 ,, 杨官显, 张静, 邹琦, 王意程, 曲常志, 姜生辉, 王楠, 陈学森山东农业大学园艺科学与工程学院/作物生物学国家重点实验室,山东泰安 271018
,, 杨官显, 张静, 邹琦, 王意程, 曲常志, 姜生辉, 王楠, 陈学森山东农业大学园艺科学与工程学院/作物生物学国家重点实验室,山东泰安 271018Molecular Mechanism of Apple MdWRKY18 and MdWRKY40 Participating in Salt Stress
XU HaiFeng ,, YANG GuanXian, ZHANG Jing, ZOU Qi, WANG YiCheng, QU ChangZhi, JIANG ShengHui, WANG Nan, CHEN XueSenCollege of Horticulture Science and Engineering, Shandong Agricultural University/State Key Laboratory of Crop Biology, Tai’an 271018, Shandong
,, YANG GuanXian, ZHANG Jing, ZOU Qi, WANG YiCheng, QU ChangZhi, JIANG ShengHui, WANG Nan, CHEN XueSenCollege of Horticulture Science and Engineering, Shandong Agricultural University/State Key Laboratory of Crop Biology, Tai’an 271018, Shandong第一联系人:
责任编辑: 李莉
收稿日期:2018-05-25接受日期:2018-07-26网络出版日期:2018-12-01
| 基金资助: |
Received:2018-05-25Accepted:2018-07-26Online:2018-12-01

摘要
关键词:
Abstract
Keywords:
PDF (1795KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
许海峰, 杨官显, 张静, 邹琦, 王意程, 曲常志, 姜生辉, 王楠, 陈学森. 苹果MdWRKY18和MdWRKY40参与盐胁迫途径分子机理研究[J]. 中国农业科学, 2018, 51(23): 4514-4521 doi:10.3864/j.issn.0578-1752.2018.23.010
XU HaiFeng, YANG GuanXian, ZHANG Jing, ZOU Qi, WANG YiCheng, QU ChangZhi, JIANG ShengHui, WANG Nan, CHEN XueSen.
0 引言
【研究意义】土壤盐渍化是对植物生长和作物产量产生不利影响的主要非生物胁迫之一[1,2],了解植物响应盐胁迫信号及抗盐胁迫反应的机制,对于提高作物抗盐能力至关重要。因此,研究WRKY家族中MdWRKY18和MdWRKY40在盐胁迫中的功能,对完善盐胁迫代谢机理具有重要意义。【前人研究进展】许多物理和化学方法能够提高植物的耐盐性,然而越来越多的研究都集中在培育新的耐盐品种,目前,耐盐机制在很多作物(玉米[3]、水稻[4,5]、小麦[6]、大豆[7]、草莓[8]、葡萄[9]和苹果[10]等)中得到广泛研究。在植物细胞中,盐胁迫会增加Na+和Cl-浓度,干扰K+浓度,导致离子毒性,影响离子稳态和代谢紊乱,植物通过自身生理和生化变化来抵抗盐胁迫[11,12],如,质膜上的Na+/H+逆转蛋白SOS1,定位于液泡膜的Na+/H+交换蛋白NHX1[13]。此外,越来越多的转录因子参与盐胁迫代谢,包括MYB家族、bHLH家族、bZIP家族和WRKY家族[14,15,16,17],其中,AtbHLH92能够影响活性氧介导的信号途径适应高盐胁迫[18],OsMYB91可以协调水稻的植物生长和耐盐性[5]。【本研究切入点】WRKY家族在植物非生物胁迫代谢调控中发挥重要作用,OsWRKY11能够提高水稻对热胁迫和干旱的耐性[19],AtWRKY57参与拟南芥中ABA和干旱胁迫的响应[20],AtWRKY25和AtWRKY33参与拟南芥中盐胁迫代谢调控[21],但苹果WRKY家族参与盐胁迫代谢的研究尚未见报道。【拟解决的关键问题】本研究通过克隆MdWRKY18和MdWRKY40,对其蛋白结构进行分析;并采用qRT-PCR及GUS分析测定该基因在盐胁迫条件下的表达,通过酵母双杂交验证其互作关系,并通过转基因验证其功能,旨在为进一步完善盐胁迫分子机理的研究提供参考。1 材料与方法
1.1 植物材料与处理
植物材料为从新疆红肉苹果(Malus sieversii f. neidzwetzkyana)中的‘塔尔阿尔玛’与‘烟富3号’(M. domestica cv. Fuji)杂种一代选育出的‘红脆2号’优株果实及‘王林’苹果愈伤组织。1.2 总RNA的提取及qRT-PCR分析
参考XU等[22]方法提取RNA及qRT-PCR分析。RNA提取试剂盒(DP432)、反转录试剂盒(KR106)、SYBR染料(FP205)均购自北京天根公司,以苹果Actin为内参,3次重复,采用2-ΔΔCT方法进行数据分析[23]。1.3 ‘王林’苹果愈伤组织的转化
用引物(MdWRKY18F:5′-gtcgacATGGACTCAA CGTGGGTGA-3′和MdWRKY18R:5′-ggatccTCATGA GTGGTCTGAAATTCTTC-3′及MdWRKY40F:5′- ccatggATGGACCATTCAGCTGCAT-3′和MdWRKY40R:5′-gctagcTTAG TAAGTATTGTGTTGAAGTATTC-3′,小写字母分别为SalⅠ、BamHⅠ、NcoⅠ和NheⅠ位点)分别扩增MdWRKY18和MdWRKY40,电泳检测,回收、连接、转化。构建重组表达载体pRI101- MdWRKY18和pCAMBIA1301-MdWRKY40。将重组质粒导入农杆菌LBA4404,得到重组农杆菌,28℃,用30 mL含50 μg·mL-1卡那霉素和50 μg·mL-1利福平的YEP液体培养基培养重组农杆菌至OD600nm=0.6,12 000 r/min离心收集菌体,用30 mL ddH2O悬浮,加入乙酰丁香酮并使其浓度为100 μmol·L-1,得到侵染液。取生长2周龄的‘王林’愈伤组织浸到侵染液中,室温振荡25 min,取愈伤组织置于含0.5 mg·L-1 6-BA+1 mg·L-1 2,4-D的MS固体培养基上,28℃暗培养2 d。随后转移到含0.5 mg·L-1 6-BA+1 mg·L-1 2,4-D+50 μg·mL-1卡那霉素+250 μg·mL-1羧苄青霉素的MS固体培养基暗培养5周左右。1.4 GUS表达载体构建、转化及GUS染色
用引物(MdWRKY18-1F:5′-tctagaACCTTACCAC ATGGCAAGTT-3′和MdWRKY18-1R:5′-ccatggTGTT GATGAAGATCAAAAGGCT-3′及MdWRKY40-1F:5′- tctagaCAGCAGAGGCTTGACAATTC-3′和MdWRKY40- 1R:5′-ccatgg CCTTTGAAAGTAACAACACTAGTTC- 3′)分别扩增MdWRKY18和MdWRKY40的启动子序列。GUS表达载体为pCAMBIA1305,构建重组表达载体及‘王林’苹果愈伤组织的转化,方法同1.3。染色液配置为100 mmol·L-1磷酸钠、10 mmol·L-1EDTA、1 mmol·L-1X-Gluc和0.1% Triton X-100。取1 g过表达MdWRKY18或MdWRKY40启动子的‘王林’愈伤置于5 mLGUS染色液中,37℃染色10—12 h。1.5 酵母双杂交试验
用引物(MdWRKY18-2F:5′-catatgATGGACTCA ACGTGGGTGA-3′和MdWRKY18-2R:5′-gtcgacTCAT GAGTGGTCTGAAATTCTTC-3′及MdWRKY40-2F:5′-catatgATGGACCATTCAGCTGCAT-3′和MdWRKY40 -2R:5′-TTA GTAAGTATTGTGTTGAAGTATTC-3′)分别扩增MdWRKY18和MdWRKY40的编码框序列。分别构建pGADT7-MdWRKY18、pGBKT7- MdWRKY18、pGADT7-MdWRKY40和pGBKT7-MdWRKY40重组载体,方法如1.3。按照YeastmakerTM Yeast Transformation System 2试剂盒(Clontech)说明书方法,将重组质粒共转化酵母Y2H感受态细胞,先在-T-L选择性培养基(-Leu/Trp,Clontech)上培养,然后将生长的细胞在-T-L-H-A选择性培养基(-Ade/- His/-Leu/-Trp,Clontech)上培养,最后用X-α-gal作为底物添加到-T-L-H-A培养基中检测β-galactosidase。1.6 数据分析
实时荧光定量分析数据用Excel 2007进行作图和标准差分析,用DPS 7.05软件(http://www.chinadps. net)进行显著性检验,显著性水平用i、ii、iii和iv来表示。2 结果
2.1 MdWRKY18和MdWRKY40蛋白结构分析
根据WRKY结构域的数量和锌指结构的特征可以将WRKY转录因子家族大体分为3类。利用SMART网站(http://smart.embl-heidelberg.de/smart/ set_mode.cgi?GENOMIC=1)对蛋白结构功能域分析,发现MdWRKY18和MdWRKY40与拟南芥AtWRKY18和AtWRKY40结构类似,都含有1个WRKY、Cx5C以及HxH结构域(图1),依据进化树、保守结构域及内含子位置将WRKY家族重新分类,MdWRKY18、MdWRKY40和AtWRKY18、AtWRKY40都属于Ⅱa+Ⅱb类[24]。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1MdWRKY18和MdWRKY40蛋白结构分析
Fig. 1Protein structure analysis of MdWRKY18 and MdWRKY40
2.2 盐胁迫诱导MdWRKY18和MdWRKY40的表达
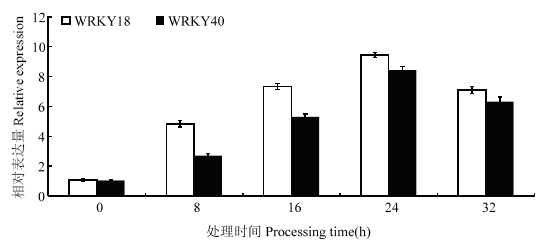
通过对生长2周龄的‘王林’愈伤组织进行150 mmol·L-1 NaCl处理(图2),结果显示,MdWRKY18和MdWRKY40的表达水平受盐胁迫诱导,在24 h达到最大,32 h略有下降;对过表达MdWRKY18和MdWRKY40启动子的‘王林’愈伤进行GUS染色,150 mmol·L-1 NaCl处理的转基因愈伤要比不处理的颜色要深(图3)图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2150 mmol·L-1 NaCl处理下MdWRKY18和MdWRKY40的表达水平
Fig. 2The expression level of MdWRKY18 and MdWRKY40 treated with 150 mmol·L-1 NaCl
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3150 mmol·L-1 NaCl处理条件下MdWRKY18和MdWRKY40启动子的GUS染色分析
Fig. 3The GUS staining analysis of MdWRKY18 and MdWRKY40 promoter treated with 150 mmol·L-1 NaCl
2.3 MdWRKY18和MdWRKY40之间蛋白相互作用分析
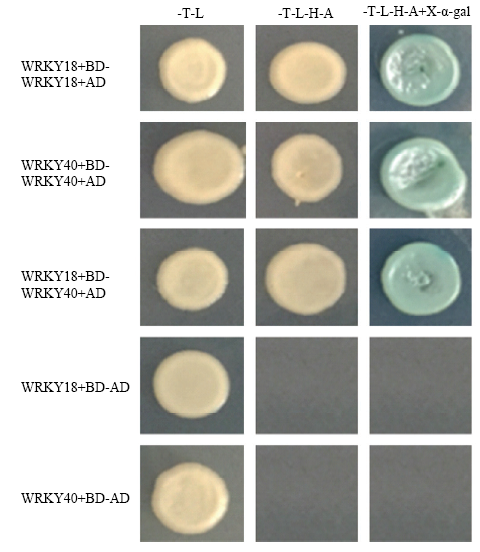
酵母双杂交结果如图4所示,WRKY18-BD和WRKY18-AD共转Y2H后,在二缺、四缺及四缺+X- α-gal均生长,说明MdWRKY18能与自身互作形成同源二聚体,并且MdWRKY18也能和MdWRKY40互作形成异源二聚体,MdWRKY40同样也可以与自身互作形成同源二聚体。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4MdWRKY18和MdWRKY40酵母双杂交分析
Fig. 4Yeast two-hybrid analysis between MdWRKY18 and MdWRKY40
2.4 MdWRKY18和MdWRKY40在‘王林’苹果愈伤中超表达后的功能分析
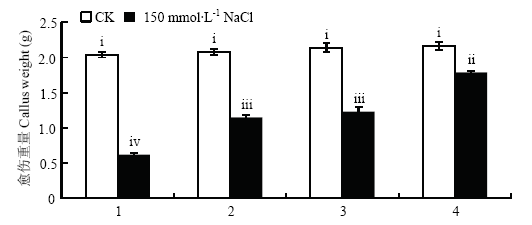
通过对‘王林’苹果愈伤的不同转基因进行处理(图5和图6),发现未处理的4种愈伤重量基本无差异,而150 mmol·L-1 NaCl处理后,过表达MdWRKY18和过表达MdWRKY40的愈伤之间重量基本无差异,但显著高于‘王林’愈伤,显著低于共表达MdWRKY18和MdWRKY40的愈伤。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5150 mmol·L-1 NaCl处理条件下过表达MdWRKY18和MdWRKY40的‘王林’愈伤形态
Fig. 5The orin callus that overexpressing MdWRKY18 and MdWRKY40 treated with 150 mmol·L-1 NaCl
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6150 mmol·L-1 NaCl处理条件下过表达MdWRKY18和MdWRKY40‘王林’愈伤的重量
1:Orin;2:OEWRKY18;3:OEWRKY40;4:OEWRKY18+OEWRKY40
Fig. 6The weight of orin callus that overexpressing MdWRKY18 and MdWRKY40 treated with 150 mmol·L-1 NaCl
2.5 4种愈伤相关基因表达水平的分析
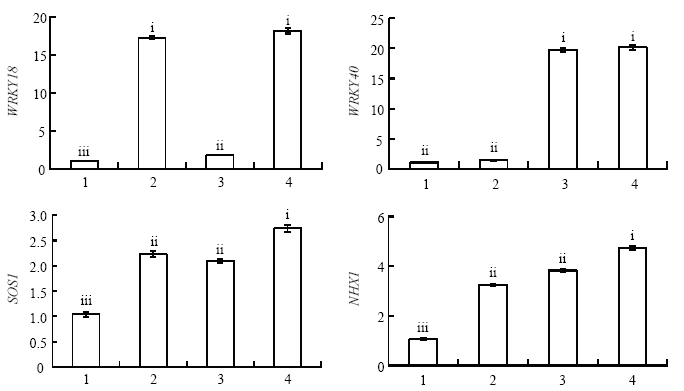
由图7可得,过表达MdWRKY18的愈伤,MdWRKY18的表达水平是‘王林’愈伤的16.58倍,但对MdWRKY40的表达水平没有影响;过表达MdWRKY40的愈伤,MdWRKY40的表达水平是‘王林’愈伤的18.08倍,但对MdWRKY18的表达水平没有影响;共表达MdWRKY18和MdWRKY40的愈伤,MdWRKY18和MdWRKY40的表达水平分别是‘王林’愈伤的17.43倍和18.47倍;与‘王林’愈伤相比,过表达MdWRKY18的愈伤,过表达MdWRKY40的愈伤以及共表达MdWRKY18和MdWRKY40的愈伤中MdSOS1和MdNHX1的表达水平均上调表达。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图74种愈伤相关基因表达水平分析
不同‘王林’苹果愈伤 Different orin apple callus
Fig. 7The expression level analysis of related gene in four types of callus
1:Orin;2:OEWRKY18;3:OEWRKY40;4:OEWRKY18+OEWRKY40
3 讨论
3.1 苹果MdWRKY18和MdWRKY40受盐胁迫诱导并提高‘王林’愈伤的盐耐性
WRKY蛋白是最近发现的一类序列特异性DNA结合转录因子,因其具有高度保守的WRKY结构域而得名[25],在拟南芥中发现了70多个WRKY蛋白,在水稻中发现了100多个WRKY蛋白[24,26]。WRKY蛋白能够调节植物发育与繁殖,酚类化合物生物合成,激素信号传导或衰老有关的多个过程[27,28,29],然而,WRKY蛋白的基础性作用在于参与植物防御信号传导以及非生物胁迫耐受性[30,31]。盐胁迫是重要的非生物胁迫之一,在大豆中,64个测试的WRKY基因中,有25个响应盐胁迫[32],并且在拟南芥,小麦和水稻等中也已经报道了类似的结果[33,34,35]。在许多植物物种中进行了WRKY蛋白响应盐胁迫的功能研究,小麦TaWRKY2、TaWRKY19、TaWRKY44和TaWRKY93能够通过增强渗透保护剂(脯氨酸和可溶性糖)积累和改善氧化应激反应来提高对盐胁迫耐受性[35,36],棉花GHWRKY34在适度盐胁迫下能够增强植物的萌发和生长,减少钠和ROS积累[37],拟南芥AtWRKY18、AtWRKY40、AtWRKY25和AtWRKY33也被报道能够响应盐胁迫诱导并且提高盐耐性[38,21]。本研究发现MdWRKY18和MdWRKY40含有1个WRKY、Cx5C及HxH结构域,属于Ⅱa+Ⅱb类WRKY蛋白,150 mmol·L-1 NaCl处理下能够增强MdWRKY18和MdWRKY40的启动子活性,诱导MdWRKY18和MdWRKY40的表达;在‘王林’愈伤分别过表达MdWRKY18和MdWRKY40以及共表达MdWRKY18和MdWRKY40时,均能够增强‘王林’愈伤的耐盐性,并且能够诱导MdSOS1和MdNHX1的表达,而MdSOS1和MdNHX1在维持胞质Na+浓度方面起着重要作用[13,39],因此MdWRKY18和MdWRKY40可能参与调控MdSOS1和MdNHX1的表达来调节‘王林’愈伤的耐盐性。3.2 MdWRKY18和MdWRKY40之间蛋白相互作用
具有调节功能的蛋白质很少单独作用,它们大多以相互作用的形式在生命系统中承担生物学功能,因此,研究蛋白质之间相互作用是理解复杂分子过程的关键步骤。在过去的研究中,发现了大量与信号转录有关的植物WRKY蛋白的相互作用蛋白,WRKY相互作用蛋白在转录中可影响WRKY转录因子与DNA的结合和转录调节活性,从而调控WRKY调节的下游基因表达。在拟南芥中,3种Ⅱ类型的WRKY蛋白(AtWRKY18、AtWRKY40和AtWRKY60)能够通过N-末端的亮氨酸拉链基序彼此相互作用[40],AtWRKY6和AtWRKY42之间也能相互作用[41],此外,通过酵母双杂交系统筛选了13个Ⅲ类型的WRKY蛋白发现,AtWRKY30、AtWRKY53、AtWRKY54和AtWRKY70之间存在显著的相互作用[42]。本研究发现MdWRKY18和MdWRKY40均能够与自身相互作用形成同源二聚体,且MdWRKY18和MdWRKY40也能够互作形成异源二聚体,与在拟南芥中研究结果一致。4 结论
盐胁迫诱导MdWRKY18和MdWRKY40表达,且MdWRKY18和MdWRKY40之间能够相互作用形成同源或异源二聚体,在‘王林’愈伤中,过表达MdWRKY18和MdWRKY40能够促进MdSOS1和MdNHX1的表达,从而提高‘王林’愈伤的耐盐性。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/j.abb.2005.10.018URLPMID:16309626 [本文引用: 1]

World population is increasing at an alarming rate and is expected to reach about six billion by the end of year 2050. On the other hand food productivity is decreasing due to the effect of various abiotic stresses; therefore minimizing these losses is a major area of concern for all nations to cope with the increasing food requirements. Cold, salinity and drought are among the major stresses, which adversely affect plants growth and productivity; hence it is important to develop stress tolerant crops. In general, low temperature mainly results in mechanical constraint, whereas salinity and drought exerts its malicious effect mainly by disrupting the ionic and osmotic equilibrium of the cell. It is now well known that the stress signal is first perceived at the membrane level by the receptors and then transduced in the cell to switch on the stress responsive genes for mediating stress tolerance. Understanding the mechanism of stress tolerance along with a plethora of genes involved in stress signaling network is important for crop improvement. Recently, some genes of calcium-signaling and nucleic acid pathways have been reported to be up-regulated in response to both cold and salinity stresses indicating the presence of cross talk between these pathways. In this review we have emphasized on various aspects of cold, salinity and drought stresses. Various factors pertaining to cold acclimation, promoter elements, and role of transcription factors in stress signaling pathway have been described. The role of calcium as an important signaling molecule in response to various stress signals has also been covered. In each of these stresses we have tried to address the issues, which significantly affect the gene expression in relation to plant physiology.
DOI:10.1016/S0076-6879(07)28024-3URL [本文引用: 1]
DOI:10.1093/aob/mcn205URLPMID:18952624 [本文引用: 1]

61 Background and Aims Corn (Zea mays) responds to salt stress via changes in gene expression, metabolism and physiology. This adaptation is achieved through the regulation of gene expression at the transcriptional and posttranscriptional levels. MicroRNAs (miRNAs) have been found to act as key regulating factors of post-transcriptional gene expression. However, little is known about the role of miRNAs in plants' responses to abiotic stresses. 61 Methods A custom 08paraflo64 microfluidic array containing release version 10.1 plant miRNA probes (http://microrna.sanger.ac.uk/) was used to discover salt stress-responsive miRNAs using the differences in miRNA expression between the salt-tolerant maize inbred line 'NC286' and the salt-sensitive maize line 'Huangzao4'. 61 Key Results miRNA microarray hybridization revealed that a total of 98 miRNAs, from 27 plant miRNA families, had significantly altered expression after salt treatment. These miRNAs displayed different activities in the salt response, and miRNAs belonging to the same miRNA family showed the same behaviour. Interestingly, 18 miRNAs were found which were only expressed in the salt-tolerant maize line, and 25 miRNAs that showed a delayed regulation pattern in the salt-sensitive line. A gene model was proposed that showed how miRNAs could regulate the abiotic stress-associated process and the gene networks coping with the stress. 61 Conclusions Salt-responsive miRNAs are involved in the regulation of metabolic, morphological and physiological adaptations of maize seedlings at the post-transcriptional level. The miRNA genotype-specific expression model might explain the distinct salt sensitivities between maize lines.
[本文引用: 1]
DOI:10.1016/j.plantsci.2015.03.023URLPMID:26025528 [本文引用: 2]

Plants have evolved a number of different mechanisms to respond and to adapt to abiotic stress for their survival. However, the regulatory mechanisms involved in coordinating abiotic stress tolerance and plant growth are not fully understood. Here, the function of OsMYB91, an R2R3-type MYB transcription factor of rice was explored. OsMYB91 was induced by abiotic stress, especially by salt stress. Analysis of chromatin structure of the gene revealed that salt stress led to rapid removal of DNA methylation from the promoter region and rapid changes of histone modifications in the locus. Plants over-expressing OsMYB91 showed reduced plant growth and accumulation of endogenous ABA under control conditions. Under salt stress, the over-expression plants showed enhanced tolerance with significant increases of proline levels and a highly enhanced capacity to scavenge active oxygen as well as the increased induction of OsP5CS1 and LOC_Os03g44130 compared to wild type, while RNAi plants were less sensitive. In addition, expression of OsMYB91 was also induced by other abiotic stresses and hormone treatment. More interestingly, SLR1, the rice homolog of Arabidopsis DELLA genes that have been shown to integrate endogenous developmental signals with adverse environmental conditions, was highly induced by OsMYB91 over-expression, while the salt-induction of SLR1 expression was impaired in the RNAi plants. These results suggested that OsMYB91 was a stress-responsive gene that might be involved in coordinating rice tolerance to abiotic stress and plant growth by regulating SLR1 expression.
DOI:10.1016/j.plantsci.2004.05.034URL [本文引用: 1]

Wheat productivity is severely affected by soil salinity mainly due to Na + toxicity to plant cells. To improve the yield performance of wheat in saline soils, we have generated transgenic wheat expressing a vacuolar Na +/H + antiporter gene AtNHX1 from Arabidopsis thaliana, to enhance the plant capacity of reducing cytosolic Na + by sequestering Na + in the vacuole. The AtNHX1 transgenic wheat lines exhibited improved biomass production at the vegetative growth stage and germination rates in severe saline conditions. A field trial revealed that the transgenic wheat lines produced higher grain yields and heavier and larger grains in the field of saline soils with the electrical conductivity values of soil saturation extracts (EC e) of 10.6 and 13.7 dS m 1. The transgenic lines accumulated a lower level of Na + and a higher level of K + in the leaves than non-transgenic plants under saline conditions (100 and 150 mM NaCl). These results indicate that the salt tolerance of wheat and grain yield in saline soils can be improved by enhancing the level of the vacuolar Na +/H + antiporter.
DOI:10.1111/j.1439-037X.2007.00288.xURL [本文引用: 1]

The effect of NaCl (610.1, 610.4 and 610.7 MPa) on some physiological parameters in six 23-day-old soya bean cultivars ( Glycine max L. Merr. namely A 3935, CX-415, Mitchell, Nazl03can, SA 88 and Türksoy) at 25, 30 and 35 °C was investigated. Salt stress treatments caused a decline in the K + /Na + ratio, plant height, fresh and dry biomass of the shoot and an increase in the relative leakage ratio and the contents of proline and Na + at all temperatures. Effects of salt stress and temperature on Chl content, Chl a / b ratio (antenna size) and qN (heat dissipation in the antenna) varied greatly between cultivars and treatments; however, in all cases approximately the same qP value was observed. It indicates that the plants were able to maintain the balance between excitation pressure and electron transport activity. Pigment content and the quantum efficiency of photosystem II exhibited significant differences that depended on the cultivar, the salt concentration and temperature. The cultivars were relatively insensitive to salt stress at 30 °C however they were very sensitive both at 25 and 35 °C. Of the cultivars tested CX-415 and SA 88 were the best performers at 25 °C compared with SA 88 and Türksoy at 35 °C.
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.scienta.2015.04.009URL [本文引用: 1]

Autotetraploid plants of apple (Malus domestica) cultivar ‘Hanfu’ and ‘Gala’ were induced from leaves by colchicine treatment. There were obvious morphological differences in the leaves of diploid and autotetraploid plants. We treated the diploid and autotetraploid apple seedlings with 200mmolL611 NaCl for 8days in 0.5× Hoagland's nutrient solution and measured the physiological characters and the expression of aquaporin genes in leaves at different times during treatment. The results showed that the response of the autotetraploid to salt stress was better than that of the diploid; the relative water content (RWC) decreased in both over time, but the RWC was higher in the autotetraploid than in the diploid for the duration of the salt stress treatment. The proline content and the accumulation of malondialdehyde (MDA) were higher in the diploid than the autotetraploid, with both increasing over the course of the treatment. We also showed that the expression of aquaporin genes (MdPIP1;1, and MdTIP1;1) decreased initially before peaking at 24h and then rapidly declining at 48h and 96h. The relative expression levels in the autotetraploid were higher than in the diploid at all sampling times, and were 4 to 10-fold higher at 24h. Our results show that autotetraploid apple possesses better salt tolerance than diploid, which may be associated with the higher levels of expression of aquaporin genes in response to salt stress. The results of our study will make a practical and theoretical contribution to the breeding of polyploid apple cultivars.
DOI:10.1111/j.1399-3054.2005.00610.xURL [本文引用: 1]

With recent advancements in DNA-chip technology, requisite software development and support and progress in related aspects of plant molecular biology, it is now possible to comprehensively analyze the expression of complete genomes. Global transcript profiling shows that in plants, salt-stress response involves simultaneous up and downregulation of a large number of genes. This analysis further suggests that apart from the transcripts that govern synthesis of osmolytes and ion transporters, two candidate systems that have attracted much of the attention thus far, transcripts encoding for proteins related to the regulation of transcriptional and translational machineries have a distinct role in salt-stress response. In particular, induction of transcripts of specific transcription factors, RNA-binding proteins, ribosomal genes, and translation initiation and elongation factors has recently been noted to be important during salt stress. There is an urgent need to examine cellular functionality of the above putative salt-tolerance-related genes emerging from the transcriptome analysis.
DOI:10.1146/annurev.arplant.51.1.463URL [本文引用: 1]
DOI:10.1104/pp.78.1.163URLPMID:16664191 [本文引用: 2]

Artificial pH gradients across tonoplast vesicles isolated from storage tissue of red beet (Beta vulgaris L.) were used to study the kinetics of a Ca2+/H+antiport across this membrane. Ca2+-dependent H+fluxes were measured by the pH-dependent fluorescence quenching of acridine orange. pH-dependent Ca2+influx was measured radiometrically. Both H+efflux and Ca2+influx displayed saturation kinetics and an identical dependence on external calcium with apparent Kmvalues of 43.9 and 41.7 micromolar, respectively. Calcium influx was unaffected by an excess of Mg2+but was inhibited by La3+> Mn2+> Cd2+. The apparent Kmfor external calcium was greatly affected (5-fold) by internal pH in the range of 6.0 to 6.5 and a transmembrane effect of internal proton binding on the affinity for external calcium is suggested.
DOI:10.1016/S1673-8527(09)60003-5URLPMID:19161942 [本文引用: 1]

Abiotic stresses cause serious crop losses. Knowledge on genes functioning in plant responses to adverse growth conditions is essential for developing stress tolerant crops. Here we report that transgenic expression of MYB15, encoding a R2R3 MYB transcription factor in Arabidopsis thaliana, conferred hypersensitivity to exogenous abscisic acid (ABA) and improved tolerance to drought and salt stresses. The promoter of MYB15 was active in not only vegetative and reproductive organs but also the guard cells of stomata. Its transcript level was substantially upregulated by ABA, drought or salt treatments. Compared with wild type (WT) control, MYB15 overexpression lines were hypersensitive to ABA in germination assays, more susceptible to ABA-elicited inhibition of root elongation, and more sensitive to ABA-induced stomatal closure. In line with the above findings, the transcript levels of ABA biosynthesis ( ABA1, ABA2), signaling ( ABI3), and responsive genes ( AtADH1, RD22, RD29B, AtEM6) were generally higher in MYB15 overexpression seedlings than in WT controls after treatment with ABA. MYB15 overexpression lines displayed improved survival and reduced water loss rates than WT control under water deficiency conditions. These overexpression lines also displayed higher tolerance to NaCl stress. Collectively, our data suggest that overexpression of MYB15 improves drought and salt tolerance in Arabidopsis possibly by enhancing the expression levels of the genes involved in ABA biosynthesis and signaling, and those encoding the stress-protective proteins.
DOI:10.1105/tpc.006130URLPMID:12509522 [本文引用: 1]

In Arabidopsis, the induction of a dehydration-responsive gene, rd22, is mediated by abscisic acid (ABA). We reported previously that MYC and MYB recognition sites in the rd22 promoter region function as cis-acting elements in the drought- and ABA-induced gene expression of rd22. bHLH- and MYB-related transcription factors, rd22BP1 (renamed AtMYC2) and AtMYB2, interact specifically with the MYC and MYB recognition sites, respectively, in vitro and activate the transcription of the 尾-glucuronidase reporter gene driven by the MYC and MYB recognition sites in Arabidopsis leaf protoplasts. Here, we show that transgenic plants overexpressing AtMYC2 and/or AtMYB2 cDNAs have higher sensitivity to ABA. The ABA-induced gene expression of rd22 and AtADH1 was enhanced in these transgenic plants. Microarray analysis of the transgenic plants overexpressing both AtMYC2 and AtMYB2 cDNAs revealed that several ABA-inducible genes also are upregulated in the transgenic plants. By contrast, a Ds insertion mutant of the AtMYC2 gene was less sensitive to ABA and showed significantly decreased ABA-induced gene expression of rd22 and AtADH1. These results indicate that both AtMYC2 and AtMYB2 proteins function as transcriptional activators in ABA-inducible gene expression under drought stress in plants.
DOI:10.1016/j.gene.2009.02.010URLPMID:19248824 [本文引用: 1]

Soil salinity severely affects plant growth and agricultural productivity. AtbZIP24 encodes a bZIP transcription factor that is induced by salt stress in Arabidopsis thaliana but suppressed in the salt-tolerant relative Lobularia maritima. Transcriptional repression of AtbZIP24 using RNA interference improved salt tolerance in A. thaliana. Under non-stress growth conditions, transgenic A. thaliana lines with decreased AtbZIP24 expression activated the expression of stress-inducible genes involved in cytoplasmic ion homeostasis and osmotic adjustment: the Na + transporter AtHKT1, the Na +/H + antiporter AtSOS1, the aquaporin AtPIP2.1, and a glutamine synthetase. In addition, candidate target genes of AtbZIP24 with functions in plant growth and development were identified such as an argonaute ( AGO1)-related protein and cyclophilin AtCYP19. The salt tolerance in transgenic plants correlated with reduced Na + accumulation in leaves. In vivo interaction of AtbZIP24 as a homodimer was shown using fluorescence energy transfer (FRET) with cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) as fused FRET pairs. Translational fusion of AtbZIP24 with GFP showed subcellular localization of the protein in nucleus and cytoplasm in plants grown under control conditions whereas in response to salt stress AtbZIP24 was preferentially targeted to the nucleus. It is concluded that AtbZIP24 is an important regulator of salt stress response in plants. The modification of transcriptional control by regulatory transcription factors provides a useful strategy for improving salt tolerance in plants.
DOI:10.1111/j.1365-313X.2010.04248.xURLPMID:20487379 [本文引用: 1]

The biological functions of WRKY transcription factors in plants have been widely studied, but their roles in abiotic stress are still not well understood. We isolated an ABA overly sensitive mutant, abo3, which is disrupted by a T-DNA insertion in At1g66600 encoding a WRKY transcription factor AtWRKY63. The mutant was hypersensitive to ABA in both seedling establishment and seedling growth. However, stomatal closure was less sensitive to ABA, and the abo3 mutant was less drought tolerant than the wild type. Northern blot analysis indicated that the expression of the ABA-responsive transcription factor ABF2/AREB1 was markedly lower in the abo3 mutant than in the wild type. The abo3 mutation also reduced the expression of stress-inducible genes RD29A and COR47, especially early during ABA treatment. ABO3 is able to bind the W-box in the promoter of ABF2in vitro. These results uncover an important role for a WRKY transcription factor in plant responses to ABA and drought stress.
[本文引用: 1]
DOI:10.1007/s00299-008-0614-xURLPMID:2018818929 [本文引用: 1]

Abstract An OsWRKY11 gene, which encodes a transcription factor with the WRKY domain, was identified as one of the genes that was induced by both heat shock and drought stresses in seedlings of rice (Oryza sativa L.). To determine if overexpression of OsWRKY11 confers heat and drought tolerance, OsWRKY11 cDNA was fused to the promoter of HSP101 of rice and introduced into a rice cultivar Sasanishiki. Overexpression of OsWRKY11 was induced by heat treatment. After heat pretreatment, the transgenic lines showed significant heat and drought tolerance, as indicated by the slower leaf-wilting and less-impaired survival rate of green parts of plants. They also showed significant desiccation tolerance, as indicated by the slower water loss in detached leaves. Our results indicate that the OsWRKY11 gene plays a role in heat and drought stress response and tolerance, and might be useful for improvement of stress tolerance.
DOI:10.1093/mp/sss080URLPMID:22930734 [本文引用: 1]

Drought is one of the most serious environmental factors that limit the productivity of agricultural crops worldwide. However, the mechanism underlying drought tolerance in plants is unclear. WRKY transcription factors are known to function in adaptation to abiotic stresses. By screening a pool of WRKY-associated T-DNA insertion mutants, we isolated a gain-of-function mutant, acquired drought tolerance (adt), showing improved drought tolerance. Under drought stress conditions, adt accumulated higher levels of ABA than wild-type plants. Stomatal aperture analysis indicated that adt was more sensitive to ABA than wild-type plants. Molecular genetic analysis revealed that a T-DNA insertion in adt led to activated expression of a WRKY gene that encodes the WRKR57 protein. Constitutive expression of WRKY57 also conferred similar drought tolerance. Consistently with the high ABA content and enhanced drought tolerance, three stress-responsive genes (RD29A, NCED3, and ABA3) were up-regulated in adt. ChIP assays demonstrated that WRKY57 can directly bind the W-box of RD29A and NCED3 promoter sequences. In addition, during ABA treatment, seed germination and early seedling growth of adt were inhibited, whereas, under high osmotic conditions, adt showed a higher seed germination frequency. In summary, our results suggested that the activated expression of WRKY57 improved drought tolerance of Arabidopsis by elevation of ABA levels. Establishment of the functions of WRKY57 will enable improvement of plant drought tolerance through gene manipulation approaches.
[本文引用: 2]
DOI:10.1007/s11103-017-0601-0URLPMID:28286910 [本文引用: 1]

Key message MdMYB16 forms homodimers and directly inhibits anthocyanin synthesis via its C-terminal EAR repressor. It weakened the inhibitory effect of MdMYB16 on anthocyanin synthesis when...
DOI:10.1006/meth.2001.1262URL [本文引用: 1]
DOI:10.1186/1471-2148-5-1URLPMID:15629062 [本文引用: 2]

pAbstract/p pBackground/p pWRKY proteins are newly identified transcription factors involved in many plant processes including plant responses to biotic and abiotic stresses. To date, genes encoding WRKY proteins have been identified only from plants. Comprehensive search for WRKY genes in non-plant organisms and phylogenetic analysis would provide invaluable information about the origin and expansion of the WRKY family./p pResults/p pWe searched all publicly available sequence data for WRKY genes. A single copy of the WRKY gene encoding two WRKY domains was identified from itGiardia lamblia/it, a primitive eukaryote, itDictyostelium discoideum/it, a slime mold closely related to the lineage of animals and fungi, and the green alga itChlamydomonas reinhardtii/it, an early branching of plants. This ancestral WRKY gene seems to have duplicated many times during the evolution of plants, resulting in a large family in evolutionarily advanced flowering plants. In rice, the WRKY gene family consists of over 100 members. Analyses suggest that the C-terminal domain of the two-WRKY-domain encoding gene appears to be the ancestor of the single-WRKY-domain encoding genes, and that the WRKY domains may be phylogenetically classified into five groups. We propose a model to explain the WRKY familys origin in eukaryotes and expansion in plants./p pConclusions/p pWRKY genes seem to have originated in early eukaryotes and greatly expanded in plants. The elucidation of the evolution and duplicative expansion of the WRKY genes should provide valuable information on their functions./p
DOI:10.1016/j.tplants.2010.02.006URL [本文引用: 1]
[本文引用: 1]
DOI:10.1105/tpc.15.00829URLPMID:26578700 [本文引用: 1]

Plant shoot branching is pivotal for developmental plasticity and crop yield. The formation of branch meristems is regulated by several key transcription factors including REGULATOR OF AXILLARY MERISTEMS1 (RAX1), RAX2, and RAX3. However, the regulatory network of shoot branching is still largely unknown. Here, we report the identification of EXCESSIVE BRANCHES1 (EXB1), which affects axillary meristem (AM) initiation and bud activity. Overexpression of EXB1 in the gain-of-function mutant exb1-D leads to severe bushy and dwarf phenotypes, which result from excessive AM initiation and elevated bud activities. EXB1 encodes the WRKY transcription factor WRKY71, which has demonstrated transactivation activities. Disruption of WRKY71/EXB1 by chimeric repressor silencing technology leads to fewer branches, indicating that EXB1 plays important roles in the control of shoot branching. We demonstrate that EXB1 controls AM initiation by positively regulating the transcription of RAX1, RAX2, and RAX3. Disruption of the RAX genes partially rescues the branching phenotype caused by EXB1 overexpression. We further show that EXB1 also regulates auxin homeostasis in control of shoot branching. Our data demonstrate that EXB1 plays pivotal roles in shoot branching by regulating both transcription of RAX genes and auxin pathways.
DOI:10.1104/pp.114.251769URLPMID:25501946 [本文引用: 1]

Abstract WRKY transcription factors (TFs) are well known for regulating plant abiotic and biotic stress tolerance. However, much less is known about how WRKY TFs affect plant-specialized metabolism. Analysis of WRKY TFs regulating the production of specialized metabolites emphasizes the values of the family outside of traditionally accepted roles in stress tolerance. WRKYs with conserved roles across plant species seem to be essential in regulating specialized metabolism. Overall, the WRKY family plays an essential role in regulating the biosynthesis of important pharmaceutical, aromatherapy, biofuel, and industrial components, warranting considerable attention in the forthcoming years. 2015 American Society of Plant Biologists. All Rights Reserved.
DOI:10.1016/j.plantsci.2015.04.014URLPMID:26025535 [本文引用: 1]

Members of the WRKY transcription factor superfamily are essential for the regulation of many plant pathways. Functional redundancy due to duplications of WRKY transcription factors, however, complicates genetic analysis by allowing single-mutant plants to maintain wild-type phenotypes. Our analyses indicate that three group I WRKY genes, OsWRKY24, -53, and -70, act in a partially redundant manner. All three showed characteristics of typical WRKY transcription factors: each localized to nuclei and yeast one-hybrid assays indicated that they all bind to W-boxes, including those present in their own promoters. Quantitative real time-PCR (qRT-PCR) analyses indicated that the expression levels of the three WRKY genes varied in the different tissues tested. Particle bombardment-mediated transient expression analyses indicated that all three genes repress the GA and ABA signaling in a dosage-dependent manner. Combination of all three WRKY genes showed additive antagonism of ABA and GA signaling. These results suggest that these WRKY proteins function as negative transcriptional regulators of GA and ABA signaling. However, different combinations of these WRKY genes can lead to varied strengths in suppression of their targets.
DOI:10.1016/j.bbagrm.2011.09.002URLPMID:21964328 [本文引用: 1]

78 WRKY gene super-family, one of the largest transcription factor gene families, has been suggested to play important roles in the regulation of transcriptional reprogramming associated with plant stress responses. 78 Modification of the expression patterns of WRKY genes and/or changes in their activity contributes to the elaboration of various signaling pathways and regulatory networks. 78 A single WRKY gene often responds to several stress factors, and then their proteins may participate in the regulation of several seemingly disparate processes as negative or positive regulators. 78 WRKY proteins function via protein–protein interaction and auto-regulation or cross-regulation exists extensively in the WRKY family.
DOI:10.1155/2015/807560URLPMID:4387944 [本文引用: 1]

WRKY proteins are emerging players in plant signaling and have been thoroughly reported to play important roles in plants under biotic stress like pathogen attack. However, recent advances in this field do reveal the enormous significance of these proteins in eliciting responses induced by abiotic stresses. WRKY proteins act as major transcription factors, either as positive or negative regulators. Specific WRKY factors which help in the expression of a cluster of stress-responsive genes are being targeted and genetically modified to induce improved abiotic stress tolerance in plants. The knowledge regarding the signaling cascade leading to the activation of the WRKY proteins, their interaction with other proteins of the signaling pathway, and the downstream genes activated by them are altogether vital for justified targeting of the WRKY genes. WRKY proteins have also been considered to generate tolerance against multiple abiotic stresses with possible roles in mediating a cross talk between abiotic and biotic stress responses. In this review, we have reckoned the diverse signaling pattern and biological functions of WRKY proteins throughout the plant kingdom along with the growing prospects in this field of research.
DOI:10.1111/j.1467-7652.2008.00336.xURLPMID:18384508 [本文引用: 1]

WRKY-type transcription factors have multiple roles in the plant defence response and developmental processes. Their roles in the abiotic stress response remain obscure. In this study, 64 GmWRKY genes from soybean were identified, and were found to be differentially expressed under abiotic stresses. Nine GmWRKY proteins were tested for their transcription activation in the yeast assay system, and five showed such ability. In a DNA-binding assay, three proteins (GmWRKY13, GmWRKY27 and GmWRKY54) with a conserved WRKYGQK sequence in their DNA-binding domain could bind to the W-box (TTGAC). However, GmWRKY6 and GmWRKY21, with an altered sequence WRKYGKK, lost the ability to bind to the W-box. The function of three stress-induced genes, GmWRKY13 , GmWRKY21 and GmWRKY54 , was further investigated using a transgenic approach. GmWRKY21- transgenic Arabidopsis plants were tolerant to cold stress, whereas GmWRKY54 conferred salt and drought tolerance, possibly through the regulation of DREB2A and STZ/Zat10 . Transgenic plants over-expressing GmWRKY13 showed increased sensitivity to salt and mannitol stress, but decreased sensitivity to abscisic acid, when compared with wild-type plants. In addition, GmWRKY13 -transgenic plants showed an increase in lateral roots. These results indicate that the three GmWRKY genes play differential roles in abiotic stress tolerance, and that GmWRKY13 may function in both lateral root development and the abiotic stress response.
DOI:10.1186/1471-2229-6-25URLPMID:1621065 [本文引用: 1]

Background Roots are an attractive system for genomic and post-genomic studies of NaCl responses, due to their primary importance to agriculture, and because of their relative structural and biochemical simplicity. Excellent genomic resources have been established for the study of Arabidopsis roots, however, a comprehensive microarray analysis of the root transcriptome following NaCl exposure is required to further understand plant responses to abiotic stress and facilitate future, systems-based analyses of the underlying regulatory networks. Results We used microarrays of 70-mer oligonucleotide probes representing 23,686 Arabidopsis genes to identify root transcripts that changed in relative abundance following 6 h, 24 h, or 48 h of hydroponic exposure to 150 mM NaCl. Enrichment analysis identified groups of structurally or functionally related genes whose members were statistically over-represented among up- or down-regulated transcripts. Our results are consistent with generally observed stress response themes, and highlight potentially important roles for underappreciated gene families, including: several groups of transporters (e.g. MATE, LeOPT1-like); signalling molecules (e.g. PERK kinases, MLO-like receptors), carbohydrate active enzymes (e.g. XTH18), transcription factors (e.g. members of ZIM, WRKY, NAC), and other proteins (e.g. 4CL-like, COMT-like, LOB-Class 1). We verified the NaCl-inducible expression of selected transcription factors and other genes by qRT-PCR. Conclusion Micorarray profiling of NaCl-treated Arabidopsis roots revealed dynamic changes in transcript abundance for at least 20% of the genome, including hundreds of transcription factors, kinases/phosphatases, hormone-related genes, and effectors of homeostasis, all of which highlight the complexity of this stress response. Our identification of these transcriptional responses, and groups of evolutionarily related genes with either similar or divergent transcriptional responses to stress, will facilitate mapping of regulatory networks and extend our ability to improve salt tolerance in plants.
DOI:10.1186/1471-2229-9-120URLPMID:19772648 [本文引用: 1]

pAbstract/p pBackground/p pThe WRKY transcription factor gene family has a very ancient origin and has undergone extensive duplications in the plant kingdom. Several studies have pointed out their involvement in a range of biological processes, revealing that a large number of itWRKY /itgenes are transcriptionally regulated under conditions of biotic and/or abiotic stress. To investigate the existence of itWRKY /itco-regulatory networks in plants, a whole gene family itWRKY/its expression study was carried out in rice (itOryza sativa/it). This analysis was extended to itArabidopsis thaliana /ittaking advantage of an extensive repository of gene expression data./p pResults/p pThe presented results suggested that 24 members of the rice itWRKY /itgene family (22% of the total) were differentially-regulated in response to at least one of the stress conditions tested. We defined the existence of nine OsitWRKY /itgene clusters comprising both phylogenetically related and unrelated genes that were significantly co-expressed, suggesting that specific sets of itWRKY /itgenes might act in co-regulatory networks. This hypothesis was tested by Pearson Correlation Coefficient analysis of the Arabidopsis itWRKY /itgene family in a large set of Affymetrix microarray experiments. itAtWRKYs /itwere found to belong to two main co-regulatory networks (COR-A, COR-B) and two smaller ones (COR-C and COR-D), all including genes belonging to distinct phylogenetic groups. The COR-A network contained several itAtWRKY /itgenes known to be involved mostly in response to pathogens, whose physical and/or genetic interaction was experimentally proven. We also showed that specific co-regulatory networks were conserved between the two model species by identifying Arabidopsis orthologs of the co-expressed itOsWRKY /itgenes./p pConclusion/p pIn this work we identified sets of co-expressed itWRKY /itgenes in both rice and Arabidopsis that are functionally likely to cooperate in the same signal transduction pathways. We propose that, making use of data from co-regulatory networks, it is possible to highlight novel clusters of plant genes contributing to the same biological processes or signal transduction pathways. Our approach will contribute to unveil gene cooperation pathways not yet identified by classical genetic analyses. This information will open new routes contributing to the dissection of WRKY signal transduction pathways in plants./p
DOI:10.1111/j.1365-3040.2012.02480.xURLPMID:22220579 [本文引用: 2]

WRKY-type transcription factors are involved in multiple aspects of plant growth, development and stress response. WRKY genes have been found to be responsive to abiotic stresses; however, their roles in abiotic stress tolerance are largely unknown especially in crops. Here, we identified stress-responsive WRKY genes from wheat (Triticum aestivum L.) and studied their functions in stress tolerance. Forty-three putative TaWRKY genes were identified and two multiple stress-induced genes, TaWRKY2 and TaWRKY19, were further characterized. TaWRKY2 and TaWRKY19 are nuclear proteins, and displayed specific binding to typical cis-element W box. Transgenic Arabidopsis plants overexpressing TaWRKY2 exhibited salt and drought tolerance compared with controls. Overexpression of TaWRKY19 conferred tolerance to salt, drought and freezing stresses in transgenic plants. TaWRKY2 enhanced expressions of STZ and RD29B, and bound to their promoters. TaWRKY19 activated expressions of DREB2A, RD29A, RD29B and Cor6.6, and bound to DREB2A and Cor6.6 promoters. The two TaWRKY proteins may regulate the downstream genes through direct binding to the gene promoter or via indirect mechanism. Manipulation of TaWRKY2 and TaWRKY19 in wheat or other crops should improve their performance under various abiotic stress conditions.
DOI:10.1016/j.bbrc.2015.06.128URLPMID:26106823 [本文引用: 1]

61A class II WRKY transcription factor, TaWRKY93 was isolated and characterized.61TaWRKY93 was induced by NaCl or abscisic acid.61TaWRKY39 increased salinity through enhancing osmotic stress tolerance.61TaWRKY93 transgenic plants showed favourable physiological parameters.61TaWRKY93 increased salt tolerance via both ABA-dependent and -independent pathways.
[本文引用: 1]
DOI:10.1186/1471-2229-10-281URLPMID:21167067 [本文引用: 1]

pAbstract/p pBackground/p pWRKY transcription factors are involved in plant responses to both biotic and abiotic stresses. Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors interact both physically and functionally in plant defense responses. However, their role in plant abiotic stress response has not been directly analyzed./p pResults/p pWe report that the three WRKYs are involved in plant responses to abscisic acid (ABA) and abiotic stress. Through analysis of single, double, and triple mutants and overexpression lines for the WRKY genes, we have shown that itWRKY18 /itand itWRKY60 /ithave a positive effect on plant ABA sensitivity for inhibition of seed germination and root growth. The same two WRKY genes also enhance plant sensitivity to salt and osmotic stress. itWRKY40/it, on the other hand, antagonizes itWRKY18 /itand itWRKY60 /itin the effect on plant sensitivity to ABA and abiotic stress in germination and growth assays. Both itWRKY18 /itand itWRKY40 /itare rapidly induced by ABA, while induction of itWRKY60 /itby ABA is delayed. ABA-inducible expression of itWRKY60 /itis almost completely abolished in the itwrky18 /itand itwrky40 /itmutants. WRKY18 and WRKY40 recognize a cluster of W-box sequences in the itWRKY60 /itpromoter and activate WRKY60 expression in protoplasts. Thus, itWRKY60 /itmight be a direct target gene of WRKY18 and WRKY40 in ABA signaling. Using a stable transgenic reporter/effector system, we have shown that both WRKY18 and WRKY60 act as weak transcriptional activators while WRKY40 is a transcriptional repressor in plant cells./p pConclusions/p pWe propose that the three related WRKY transcription factors form a highly interacting regulatory network that modulates gene expression in both plant defense and stress responses by acting as either transcription activator or repressor./p
DOI:10.1007/s11240-017-1283-7URL [本文引用: 1]

Exposure to high salt concentrations is one of the main stresses adversely affecting crop growth, development, and yield. In this study, a putative salt stress-responsive gene ( MdMYB4 ) encoding an R2R3-MYB transcription factor was cloned and characterized. This gene belongs to the SG2 subgroup, and its C-terminal contains a conserved SG2 motif. The relative MdMYB4 expression levels in Malus 65×65 domestica Borkh. ‘Orin’ calli cultured under various NaCl concentrations were analyzed. Saline conditions upregulated the expression of MdMYB4 , and the effects of the encoded transcription factor on salt tolerance were determined in MdMYB4- overexpressing (OE-MYB4) ‘Orin’ calli. The OE-MYB4 calli grew significantly better than the wild-type calli under salt stress conditions. Additionally, an analysis of the cis -acting elements in the MdNHX1 and MdSOS1 promoters revealed MYB-binding sites. However, the electrophoretic mobility shift assay results indicated that MdMYB4 can bind only to the MdNHX1 promoter. Moreover, the MdNHX1 expression levels were higher in OE-MYB4 calli than in wild-type calli at 150, 200, and 25002mM NaCl. In contrast, there were no significant changes to the MdSOS1 expression levels in OE-MYB4 calli. In conclusion, MdMYB4 is an R2R3-MYB transcription factor that mediates salt tolerance by activating the MdNHX1 promoter. Our findings may provide a theoretical basis for enriching the genetic resources associated with stress tolerance and for improving stress resistance in apple cultivars.
DOI:10.1105/tpc.105.037523URLPMID:16603654 [本文引用: 1]

Limited information is available about the roles of specific WRKY transcription factors in plant defense. We report physical and functional interactions between structurally related and pathogen-induced WRKY18, WRKY40, and WRKY60 transcription factors in Arabidopsis thaliana. The three WRKY proteins formed both homocomplexes and heterocomplexes and DNA binding activities were significantly shifted depending on which WRKY proteins were present in these complexes. Single WRKY mutants exhibited no or small alterations in response to the hemibiotrophic bacterial pathogen Pseudomonas syringae and the necrotrophic fungal pathogen Botrytis cinerea. However, wrky18 wrky40 and wrky18 wrky60 double mutants and the wrky18 wrky40 wrky60 triple mutant were substantially more resistant to P. syringae but more susceptible to B. cinerea than wild-type plants. Thus, the three WRKY proteins have partially redundant roles in plant responses to the two distinct types of pathogens, with WRKY18 playing a more important role than the other two. The contrasting responses of these WRKY mutants to the two pathogens correlated with opposite effects on pathogen-induced expression of salicylic acid-regulated PATHOGENESIS-RELATED1 and jasmonic acid-regulated PDF1.2. While constitutive expression of WRKY18 enhanced resistance to P. syringae, its coexpression with WRKY40 or WRKY60 made plants more susceptible to both P. syringae and B. cinerea. These results indicate that the three WRKY proteins interact both physically and functionally in a complex pattern of overlapping, antagonistic, and distinct roles in plant responses to different types of microbial pathogens.
DOI:10.1105/tpc.108.064980URLPMID:19934380 [本文引用: 1]

Arabidopsis thaliana WRKY family comprises 74 members and some of them are involved in plant responses to biotic and abiotic stresses. This study demonstrated that WRKY6 is involved in Arabidopsis responses to low-Pi stress through regulating PHOSPHATE1 (PH01) expression. WRKY6 overexpression lines, similar to the pho1 mutant, were more sensitive to low Pi stress and had lower Pi contents in shoots compared with wild-type seedlings and the wrkyS-1 mutant. Immunoprecipitation assays demonstrated that WRKY6 can bind to two W-boxes of the PHO1 promoter. RNA gel blot and -glucuronidase activity assays showed that PHO1 expression was repressed in WRKY6-overexpressing lines and enhanced in the wrky6-1 mutant. Low Pi treatment reduced WRKY6 binding to the PHO1 promoter, which indicates that PHO1 regulation by WRKY6 is Pi dependent and that low Pi treatment may release inhibition of PHO1 expression. Protein gel blot analysis showed that the decrease in WRKY6 protein induced by low Pi treatment was inhibited by a 26S proteosome inhibitor, MG132, suggesting that low Pi-induced release of PHO1 repression may result from 26S proteosome-mediated proteolysis. In addition, WRKY42 also showed binding to W-boxes of the PHO1 promoter and repressed PHO1 expression. Our results demonstrate that WRKY6 and WRKY42 are involved in Arabidopsis responses to low Pi stress by regulation of PHO1 expression.
[本文引用: 1]
