 ,南京农业大学园艺学院,南京 210095
,南京农业大学园艺学院,南京 210095vvi-miR160s in Mediating VvARF18 Response to Gibberellin Regulation of Grape Seed Development
BAI YunHe, WANG WenRan, DONG TianYu, GUAN Le, SU ZiWen, JIA HaiFeng, FANG JingGui, WANG Chen ,College of Horticulture, Nanjing Agricultural University, Nanjing 210095
,College of Horticulture, Nanjing Agricultural University, Nanjing 210095通讯作者:
责任编辑: 赵伶俐
收稿日期:2019-09-5接受日期:2019-12-25网络出版日期:2020-05-16
| 基金资助: |
Received:2019-09-5Accepted:2019-12-25Online:2020-05-16
作者简介 About authors
白云赫,E-mail:2017104028@njau.edu.cn。

摘要
关键词:
Abstract
Keywords:
PDF (6560KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
白云赫, 王文然, 董天宇, 管乐, 宿子文, 贾海锋, 房经贵, 王晨. vvi-miR160s介导VvARF18应答赤霉素调控葡萄种子的发育[J]. 中国农业科学, 2020, 53(9): 1890-1903 doi:10.3864/j.issn.0578-1752.2020.09.015
BAI YunHe, WANG WenRan, DONG TianYu, GUAN Le, SU ZiWen, JIA HaiFeng, FANG JingGui, WANG Chen.
0 引言
【研究意义】MicroRNA(miRNA)是一类长度约为19—24个核苷酸的非编码RNA序列,其主要通过特异性切割或抑制靶mRNA的翻译调节靶基因的表达[1,2],在植物的多种生理代谢过程中起着重要的调节作用[3],如信号转导[4]、生长发育[5]和非生物胁迫[6]等。植物中已经鉴定了许多miRNA家族,其中microR160家族比较保守,其在调节植物形态[7],增强植物抗性[8],调节花和胚胎的发育[9,10],影响植物体内激素含量的变化[11]等过程中起着重要作用。到目前为止,miR160及其靶基因功能的研究主要集中在营养生长和生殖生长方面,笔者课题组前期研究发现葡萄miR160(vvi-miR160s)及其靶基因可能参与单性结实发育的调控,而对浆果发育,尤其是在果核发育方面尚未见报道。因此,开展miR160及其靶基因对葡萄果核发育的调控机制研究,对阐释葡萄果实核发育机制以及无核育种具有重要意义。【前人研究进展】研究发现miR160主要靶向生长素响应因子ARF(Auxin response factor,ARF)转录因子,其主要调控生长素响应基因的表达[12],能够与生长素基因ARF启动子中的TGTCTC序列特异性结合,激活或抑制相关基因的表达[13]。目前,ARF家族已经在番茄、拟南芥、苹果、柑橘、大豆、黄瓜、谷子、白菜等作物中得到鉴定[14]。番茄miR160a通过抑制靶基因SlARF10,影响叶片生长[15];拟南芥miR160负调控靶基因ARF17,参与生长素信号转导,影响植株的营养生长与生殖生长[11]。拟南芥miR160还可负调控靶基因AtARF10和AtARF16,通过ABA途径参与种子的萌发和休眠[16]。近期研究发现,ARF是生长素与赤霉素信号途径的交互作用因子。通过降低番茄SlARF7 mRNA水平可能影响GA信号通路,且SlARF7仅调控番茄果实坐果及其相关的生长素信号通路[17]。例如,在拟南芥中,生长素处理导致AUX/IAA和ARF蛋白可直接调节GA代谢基因的表达,并且该结果与生长素过表达突变体yucca相同[18]。此外,拟南芥胚珠受精可引发生长素介导的GA生物合成[19]。尽管如此,有关ARFs调控植物种子发育的研究尚未见报道。葡萄(Vitis. L)是世界上重要的经济作物之一,其栽培面积和产量仅次于柑橘,位居世界第二。葡萄是赤霉素(GA)高度敏感型果树,外源GA处理极易诱导葡萄无核或残核;另一方面,GA也可调控葡萄种子发育从而影响果实大小。尽管GA被广泛应用于葡萄无核生产或种子发育调控,但其分子机制尚不清晰。【本研究切入点】基因组测序使葡萄基因组中vvi-miR160s成熟体精确序列的鉴定及进化分析成为可能,但其对靶基因VvARF18的裂解作用,以及它们在GA介导的葡萄果实发育过程中的时空表达模式尚无报道。【拟解决的关键问题】鉴定‘魏可’葡萄中vvi-miR160家族成员的精确序列,预测并验证其靶基因,分析其序列特征及其潜在功能,鉴定它们在葡萄种子发育中的时空表达特征及其应答GA的模式,为进一步认识vvi-miR160s及其靶基因VvARF18在种子发育过程中的作用提供重要信息,同时也为GA诱导果实无核过程提供一定的理论依据。1 材料与方法
试验于2017年在江苏省农业科学院葡萄科研基地进行。1.1 试验材料
以5年生欧亚种葡萄‘魏可’为材料。选取生长势基本一致的花序,根据前人研究结果,以50 mg∙L-1赤霉素溶液(含0.2%吐温),于盛花前9 d浸沾花穗30 s,以清水为对照,分别在花后2、5、10、20、30和40 d采集样品,一部分用于体式显微镜观察,另一部分分离果皮、果肉、种子,冻存于液氮后于-80℃保存备用。1.2 试验方法
1.2.1 vvi-miR160s成熟体序列克隆鉴定 在miRBase 22.1数据库(http://www.mirbase.org/)中搜索不同物种miR160s的成熟体序列,基于葡萄miR160s成熟体序列,利用miR-RACE在‘魏可’种子中克隆其成熟体序列,测序鉴定其精准序列[20],并在葡萄基因组中比对葡萄miR160s前体基因(VvMIR160s)。使用在线软件Primer3 Input(http:// bioinfo.ut.ee/primer3-0.4.0/)设计VvMIR160s引物(表1),以‘魏可’葡萄总DNA为模板进行扩增(95℃预变性5 min;95℃变性30 s,58℃退火30 s,72℃延伸30 s,36个循环;72℃延伸5 min),产物经电泳回收,送公司测序。利用在线软件RNA Folding Form(http://unafold.rna.albany.edu/?q=mfold/RNA-Folding-Form)分析前体茎环结构;利用软件MapInsepet进行染色体位置的绘制。Table 1
表1
表1VvMIR160s PCR扩增引物序列
Table 1
| 基因名称 Gene name | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| VvMIR160a | ACACCTCCTAAAATCATTGTCTG | CTTGTGACATGAATATGGTGCG |
| VvMIR160b | CTATGTATTTGTCTTGTTCTGATTGAA | TGAATGGTCACAGTTCTTTGG |
| VvMIR160c | GGCCTGGCCTCTATAAATATCA | AATCGACCCACAATCAAACC |
| VvMIR160d | GATGTGGTGCTTCGCCAAT | ATGTGGGTTTTCTAAATGCCTAACC |
| VvMIR160e | CACTCACTCACACCCTTCC | ATATTATATTCTCTCTGCAGCCAAG |
新窗口打开|下载CSV
1.2.2 vvi-miR160s靶基因预测及生物信息学分析 运用在线软件PSRNA Target(http://plantgrn.noble.org/ v1_psRNATarget/#)预测vvi-miR160s的靶基因。运用在线软件ExPASy(http://web.expasy.org/protparam/)分析VvARF18蛋白的分子式、分子质量、等电点、氨基酸含量、不稳定指数、亲疏水性;在线软件NCBI Conserved Domain Serch(https://www.ncbi.nlm.nih. gov/Structure/cdd/wrpsb.cgi)分析VvARF18蛋白的保守结构域;在线软件NLS Mapper(http://nls-mapper. iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi)进行核定位信号分析;Genscript(https://www.genscript.com/ wolf-psort.html)进行亚细胞定位分析。利用NCBI- BLAST寻找VvARF18蛋白的同原序列,并用MEGA 6.0 构建系统进化树。利用在线软件GSDS(http:// gsds.cbi.pku.edu.cn/)进行不同物种基因结构构建;在线软件MEME(http://meme-suite.org/tools/meme)进行不同物种作用元件motif分析;在线软件PlantCARE(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)进行启动子作用元件分析。
1.2.3 VvARF18启动子顺式作用元件分析 在Grape Genome Browser(http://www.genoscope.cns.fr/externe/ GenomeBrowser/Vitis/)数据库中寻找VvARF18转录起始位置上游1 500 bp序列,即VvARF18启动子序列,将启动子序列放入在线数据库Plant CARE中,寻找相对应的作用元件。
1.2.4 vvi-miR160s及其靶基因在葡萄果核发育过程中的表达分析 采用CTAB法提取葡萄种子总RNA,并使用abm试剂盒反转录为cDNA。根据定量引物设计的原则,利用Primer3 Input(http://bioinfo.ut. ee/primer3-0.4.0/)设计vvi-miR160家族以及VvARF18的定量引物(表2)。
Table 2
表2
表2vvi-miR160s及VvARF18定量引物序列
Table 2
| 基因名称 Gene name | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| vvi-miR160a | TGACCTTTGTGCTTCAGTGG | GCTATCTGGGTTGACCTCCA |
| vvi-miR160b | TTCTGCAGGAGATGGAGCTT | AGTGTTTCGCCTGCTTGACT |
| vvi-miR160c | CCACATTCCGTGACCTTTCT | GCACAACCCATTTCACCTTT |
| vvi-miR160d | CGCCAATGCAGGAAATTTAT | GGGAGCCAGGCATGTAAGTA |
| vvi-miR160e | CTGTATGCCATTTGCAGAGC | GGGGGAGAAGATTGAAGAGG |
| VvARF18 | CTGAACACGCCTATGGGAAT | CCGTTTCACCCTCAGTGTTT |
新窗口打开|下载CSV
按照SYPBR Premix ExTaqTM试剂盒(宝生物工程有限公司)指导,采用实时定量PCR(real-time quantitative PCR,qRT-PCR)检测基因的相对表达量。扩增体系为20 μL(1 μL cDNA,上、下游引物各0.4 μL,10 μL 2×TransStart Tip GreenqPCR SuperMix,8.2 μL ddH20)。反应程序为:94℃ 30 s;94℃5 s,60℃15 s,72℃ 10 s,40个循环。葡萄VvACTIN(XM002273532)为内参基因,进行3次生物学重复。采用ΔΔCT法分析数据。
1.3 数据分析
使用Excel 2017进行数据整理,SPSS 22.0进行方差分析,使用Origin 9软件进行绘图。2 结果
2.1 GA处理对‘魏可’葡萄果核发育的影响
本试验通过花前赤霉素(GA)处理发现,花后2 d时,对照组和GA处理组均有胚珠,但是对照比GA处理组的胚珠大;到花后5 d时,对照组可明显观察到胚珠,而GA处理组胚珠发育不良,逐渐退化、缩小;在花后10、20、30和40 d中,对照组中胚珠逐渐发育成成熟种子,而GA处理组胚珠退化、消失,导致果实无核,仅可以观察到木质化的细线(图1-A)。除此之外,GA处理还增大果实纵经、减小果实横径,增大果形指数(图1-B)。以上结果表明,GA可以抑制葡萄果实胚珠及种子的发育,从而诱导果实无核,同时影响果实形状。图1
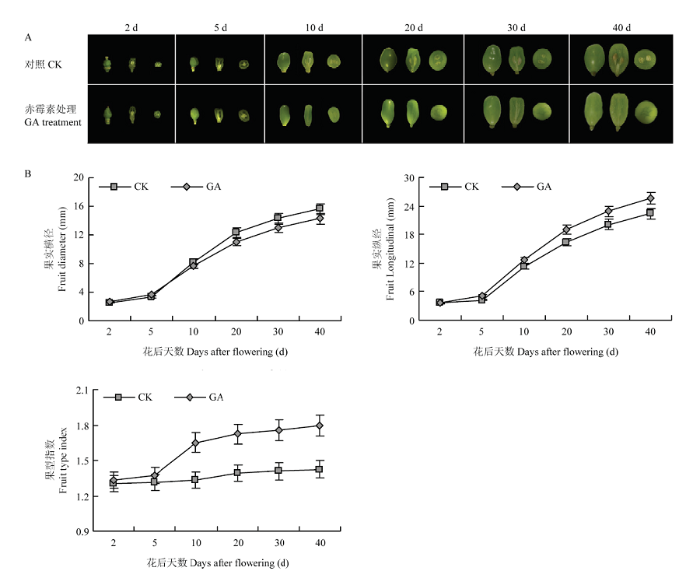 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1GA处理对‘魏可’葡萄果实发育的影响
Fig. 1Effect of GA treatment on fruit development of Wink grape
2.2 vvi-miR160s序列鉴定及前体基因扩增
采用miR-RACE技术,鉴定葡萄中vvi-miR160s成员的精确序列。vvi-miR160s成员的3′和5′-miR- RACE的产物分别为88 bp和62 bp(图2-A),且该序列与miRBase22.1数据库中所上传的序列一致。以‘魏可’葡萄总DNA为模板,对vvi-miR160s的前体基因进行扩增。经凝胶电泳后获得5条约501 bp的特异性条带,并且没有非特异性杂带。经纯化、测序后发现,其前体基因序列均为501 bp(图2-B)。图2
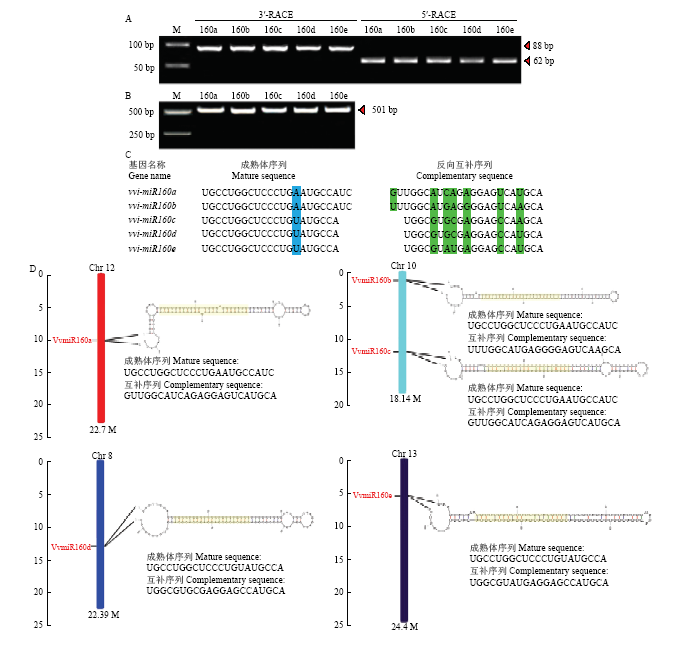 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2vvi-miR160s成熟体序列分析和染色体分布
Fig. 2Sequence analysis and chromosome distribution of mature vvi-miR160s
其中,vvi-miR160a/b成熟体序列为23 bp,而vvi-miR160c/d/e的序列长度为21 bp;vvi-miR160s成熟体序列保守程度较高,其中vvi-miR160a、b相同,vvi-miR160c、d、e序列相同;而vvi-miR160s互补序列的保守度较低(图2-C)。vvi-miR160s成员分别分布在染色体12(vvi-miR160a)、染色体10(vvi-miR160b和vvi-miR160c)、染色体8(vvi-miR160d)和染色体13(vvi-miR160e)上,其中vvi-miR160a和b成熟体序列相同,但却分布在不同染色体上,而vvi-miR160b和c成熟体序列不同,却分布在相同的染色体上,表明成熟体序列保守的miRNA在染色体上位置并不一定保守;通过在线软件RNA Folding Form分析可知,vvi-miR160s前体序列均可形成稳定的茎环结构,这为VvmiRNAs稳定存在提供了有力的证据(图2-D)。
2.3 VvMIR160s前体基因的进化分析与成熟体序列比对
利用MEGA6.0软件对拟南芥(Arabidopsis thaliana)、烟草(Nicotiana tabacum)、番茄(Solanum lycopersicum)、毛果杨(Populus trichocarpa)、水稻(Oryza sativa)等17种植物的miR160家族进行进化关系分析以及成熟体序列比对(图3)。结果表明,不同物种miR160家族成员数量不同,小麦(Triticum aestivum)、棉花(Gossypium herbaceum)、花生(Arachis hypogaea)、向日葵(Helianthus annuus)等物种中miR160家族成员仅有1个,而水稻(6个)(Oryza sativa)、烟草(4个)(Nicotiana tabacum)、高粱(6个)(Sorghum bicolor)、苹果(5个)(malus domestica)、毛果杨(8个)(Populus trichocarpa)等物种的miR160家族成员较多;除vvi-miR160a/b外,其余物种miR160家族成员成熟体序列为21个碱基。并且从进化图谱和成熟体序列比对中发现,vvi-miR160c/d/e与小麦、草莓(Fragaria vesca)、玉米(Zea mays)、高粱、水稻等物种的亲缘关系较进,且成熟体序列相同;与向日葵、柑橘(Citurs sinensis)等物种的亲缘关系较远,且成熟体序列间存在差异,而vvi-miR160a/b与毛果杨ptc-miR160e/ f/g/h、烟草nta-miR160d、甜瓜cme-miR160d亲缘关系较进,且成熟体序列相同,但值得注意的是,vvi-miR160s成员与棉花成熟体序列的差异较大,仅有9个碱基完全匹配。综上所述,尽管不同物种miR160家族成员保守性较高,但仍存在个别物种间miR160家族成员存在一定的差异,表明其序列存在较高保守性的同时也具有一定的进化多样性。图3
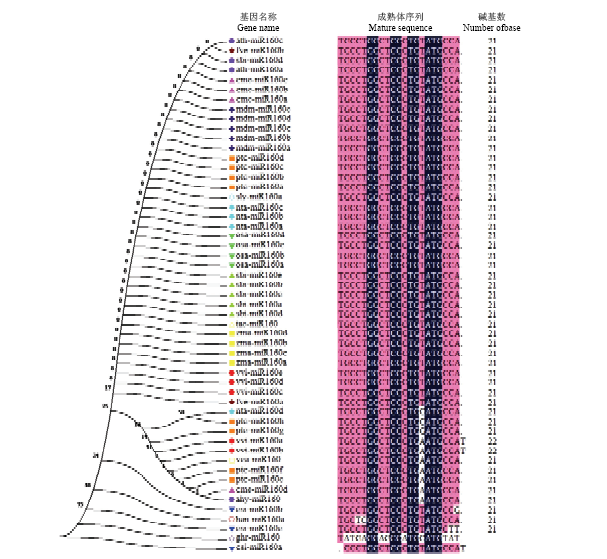 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3microR160s进化分析和成熟体序列比对
Vv:葡萄Vvtis vinifera;Ha:向日葵Helianthus annuus L;Zm:玉米Zea mays;Ta:小麦Triticum aestivum;Sb:高粱Sorghum bicolor;Os:水稻Oryza sativa;Nt:烟草Nicotiana tabacum;Sl:番茄Solanum lycopersicum;Pt:毛果杨Populus trichocarpa;Cs:柑橘Citurs sinensis;Md:苹果Malus domestica;Gh:棉花Gossypium herbaceum;Cm:甜瓜Cucumis melo;At:拟南芥Arabidopsis thaliana;Ah:花生Arachis hypogaea;Fv:草莓Fragaria vesca
Fig. 3MicroR160s evolution analysis and mature sequence alignment
2.4 vvi-miR160s靶基因的预测
利用在线软件psRNATarget预测vvi-miR160s的靶基因(图4)。结果发现,vvi-miR160家族不同成员均有相同的靶基因VvARF18(VIT213s0019g04380)、VvARF17(VIT218s0001g04180)、VvARF18-like(VIT208s0040g01810)。其中,vvi-miR160s与VvARF18的错配率为2.5,与VvARF17和VvARF8-like的错配率为3。鉴于前期已有vvi-miR160s靶基因VvARF17和VvARF8-like的研究[21],因此,本研究重点对错配率最低的靶基因VvARF18开展研究,分析vvi-miR160s介导VvARF18的调控作用。图4
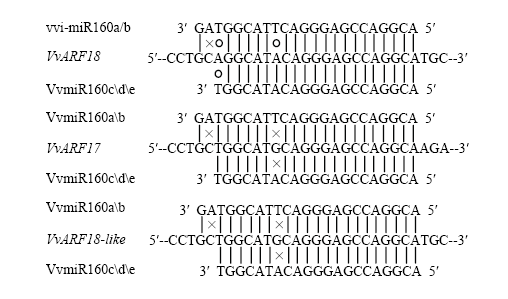 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4vvi-miR160s与靶基因互补错配
×:错配为1,○:错配为0.5
Fig. 4vvi-miR160s and target gene complementary mismatch
×:Mismatch is 1, ○: Mismatch is 0.5
2.5 靶基因VvARF18的序列及潜在功能分析
2.5.1 VvARF18序列结构分析 VvARF18共编码683个氨基酸。VvARF18蛋白分子式为C3327H5169N939O1003S28,分子质量为75 268.01 Da,理论等电点pI为6.43;丙氨酸的含量最高为9.5%,其次为亮氨酸占8.9%,最低为色氨酸,仅1.6%;不稳定指数48.84,为不稳定蛋白(40以下为稳定蛋白),总平均输水指数为-0.388,表明其为亲水性蛋白。VvARF18蛋白有3个结构域,分别为B3结构域(117—218位氨基酸)、生长素响应结构域(279—362位氨基酸)和AUX-IAA结构域(604—681位氨基酸)。结构分析发现,蛋白的第398—411位存在核定位信号(PPRKKLRLQQQSEF)(图5-A)。亚细胞定位预测结果也表明,其主要定位于细胞核上。图5
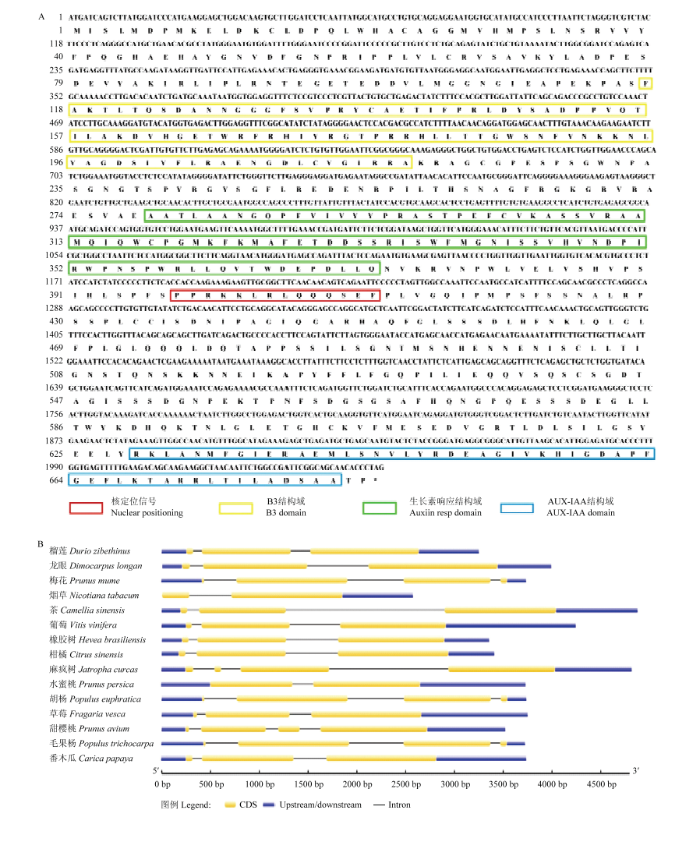 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5ARF序列结构分析
Fig. 5ARF gene sequence structure analysis
通过基因结构分析发现,不同植物中ARF18的内含子数量从2—4个不等(图5-B),其中葡萄与榴莲(Durio zibethinus)、龙眼(Dimocarpus longan)、番木瓜(Carica papaya)、草莓、茶(Camellia sinensis)、橡胶树(Hevea brasiliensis)基因结构相似(均含有2个内含子、3个外显子),且每个外显子长度和分布相似,而内含子长度差异较大。特殊的是,烟草ARF18没有5′ UTR序列。这些结果表明,ARF18结构较为保守,但在进化的过程中表现出一定的多样性。
2.5.2 ARF18蛋白进化分析 利用NCBI数据库,对葡萄ARF18蛋白序列进行同源比较,使用MEGA6.0软件构建系统进化树(图6)。结果发现,VvARF18蛋白与茶、烟草、梅花等物种的亲缘关系较进,与毛白杨、番木瓜、甜樱桃、草莓等物种亲缘关系较远。
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6ARF18蛋白进化分析
Fig. 6ARF18 protein evolution analysis and gene structure analysis
2.5.3 ARF18蛋白作用元件分析 利用在线软件MEME对ARF18蛋白进行作用元件motif分析(图7)。结果表明,VvARF18蛋白和其他14个物种ARF蛋白作用元件的数量和排列顺序完全相同,且这15个motif作用元件保守性也非常高,表明VvARF18蛋白与其他物种氨基酸序列保守性较高,功能相似。
图7
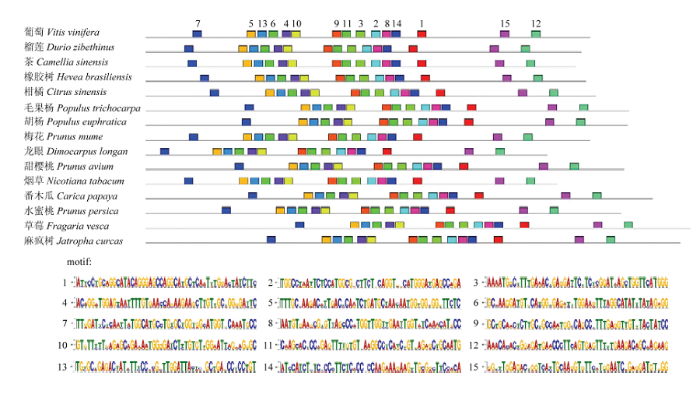 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7ARF18蛋白作用元件
Fig. 7ARF18 protein action element
2.6 VvARF18启动子顺式作用元件分析
利用在线软件Plant CARE对VvARF18启动子分析发现,在VvARF18启动子中,除了含有基本的作用元件CAAT-box、TATA-box外,其余作用元件可以分为4类:光响应作用元件如3-AF1 binding site、AE-box、Box 4、G-box、GT1-motif;胁迫相关作用元件如厌氧诱导元件(ARE)、低温诱导元件(LRT);而MYBHv1结合位点(CCAAT-box)和MYB参与干旱诱导位点(MBS);在激素类响应元件中,有较多的激素类型,如脱落酸响应元件(ABRE)、生长素响应元件(AuxRR-core)、茉莉酸甲酯响应元件(CGTCA-motif和TGACG-motif)以及赤霉素响应元件(P-box)(表3),由此推测VvARF18可能参与不同激素信号通路。Table 3
表3
表3VvARF18基因启动子顺式作用元件
Table 3
| 元件类型 Component type | 相关元件 Related component | 数量 Number | 功能注释 Functional comment |
|---|---|---|---|
| 光响应元件 Photoresponsive element | 3-AF1 binding site | 1 | 光响应元件Photoresponsive element |
| AE-box | 1 | 光反应元件Photoresponsive element | |
| Box 4 | 7 | 光响应元件Photoresponsive element | |
| G-box | 2 | 光响应元件Photoresponsive element | |
| GT1-motif | 6 | 光响应元件Photoresponsive element | |
| 激素响应元件 Hormone response element | ABRE | 2 | 脱落酸响应元件Abscisic acid response element |
| AuxRR-core | 1 | 生长素响应元件Auxin response element | |
| CGTCA-motif | 1 | 茉莉酸甲酯响应元件Methyl jasmonate response element | |
| TGACG-motif | 1 | 茉莉酸甲酯响应元件Methyl jasmonate response element | |
| P-box | 1 | 赤霉素响应元件Gibberellin response element | |
| 胁迫相关元件 Stress related component | ARE | 3 | 厌氧诱导元件Anaerobic inducing element |
| LRT | 1 | 低温诱导元件Low temperature inducing element | |
| 结合位点 Bonding component | CCAAT-box | 1 | MYBHv1结合位点MYBHv1 binding site |
| MBS | 2 | MYB参与干旱诱导位点MYB participates in drought induction sites |
新窗口打开|下载CSV
2.7 vvi-miR160s及其靶基因裂解验证
鉴于植物miRNA主要通过介导靶基因的裂解调控靶基因的表达,本研究利用5′-RLM-RACE和3′-PPM- RACE技术验证vvi-miR160s的靶基因VvARF18。RLM-RACE结果表明(图8),vvi-miR160a/b和vvi- miR160c/d/e均可裂解VvARF18,裂解位点位于miRNA 5′端的第10位,且裂解频度为9/17;PPM-RACE结果进一步证实,vvi-miR160s均可裂解靶基因VvARF18,其裂解位点相同于RLM-RACE结果,裂解频度为3/20,显著低于RLM-RACE结果,这可能是由于5′末端的裂解产物比3′末端产物更容易降解。该试验结果证明了VvARF18是vvi-miR160s家族真实的靶基因。图8
图8vvi-miR160s及其靶基因裂解验证
Fig. 8vvi-miR160s and its target gene fragmentation validation
2.8 vvi-miR160s和VvARF18在葡萄种子发育过程中的时空表达模式及其相关性
在葡萄种子发育过程中,vvi-miR160a/b的表达量在花后2—10 d呈上升趋势,而在20—40 d时,表达量较低,几乎不表达;vvi-miR160c/d/e的表达量在种子发育的整个时期呈先下降后上升的变化趋势,并且在花后20 d时,表达量最低;而VvARF18在种子发育的过程中呈先上升后下降的变化趋势,并在花后10 d表达量最高(图9-A)。图9
图9vvi-miR160s:VvARF18时空表达模式及其相关性
Fig. 9vvi-miR160s: VvARF18 spatiotemporal expression pattern and its correlation
对比vvi-miR160s和VvARF18的表达模式,发现vvi-miR160c/d/e与靶基因VvARF18呈现比较显著的负相关,而vvi-miR160a/b与VvARF18之间相关性较差(图9-B),表明在葡萄种子发育过程中,vvi-miR160家族可能主要通过vvi-miR160c/d/e介导VvARF18起调控作用,而vvi-miR160a/b在葡萄种子发育过程的调控作用可能较弱。
2.9 vvi-miR160s和VvARF18响应GA的时空表达模式及其相关性
通过外源赤霉素处理,研究vvi-miR160s及其靶基因VvARF18对葡萄种子发育的影响(图10-A)。结果表明,赤霉素处理使vvi-miR160a/b的表达呈上升趋势,尤其在花后20—40 d,表达差异达到极显著水平;相比之下,赤霉素处理对vvi-miR160c/d/e表达的影响并不太显著。有趣的是,赤霉素处理却显著上调了靶基因VvARF18在花后2—5 d葡萄胚珠中的表达,而在花后10—40 d,VvARF18的表达却被显著下调。对比赤霉素处理后vvi-miR160s与VvARF18表达水平的相关性(图10-B),发现在赤霉素处理诱导葡萄种子败育过程中,赤霉素处理诱导了vvi-miR160a/b与VvARF18表达水平的负相关,却减弱了vvi-miR160c/d/e与VvARF18表达水平的负相关,表明GA增强了vvi-miR160a/b对VvARF18的负调控作用,却减弱了vvi-miR160c/d/e对VvARF18的负调控作用,同时也表明在vvi-miR160家族中,vvi-miR160a/b可能是主要应答GA介导葡萄种子败育的调控因子。图10
图10vvi-miR160s和VvARF18响应GA在葡萄种子发育过程中的时空表达分析
Fig. 10vvi-miR160s and VvARF18 respond to GA regulation of grapevine nuclear development
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1073/pnas.0405570102URLPMID:15738428 [本文引用: 1]

In mammalian cells, the nuclear export receptor, Exportin 5 (Exp5), exports pre-microRNAs (pre-miRNAs) as well as tRNAs into the cytoplasm. In this study, we examined the function of HASTY (HST), the Arabidopsis ortholog of Exp5, in the biogenesis of miRNAs and tRNAs. In contrast to mammals, we found that miRNAs exist as single-stranded 20- to 21-nt molecules in the nucleus in Arabidopsis. This observation is consistent with previous studies indicating that proteins involved in miRNA biogenesis are located in the nucleus in Arabidopsis. Although miRNAs exist in the nucleus, a majority accumulate in the cytoplasm. Interestingly, loss-of-function mutations in HST reduced the accumulation of most miRNAs but had no effect on the accumulation of tRNAs and endogenous small interfering RNAs, or on transgene silencing. In contrast, a mutation in PAUSED (PSD), the Arabidopsis ortholog of the tRNA export receptor, Exportin-t, interfered with the processing of tRNA-Tyr but did not affect the accumulation or nuclear export of miRNAs. These results demonstrate that HST and PSD do not share RNA cargos in nuclear export and strongly suggest that there are multiple nuclear export pathways for these small RNAs in Arabidopsis.
DOI:10.1126/science.1159151URLPMID:18483398 [本文引用: 1]

High complementarity between plant microRNAs (miRNAs) and their messenger RNA targets is thought to cause silencing, prevalently by endonucleolytic cleavage. We have isolated Arabidopsis mutants defective in miRNA action. Their analysis provides evidence that plant miRNA-guided silencing has a widespread translational inhibitory component that is genetically separable from endonucleolytic cleavage. We further show that the same is true of silencing mediated by small interfering RNA (siRNA) populations. Translational repression is effected in part by the ARGONAUTE proteins AGO1 and AGO10. It also requires the activity of the microtubule-severing enzyme katanin, implicating cytoskeleton dynamics in miRNA action, as recently suggested from animal studies. Also as in animals, the decapping component VARICOSE (VCS)/Ge-1 is required for translational repression by miRNAs, which suggests that the underlying mechanisms in the two kingdoms are related.
[本文引用: 1]
DOI:10.1515/jib-2017-0002URLPMID:28637931 [本文引用: 1]

Increase in soil salt causes osmotic and ionic stress to plants, which inhibits their growth and productivity. Rice production is also hampered by salinity and the effect of salt is most severe at the seedling and reproductive stages. Salainity tolerance is a quantitative property controlled by multiple genes coding for signaling molecules, ion transporters, metabolic enzymes and transcription regulators. MicroRNAs are key modulators of gene-expression that act at the post-transcriptional level by translation repression or transcript cleavage. They also play an important role in regulating plant's response to salt-stress. In this work we adopted the approach of comparative and integrated data-mining to understand the miRNA-mediated regulation of salt-stress in rice. We profiled and compared the miRNA regulations using natural varieties and transgenic lines with contrasting behaviors in response to salt-stress. The information obtained from sRNAseq, RNAseq and degradome datasets was integrated to identify the salt-deregulated miRNAs, their targets and the associated metabolic pathways. The analysis revealed the modulation of many biological pathways, which are involved in salt-tolerance and play an important role in plant phenotype and physiology. The end modifications of the miRNAs were also studied in our analysis and isomiRs having a dynamic role in salt-tolerance mechanism were identified.
DOI:10.1016/S0070-2153(10)91012-0URLPMID:20705188 [本文引用: 1]

microRNAs (miRNAs) are small approximately 21-nucleotide RNAs that function posttranscriptionally to regulate gene activity. miRNAs function by binding to complementary sites in target genes causing mRNA degradation and/or translational repression of the target. Since the discovery of miRNAs in plants in 2002 much has been learned about the function of these small regulatory RNAs. miRNAs function broadly to control many aspects of plant biology and plant development. This review focuses on the role of miRNAs in flower development. miRNAs function throughout flower development, from the earliest stages (floral induction) to very late stages (floral organ cell type specification). miRNAs such as miR156 and miR172 play a key role in vegetative phase change and in the vegetative to reproductive transition in both Arabidopsis and maize. miR172 in Arabidopsis and maize and miR169 in Petunia and Antirrhinum function to control floral organ identity fate during the early stages of flower development by regulating the spatial boundaries of expression of target genes. miR164, miR319, miR159, and miR167 function to specify particular cell types during later stages of flower development. Although much has been learned about the role of miRNAs in flower development in the last 8 years, many challenges remain to fully elucidate the function of these important regulatory molecules.
DOI:10.1186/s12864-016-3372-0URLPMID:28068898 [本文引用: 1]

Drought stress is one of the most severe problem limited agricultural productivity worldwide. It has been reported that plants response to drought-stress by sophisticated mechanisms at both transcriptional and post-transcriptional levels. However, the precise molecular mechanisms governing the responses of tobacco leaves to drought stress and water status are not well understood. To identify genes and miRNAs involved in drought-stress responses in tobacco, we performed both mRNA and small RNA sequencing on tobacco leaf samples from the following three treatments: untreated-control (CL), drought stress (DL), and re-watering (WL).
DOI:10.1104/pp.113.220699URL [本文引用: 1]

Symbiotic root nodules in leguminous plants result from interaction between the plant and nitrogen-fixing rhizobia bacteria. There are two major types of legume nodules, determinate and indeterminate. Determinate nodules do not have a persistent meristem, while indeterminate nodules have a persistent meristem. Auxin is thought to play a role in the development of both these types of nodules. However, inhibition of rootward auxin transport at the site of nodule initiation is crucial for the development of indeterminate nodules but not determinate nodules. Using the synthetic auxin-responsive DR5 promoter in soybean ( Glycine max), we show that there is relatively low auxin activity during determinate nodule initiation and that it is restricted to the nodule periphery subsequently during development. To examine if and what role auxin plays in determinate nodule development, we generated soybean composite plants with altered sensitivity to auxin. We overexpressed microRNA393 to silence the auxin receptor gene family, and these roots were hyposensitive to auxin. These roots nodulated normally, suggesting that only minimal/reduced auxin signaling is required for determinate nodule development. We overexpressed microRNA160 to silence a set of repressor auxin response factor transcription factors, and these roots were hypersensitive to auxin. These roots were not impaired in epidermal responses to rhizobia but had significantly reduced nodule primordium formation, suggesting that auxin hypersensitivity inhibits nodule development. These roots were also hyposensitive to cytokinin and had attenuated expression of key nodulation-associated transcription factors known to be regulated by cytokinin. We propose a regulatory feedback loop involving auxin and cytokinin during nodulation.
DOI:10.1016/j.jplph.2014.09.006URLPMID:25462963 [本文引用: 1]

Cassava is a starchy root crop for food and industrial applications in many countries around the world. Among the factors that affect cassava production, diseases remain the major cause of yield loss. Cassava anthracnose disease is caused by the fungus Colletotrichum gloeosporioides. Severe anthracnose attacks can cause tip die-backs and stem cankers, which can affect the availability of planting materials especially in large-scale production systems. Recent studies indicate that plants over- or under-express certain microRNAs (miRNAs) to cope with various stresses. Understanding how a disease-resistant plant protects itself from pathogens should help to uncover the role of miRNAs in the plant immune system. In this study, the disease severity assay revealed different response to C. gloeosporioides infection in two cassava cultivars. Quantitative RT-PCR analysis uncovered the differential expression of the two miRNAs and their target genes in the two cassava cultivars that were subjected to fungal infection. The more resistant cultivar revealed the up-regulation of miR160 and miR393, and consequently led to low transcript levels in their targets, ARF10 and TIR1, respectively. The more susceptible cultivar exhibited the opposite pattern. The cis-regulatory elements relevant to defense and stress responsiveness, fungal elicitor responsiveness and hormonal responses were the most prevalent present in the miRNAs gene promoter regions. The possible dual role of these specific miRNAs and their target genes associated with cassava responses to C. gloeosporioides is discussed. This is the first study to address the molecular events by which miRNAs which might play a role in fungal-infected cassava. A better understanding of the functions of miRNAs target genes should greatly increase our knowledge of the mechanism underlying susceptibility and lead to new strategies to enhance disease tolerance in this economically important crop.
DOI:10.1111/j.1365-313X.2010.04164.xURLPMID:20136729 [本文引用: 1]

MicroRNAs (miRNAs) have emerged as key regulators of gene expression at the post-transcriptional level in both plants and animals. However, the specific functions of MIRNAs (MIRs) and the mechanisms regulating their expression are not fully understood. Previous studies showed that miR160 negatively regulates three genes that encode AUXIN RESPONSE FACTORs (ARF10, -16, and -17). Here, we characterized floral organs in carpels (foc), an Arabidopsis mutant with a Ds transposon insertion in the 3' regulatory region of MIR160a. foc plants exhibit a variety of intriguing phenotypes, including serrated rosette leaves, irregular flowers, floral organs inside siliques, reduced fertility, aberrant seeds, and viviparous seedlings. Detailed phenotypic analysis showed that abnormal cell divisions in the basal embryo domain and suspensor led to diverse defects during embryogenesis in foc plants. Further analysis showed that the 3' region was required for the expression of MIR160a. The accumulation of mature miR160 was greatly reduced in foc inflorescences. In addition, the expression pattern of ARF16 and -17 was altered during embryo development in foc plants. foc plants were also deficient in auxin responses. Moreover, auxin was involved in regulating the expression of MIR160a through its 3' regulatory region. Our study not only provides insight into the molecular mechanism of embryo development via MIR160a-regulated ARFs, but also reveals the mechanism regulating MIR160a expression.
DOI:10.3389/fpls.2017.02024URLPMID:29321785 [本文引用: 2]

MicroRNAs are non-coding small RNA molecules that are involved in the post-transcriptional regulation of the genes that control various developmental processes in plants, including zygotic embryogenesis (ZE). miRNAs are also believed to regulate somatic embryogenesis (SE), a counterpart of the ZE that is induced in vitro in plant somatic cells. However, the roles of specific miRNAs in the regulation of the genes involved in SE, in particular those encoding transcription factors (TFs) with an essential function during SE including LEAFY COTYLEDON2 (LEC2), remain mostly unknown. The aim of the study was to reveal the function of miR165/166 and miR160 in the LEC2-controlled pathway of SE that is induced in in vitro cultured Arabidopsis explants.In ZE, miR165/166 controls the PHABULOSA/PHAVOLUTA (PHB/PHV) genes, which are the positive regulators of LEC2, while miR160 targets the AUXIN RESPONSE FACTORS (ARF10, ARF16, ARF17) that control the auxin signaling pathway, which plays key role in LEC2-mediated SE. We found that a deregulated expression/function of miR165/166 and miR160 resulted in a significant accumulation of auxin in the cultured explants and the spontaneous formation of somatic embryos. Our results show that miR165/166 might contribute to SE induction via targeting PHB, a positive regulator of LEC2 that controls embryogenic induction via activation of auxin biosynthesis pathway (Wójcikowska et al., 2013). Similar to miR165/166, miR160 was indicated to control SE induction through auxin-related pathways and the negative impact of miR160 on ARF10/ARF16/ARF17 was shown in an embryogenic culture. Altogether, the results suggest that the miR165/166- and miR160-node contribute to the LEC2-mediated auxin-related pathway of embryogenic transition that is induced in the somatic cells of Arabidopsis. A model summarizing the suggested regulatory interactions between the miR165/166-PHB and miR160-ARF10/ARF16/ARF17 nodes that control SE induction in Arabidopsis was proposed.
DOI:10.1105/tpc.105.031716URLPMID:15829600 [本文引用: 2]

The phytohormone auxin plays critical roles during plant growth, many of which are mediated by the auxin response transcription factor (ARF) family. MicroRNAs (miRNAs), endogenous 21-nucleotide riboregulators, target several mRNAs implicated in auxin responses. miR160 targets ARF10, ARF16, and ARF17, three of the 23 Arabidopsis thaliana ARF genes. Here, we describe roles of miR160-directed ARF17 posttranscriptional regulation. Plants expressing a miRNA-resistant version of ARF17 have increased ARF17 mRNA levels and altered accumulation of auxin-inducible GH3-like mRNAs, YDK1/GH3.2, GH3.3, GH3.5, and DFL1/GH3.6, which encode auxin-conjugating proteins. These expression changes correlate with dramatic developmental defects, including embryo and emerging leaf symmetry anomalies, leaf shape defects, premature inflorescence development, altered phyllotaxy along the stem, reduced petal size, abnormal stamens, sterility, and root growth defects. These defects demonstrate the importance of miR160-directed ARF17 regulation and implicate ARF17 as a regulator of GH3-like early auxin response genes. Many of these defects resemble phenotypes previously observed in plants expressing viral suppressors of RNA silencing and plants with mutations in genes important for miRNA biogenesis or function, providing a molecular rationale for phenotypes previously associated with more general disruptions of miRNA function.
DOI:10.1105/tpc.008417URLPMID:12566590 [本文引用: 1]

Auxin response factors (ARFs) are transcription factors that bind to TGTCTC auxin response elements in promoters of early auxin response genes. ARFs have a conserved N-terminal DNA binding domain (DBD) and in most cases a conserved C-terminal dimerization domain (CTD). The ARF CTD is related in amino acid sequence to motifs III and IV found in Aux/IAA proteins. Just C terminal to the DBD, ARFs contain a nonconserved region referred to as the middle region (MR), which has been proposed to function as a transcriptional repression or activation domain. Results with transfected protoplasts reported here show that ARFs with Q-rich MRs function as activators, whereas most, if not all other ARFs, function as repressors. ARF DBDs alone are sufficient to recruit ARFs to their DNA target sites, and auxin does not influence this recruitment. ARF MRs alone function as activation or repression domains when targeted to reporter genes via a yeast Gal4 DBD, and auxin does not influence the potency of activation or repression. ARF CTDs, along with a Q-rich MR, are required for an auxin response whether the MRs plus CTDs are recruited to a promoter by an ARF DBD or by a Gal4 DBD. The auxin response is mediated by the recruitment of Aux/IAA proteins to promoters that contain a DNA binding protein with a Q-rich MR and an attached CTD.
DOI:10.1038/35104500URLPMID:11713520 [本文引用: 1]

The plant hormone auxin is central in many aspects of plant development. Previous studies have implicated the ubiquitin-ligase SCF(TIR1) and the AUX/IAA proteins in auxin response. Dominant mutations in several AUX/IAA genes confer pleiotropic auxin-related phenotypes, whereas recessive mutations affecting the function of SCF(TIR1) decrease auxin response. Here we show that SCF(TIR1) is required for AUX/IAA degradation. We demonstrate that SCF(TIR1) interacts with AXR2/IAA7 and AXR3/IAA17, and that domain II of these proteins is necessary and sufficient for this interaction. Further, auxin stimulates binding of SCF(TIR1) to the AUX/IAA proteins, and their degradation. Because domain II is conserved in nearly all AUX/IAA proteins in Arabidopsis, we propose that auxin promotes the degradation of this large family of transcriptional regulators, leading to diverse downstream effects.
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s11103-012-9883-4URL [本文引用: 1]

Auxin response factors (ARFs) are plant transcription factors that activate or repress the expression of auxin-responsive genes and accordingly, play key roles in auxin-mediated developmental processes. Here we identified and characterized the Solanum lycopersicum (tomato) ARF10 homolog (SlARF10), demonstrated that it is post-transcriptionally regulated by Sl-miR160, and investigated the significance of this regulation for tomato development. In wild-type tomato, SlARF10 is primarily expressed in the pericarp of mature and ripened fruit, showing an expression profile complementary to that of Sl-miR160. Constitutive expression of wild-type SlARF10 did not alter tomato development. However, transgenic tomato plants that constitutively expressed the Sl-miR160a-resistant version (mSlARF10) developed narrow leaflet blades, sepals and petals, and abnormally shaped fruit. During compound leaf development, mSlARF10 accumulation specifically inhibited leaflet blade outgrowth without affecting other auxin-driven processes such as leaflet initiation and lobe formation. Moreover, blade size was inversely correlated with mSlARF10 transcript levels, strongly implying that the SlARF10 protein, which was localized to the nucleus, can function as a transcriptional repressor of leaflet lamina outgrowth. Accordingly, known auxin-responsive genes, which promote cell growth, were downregulated in shoot apices that accumulated increased mSlARF10 levels. Taken together, we propose that repression of SlARF10 by Sl-miR160 is essential for auxin-mediated blade outgrowth and early fruit development.
DOI:10.1111/j.1365-313X.2007.03218.xURLPMID:17672844 [本文引用: 1]

AUXIN RESPONSE FACTORS (ARFs) are transcription factors involved in auxin signal transduction during many stages of plant growth development. ARF10, ARF16 and ARF17 are targeted by microRNA160 (miR160) in Arabidopsis thaliana. Here, we show that negative regulation of ARF10 by miR160 plays important roles in seed germination and post-germination. Transgenic plants expressing an miR160-resistant form of ARF10, which has silent mutations in the miRNA target site (termed mARF10), exhibited developmental defects such as serrated leaves, curled stems, contorted flowers and twisted siliques. These phenotypes were not observed in wild-type plants or plants transformed with the targeted ARF10 gene. During sensu stricto germination and post-germination, mARF10 mutant seeds and plants were hypersensitive to ABA in a dose-dependent manner. ABA hypersensitivity was mimicked in wild-type plants by exogenous auxin. In contrast, overexpression of MIR160 (35S:MIR160) resulted in reduced sensitivity to ABA during germination. Transcriptome analysis of germinating ARF10 and mARF10 seeds indicated that typical ABA-responsive genes expressed during seed maturation were overexpressed in germinating mARF10 seeds. These results indicate that negative regulation of ARF10 by miR160 plays a critical role in seed germination and post-embryonic developmental programs, at least in part by mechanisms involving interactions between ARF10-dependent auxin and ABA pathways.
DOI:10.1093/jxb/erq293URLPMID:20937732 [本文引用: 1]

Transgenic tomato plants (Solanum lycopersicum L.) with reduced mRNA levels of AUXIN RESPONSE FACTOR 7 (SlARF7) form parthenocarpic fruits with morphological characteristics that seem to be the result of both increased auxin and gibberellin (GA) responses during fruit growth. This paper presents a more detailed analysis of these transgenic lines. Gene expression analysis of auxin-responsive genes show that SlARF7 may regulate only part of the auxin signalling pathway involved in tomato fruit set and development. Also, part of the GA signalling pathway was affected by the reduced levels of SlARF7 mRNA, as morphological and molecular analyses display similarities between GA-induced fruits and fruits formed by the RNAi SlARF7 lines. Nevertheless, the levels of GAs were strongly reduced compared with that in seeded fruits. These findings indicate that SlARF7 acts as a modifier of both auxin and gibberellin responses during tomato fruit set and development.
DOI:10.1104/pp.106.084871URLPMID:16905669 [本文引用: 1]

Auxin and gibberellins (GAs) overlap in the regulation of multiple aspects of plant development, such as root growth and organ expansion. This coincidence raises questions about whether these two hormones interact to regulate common targets and what type of interaction occurs in each case. Auxins induce GA biosynthesis in a range of plant species. We have undertaken a detailed analysis of the auxin regulation of expression of Arabidopsis (Arabidopsis thaliana) genes encoding GA 20-oxidases and GA 3-oxidases involved in GA biosynthesis, and GA 2-oxidases involved in GA inactivation. Our results show that auxin differentially up-regulates the expression of various genes involved in GA metabolism, in particular several AtGA20ox and AtGA2ox genes. Up-regulation occurred very quickly after auxin application; the response was mimicked by incubations with the protein synthesis inhibitor cycloheximide and was blocked by treatments with the proteasome inhibitor MG132. The effects of auxin treatment reflect endogenous regulation because equivalent changes in gene expression were observed in the auxin overproducer mutant yucca. The results suggest direct regulation of the expression of GA metabolism genes by Aux/IAA and ARF proteins. The physiological relevance of this regulation is supported by the observation that the phenotype of certain gain-of-function Aux/IAA alleles could be alleviated by GA application, which suggests that changes in GA metabolism mediate part of auxin action during development.
DOI:10.1111/j.1365-313X.2008.03781.xURLPMID:19207215 [本文引用: 2]

Fruit development is usually triggered by ovule fertilization, and it requires coordination between seed development and the growth and differentiation of the ovary to host the seeds. Hormones are known to synchronize these two processes, but the role of each hormone, and the mechanism by which they interact, are still unknown. Here we show that auxin and gibberellins (GAs) act in a hierarchical scheme. The synthetic reporter construct DR5:GFP showed that fertilization triggered an increase in auxin response in the ovules, which could be mimicked by blocking polar auxin transport. As the application of GAs did not affect auxin response, the most likely sequence of events after fertilization involves auxin-mediated activation of GA synthesis. We have confirmed this, and have shown that GA biosynthesis upon fertilization is localized specifically in the fertilized ovules. Furthermore, auxin treatment caused changes in the expression of GA biosynthetic genes similar to those triggered by fertilization, and also restricted to the ovules. Finally, GA signaling was activated in ovules and valves, as shown by the rapid downregulation of the fusion protein RGA-GFP after pollination and auxin treatment. Taken together, this evidence suggests a model in which fertilization would trigger an auxin-mediated promotion of GA synthesis specifically in the ovule. The GAs synthesized in the ovules would be then transported to the valves to promote GA signaling and thus coordinate growth of the silique.
DOI:10.1111/j.1399-3054.2011.01481.xURLPMID:21496033 [本文引用: 1]

MicroRNAs (miRNAs) are a class of non-coding RNA molecules which have significant gene regulation roles in organisms. The advent of new high-throughput sequencing technologies has enabled the discovery of novel miRNAs. Although there are two recent reports on high-throughput sequencing analysis of small RNA libraries from different organs of two wine grapevine varieties, there was a significant divergence in the number and kinds of miRNAs sequenced in these studies. More sequencing of small RNA libraries is still important for the discovery of novel miRNAs in grapevine. In this study, a total of 130 conserved grapevine Vitis vinifera miRNA (Vv-miRNA) belonging to 28 Vv-miRNA families were validated, other 80 unconserved Vv-miRNAs including 72 novel potential and 8 known but unconserved ones were found. Fifty-two (52.5%) of these 80 unconserved Vv-miRNAs exhibited differential poly(A)-tailed reverse transcriptase-polymerase chain reaction expression profiles in various grapevine tissues that could further confirm their existence in grapevine, among which 20 were expressed only in grapevine berries, indicating a degree of fruit-specificity. One hundred thirty target genes for 56 unconserved miRNAs could be predicted. The locations of these potential target genes on grapevine chromosomes and their complementary levels with the corresponding miRNAs were also analyzed. These results point to a regulatory role of miRNAs in grapevine berry development and response to various environments.
DOI:10.1186/s12870-019-1719-9URLPMID:30898085 [本文引用: 3]

Grape (Vitis vinifera) is highly sensitive to gibberellin (GA), which effectively induce grape parthenocarpy. Studies showed that miR160s and their target AUXIN RESPONSIVE FACTOR (ARF) responding hormones are indispensable for various aspects of plant growth and development, but their functions in GA-induced grape parthenocarpy remain elusive.
[本文引用: 1]
DOI:10.1371/journal.pone.0080044URLPMID:24224035 [本文引用: 1]

The phytohormone gibberellic acid (GA3) is widely used in the table grape industry to induce seedlessness in seeded varieties. However, there is a paucity of information concerning the mechanisms by which GAs induce seedlessness in grapes.
DOI:10.1093/jxb/ert453URLPMID:24474808 [本文引用: 1]

The onset of flowering in plants is regulated by complex gene networks that integrate multiple environmental and endogenous cues to ensure that flowering occurs at the appropriate time. This is achieved by precise control of the expression of key flowering genes at both the transcriptional and post-transcriptional level. In recent years, a class of small non-coding RNAs, called microRNAs (miRNAs), has been shown to regulate gene expression in a number of plant developmental processes and stress responses. MiRNA-based biotechnology, which harnesses the regulatory functions of such endogenous or artificial miRNAs, therefore represents a highly promising area of research. In this review, the process of plant miRNA biogenesis, their mode of action, and multiple regulatory functions are summarized. The roles of the miR156, miR172, miR159/319, miR390, and miR399 families in the flowering time regulatory network in Arabidopsis thaliana are discussed in depth.
DOI:10.1016/j.ydbio.2013.05.009URLPMID:23707900 [本文引用: 1]

MicroRNAs (miRNAs) are post-transcriptional regulators of growth and development in both plants and animals. Flowering is critical for the reproduction of angiosperms. Flower development entails the transition from vegetative growth to reproductive growth, floral organ initiation, and the development of floral organs. These developmental processes are genetically regulated by miRNAs, which participate in complex genetic networks of flower development. A survey of the literature shows that miRNAs, their specific targets, and the regulatory programs in which they participate are conserved throughout the plant kingdom. This review summarizes the role of miRNAs and their targets in the regulation of gene expression during the floral developmental phase, which includes the floral transition stage, followed by floral patterning, and then the development of floral organs. The conservation patterns observed in each component of the miRNA regulatory system suggest that these miRNAs play important roles in the evolution of flower development.
DOI:10.1242/dev.01206URLPMID:15226253 [本文引用: 1]

Floral initiation and floral organ development are both regulated by the phytohormone gibberellin (GA). For example, in short-day photoperiods, the Arabidopsis floral transition is strongly promoted by GA-mediated activation of the floral meristem-identity gene LEAFY. In addition, anther development and pollen microsporogenesis depend on GA-mediated opposition of the function of specific members of the DELLA family of GA-response repressors. We describe the role of a microRNA (miR159) in the regulation of short-day photoperiod flowering time and of anther development. MiR159 directs the cleavage of mRNA encoding GAMYB-related proteins. These proteins are transcription factors that are thought to be involved in the GA-promoted activation of LEAFY, and in the regulation of anther development. We show that miR159 levels are regulated by GA via opposition of DELLA function, and that both the sequence of miR159 and the regulation of miR159 levels by DELLA are evolutionarily conserved. Finally, we describe the phenotypic consequences of transgenic over-expression of miR159. Increased levels of miR159 cause a reduction in LEAFY transcript levels, delay flowering in short-day photoperiods, and perturb anther development. We propose that miR159 is a phytohormonally regulated homeostatic modulator of GAMYB activity, and hence of GAMYB-dependent developmental processes.
[本文引用: 2]
[本文引用: 2]
DOI:10.1007/s00438-018-1462-1URLPMID:29943289 [本文引用: 1]

SPL is a plant-specific transcription factor family. Many researchers reported that SPL members targeted by miR156s could play crucial roles in the modulation of plant growth and development. Although there are similar reports on grapes, till now little is known about grape berry development and ripening. To gain more insight into how grape miR156s (Vv-miR156s) modulated the above given processes of grape berries by mediating their target gene Vv-SPLs, here we identified the precise sequences of Vv-miR156s in 'Giant Rose' grape berries, predicted their potential targets, and revealed that the matching degree of various Vv-miR156: Vv-SPL pairs exhibited some discrepancy, implying the divergence of their interaction. Subsequently, we also discovered similar motifs such as ABRE, CGTCA and ERE, which are more specific to berry development and ripening, within the promoters of both Vv-MIR156s and Vv-SPLs. With berry development and ripening, meanwhile, Vv-miR156a, b/c/d, e and f/g/i exhibited an overall increasing expression trend, while their targets showed opposite trends at the corresponding stages. Additionally, exogenous ABA and NAA application promoted or curbed the expression of Vv-miR156s to some extent, before grape berry ripening stage. The cleavage products, sites and frequencies of Vv-miR156a, b/c/d, e, f/g/i and their respective targets (Vv-SPL2, 9, 10, 16) during grape berry development and ripening process were validated by our developed PPM-RACE and modified RLM-RACE together with qRT-PCR, which demonstrated that Vv-miR156s can be involved in the modulation of grape berry development and ripening process by mediating the expression of Vv-SPL2, 9, 10, 16. Our findings lay an important foundation for further recognizing their functions in grape berries, and enrich the knowledge of the regulatory mechanism of miRNA-mediated grape berry development and ripening.
DOI:10.1111/j.1399-3054.2010.01411.xURLPMID:20875055 [本文引用: 1]

Thirty-one potential miRNAs that belong to 16 miRNA families were discovered from more than 324 000 EST sequences of apple (Malus domestica). In addition, precise sequences, especially terminal nucleotides of the 16 apple miRNAs (mdo-miRNAs) in 16 families were validated by miR-RACE, a newly developed method for the determination of the potential miRNAs predicted computationally. The expression of these 16 microRNAs could be detected in apple young leaf, old leaf, young stem, flower bud, flower and developing fruits by quantitative RT-PCR (qRT-PCR) and some of them showed tissue-specific expression. Fifty-six potential targets were identified for the 16 apple miRNAs, most of which were transcription factors that play important roles in apple development. Twelve target genes were experimentally verified by qRT-PCR, with some exhibiting different expression trends from their corresponding microRNAs, indicating the cleavage mode of miRNAs on their target genes.
DOI:10.1007/s00425-009-0971-xURLPMID:19585144 [本文引用: 1]

MicroRNAs (miRNAs) are a class of non-protein-coding small RNAs. Considering the conservation of many miRNA genes in different plant genomes, the identification of miRNAs from non-model organisms is both practicable and instrumental in addressing miRNA-guided gene regulation. Citrus is an important staple fruit tree, and publicly available expressed sequence tag (EST) database for citrus are increasing. However, until now, little has been known about miRNA in citrus. In this study, 27 known miRNAs from Arabidopsis were searched against citrus EST databases for miRNA precursors, of which 13 searched precursor sequences could form fold-back structures similar with those of Arabidopsis. The ubiquitous expression of those 13 citrus microRNAs and other 13 potential citrus miRNAs could be detected in citrus leaf, young shoot, flower, fruit and root by northern blotting, and some of them showed differential expression in different tissues. Based on the fact that miRNAs exhibit perfect or nearly perfect complementarity with their target sequences, a total of 41 potential targets were identified for 15 citrus miRNAs. The majority of the targets are transcription factors that play important roles in citrus development, including leaf, shoot, and root development. Additionally, some other target genes appear to play roles in diverse physiological processes. Four target genes have been experimentally verified by detection of the miRNA-mediated mRNA cleavage in Poncirus trifoliate. Overall, this study in the identification and characterization of miRNAs in citrus can initiate further study on citrus miRNA regulation mechanisms, and it can help us to know more about the important roles of miRNAs in citrus.
DOI:10.1371/journal.pone.0070959URLPMID:23990918 [本文引用: 1]

In non-climacteric fruits, the respiratory increase is absent and no phytohormone is appearing to be critical for their ripening process. They must remain on the parent plant to enable full ripening and be picked at or near the fully ripe stage to obtain the best eating quality. However, huge losses often occur for their quick post-harvest senescence. To understanding the complex mechanism of non-climacteric fruits post-harvest senescence, we constructed two small RNA libraries and one degradome from strawberry fruit stored at 20°C for 0 and 24 h. A total of 88 known and 1224 new candidate miRNAs, and 103 targets cleaved by 19 known miRNAs families and 55 new candidatemiRNAs were obtained. These targets were associated with development, metabolism, defense response, signaling transduction and transcriptional regulation. Among them, 14 targets, including NAC transcription factor, Auxin response factors (ARF) and Myb transcription factors, cleaved by 6 known miRNA families and 6 predicted candidates, were found to be involved in regulating fruit senescence. The present study provided valuable information for understanding the quick senescence of strawberry fruit, and offered a foundation for studying the miRNA-mediated senescence of non-climacteric fruits.
DOI:10.1186/1471-2164-15-111URL [本文引用: 1]
DOI:10.1093/jhered/esw030URLPMID:27660497 [本文引用: 1]

miR156 regulates the expression of its target SPL (PROMOTER BINDING-LIKE) genes during flower and fruit development, diverse developmental stage transitions, especially from vegetative to reproductive growth phases, by cleaving the target mRNA SPL of one plant-specific transcription factor. However, systematic reports on grapevine have yet to be presented. Here, the precise sequence of miR156 (vvi-miR156b/c/d) in grapevine "Takatsuma" was cloned with a previously cloned grapevine SPL (Vv-SPL9). Expression profiles in 18 grapevine tissues were identified through stem-loop RT-PCR. The interaction mode between vvi-miR156b/c/d and Vv-SPL9 was further validated by detecting the cleavage site and cleavage products of 3'- and 5'-ends via an integrated approach of 5'-RLM-RACE (RNA ligase-mediated 5'-rapid amplification of cDNA ends), 3'-PPM-RACE (poly(A) polymerase-mediated 3'-rapid amplification of cDNA ends), and qRT-PCR (real time reverse transcriptase-polymerase chain reaction). The variation in their cleavage roles in the whole growth stage of grapevine was also systematically investigated. Results showed that vvi-miR156b/c/d exhibited typical temporal-spatial-specific expression levels. The expression levels were higher in vegetative organs, such as leaf, than in reproductive organs, such as tendrils, flowers, and berries. A significant variation was observed during vegetative-to-reproductive transition. The expression patterns of Vv-SPL9 showed the opposite trends with those of vvi-miR156b. We confirmed that the cleavage site was at the 10th site of vvi-miR156b/c/d complementary to Vv-SPL9 in "Takatsuma" grapevine. We also identified the temporal-spatial variation of the cleavage products. This variation can indicate the regulatory function of miR156 on SPL in grapevines. Our findings provide further insights into the functions of vvi-miR156b/c/d and its target Vv-SPL9, and also help enrich our knowledge of small RNA-mediated regulation in grapevine.
DOI:10.1016/j.scib.2017.03.013URL [本文引用: 1]
DOI:10.1038/srep35675URLPMID:27762296 [本文引用: 1]

Recently, our transcriptomic analysis has identified some functional genes responsible for oil biosynthesis in developing SASK, yet miRNA-mediated regulation for SASK development and oil accumulation is poorly understood. Here, 3 representative periods of 10, 30 and 60 DAF were selected for sRNA sequencing based on the dynamic patterns of growth tendency and oil content of developing SASK. By miRNA transcriptomic analysis, we characterized 296 known and 44 novel miRNAs in developing SASK, among which 36 known and 6 novel miRNAs respond specifically to developing SASK. Importantly, we performed an integrated analysis of mRNA and miRNA transcriptome as well as qRT-PCR detection to identify some key miRNAs and their targets (miR156-SPL, miR160-ARF18, miR164-NAC1, miR171h-SCL6, miR172-AP2, miR395-AUX22B, miR530-P2C37, miR393h-TIR1/AFB2 and psi-miRn5-SnRK2A) potentially involved in developing response and hormone signaling of SASK. Our results provide new insights into the important regulatory function of cross-talk between development response and hormone signaling for SASK oil accumulation.
[本文引用: 1]
DOI:10.1111/j.1365-313X.2010.04164.xURLPMID:20136729 [本文引用: 1]

MicroRNAs (miRNAs) have emerged as key regulators of gene expression at the post-transcriptional level in both plants and animals. However, the specific functions of MIRNAs (MIRs) and the mechanisms regulating their expression are not fully understood. Previous studies showed that miR160 negatively regulates three genes that encode AUXIN RESPONSE FACTORs (ARF10, -16, and -17). Here, we characterized floral organs in carpels (foc), an Arabidopsis mutant with a Ds transposon insertion in the 3' regulatory region of MIR160a. foc plants exhibit a variety of intriguing phenotypes, including serrated rosette leaves, irregular flowers, floral organs inside siliques, reduced fertility, aberrant seeds, and viviparous seedlings. Detailed phenotypic analysis showed that abnormal cell divisions in the basal embryo domain and suspensor led to diverse defects during embryogenesis in foc plants. Further analysis showed that the 3' region was required for the expression of MIR160a. The accumulation of mature miR160 was greatly reduced in foc inflorescences. In addition, the expression pattern of ARF16 and -17 was altered during embryo development in foc plants. foc plants were also deficient in auxin responses. Moreover, auxin was involved in regulating the expression of MIR160a through its 3' regulatory region. Our study not only provides insight into the molecular mechanism of embryo development via MIR160a-regulated ARFs, but also reveals the mechanism regulating MIR160a expression.
DOI:10.1111/tpj.13127URLPMID:26800988 [本文引用: 1]

Plant microRNAs play vital roles in auxin signaling via the negative regulation of auxin response factors (ARFs). Studies have shown that targeting of ARF10/16/17 by miR160 is indispensable for various aspects of development, but its functions in the model crop tomato (Solanum lycopersicum) are unknown. Here we knocked down miR160 (sly-miR160) using a short tandem target mimic (STTM160), and investigated its roles in tomato development. Northern blot analysis showed that miR160 is abundant in developing ovaries. In line with this, its down-regulation perturbed ovary patterning as indicated by the excessive elongation of the proximal ends of mutant ovaries and thinning of the placenta. Following fertilization, these morphological changes led to formation of elongated, pear-shaped fruits reminiscent of those of the tomato ovate mutant. In addition, STTM160-expressing plants displayed abnormal floral organ abscission, and produced leaves, sepals and petals with diminished blades, indicating a requirement for sly-miR160 for these auxin-mediated processes. We found that sly-miR160 depletion was always associated with the up-regulation of SlARF10A, SlARF10B and SlARF17, of which the expression of SlARF10A increased the most. Despite the sly-miR160 legitimate site of SlARF16A, its mRNA levels did not change in response to sly-miR160 down-regulation, suggesting that it may be regulated by a mechanism other than mRNA cleavage. SlARF10A and SlARF17 were previously suggested to function as inhibiting ARFs. We propose that by adjusting the expression of a group of ARF repressors, of which SlARF10A is a primary target, sly-miR160 regulates auxin-mediated ovary patterning as well as floral organ abscission and lateral organ lamina outgrowth.
