吸取1 μL浓度为105 cfu/mL柑橘溃疡病菌悬液接种到伤口,同时对照组接种无菌水。运用软件Image J V1.47T统计病斑面积(lesion area,LA),按照病斑面积大小将病情分为6个等级,0级(LA<0.3 mm2),1级(0.3 mm2<LA≤0.6 mm2),3级(0.6 mm2<LA≤0.9 mm2),5级(0.9 mm2<LA≤1.2 mm2),7级(1.2 mm2<LA≤1.5 mm2),9级(LA>1.5 mm2),根据公式计算病情指数(disease index,DI),DI=100×Σ[(各级病斑数×相应级数值)/(病斑总数×最大级数)]。根据病情指数进行抗病性分级,病情指数≤20为免疫,20—40之间为高抗,40—60之间为中抗,60—80之间为中感,80—100为高感。1.2.2 SNP分型 采用EASY spin植物DNA快速提取试剂盒(购自北京艾德莱公司)提取叶片DNA。14个SNP位点基因分型的扩增引物(表1)由英维捷基(上海)贸易有限公司合成。PCR反应采用20 μL体系:13.4 μL H2O,0.3 μL 10 mmol·L-1上、下游引物,2 μL 10×buffer,1.6 μL 50 ng DNA,10 mmol·L-1 dNTP,0.8 U DNA Taq酶。反应条件:94℃预变性3 min;94℃ 30 s,58℃ 30 s,72℃ 1 min,共30个循环;72℃延伸5 min。在高分辨率熔解曲线(HRM)仪LightScanner 96(Idaho Technology Inc.)上进行样品SNP分型。
Table 1
表1
表1SNP位点及高分辨率熔解曲线分型PCR引物
Table 1
| SNP编号 SNP number | SNP位点 SNP loci | 正向引物序列 Forward primer sequence (5′-3′) | 反向引物序列 Reverse primer sequence (5′-3′) |
|---|---|---|---|
| HP02 | GGTTTTT GGTGTTT | AGAGCTGCCTTAGTGGTGTC | CAGTCTCTACTGCCCCGTTC |
| HP17 | ATATCCC ATAGCCC | ACCAGTTACTCTCCGTCCGA | TTAGGGTTTGGCGAGGACG |
| HP18 | GATCGAA GATTGAA | CAGAACATTATCCGGCATGGG | CCTGAATGAGAGGTGCTGATTT |
| HP31 | GCCATAT GCCGTAT | TGGCAGAATGTTGGGTAGGG | CCACTCAAGAGCCGCCTTAT |
| HP40 | ACGCTCA ACGTTCA | ATCCTCCTTCTCCCGTCACC | TGGGTTTGGTCATGTCCTTCC |
| HP42 | ATTCTCC ATTTTCC | ATCAAGTAACACACTCCCTTTACG | TGAGACTTTATATTTGTGATTGCGA |
| HP54 | TGCCCTT TGCTCTT | GGGTTCCCCGTTTCAGATGT | CCACCTCCGATGGAAAGAAGT |
| HP75 | TTACTCC TTATTCC | TCCTTCTGCCCGGATGATGA | TCCTTCTGCCCGGATGATGA |
| HP85 | TTGCCC TTGTCC | CACAAGTCACAAGCCGCAAT | AAGTACGATTGCCCGAGGTC |
| HP87 | ACAGTAG ACATTAG | ATCAATGGGGTCACCAACCC | ATTGGTGGTGGCGTTAGTCC |
| HP108 | CATACCAA CATATCAA | TGAAAGTGAACTTCAACGATGC | TCGTTGAGGAGAGGTCCAAGA |
| HP127 | CTCTTTCC CTCTCTCC | TCGGCTTTTGTCGCGTTTAC | TAGCTGACACGACACCGTTT |
| HP128 | ATCCCTTT ATCACTT | TGGATCCCACCATGGAATTGT | AGCGATGAGTTTAAGGAGAAGGG |
| HP170 | AACTCGA AACCCGA | ATGGATCCTTTTGAGAGAAGAAAACTGT | TGTCATTCTTGCCTTTTCCTTTCT |
新窗口打开|下载CSV
1.2.3 CsCDPK生物信息学分析 从甜橙基因组数据库(http://citrus.hzau.edu.cn/orange/)下载CsCDPK的核苷酸序列。利用Vector NTI软件寻找开放阅读框(ORF),翻译出氨基酸序列;利用在线软件预测氨基酸序列的分子量、理论等电点(https://web.expasy.org/ protparam/)[21]、基因内含子、外显子数及ORF范围(http://gsds.cbi.pku.edu.cn/)、基因的功能结构(http:// pfam.xfam.org/)、跨膜结构域(http://www.cbs.dtu.dk/ services/TMHMM/)。
1.2.4 CsCDPK诱导表达分析 以柑橘溃疡病敏感品种‘冰糖橙’、中抗品种‘新生系3号椪柑’和高抗品种‘金弹’为试材,每个品种选3株树,分别从每株的树冠外围选取24片完全展开且成熟度一致的春梢叶片混合为1个样品,设置3个生物学重复。叶片清洗干净后用75%酒精进行表面消毒,无菌水清洗3次,置于无菌培养皿中;用注射器将105 cfu/mL的溃疡病菌悬液注射入叶片下表皮,每个样品均处理18片叶;用浸有10 μmol·L-1水杨酸(SA)溶液、100 μmol·L-1茉莉酸甲酯(MeJA)溶液和100 μmol·L-1脱落酸(ABA)溶液的无菌脱脂棉覆盖不同品种叶片的叶柄部位。分别在处理后0、6、12、24、48、72 h从4种处理的样品中随机选取3片叶用刀片将处理部位切取下来混合,以无菌水处理的各品种为对照。用EASY spin 植物RNA快速提取试剂盒(购自北京艾德莱公司)提取总RNA[22], Prime ScriptTM RT reagent Kit 反转录试剂盒(TaKaRa公司)将RNA反转录成cDNA。特异性扩增引物(表2)由英维捷基(上海)贸易有限公司合成。以actin为内参基因,采用实时荧光定量PCR(qRT-PCR)技术分析CsCDPK的相对表达量。采用20 μL反应体系:10 μL 2×SYBRG PCR Master Mix,5 μL cDNA,各0.5 μL 10 mmol·L-1引物,4 μL H2O。扩增反应在7500 Fast Real-Time PCR System(Applied Biosystem)上进行,反应程序:95℃ 10 min;95℃ 15 s;60℃ 1 min;共40个循环。
Table 2
表2
表2实时荧光定量PCR引物
Table 2
| 基因Gene | 上游引物序列Upstream primer sequence (5′-3′) | 下游引物序列Downstream primer sequence (5′-3′) |
|---|---|---|
| CsCDPK | CTACCCGGTTTGCCTTGCTA | CAGCTCTGTTGCCTGACTGA |
| actin | CATCCCTCAGCACCTTCC | CCAACCTTAGCACTTCTCC |
新窗口打开|下载CSV
1.2.5 数据分析 根据SNP基因型进行赋值,A/A和T/T赋值为-1,G/G和C/C赋值为1,杂合型均赋值为0。运用DPS软件分析供试材料溃疡病抗性表型(病斑面积和病情指数)和SNP基因型的相关性;运用Excel软件采用ΔΔCT法计算基因相对表达量。
2 结果
2.1 F1群体溃疡病抗性鉴定
采用针刺接种法对143个供试材料进行溃疡病抗性离体鉴定,接菌10 d后拍照统计计算病斑面积,以敏感品种‘冰糖橙’和高抗品种‘金弹’作对照,根据病斑面积大小分级,计算材料的病情指数。结果显示,杂交F1代群体病斑面积在0.75—3.29 mm2,而‘金弹’病斑面积为0.5 mm2,‘冰糖橙’病斑面积为3.69 mm2;杂交F1代群体的病情指数在30.2—100,对照‘金弹’的病情指数为11.1,‘冰糖橙’为100。发病程度较低的杂交后代,其亲本至少有1个为耐病性品种,母本耐病性强的居多。但母本对溃疡病敏感而父本耐病性强的杂交组合其F1代表现耐病性强的也较多,如‘沙田柚’♀ב梁平柚’♂、‘沃柑’♀ב南丰蜜橘’♂等杂交组合,其F1代中抗性表型为中抗以上的占近1/3。根据病情指数分级,143个供试材料中对溃疡病抗性表现为免疫的1个(‘金弹’),高抗17个,中抗31个,中感38个,高感的56个。2.2 SNP分型及相关性分析
采用HRM技术对143个供试材料进行SNP分型。结果显示,验证的14个SNP位点在供试材料间均有多态性,可分为3种不同的基因型,2种纯合型和1种杂合型。如图1为1个SNP位点(HP31)的HRM分型结果,143个供试材料被准确地区分为A/A、G/G和A/G 3种基因型。图1
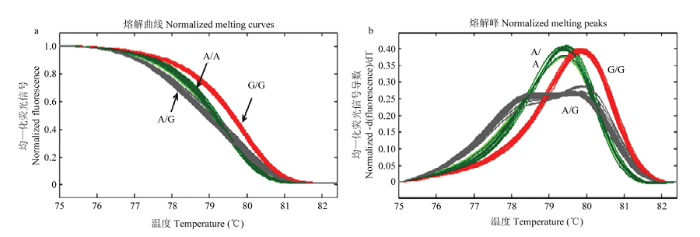 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1供试材料(SNP HP31)分型结果
Fig. 1Melting curve and genotyping of locus 31
根据SNP分型结果对143个供试材料SNP位点的基因型进行赋值,运用DPS软件对供试材料的基因型值和病斑面积及病情指数进行简单相关和典型相关分析。简单相关分析结果表明,多个SNP位点基因型与病斑面积及病情指数相关性显著。而典型相关显著性检验结果显示,供试材料SNP基因型与病斑面积相关系数为0.4849,P=0.0016<0.05,相关性显著(表3);SNP基因型与病情指数相关系数0.3796,P=0.088>0.05,两者不相关。典型相关分析结果显示,与病斑面积相关系数绝对值>0.2的SNP位点5个,分别是HP31、HP42、HP85、HP87、HP170(表4),表明这5个SNP与柑橘溃疡病病斑面积相关性较高,可作为柑橘溃疡病耐性材料筛选的候选SNP标记。其中HP31为CsCDPK编码区靠近5′ UTR端的SNP。
Table 3
表3
表3典型相关显著性检验结果
Table 3
| 相关系数 Correlation coefficient | Wilks统计量 Wilks Lambda | 卡方值 Chi-Square | 自由度 Degree of freedom (df) | P值 P value | |
|---|---|---|---|---|---|
| 病斑面积Lesion area | 0.4849 | 0.6547 | 55.2766 | 28 | 0.0016 |
| 病情指数Disease index | 0.3796 | 0.8559 | 20.2999 | 13 | 0.088 |
新窗口打开|下载CSV
Table 4
表4
表4典型相关系数矩阵
Table 4
| HP 02 | HP 17 | HP 18 | HP 31 | HP 40 | HP 42 | HP 54 | HP 75 | HP 85 | HP 87 | HP 108 | HP 127 | HP 128 | HP 170 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 病斑面积 Lesion area | 0.13 | 0.12 | -0.01 | -0.30 | -0.16 | -0.30 | 0.01 | 0.07 | 0.24 | -0.28 | -0.14 | -0.04 | 0.11 | 0.20 |
| 病情指数 Disease index | 0.09 | 0.05 | 0.04 | -0.23 | -0.12 | -0.23 | 0.06 | 0.01 | 0.27 | -0.24 | -0.15 | -0.02 | 0.01 | 0.24 |
新窗口打开|下载CSV
2.3 CsCDPK生物信息学分析
CsCDPK位于基因组第4号染色体上,编号为Cs4g10370,全长5 162 bp(图2-A)。结构分析发现该基因由13个外显子组成(图2-B)。特定结构域分析发现蛋白激酶结构域(PKinase)位于1—236个氨基酸的位置;此外还含有4个螺旋-环-螺旋结构域(EF hand)(图2-C)。对CsCDPK进行跨膜区分析,发现在160—180个氨基酸的位置存在20个氨基酸的跨膜区,1—159是在胞内区,181—615为胞外区(图2-D、2-E)。图2
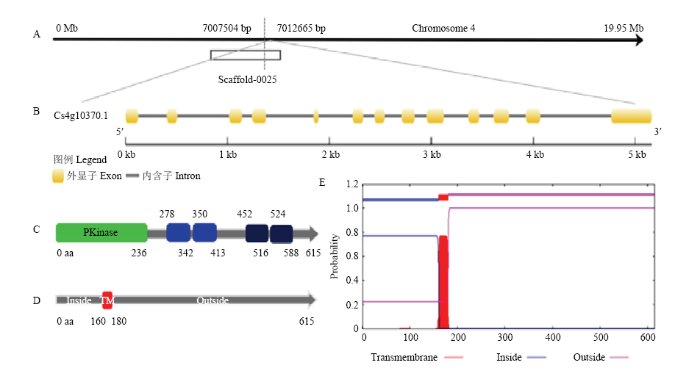 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2CsCDPK 生物信息学分析
A:染色体定位Chromosome localization;B:基因结构Gene structure;C:功能结构域Functional domain;D:跨膜结构Transmembrane structure;E:跨膜区预测Transmembrane domain prediction
Fig. 2Bioinformatics analysis of CsCDPK
2.4 CsCDPK诱导表达分析
CsCDPK受溃疡病菌诱导表达,接种后6、12、24、48和72 h,3个品种中基因的相对表达量基本呈逐渐升高再降低的趋势,且均在处理后48 h相对表达量达到最高。接种后6 h,‘金弹’中CsCDPK相对表达量为0.7,而接种后48 h其相对表达量升至7.0;‘冰糖橙’和‘新生系3号椪柑’接种后48 h,基因相对表达量分别升至4.0和1.8,但与接种后6 h相比,表达量升高的幅度不明显;接种后12 h,在高抗品种‘金弹’中CsCDPK相对表达量为对照的3倍,而此时‘新生系3号椪柑’和‘冰糖橙’中该基因相对表达量与对照无明显差异。说明高抗品种‘金弹’可更早地响应溃疡病菌胁迫,并且‘金弹’中响应胁迫的分子信号可迅速地调控CsCDPK大量表达(图3-A)。图3
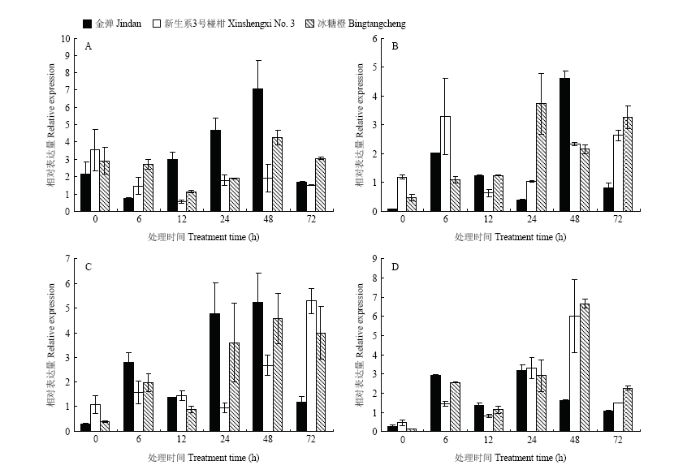 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3溃疡病菌和激素诱导处理3个柑橘品种后不同时间CsCDPK相对表达量
A:溃疡病菌诱导处理Xcc induction treatment;B:茉莉酸甲酯诱导处理MeJA induction treatment;C:水杨酸诱导处理SA induction treatment;D:脱落酸诱导处理ABA induction treatment
Fig. 3CsCDPK relative expression in 3 citrus varieties induced by Xcc and exogenous hormones
利用MeJA诱导处理‘金弹’‘新生系3号椪柑’‘冰糖橙’后,CsCDPK相对表达量随时间变化趋势与溃疡病菌处理类似。在处理后6 h CsCDPK相对表达量相对较高,但处理后12 h与对照无明显差异;诱导处理后12—48 h,其相对表达量则呈逐渐升高的趋势(图3-B)。SA诱导处理后,各品种CsCDPK相对表达量变化均呈波动趋势,‘金弹’在处理后24 h和48 h的相对表达量均在5倍左右,但72 h后表达量与对照无明显差异;‘新生系3号椪柑’和‘冰糖橙’CsCDPK相对表达量分别在72 h和48 h达到最高(图3-C)。ABA诱导处理后,‘金弹’中CsCDPK相对表达量变化趋势与溃疡病菌处理的变化趋势大致相反,处理后6、24 h CsCDPK的表达量相对较高,而处理后48 h其表达量较低;‘新生系3号椪柑’和 ‘冰糖橙’在处理后48 h CsCDPK相对表达量达到最高(图3-D)。
3 讨论
柑橘溃疡病被国内外列为检疫性病害,给世界柑橘产业带来了巨大的经济损失。栽培耐病品种在一定程度上可减轻溃疡病对柑橘生产的影响,目前生产上已有一些对溃疡病耐性较强的品种资源,如‘金柑’‘椪柑’‘南丰蜜橘’及其杂种后代。利用抗性资源进行品种改良是柑橘育种的一个重要方向。近年来,有研究者通过基因编辑技术修饰特定基因位点提高甜橙对溃疡病的抗性[23],其实质是改变敏感品种中特定基因的遗传多态性。因此,利用含有抗性基因(纯合型或杂合型)的品种资源作亲本,采用常规杂交育种方法,经过遗传重组后也可能得到抗性提高的材料。但传统的柑橘育种及溃疡病抗性鉴定方法筛选效率不高,费时费力,若采用杂交育种法与抗性分子标记辅助选择相结合,将大大提筛选效率,加快育种进程。目前,在果树中研究性状关联SNP的报道不少,例如利用桃果肉软化性状相关基因SNP鉴别桃果肉软化程度不同品种[24],木瓜果皮红色和黄色相关SNP[25],利用SNP区分杏甜味和苦味基因型[26]等。本研究在前期通过关联分析发掘的14个与柑橘溃疡病耐/感性关联SNP基础上,利用耐/感品种杂交后代进一步验证这14个SNP与柑橘溃疡病耐/感性的相关性。结果发现,其中至少5个SNP与柑橘的溃疡病病斑面积存在显著相关性,这些SNP的获得为今后柑橘溃疡病抗性材料的筛选鉴定打下了基础。5个与柑橘溃疡病耐性存在显著相关性的SNP,其中1个为CsCDPK编码区的单核苷酸变异。CDPK是一种重要的调节蛋白[27],1987年HARMON 等在豌豆中首次发现CDPK[28], 经过几十年不断研究,发现CDPK广泛存在于植物、绿藻及顶复门原虫等生物中,但不存在于哺乳动物中[29]。CDPK通过识别胞内Ca2+信号,在植物茎和根发育、花粉管生长、气孔开闭、激素信号传导及响应逆境胁迫过程中发挥关键作用[30]。在拟南芥中AtCDPK1(CPK10)和AtCDPK2(CPK11)响应干旱和高盐度诱导[31]。水稻OsCPK12突变体可导致叶片的过早衰老,OsCPK12的过量表达导致其生育期延迟[32]。CDPK在植物激素信号转导中也具有重要作用,CDPK12在拟南芥中普遍表达并定位于细胞质和细胞核[33],CPK12 RNAi株系在种子萌发过程和萌发后的生长中表现出对ABA的极度敏感性[34]。BOTELLA等[35]发现生长素处理能够使绿豆插条CDPK上调表达;BREVIARIO等[36]在ABA信号研究中发现,ABA处理能够抑制水稻胚芽鞘的伸长,同时也抑制了水稻CDPK cDNA(OsCDPK11)的mRNA表达。植物能够成功抵御微生物的侵染,主要是依靠早期对病原体的识别,从而诱导多种信号传导途径以启动多元防御反应。在基因间反应中抑制病原体则需要植物抗性基因,使得携带相应无毒基因的病原体产生抗体,ROMEIS等[37]在转基因烟草植株研究中发现CDPK参与Cf-9(番茄的抗真菌病原体Cladosporium fulvum基因)和AVR-9(Cf-9对应的无毒性基因)介导植物防御反应信号的转导,AVR-9可引起NtCDPK2磷酸化从而使其活化。近年的研究表明CDPK在从枝菌根、根瘤菌-宿主植物共生体系中起重要的调控作用,如OsCDPK18上调表达参与水稻菌根早期共生[38]。大豆中CDPK通过磷酸化调控根瘤特异蛋白Nodulin26,从而参与调控根瘤的发生[39]。本研究对柑橘中与溃疡病抗性相关SNP的CsCDPK进行生物信息学分析,发现CsCDPK为一个跨膜蛋白,预测其功能可能与信号转导有关。利用溃疡病菌和3种激素对3个耐病性不同品种进行诱导处理,分析该基因的表达量变化,结果发现CsCDPK受溃疡病菌和SA、MeJA和ABA诱导表达,但在不同品种中表达模式不同。SA、MeJA和ABA在植物对逆境胁迫的应答过程中起到至关重要的作用,是植物应答逆境胁迫的重要调控因子。因此,笔者推测CsCDPK可能参与柑橘对溃疡病及其他非生物逆境胁迫信号转导过程的调控。
4 结论
验证的14个SNP位点中,5个SNP与柑橘的溃疡病病斑面积显著相关,可作为柑橘溃疡病耐性筛选的SNP标记。柑橘钙依赖性蛋白激酶基因(CsCDPK)受溃疡病菌和SA、MeJA及ABA诱导表达,该基因可能在柑橘应答溃疡病等逆境胁迫过程中发挥作用。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
//
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/nmeth0508-447URL [本文引用: 1]
DOI:10.1016/j.scienta.2010.03.023URL [本文引用: 1]
DOI:10.1128/AEM.03043-10URLPMID:21515725 [本文引用: 1]

Copper sprays have been widely used for control of endemic citrus canker caused by Xanthomonas citri subsp. citri in citrus-growing areas for more than 2 decades. Xanthomonas alfalfae subsp. citrumelonis populations were also exposed to frequent sprays of copper for several years as a protective measure against citrus bacterial spot (CBS) in Florida citrus nurseries. Long-term use of these bactericides has led to the development of copper-resistant (Cu(r)) strains in both X. citri subsp. citri and X. alfalfae subsp. citrumelonis, resulting in a reduction of disease control. The objectives of this study were to characterize for the first time the genetics of copper resistance in X. citri subsp. citri and X. alfalfae subsp. citrumelonis and to compare these organisms to other Cur bacteria. Copper resistance determinants from X. citri subsp. citri strain A44(pXccCu2) from Argentina and X. alfalfae subsp. citrumelonis strain 1381(pXacCu2) from Florida were cloned and sequenced. Open reading frames (ORFs) related to the genes copL, copA, copB, copM, copG, copC, copD, and copF were identified in X. citri subsp. citri A44. The same ORFs, except copC and copD, were also present in X. alfalfae subsp. citrumelonis 1381. Transposon mutagenesis of the cloned copper resistance determinants in pXccCu2 revealed that copper resistance in X. citri subsp. citri strain A44 is mostly due to copL, copA, and copB, which are the genes in the cloned cluster with the highest nucleotide homology (>= 92%) among different Cur bacteria.
[本文引用: 1]
[本文引用: 1]
DOI:10.1139/g97-792URLPMID:18464859 [本文引用: 1]

Twelve new dominant randomly amplified polymorphic DNA (RAPD) fragments associated with a single dominant gene for resistance to citrus tristeza virus (CTV) were identified using bulked segregant analysis of an intergeneric backcross family. These and eight previously reported RAPDs were mapped in the resistance gene (Ctv) region; the resulting localized linkage map spans about 32 cM, with nine close flanking markers within 2.5 cM of Ctv. Seven of 20 RAPD fragments linked with the resistance gene were cloned and sequenced, and their sequences were used to design longer primers to develop sequence characterized amplified region (SCAR) markers that can be utilized reliably in marker-assisted selection, high-resolution mapping, and map-based cloning of the resistance gene. All seven cloned RAPDs were converted successfully into SCARs by redesigning primers, optimizing PCR parameters (especially the annealing temperature), or digesting amplification products with restriction enzymes. Four of the seven remained dominant markers, displaying presence-absence polymorphism patterns; the other three detected restriction site changes or length variations and thus were transformed into codominant markers. Two genomic regions rich in variability were also detected by two codominant SCAR markers.
DOI:10.1007/s001220051382URLPMID:12582928 [本文引用: 1]

Eleven RAPD markers linked to a gene region conferring resistance to citrus nematodes in an intergen-eric backcross family were identified. Two sequence- characterized amplified region markers linked to a citrus tristeza virus resistance gene and one selected resistance gene candidate marker were evaluated for their association with citrus nematode resistance. A nematode-susceptible citrus hybrid, LB6-2 [Clementine mandarin (Citrus reticulata)×Hamlin orange (C. sinensis)], was crossed with the citrus nematode-resistant hybrid Swingle citrumelo (C. paradisi×Poncirus trifoliata) to produce 62 hybrids that were reproduced by rooted cuttings. The plants were grown in a greenhouse and inoculated with nematodes isolated from infected field trees. The hybrids segregated widely for this trait in a continuous distribution, suggesting possible polygenic control of the resistance. Bulked segregant analysis was used to identify markers associated with resistance by bulking DNA samples from individuals at the phenotypic distribution extremes. Linkage relationships were established by the inheritance of the markers in the entire population. A single major gene region that contributes to nematode resistance was identified. The resistance was inherited in this backcross family from the grandparent Poncirus trifoliata as a single dominant gene. QTL analysis revealed that 53.6% of the phenotypic variance was explained by this major gene region. The existence of other resistance-associated loci was suggested by the continuous phenotypic distribution and the fact that some moderately susceptible hybrids possessed the resistance-linked markers. The markers may be useful in citrus rootstock breeding programs if it can be demonstrated that they are valid in other genetic backgrounds.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S1360-1385(98)01240-0URL [本文引用: 1]
DOI:10.1016/j.aquaculture.2004.05.027URL [本文引用: 1]
DOI:10.1007/s00122-018-3054-1URLPMID:29352323 [本文引用: 1]

Genotyping of disease resistance to bacterial wilt in tomato by a genome-wide SNP analysis Bacterial wilt caused by Ralstonia pseudosolanacearum is one of the destructive diseases in tomato. The previous studies have identified Bwr-6 (chromosome 6) and Bwr-12 (chromosome 12) loci as the major quantitative trait loci (QTLs) contributing to resistance against bacterial wilt in tomato cultivar 'Hawaii7996'. However, the genetic identities of two QTLs have not been uncovered yet. In this study, using whole-genome resequencing, we analyzed genome-wide single-nucleotide polymorphisms (SNPs) that can distinguish a resistant group, including seven tomato varieties resistant to bacterial wilt, from a susceptible group, including two susceptible to the same disease. In total, 5259 non-synonymous SNPs were found between the two groups. Among them, only 265 SNPs were located in the coding DNA sequences, and the majority of these SNPs were located on chromosomes 6 and 12. The genes that both carry SNP(s) and are near Bwr-6 and Bwr-12 were selected. In particular, four genes in chromosome 12 encode putative leucine-rich repeat (LRR) receptor-like proteins. SNPs within these four genes were used to develop SNP markers, and each SNP marker was validated by a high-resolution melting method. Consequently, one SNP marker, including a functional SNP in a gene, Solyc12g009690.1, could efficiently distinguish tomato varieties resistant to bacterial wilt from susceptible varieties. These results indicate that Solyc12g009690.1, the gene encoding a putative LRR receptor-like protein, might be tightly linked to Bwr-12, and the SNP marker developed in this study will be useful for selection of tomato cultivars resistant to bacterial wilt.
DOI:10.1111/pbr.2007.126.issue-5URL [本文引用: 1]
DOI:10.1139/g06-096URLPMID:17213905 [本文引用: 1]

Apple scab, caused by the fungus Venturia inaequalis, is the major production constraint in temperate zones with humid springs. Normally, its control relies on frequent and regular fungicide applications. Because this control strategy has come under increasing criticism, major efforts are being directed toward the breeding of scab-resistant apple cultivars. Modern apple breeding programs include the use of molecular markers, making it possible to combine several different scab-resistance genes in 1 apple cultivar (pyramiding) and to speed up the breeding process. The apple scab-resistance gene Vb is derived from the Siberian crab apple 'Hansen's baccata #2', and is 1 of the 6 &quot;historical&quot; major apple scab-resistance genes (Vf, Va, Vr, Vbj, Vm, and Vb). Molecular markers have been published for all these genes, except Vr. In testcross experiments conducted in the 1960s, it was reported that Vb segregated independently from 3 other major resistance genes, including Vf. Recently, however, Vb and Vf have both been mapped on linkage group 1, a result that contrasts with the findings from former testcross experiments. In this study, simple sequence repeat (SSR) markers were used to identify the precise position of Vb in a cross of 'Golden Delicious' (vbvb) and 'Hansen's baccata #2' (Vbvb). A genome scanning approach, a fast method already used to map apple scab-resistance genes Vr2 and Vm, was used, and the Vb locus was identified on linkage group 12, between the SSR markers Hi02d05 and Hi07f01. This finding confirms the independent segregation of Vb from Vf. With the identification of SSR markers linked to Vb, another major apple scab-resistance gene has become available; breeders can use it to develop durable resistant cultivars with several different resistance genes.
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1007/s00122-013-2202-xURLPMID:24072208 [本文引用: 1]

Improved efficacy and durability of powdery mildew resistance can be enhanced via knowledge of the genetics of resistance and susceptibility coupled with the development of high-resolution maps to facilitate the stacking of multiple resistance genes and other desirable traits. We studied the inheritance of powdery mildew (Erysiphe necator) resistance and susceptibility of wild Vitis rupestris B38 and cultivated V. vinifera 'Chardonnay', finding evidence for quantitative variation. Molecular markers were identified using genotyping-by-sequencing, resulting in 16,833 single nucleotide polymorphisms (SNPs) based on alignment to the V. vinifera 'PN40024' reference genome sequence. With an average density of 36 SNPs/Mbp and uniform coverage of the genome, this 17K set was used to identify 11 SNPs on chromosome 7 associated with a resistance locus from V. rupestris B38 and ten SNPs on chromosome 9 associated with a locus for susceptibility from 'Chardonnay' using single marker association and linkage disequilibrium analysis. Linkage maps for V. rupestris B38 (1,146 SNPs) and 'Chardonnay' (1,215 SNPs) were constructed and used to corroborate the 'Chardonnay' locus named Sen1 (Susceptibility to Erysiphe necator 1), providing the first insight into the genetics of susceptibility to powdery mildew from V. vinifera. The identification of markers associated with a susceptibility locus in a V. vinifera background can be used for negative selection among breeding progenies. This work improves our understanding of the nature of powdery mildew resistance in V. rupestris B38 and 'Chardonnay', while applying next-generation sequencing tools to advance grapevine genomics and breeding.
DOI:10.1128/AEM.67.5.2263-2269.2001URLPMID:11319110 [本文引用: 1]

Xylella fastidiosa is a gram-negative, xylem-limited bacterium affecting economically important crops (e.g., grapevine, citrus, and coffee). The citrus variegated chlorosis (CVC) strain of X. fastidiosa is the causal agent of this severe disease of citrus in Brazil and represents the first plant-pathogenic bacterium for which the genome sequence was determined. Plasmids for the CVC strain of X. fastidiosa were constructed by combining the chromosomal replication origin (oriC) of X. fastidiosa with a gene which confers resistance to kanamycin (Kan(r)). In plasmid p16KdAori, the oriC fragment comprised the dnaA gene as well as the two flanking intergenic regions, whereas in plasmid p16Kori the oriC fragment was restricted to the dnaA-dnaN intergenic region, which contains dnaA-box like sequences and AT-rich clusters. In plasmid p16K, no oriC sequence was present. In the three constructs, the promoter region of one of the two X. fastidiosa rRNA operons was used to drive the transcription of the Kan(r) gene to optimize the expression of kanamycin resistance in X. fastidiosa. Five CVC X. fastidiosa strains, including strain 9a5c, the genome sequence of which was determined, and two strains isolated from coffee, were electroporated with plasmid p16KdAori or p16Kori. Two CVC isolates, strains J1a12 and B111, yielded kanamycin-resistant transformants when electroporated with plasmid p16KdAori or p16Kori but not when electroporated with p16K. Southern blot analyses of total DNA extracted from the transformants revealed that, in all clones tested, the plasmid had integrated into the host chromosome at the promoter region of the rRNA operon by homologous recombination. To our knowledge, this is the first report of stable transformation in X. fastidiosa. Integration of oriC plasmids into the X. fastidiosa chromosome by homologous recombination holds considerable promise for functional genomics by specific gene inactivation.
DOI:10.1007/s10681-009-9950-3URL [本文引用: 1]

Leprosis, caused by citrus leprosis virus (CiLV) and transmitted by the tenuipalpid mite Brevipalpus phoenicis, is one of the most important viruses of citrus in the Americas. Sweet oranges (Citrus sinensis L. Osb.) are highly susceptible to CiLV, while mandarins (C. reticulata Blanco) and some of their hybrids have higher tolerance or resistance to this disease. The mechanisms involved in the resistance and its inheritance are still largely unknown. To study the quantitative trait loci (QTL; quantitative trait loci) associated with the resistance to CiLV, progeny analyses were established with 143 hybrid individuals of ‘Pêra’ sweet orange (C. sinensis L. Osb.) and ‘Murcott’ tangor (C. reticulata Blanco×C. sinensis L. Osb.) from controlled crossings. Disease assessment of the hybrid individuals was conducted by infesting the plants with viruliferous mites in the field. The experiment consisted of a randomized completely block design with ten replicates. The evaluated phenotypic traits were incidence and severity of the disease on leaves and branches, for a period of 3years. The MapQTL™ v.4.0 software was used for the identification and location of possible QTL associated with resistance to CiLV on a genetic map obtained from 260 AFLP and 5 RAPD markers. Only consistent QTLs from different phenotypic traits and years of evaluation, with the critical LOD scores to determine the presence or absence of each QTL calculated through the random permutation test, were considered. A QTL was observed and had a significant effect on the phenotypic variation, ranging from 79.4 to 84% depending on which trait (incidence or severity) was assessed. This suggests that few genes are involved in the genetic resistance of citrus to CiLV.
[本文引用: 1]
DOI:10.1093/nar/gks400URLPMID:22661580 [本文引用: 1]

ExPASy (http://www.expasy.org) has worldwide reputation as one of the main bioinformatics resources for proteomics. It has now evolved, becoming an extensible and integrative portal accessing many scientific resources, databases and software tools in different areas of life sciences. Scientists can henceforth access seamlessly a wide range of resources in many different domains, such as proteomics, genomics, phylogeny/evolution, systems biology, population genetics, transcriptomics, etc. The individual resources (databases, web-based and downloadable software tools) are hosted in a 'decentralized' way by different groups of the SIB Swiss Institute of Bioinformatics and partner institutions. Specifically, a single web portal provides a common entry point to a wide range of resources developed and operated by different SIB groups and external institutions. The portal features a search function across 'selected' resources. Additionally, the availability and usage of resources are monitored. The portal is aimed for both expert users and people who are not familiar with a specific domain in life sciences. The new web interface provides, in particular, visual guidance for newcomers to ExPASy.
[本文引用: 1]
[本文引用: 1]
DOI:10.1111/pbi.12733URLPMID:28371200 [本文引用: 1]

Citrus canker, caused by Xanthomonas citri subsp. citri (Xcc), is severely damaging to the global citrus industry. Targeted editing of host disease-susceptibility genes represents an interesting and potentially durable alternative in plant breeding for resistance. Here, we report improvement of citrus canker resistance through CRISPR/Cas9-targeted modification of the susceptibility gene CsLOB1 promoter in citrus. Wanjincheng orange (Citrus sinensis Osbeck) harbours at least three copies of the CsLOB1G allele and one copy of the CsLOB1- allele. The promoter of both alleles contains the effector binding element (EBEPthA4 ), which is recognized by the main effector PthA4 of Xcc to activate CsLOB1 expression to promote citrus canker development. Five pCas9/CsLOB1sgRNA constructs were designed to modify the EBEPthA4 of the CsLOB1 promoter in Wanjincheng orange. Among these constructs, mutation rates were 11.5%-64.7%. Homozygous mutants were generated directly from citrus explants. Sixteen lines that harboured EBEPthA4 modifications were identified from 38 mutant plants. Four mutation lines (S2-5, S2-6, S2-12 and S5-13), in which promoter editing disrupted CsLOB1 induction in response to Xcc infection, showed enhanced resistance to citrus canker compared with the wild type. No canker symptoms were observed in the S2-6 and S5-13 lines. Promoter editing of CsLOB1G alone was sufficient to enhance citrus canker resistance in Wanjincheng orange. Deletion of the entire EBEPthA4 sequence from both CsLOB1 alleles conferred a high degree of resistance to citrus canker. The results demonstrate that CRISPR/Cas9-mediated promoter editing of CsLOB1 is an efficient strategy for generation of canker-resistant citrus cultivars.
DOI:10.1111/j.1469-8137.2006.01763.xURLPMID:16866939 [本文引用: 1]

The changes in endopolygalacturonase (endo-PG) levels and endo-PG expression in nonmelting flesh (NMF) and melting flesh (MF) peach fruits (Prunus persica) during softening were studied. The endo-PG gene was analysed to identify polymorphisms exploitable for early marker-assisted selection (MAS) of flesh texture. The role of endo-PG in softening was assessed by western and northern blotting and by biochemical analyses. Polymorphisms in the endo-PG gene were revealed by reverse transcription-polymerase chain reaction (RT-PCR) and sequencing. An endo-PG protein was detected in both NMF and MF fruits. The levels of this endo-PG protein were higher and increased with softening in MF fruits, but remained lower and were constant in NMF fruits. The different levels of endo-PG appeared to be caused by the differential expression of an endo-PG gene, whose open-reading frame (ORF) showed five single nucleotide polymorphisms (SNPs) in NMF 'Oro A' compared with MF 'Bolero'. One of these SNPs allowed us to determine the allelic configuration at the melting flesh (M) locus and also seemed to be exploitable for early MAS in other NMF/MF phenotypes. The NMF phenotype does not seem to be caused by a large deletion of the endo-PG gene.
DOI:10.1093/jxb/erp284URLPMID:19887502 [本文引用: 1]

The colour of papaya fruit flesh is determined largely by the presence of carotenoid pigments. Red-fleshed papaya fruit contain lycopene, whilst this pigment is absent from yellow-fleshed fruit. The conversion of lycopene (red) to beta-carotene (yellow) is catalysed by lycopene beta-cyclase. This present study describes the cloning and functional characterization of two different genes encoding lycopene beta-cyclases (lcy-beta1 and lcy-beta2) from red (Tainung) and yellow (Hybrid 1B) papaya cultivars. A mutation in the lcy-beta2 gene, which inactivates enzyme activity, controls lycopene production in fruit and is responsible for the difference in carotenoid production between red and yellow-fleshed papaya fruit. The expression level of both lcy-beta1 and lcy-beta2 genes is similar and low in leaves, but lcy-beta2 expression increases markedly in ripe fruit. Isolation of the lcy-beta2 gene from papaya, that is preferentially expressed in fruit and is correlated with fruit colour, will facilitate marker-assisted breeding for fruit colour in papaya and should create possibilities for metabolic engineering of carotenoid production in papaya fruit to alter both colour and nutritional properties.
DOI:10.3390/genes9080385URLPMID:30065184 [本文引用: 1]

The bitterness and toxicity of wild-type seeds of Prunoideae is due to the cyanogenic glucoside amygdalin. In cultivated almond (Prunus dulcis (Mill.) D.A. Webb), a dominant mutation at the Sk locus prevents amygdalin accumulation and thus results in edible sweet kernels. Here, we exploited sequence similarity and synteny between the genomes of almond and peach (Prunus persica (L.) Batsch) to identify cleaved amplified polymorphic sequence (CAPS) molecular markers linked to the Sk locus. A segregant F? population was used to map these markers on the Sk genomic region, together with Sk-linked simple sequence repeat (SSR) markers previously described. Molecular fingerprinting of a cultivar collection indicated the possibility to use CAPS polymorphisms identified in this study in breeding programs arising from different parental combinations. Overall, we highlight a set of codominant markers useful for early selection of sweet kernel genotypes, an aspect of primary importance in almond breeding. In addition, by showing collinearity between the physical map of peach and the genetic map of almond with respect to the Sk genomic region, we provide valuable information for further marker development and Sk positional cloning.
DOI:10.1111/nph.16088URLPMID:31369160 [本文引用: 1]

Calcium is a ubiquitous second messenger that mediates plant responses to developmental and environmental cues. Calcium-dependent protein kinases (CDPKs) are key actors of plant signaling that convey calcium signals into physiological responses by phosphorylating various substrates including ion channels, transcription factors and metabolic enzymes. This large diversity of targets confers pivotal roles of CDPKs in shoot and root development, pollen tube growth, stomatal movements, hormonal signaling, transcriptional reprogramming and stress tolerance. On the one hand, specificity in CDPK signaling is achieved by differential calcium sensitivities, expression patterns, subcellular localizations and substrates. On the other hand, CDPKs also target some common substrates to ensure key cellular processes indispensable for plant growth and survival in adverse environmental conditions. In addition, the CDPK-related protein kinases (CRKs) might be closer to some CDPKs than previously anticipated and could contribute to calcium signaling despite their inability to bind calcium. This review highlights the regulatory properties of Arabidopsis CDPKs and CRKs that coordinate their multifaceted functions in development, immunity and abiotic stress responses.
DOI:10.1104/pp.83.4.830URLPMID:16665348 [本文引用: 1]

A calcium-dependent protein kinase activity from suspension-cultured soybean cells (Glycine max L. Wayne) was shown to be dependent on calcium but not calmodulin. The concentrations of free calcium required for half-maximal histone H1 phosphorylation and autophosphorylation were similar ( approximately 2 micromolar). The protein kinase activity was stimulated 100-fold by &gt;/=10 micromolar-free calcium. When exogenous soybean or bovine brain calmodulin was added in high concentration (1 micromolar) to the purified kinase, calcium-dependent and -independent activities were weakly stimulated (&lt;/=2-fold). Bovine serum albumin had a similar effect on both activities. The kinase was separated from a small amount of contaminating calmodulin by sodium dodecyl sulfate polyacrylamide gel electrophoresis. After renaturation the protein kinase autophosphorylated and phosphorylated histone H1 in a calcium-dependent manner. Following electroblotting onto nitrocellulose, the kinase bound (45)Ca(2+) in the presence of KCl and MgCl(2), which indicates that the kinase itself is a high-affinity calcium-binding protein. Also, the mobility of one of two kinase bands in SDS gels was dependent on the presence of calcium. Autophosphorylation of the calmodulin-free kinase was inhibited by the calmodulin-binding compound N-(6-aminohexyl)-5-chloro-1-naphthalene sulfonamide (W-7), showing that the inhibition of activity by W-7 is independent of calmodulin. These results show that soybean calcium-dependent protein kinase represents a new class of protein kinase which requires calcium but not calmodulin for activity.
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.3390/ijms18112436URLPMID:29156607 [本文引用: 1]

Calcium-dependent protein kinases (CPKs/CDPKs) are Ca2+-sensors that decode Ca2+ signals into specific physiological responses. Research has reported that CDPKs constitute a large multigene family in various plant species, and play diverse roles in plant growth, development, and stress responses. Although numerous CDPKs have been exhaustively studied, and many of them have been found to be involved in plant hormone biosynthesis and response mechanisms, a comprehensive overview of the manner in which CDPKs participate in phytohormone signaling pathways, regulating nearly all aspects of plant growth, has not yet been undertaken. In this article, we reviewed the structure of CDPKs and the mechanism of their subcellular localization. Some CDPKs were elucidated to influence the intracellular localization of their substrates. Since little work has been done on the interaction between CDPKs and cytokinin signaling pathways, or on newly defined phytohormones such as brassinosteroids, strigolactones and salicylic acid, this paper mainly focused on discussing the integral associations between CDPKs and five plant hormones: auxins, gibberellins, ethylene, jasmonates, and abscisic acid. A perspective on future work is provided at the end.
DOI:10.1007/BF00286684URLPMID:8078458 [本文引用: 1]

Two cDNA clones, cATCDPK1 and cATCDPK2, encoding Ca(2+)-dependent, calmodulin-independent protein kinases (CDPK) were cloned from Arabidopsis thaliana and their nucleotide sequences were determined. Northern blot analysis indicated that the mRNAs corresponding to the ATCDPK1 and ATCDPK2 genes are rapidly induced by drought and high-salt stress but not by low-temperature stress or heat stress. Treatment of Arabidopsis plants with exogenous abscisic acid (ABA) had no effect on the induction of ATCDPK1 or ATCDPK2. These findings suggest that a change in the osmotic potential of the environment can serve as a trigger for the induction of ATCDPK1 and ATCDPK2. Putative proteins encoded by ATCDPK1 and ATCDPK2 which contain open reading frames of 1479 and 1488 bp, respectively, are designated ATCDPK1 and ATCDPK2 and show 52% identity at the amino acid sequence level. ATCDPK1 and ATCDPK2 exhibit significant similarity to a soybean CDPK (51% and 73%, respectively). Both proteins contain a catalytic domain that is typical of serine/threonine protein kinases and a regulatory domain that is homologous to the Ca(2+)-binding sites of calmodulin. Genomic Southern blot analysis suggests the existence of a few additional genes that are related to ATCDPK1 and ATCDPK2 in the Arabidopsis genome. The ATCDPK2 protein expressed in Escherichia coli was found to phosphorylate casein and myelin basic protein preferentially, relative to a histone substrate, and required Ca2+ for activation.
DOI:10.3389/fpls.2019.00052URLPMID:30778363 [本文引用: 1]

Premature leaf senescence affects plant yield and quality, and numerous researches about it have been conducted until now. In this study, we identified an early senescent mutant es4 in rice (Oryza sativa L.); early senescence appeared approximately at 60 dps and became increasingly senescent with the growth of es4 mutant. We detected that content of reactive oxygen species (ROS) and malondialdehyde (MDA), as well as activity of superoxide dismutase (SOD) were elevated, while chlorophyll content, soluble protein content, activity of catalase (CAT), activity of peroxidase (POD) and photosynthetic rate were reduced in the es4 mutant leaves. We mapped es4 in a 33.5 Kb physical distance on chromosome 4 by map-based cloning. Sequencing analysis in target interval indicated there was an eight bases deletion mutation in OsCPK12 which encoded a calcium-dependent protein kinase. Functional complementation of OsCPK12 in es4 completely restored the normal phenotype. We used CRISPR/Cas9 for targeted disruption of OsCPK12 in ZH8015 and all the mutants exhibited the premature senescence. All the results indicated that the phenotype of es4 was caused by the mutation of OsCPK12. Overexpression of OsCPK12 in ZH8015 enhanced the net photosynthetic rate (Pn) and chlorophyll content. OsCPK12 was mainly expressed in green organs. The results of qRT-PCR analysis showed that the expression levels of some key genes involved in senescence, chlorophyll biosynthesis, and photosynthesis were significantly altered in the es4 mutant. Our results demonstrate that the mutant of OsCPK12 triggers the premature leaf senescence; however, the overexpression of OsCPK12 may delay its growth period and provide the potentially positive effect on productivity in rice.
DOI:10.1111/j.1469-8137.2011.03793.xURLPMID:21692804 [本文引用: 1]

? Ca(2+) -dependent protein kinase (CDPK) is believed to be involved in abscisic acid (ABA) signaling, and several members of the Arabidopsis CDPK superfamily have been identified as positive ABA signaling regulators, but it remains unknown if CDPK negatively regulates ABA signaling. ? Here, we investigated the function of an Arabidopsis (Arabidopsis thaliana) CDPK, CPK12, in ABA signaling pathway. ? We generated Arabidopsis CPK12-RNAi lines, and observed that downregulation of CPK12 resulted in ABA hypersensitivity in seed germination and post-germination growth, and altered expression of a set of ABA-responsive genes. Expression assay showed that CPK12 was ubiquitously expressed and localized to both cytosol and nucleus. Biochemical assays showed that CPK12 interacted with, phosphorylated and stimulated a type 2C protein phosphatase ABI2, and phosphorylated two ABA-responsive transcription factors (ABF1 and ABF4) in vitro. ? Our findings show that the Arabidopsis CPK12 is a negative ABA-signaling regulator in seed germination and post-germination growth, suggesting that different members of the CDPK family may constitute a regulation loop by functioning positively and negatively in ABA signal transduction.
DOI:10.4161/psb.6.11.17954URLPMID:22041934 [本文引用: 1]

Ca2+ is believed to be a critical second messenger in ABA signal transduction. Ca2+-dependent protein kinases (CDPKs) are the best characterized Ca2+ sensors in plants. Recently, we identified an Arabidopsis CDPK member CPK12 as a negative regulator of ABA signaling in seed germination and post-germination growth, which reveals that different members of the CDPK family may constitute a regulation loop by functioning positively and negatively in ABA signal transduction. We observed that both RNA interference and overexpression of CPK12 gene resulted in ABA-hypersensitive phenotypes in seed germination and post-germination growth, suggesting a high complexity of the CPK12-mediated ABA signaling pathway. CPK12 stimulates a negative ABA-signaling regulator (ABI2) and phosphorylates two positive ABA-signaling regulators (ABF1 and ABF4), which may partly explain the ABA hypersensitivity induced by both downregulation and upregulation of CPK12 expression. Our data indicate that CPK12 appears to function as a balancer in ABA signal transduction in Arabidopsis.
DOI:10.1007/BF00019547URLPMID:8704124 [本文引用: 1]

Protein kinases are important in eukaryotic signal transduction pathways. In this study we designed degenerate oligonucleotides corresponding to two conserved regions of protein kinases and using the polymerase chain reaction (PCR) have amplified a 141 bp fragment of DNA from mungbeans (Vigna radiata Rwilcz cv. Berken). Sequence analysis of the PCR products indicates that they encode several putative protein kinases with respect to their identity with other known plant protein kinases. Using one of the six fragments (CPK3-8), we isolated a 2022 bp cDNA (VrCDPK-1) from a Vigna radiata lambda gt11 library. VrCDPK-1 has a 96 bp 5'-untranslated region and a 465 bp 3'-untranslated region and an open reading frame of 1461 bp. VrCDPK-1 contains all of the conserved regions commonly found in calcium dependent protein kinases (CDPK). VrCDPK-1 shares 24 to 89% sequence identity with previously reported sequences for plant CDPKs at the protein level. Southern analysis revealed the presence of several copies of the CDPK gene. VrCDPK-1 expression was stimulated when mungbean cuttings were treated with CaCl2, while treatment with MgCl2 had no effect. We are reporting for the first time a CDPK gene in mungbean which is inducible by mechanical strain. Cuttings treated with indole-3-acetic acid (IAA) or subjected to salt stress showed an increase in VrCDPK-1 expression. There was a dramatic stimulation in VrCDPK-1 expression 6 h after cuttings were treated with cycloheximide.
DOI:10.1007/BF00037023URLPMID:7766885 [本文引用: 1]

We have isolated, from a cDNA library constructed from rice coleoptiles, two sequences, OSCPK2 and OSCPK11, that encode for putative calcium-dependent protein kinase (CDPK) proteins. OSCPK2 and OSCPK11 cDNAs are related to SPK, another gene encoding a rice CDPK that is specifically expressed in developing seeds [20]. OSCPK2 and OSCPK11-predicted protein sequences are 533 and 542 amino acids (aa) long with a corresponding molecular mass of 59436 and 61079 Da respectively. Within their polypeptide chain, they all contain those conserved features that define a plant CDPK; kinase catalytic sequences are linked to a calmodulin-like regulatory domain through a junction region. The calmodulin-like regulatory domain of the predicted OSCPK2 protein contains 4 EF-hand calcium-binding sites while OSCPK11 has conserved just one canonical EF-hand motif. In addition, OSCPK2- and OSCPK11-predicted proteins contain, at their N-terminal region preceding the catalytic domain, a stretch of 80 or 74 residues highly rich in hydrophilic amino acids. Comparison of the NH2-terminal sequence of all three rice CDPKs so far identified (OSCPK2, OSCPK11 and SPK) indicates the presence of a conserved MGxxC(S/Q)xxT motif that may define a consensus signal for N-myristoylation. OSCPK2 and OSCPK11 proteins are both encoded by a single-copy gene and their polyadenylated transcripts are 2.4 and 3.5 kb long respectively. OSCPK2 and OSCPK11 mRNAs are equally abundant in rice roots and coleoptiles. A 12 h white light treatment of the coleoptiles reduces the amount of OSCPK2 mRNA with only a slight effect on the level of OSCPK11 transcript. With anoxic treatments, OSCPK2 mRNA level declined significantly and promptly while the amount of OSCPK11 transcript remained constant.
DOI:10.1105/tpc.12.5.803URLPMID:10810151 [本文引用: 1]

In the Cf-9/Avr9 gene-for-gene interaction, the Cf-9 resistance gene from tomato confers resistance to the fungal pathogen Cladosporium fulvum, which expresses the corresponding pathogen-derived avirulence product Avr9. To understand R gene function and dissect the signaling mechanisms involved in the induction of plant defenses, we studied Cf-9/Avr9-dependent activation of protein kinases in transgenic Cf9 tobacco cell cultures. Using a modified in-gel kinase assay with histone as substrate, we identified a membrane-bound, calcium-dependent protein kinase (CDPK) that showed a shift in electrophoretic mobility from 68 to 70 kD within 5 min after Avr9 elicitor was added. This transition from the nonelicited to the elicited CDPK form was caused by a phosphorylation event and was verified when antibodies to CDPK were used for protein gel blot analysis. In addition, the interconversion of the corresponding CDPK forms could be induced in vitro in both directions by treatment with either phosphatase or ATP. In vitro protein kinase activity toward syntide-2 or histone with membrane extracts or gel-purified enzyme was dependent on Ca(2)+ content and was compromised by the calmodulin antagonist N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide (W-7) but not by its inactive isoform N-(6-aminohexyl)-1-naphthalenesulfonamide. In these assays, the CDPK activity in elicited samples, reflecting predominantly the phosphorylated 70-kD CDPK form, was greater than in nonelicited samples. Thus, Avr9/Cf-9-dependent phosphorylation and subsequent transition from the nonelicited to the elicited form correlate with the activation of a CDPK isoform after in vivo stimulation. Because that transition was not inhibited by W-7, the in vivo CDPK activation probably is not the result of autophosphorylation. Studies with pharmacological inhibitors indicated that the identified CDPK is independent of or is located upstream from a signaling pathway that is required for the Avr9-induced active oxygen species.
DOI:10.1186/1471-2229-11-90URLPMID:21595879 [本文引用: 1]

The arbuscular mycorrhizal (AM) symbiosis consists of a mutualistic relationship between soil fungi and roots of most plant species. This association provides the arbuscular mycorrhizal fungus with sugars while the fungus improves the uptake of water and mineral nutrients in the host plant. Then, the establishment of the arbuscular mycorrhizal (AM) symbiosis requires the fine tuning of host gene expression for recognition and accommodation of the fungal symbiont. In plants, calcium plays a key role as second messenger during developmental processes and responses to environmental stimuli. Even though calcium transients are known to occur in host cells during the AM symbiosis, the decoding of the calcium signal and the molecular events downstream are only poorly understood.
DOI:10.1021/bi00152a035URLPMID:1390682 [本文引用: 1]

Nodulin 26 is a nodule-specific protein that is associated with the symbiosome membrane of soybean root nodules. Nodulin 26 is an endogenous substrate for a novel calcium-dependent protein kinase (CDPK) of soybean root nodules. By phosphopeptide mapping of endoproteinase Lys-C-digested nodulin 26 and automated and manual peptide sequence analyses, we have identified the site on nodulin 26 phosphorylated by CDPK. We have also established that the phosphorylation site of nodulin 26 is identical to the phosphorylation site of CK-15, a synthetic peptide with the carboxyl-terminal sequence of nodulin 26. The phosphorylation of nodulin 26 occurs at position Ser262, and the phosphorylation of CK-15 occurs at the analogous position, Ser,6 in vitro. Thus, the CK-15 sequence apparently contains sufficient structural features of the phosphorylation site of nodulin 26 to be recognized by CDPK. On the basis of peptide mapping analysis of nodulin 26 from nodules that are metabolically labeled with [32P]phosphate, it appears that the site of nodulin 26 that is phosphorylated in vitro is also labeled in vivo. The data indicate that the carboxyl terminus of nodulin 26 is phosphorylated by CDPK and provide initial sequence data for the phosphorylation site of an endogenous substrate for a plant CDPK.
