 ,, 徐文娟
,, 徐文娟 ,, 朱春红, 陶志云, 宋卫涛, 章双杰, 李慧芳
,, 朱春红, 陶志云, 宋卫涛, 章双杰, 李慧芳 ,江苏省家禽科学研究所,江苏扬州 225125
,江苏省家禽科学研究所,江苏扬州 225125RNA-seq Analysis on Development Arrest of Duck Pectoralis Muscle During Semi-Late Embryonic Period
LIU HongXiang ,, XU WenJuan
,, XU WenJuan ,, ZHU ChunHong, TAO ZhiYun, SONG WeiTao, ZHANG ShuangJie, LI HuiFang
,, ZHU ChunHong, TAO ZhiYun, SONG WeiTao, ZHANG ShuangJie, LI HuiFang ,Jiangsu Institute of Poultry Sciences, Yangzhou 225125, Jiangsu
,Jiangsu Institute of Poultry Sciences, Yangzhou 225125, Jiangsu通讯作者:
第一联系人:
收稿日期:2017-08-3接受日期:2018-09-12网络出版日期:2018-11-16
| 基金资助: |
Received:2017-08-3Accepted:2018-09-12Online:2018-11-16

摘要
关键词:
Abstract
Keywords:
PDF (1210KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
刘宏祥, 徐文娟, 朱春红, 陶志云, 宋卫涛, 章双杰, 李慧芳. 鸭胚胎发育中后期胸肌发育阻滞的RNA-seq分析[J]. 中国农业科学, 2018, 51(22): 4373-4386 doi:10.3864/j.issn.0578-1752.2018.22.015
LIU HongXiang, XU WenJuan, ZHU ChunHong, TAO ZhiYun, SONG WeiTao, ZHANG ShuangJie, LI HuiFang.
0 引言
【研究意义】动物肌肉的生长发育大体可以分为两个阶段:出雏(生)前的胚胎期阶段和出雏(生)后阶段。动物肌纤维数量在出雏(生)前就已经固定[1,2,3],后期肌肉量的增加主要由于肌纤维的肥大,因此禽类上市日龄时的产肉量取决于胚胎中后期肌纤维的数量。胸肌是禽类肌肉的重要组成部分,因此研究禽类胚胎期胸肌发育的调控机制,挖掘影响肌纤维发育的关键基因,对育种实践中禽类产肉量的选育具有重要指导意义。【前人研究进展】动物在胚胎发育期间,成肌细胞增殖、分化成为多核的肌管,最终形成成熟的肌纤维,此过程涉及肌纤维的增生和肥大[2]。哺乳动物胚胎期胸肌重量持续增长[4],而禽类胚胎期胸肌发育过程较为复杂,表现为胚胎前期发育迅速而中后期发育迟缓甚至萎缩。MOORE等[5]报道火鸡胸肌肌纤维横截面积在胚胎后期减小。CHEN等[6,7]对鸭的研究发现,鸭胸肌重量和肌纤维横截面积从22胚龄到出雏期呈减小趋势。本课题组前期研究发现高邮鸭和金定鸭胚胎期21胚龄后胸肌重量没有增加[8]。这可能与此时胸肌肌纤维主要表现为融合,而非肥大有关[7],但具体分子机制还未完全了解。近年来,随着鸭基因组测序的完成[9](基因组草图见http://www.ensembl.org/Anas_platyrhynchos/Info/ Index),以及高通量测序、分析技术的发展,RNA-seq分析鸭不同发育时期基因mRNA表达水平变化情况及其调控机制成为可能。【本研究切入点】鸭胚胎中后期胸肌发育阻滞现象已有多方报道,但具体分子机制还未完全明晰。【拟解决的关键问题】高邮鸭和金定鸭分别为中国地方体型差异较大的品种,其肌肉生长具有明显的表型差异,这为研究肌肉生长发育的分子机制提供了良好的素材。本研究拟通过RNA-seq技术方法,探讨胚胎发育中后期高邮鸭、金定鸭的胸肌转录组表达差异情况,为深入了解此阶段胸肌生长发育的分子机制奠定基础。1 材料与方法
1.1 试验动物及样品采集
本试验研究对象为中国地方品种鸭——蛋肉兼用型品种高邮鸭和蛋用型品种金定鸭,于2014年饲养于江苏省高邮鸭集团。选择正常的高邮鸭和金定鸭种蛋(重量接近)各10枚于微电脑全自动孵化器(山东德州,科裕孵化机)中孵化,21胚龄和27胚龄时随机挑选高邮鸭、金定鸭胚蛋各3枚。解剖胚胎,采集右侧胸大肌样品,迅速置于液氮中冻存。1.2 试验方法
1.2.1 总RNA提取与质量鉴定 使用Trizol试剂(Invitrogen)从约30 mg胸大肌样品中提取总RNA,使用1%甲醛凝胶对RNA样品进行电泳,鉴定RNA完整性和降解情况;使用Qubit RNA试剂盒(Invitrogen)测定RNA浓度;使用Nano Photometer分光光度计(德国Implen)检测RNA纯度。1.2.2 测序文库的构建 RNA检测合格后,将mRNA打断成短片段并反转录成cDNA,选择合适大小的片段进行PCR富集得到最终的cDNA文库。使用qRT-PCR方法对文库的有效浓度进行准确定量(文库有效浓度>2 nM),以保证文库质量。
1.2.3 Illumina测序 库检合格后,交由北京诺禾致源公司使用HiseqTM2000测序仪(Illumina)进行转录组测序。
1.2.4 qRT-PCR验证 根据转录组测序结果,挑选16个差异表达基因进行qRT-PCR验证。使用Primer 3在线工具设计引物(http://fokker.wi.mit.edu/primer3/ input.htm)(所选基因和设计的引物见表1)。
Table 1
表1
表1用于qRT-PCR验证的基因及其引物
Table 1
| 基因 Gene | 上游引物 Up-primer | 下游引物 Lower-primer | 产物长度 Product length (bp) | 退火温度 Temperature (℃) |
|---|---|---|---|---|
| F16P1 | CCCTAAAGGAAAGCTGAGAC | CTCGGGCACTATATCCAGTA | 115 | 60 |
| COL1A1 | GCCAACGAAATCGAGATCAG | CGTCTTTGTCGTCTTGTACT | 126 | 60 |
| WIPI1 | GAAATCCCAGATGTTTACATCG | TCCGTCCCTTTCTTGAAGT | 113 | 60 |
| ARMC3 | TTGGCATTGGCTGTAGTTTAG | TACTCTTGAGGAGGAAGCAGAA | 115 | 60 |
| CEA20 | TCTCCGAGCCAGAAATCC | TCTTCCACCAGTACACGTC | 105 | 60 |
| GLIS3 | ATGACGCAGAGACAAACT | TTGTGTTGTACGTTCTTCTGAG | 188 | 60 |
| SRSF4 | CTCTTACTCCAGAAGCCGA | GATCTACTCTTGGAGCGAC | 105 | 60 |
| LAMC3 | GAGGCCCAGAAGAAGATCA | ACAACCTGTGCCCTCTTA | 131 | 60 |
| TNNI1 | CTGCACGAGAAGGTTGAG | GCAGGTCAAGCACTTTGAT | 109 | 60 |
| TNNT2 | TCTCCAACATGCTGCATT | CTGAGGTGGTCGATGTTC | 130 | 60 |
| TNNC1 | ATGCTGCTTTAACTGGAATG | CACCACAGGGTGGAAATC | 179 | 60 |
| MYOZ1 | CAGAAGATTCAGTCTCACAAGT | AGGTCTTTATCCAAGCCAC | 103 | 60 |
| MYBPH | TCATGGGCAACACCTACTC | GGATCTTCTCTGGCTGGTAA | 129 | 60 |
| COEA1 | CTTCCTGCCAGCAATTAC | AGCCAAACACTTCCATCATA | 134 | 60 |
| KCRS | GACCAGTGCATCCAAACC | TCTCAGCAAACACCTCGTAG | 103 | 60 |
| DEP1A | ATCTGTTGTTTGTTGCTTCC | TGTATCATCAAAGAGCGTGTG | 134 | 60 |
| ACTB* | TGAGAGTAGCCCCTGAGGAGCAC | TAACACCATCACCAGACTCCATCAC | 198 | 60 |
| GAPDH* | CTTCGGAATAGGGAGGAGAC | CGGAGATGATGACACGCTTA | 131 | 60 |
新窗口打开|下载CSV
1.3 测序数据的处理分析
1.3.1 测序数据的处理 为保证信息分析质量,对原始序列(Raw reads)进行过滤去杂(去除测序接头序列、重复冗余序列、低质量序列)得到高质量的序列数据(clean reads)。采用TopHat2方法[10]将clean reads与鸭参考基因组进行比对。1.3.2 差异基因的筛选 本试验每个胚龄点都有3个生物学重复。通过DESeq软件包,对前步得到的clean reads进行标准化,并评估read count的分布,进行BH检验(Benjamini和Hochberg于1995年提出的控制假阳性结果错误率的多重比较方法[11]),最终得到不同组别之间的差异基因比较结果。
1.3.3 mRNA差异表达基因GO分类和KEGG富集分析 使用R语言中的goseq包[12]将mRNA差异表达基因序列与GO(gene ontology)数据库进行比对分析,获得GO功能注释;将mRNA差异表达基因与KEGG(kyoto encyclopedia of genes and genomes)数据库进行BLASTX比对获得mRNA差异表达基因相对应的Pathway注释信息。
2 结果
2.1 RNA-seq数据总体评价
本研究对高邮鸭、金定鸭21胚龄和27胚龄胸大肌组织转录组测序,高邮鸭和金定鸭两个时间点均获得了43+ M条的reads,总碱基数5+ Gb,Q20接近95%,Q30接近90%(表2),说明RNA-seq测序结果可靠,可用于后续分析。高邮鸭、金定鸭21胚龄和27胚龄样品比对到参考基因组上的reads均达到总reads数的60%左右(表3),比对率均较高。Table 2
表2
表2测序数据评估统计
Table 2
| 品种 Breeds | 样品 Sample | 总碱基数(Gbp)Total nucleotides | Q20(%) | Q30(%) | GC(%) |
|---|---|---|---|---|---|
| 高邮鸭 GY | 21胚龄 21 ed | 5.45 | 94.42 | 89.55 | 51.47 |
| 27胚龄 27 ed | 6.25 | 94.34 | 89.46 | 51.89 | |
| 金定鸭 JD | 21胚龄 21 ed | 6.16 | 94.28 | 89.37 | 50.11 |
| 27胚龄 27 ed | 5.79 | 94.33 | 89.37 | 51.42 |
新窗口打开|下载CSV
Table 3
表3
表3Reads与参考基因组比对情况
Table 3
| 项目 Statistical content | 品种 Breeds | 21胚龄 21 ed | 27胚龄 27 ed | ||
|---|---|---|---|---|---|
| 数量 Number | 百分比 Percentage | 数量 Number | 百分比 Percentage | ||
| 总读段数 Total reads | 高邮鸭/GY | 43570695 | - | 50059412 | - |
| 金定鸭/JD | 49264929 | - | 46361619 | - | |
| 总比对数 Total mapped | 高邮鸭/GY | 26927881 | 61.80 | 29480876 | 58.89 |
| 金定鸭/JD | 31722102 | 64.39 | 27119296 | 58.50 | |
| 多次比对数 Multiple mapped | 高邮鸭/GY | 560376 | 1.29 | 668497 | 1.33 |
| 金定鸭/JD | 636927 | 1.29 | 603064 | 1.30 | |
| 唯一比对数 Uniquely mapped | 高邮鸭/GY | 26367505 | 60.51 | 28812378 | 57.56 |
| 金定鸭/JD | 31085175 | 63.10 | 26516232 | 57.19 | |
新窗口打开|下载CSV
2.2 RNA-seq水平相关性检查
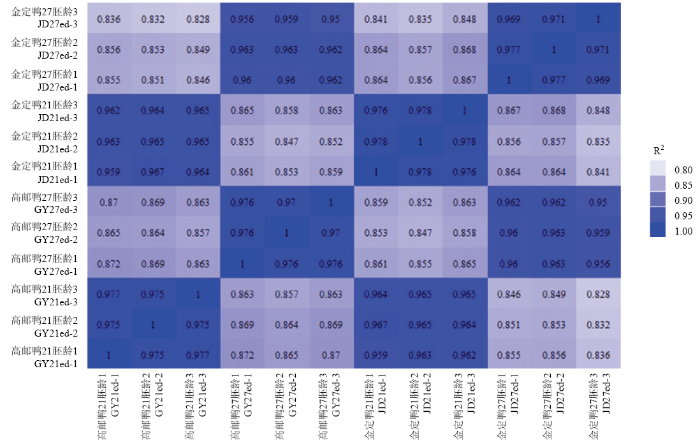
使用皮尔逊相关系数对样品进行相关性分析。同一时间点、品种内的3个生物学重复的相关系数平方(R2)均大于0.95,同一时间点不同品种之间的相似性高于同一品种不同时间点之间的相似性(图1)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1样品间RNA-seq水平相关性检查
Fig. 1correlation check on RNA-seq level between samples
2.3 基因mRNA表达聚类图
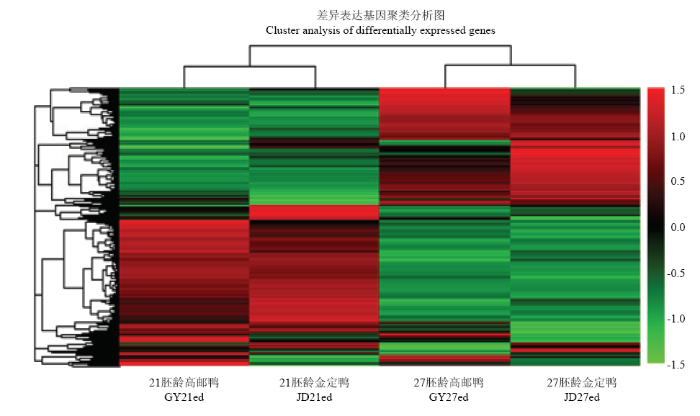
根据高邮鸭、金定鸭21胚龄、27胚龄各基因RPKM进行层次聚类分析。同一时间点不同品种之间的表达模式相近,而同一品种不同时间点的表达模式差异较大(图2)。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2基因mRNA差异表达聚类图
Fig. 2Cluster analysis of mRNA differentially expressed genes
2.4 mRNA差异表达基因的筛选
表4显示了21胚龄和27胚龄样品的RPKM值。21胚龄高邮鸭、金定鸭胸大肌组织高表达基因较多,RPKM值大于3的基因均占到60%左右。Table 4
表4
表4不同胚龄胸大肌RPKM值
Table 4
| 品种 Breeds | RPKM值 RPKM value | 0-1 | 1-3 | 3-15 | 15-60 | > 60 | 总计 Total |
|---|---|---|---|---|---|---|---|
| 高邮鸭 GY | 21胚龄 21 ed | 5197(27.22%) | 2114(11.07%) | 5245(27.47%) | 4711(24.67%) | 1828(9.57%) | 19095 |
| 27胚龄 27 ed | 5757(30.15%) | 2186(11.45%) | 5053(26.46%) | 4310(22.57%) | 1789(9.37%) | 19095 | |
| 金定鸭 JD | 21胚龄 21 ed | 5144(26.93%) | 2152(11.27%) | 5254(27.51%) | 4744(24.84%) | 1802(9.44%) | 19095 |
| 27胚龄 27 ed | 5884(30.81%) | 2166(11.34%) | 4894(25.63%) | 4356(22.81%) | 1796(9.40%) | 19095 |
新窗口打开|下载CSV
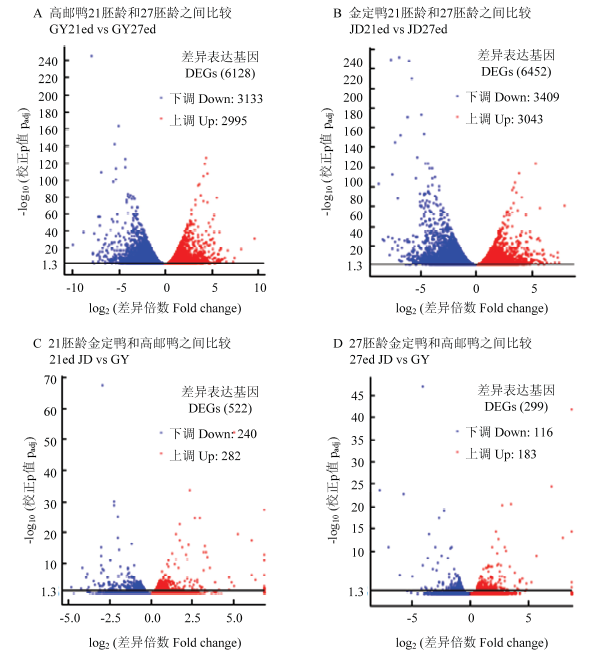
高邮鸭和金定鸭不同时间点之间的mRNA差异表达基因均较多。高邮鸭21胚龄和27胚龄之间、金定鸭21胚龄和27胚龄之间的mRNA显著差异表达基因数量均超过6 000个。21胚龄金定鸭和高邮鸭mRNA显著差异表达基因、27胚龄金定鸭和高邮鸭mRNA显著差异表达基因分别为522个和299个(图3),明显少于品种内不同时间点之间的mRNA差异表达基因数量。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3mRNA差异表达基因火山图
纵坐标1.3以上表示padj<0.05
Fig. 3Valcano chart of mRNA differentially expressed genes
Numbers higher than 1.3 in Y-axis indicate padj<0.05
2.5 qRT-PCR验证
从差异表达分析结果中挑选出16个基因进行qRT-PCR验证。结果显示,所选择的16个基因qRT-PCR结果(??Ct值)与RNA-seq结果(RPKM比值)具有较强的相关性(R > 0.70)(表5),表明RNA-seq结果可靠。Table 5
表5
表5qRT-PCR实验得到的基因表达??Ct值与RPKM比值之间的相关
Table 5
| 基因名 Gene symbol | 相关系数 Correlation coefficient | 描述 Description |
|---|---|---|
| F16P1 | 0.81 | 果糖-1,6-二磷酸酶1 Fructose-1,6-bisphosphatase 1 |
| COL1A1 | 0.79 | 胶原蛋白α-1(I)链 Collagen alpha-1(I) chain |
| WIPI1 | 0.87 | WD重复区域磷酸肌醇互作蛋白1 WD repeat domain phosphoinositide-interacting protein 1 |
| ARMC3 | 0.88 | Armadillo重复蛋白3 Armadillo repeat-containing protein 3 |
| CEA20 | 0.89 | 癌胚抗原相关细胞粘附因子20 Carcinoembryonic antigen-related cell adhesion molecule 20 |
| GLIS3 | 0.75 | 锌指蛋白GLIS3 Zinc finger protein GLIS3 |
| SRSF4 | 0.79 | 富含丝氨酸/精氨酸剪切因子 Serine/arginine-rich splicing factor 4 |
| LAMC3 | 0.82 | 层连蛋白亚基γ3 Laminin subunit gamma-3 |
| TNNI1 | 0.96 | 慢肌中肌钙蛋白1 Troponin 1, slow skeletal muscle |
| TNNT2 | 0.81 | 心肌型肌钙蛋白2 Troponin T2, Cardiac Type |
| TNNC1 | 0.70 | 慢肌和心肌中肌钙蛋白C Troponin C, slow skeletal and cardiac muscles |
| MYOZ1 | 0.76 | Myozenin蛋白1 Myozenin-1 |
| MYBPH | 0.72 | Myosin结合蛋白H Myosin-binding protein H |
| COEA1 | 0.87 | 胶原蛋白α-1(XIV)链 Collagen alpha-1(XIV) chain |
| KCRS | 0.82 | 线粒体中S型肌酸激酶 Creatine kinase S-type, mitochondrial |
| DEP1A | 0.75 | 含DEP区域蛋白1A DEP domain-containing protein 1A |
新窗口打开|下载CSV
2.6 骨骼肌相关基因差异表达情况
挑选出与骨骼肌生长发育相关的关键基因IGF1、MUSTN1、MyoD1、MyoG和MSTN,发现在21胚龄和27胚龄两个时间点,高邮鸭和金定鸭之间IGF1、MUSTN1、MSTN、MyoD1 mRNA表达水平均没有显著变化。两个品种21胚龄到27胚龄IGF1、MyoG均显著下调,MUSTN1、MyoD1均显著上调,MSTN均没有显著变化(表6)。Table 6
表6
表6骨骼肌相关基因mRNA表达水平变化
Table 6
| 基因登录号 GeneID | 基因名 Gene symbol | 比较对象 Objects compared | 前一时间 Before | 后一时间 After | log2(差异倍数) log2(FoldChange) | 校正p值 padj | 原始p值 pval | 上/下调 Up or down |
|---|---|---|---|---|---|---|---|---|
| ENSAPLG00000010676 | IGF1 | 高邮鸭21、27胚龄 GY21v27 | 95.09 | 25.38 | -1.91 | 1.41E-07 | 1.23E-08 | 下调Down |
| 金定鸭21、27胚龄 JD21v27 | 95.24 | 25.00 | -1.93 | 2.02E-07 | 1.91E-08 | 下调Down | ||
| ENSAPLG00000004095 | MUSTN1 | 高邮鸭21、27胚龄 GY21v27 | 2652.05 | 19892.66 | 2.91 | 6.81E-11 | 3.73E-12 | 上调Up |
| 金定鸭21、27胚龄 JD21v27 | 2066.24 | 11844.22 | 2.52 | 1.86E-33 | 1.78E-35 | 上调Up | ||
| ENSAPLG00000012230 | MSTN | 高邮鸭21、27胚龄 GY21v27 | 1281.81 | 825.70 | -0.63 | 0.53 | 0.34 | 下调Down |
| 金定鸭21、27胚龄 JD21v27 | 967.86 | 483.89 | -1.00 | 0.08 | 0.03 | 下调Down | ||
| ENSAPLG00000005673 | MyoD1 | 高邮鸭21、27胚龄 GY21v27 | 3800.16 | 6924.07 | 0.87 | 3.39E-04 | 5.70E-05 | 上调Up |
| 金定鸭21、27胚龄 JD21v27 | 4074.60 | 6240.65 | 0.62 | 0.01 | 2.65E-03 | 上调Up | ||
| ENSAPLG00000001996 | MyoG | 高邮鸭21、27胚龄 GY21v27 | 870.47 | 23.75 | -5.20 | 1.39E-100 | 9.27E-104 | 下调Down |
| 金定鸭21、27胚龄 JD21v27 | 864.99 | 51.91 | -4.06 | 2.58E-73 | 5.01E-76 | 下调Down |
新窗口打开|下载CSV
2.7 mRNA差异表达基因GO富集分析
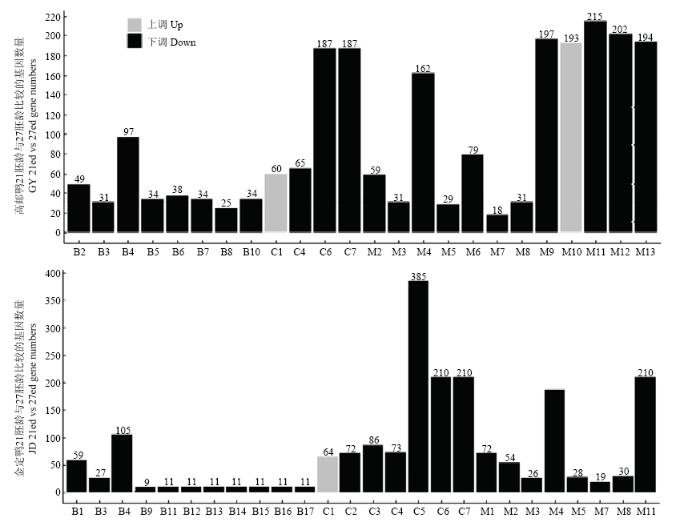
分别对高邮鸭、金定鸭21胚龄和27胚龄胸大肌组织的上调差异基因、下调差异基因进行GO(Gene Ontology)分析,高邮鸭和金定鸭分别富集到24个(上调和下调分别2个和22个)和26个显著GO条目(上调和下调分别1个和25个(图4)。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4高邮鸭及金定鸭21胚龄和27胚龄mRNA差异表达基因GO功能分类图
B1:细胞成分运动(cellular component movement),B2:基于微管的过程(microtubule-based process),B3:基于微管的运动(microtubule-based movement),B4:细胞周期(cell cycle),B5:分解代谢过程的正向调控(positive regulation of catabolic process),B6:自噬调控(regulation of autophagy),B7:自噬正调控(positive regulation of autophagy),B8:细胞膜组织(cellular membrane organization),B9:(evasion or tolerance of host defenses by virus),B10:细胞分解过程的正向调控(positive regulation of cellular catabolic process),B11:宿主防御逃逸(avoidance of host defenses),B12:宿主防御回避或耐受(evasion or tolerance of host defenses),B13:其它共生互作生物的防御逃逸avoidance of defenses of other organism involved in symbiotic interaction),B14:其他共生互作生物的防御回避或耐受(evasion or tolerance of defenses of other organism involved in symbiotic interaction),B15:其他共生互作生物的防御响应(response to defenses of other organism involved in symbiotic interaction),B16:对宿主防御的响应(response to host defenses),B17:对宿主的响应(response to host)
C1:线粒体(mitochondrion),C2:核糖体(ribosome),C3:核糖核蛋白复合物(ribonucleoprotein complex),C4:细胞外基质(extracellular matrix),C5:大分子复合物(macromolecular complex),C6:非膜结合细胞器(non-membrane-bounded organelle),C7:胞内非膜结合细胞器(intracellular non-membrane-bounded organelle)
M1:核糖体结构成分(structural constituent of ribosome),M2:肌动活性(motor activity),M3:微管肌动活性(microtubule motor activity),M4:结构分子活性(structural molecule activity),M5:微管结合(microtubule binding),M6:细胞支架蛋白结合(cytoskeletal protein binding),M7:DNA依赖的ATP酶活性(DNA-dependent ATPase activity),M8:微管蛋白结合(tubulin binding),M9:焦磷酸酶活性(pyrophosphatase activity),M10:氧化还原酶活性(oxidoreductase activity),M11:作用于酸酐的水解酶活性(hydrolase activity, acting on acid anhydrides),M12:作用于含磷酸酐的水解酶活性(hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides),M13:核苷三磷酸酶活性(nucleoside-triphosphatase activity)
Fig. 4GO function classification of mRNA differentially expressed genes of GY and JD between 21 ed and 27 ed
两个品种富集到13个共有GO条目(表7),其中,C1(GO:0005739,mitochondrion)为上调基因所富集,与线粒体能量代谢有关;其余均由下调基因所富集,主要与细胞周期、DNA复制有关。每个共有GO条目下的共有基因数量见表7。
Table 7
表7
表7高邮鸭、金定鸭21胚龄和27胚龄之间显著富集的共有GO分类
Table 7
| GO类型 GO type | GO条目号 GO terms | GO描述 GO description | 标记 Mark | 共有基因数 Number of shared genes | 上/下调 Up or down |
|---|---|---|---|---|---|
| 生物过程 Biological process | GO:0007018 | 基于微管的运动 microtubule-based movement | B3 | 24 | 下调Down |
| GO:0007049 | 细胞周期 cell cycle | B4 | 86 | 下调Down | |
| 细胞组分 Cellular component | GO:0005739 | 线粒体 mitochondrion | C1 | 53 | 上调Up |
| GO:0031012 | 细胞外基质 extracellular matrix | C4 | 57 | 下调Down | |
| GO:0043228 | 无膜细胞器 non-membrane-bounded organelle | C6 | 161 | 下调Down | |
| GO:0043232 | 细胞内无膜细胞器 intracellular non-membrane-bounded organelle | C7 | 161 | 下调Down | |
| 分子功能 Molecular function | GO:0003774 | 马达运动 motor activity | M2 | 48 | 下调Down |
| GO:0003777 | 微管马达运动 microtubule motor activity | M3 | 24 | 下调Down | |
| GO:0005198 | 结构分子活性 structural molecule activity | M4 | 137 | 下调Down | |
| GO:0008017 | 微管结合 microtubule binding | M5 | 25 | 下调Down | |
| GO:0008094 | DNA依赖的ATP酶活性 DNA-dependent ATPase activity | M7 | 17 | 下调Down | |
| GO:0015631 | 微管蛋白结合 tubulin binding | M8 | 26 | 下调Down | |
| GO:0016817 | 作用于酸酐的水解酶活性hydrolase activity, acting on acid anhydrides | M11 | 178 | 下调Down |
新窗口打开|下载CSV
2.8 mRNA差异表达基因KEGG富集分析
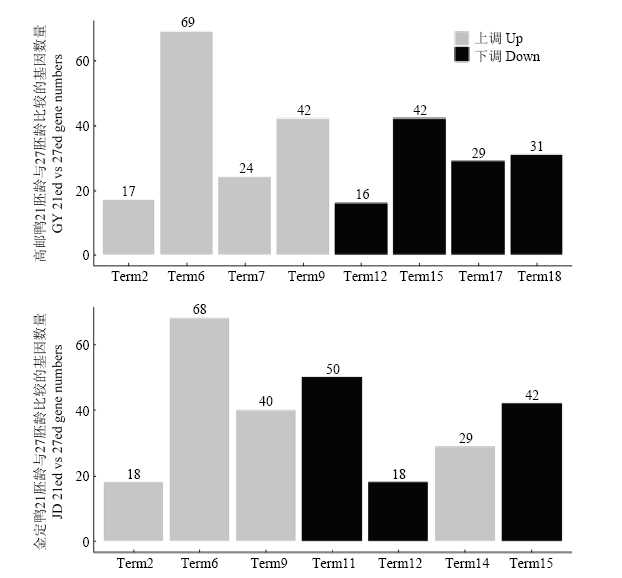
本试验分别对高邮鸭、金定鸭21胚龄和27胚龄胸大肌组织的上调差异基因、下调差异基因进行KEGG分析,高邮鸭和金定鸭分别富集到8个和7个显著KEGG通路(图5)。高邮鸭上调基因富集到4个显著KEGG通路,下调基因富集到4个显著KEGG通路;金定鸭上调基因富集到4个显著KEGG通路,下调基因富集到3个显著KEGG通路。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5高邮鸭及金定鸭21胚龄和27胚龄mRNA差异表达基因KEGG功能分类图
Term2:柠檬酸循环(三羧酸循环)(Citrate cycle (TCA cycle)),Term6:氧化磷酸化(Oxidative phosphorylation),Term7:缬氨酸、亮氨酸和异亮氨酸降解(Valine, leucine and isoleucine degradation),Term9:碳新陈代谢(Carbon metabolism),Term11:核糖体(Ribosome),Term12:DNA复制(DNA replication),Term14:PPAR信号途径(PPAR signaling pathway),Term15:细胞周期(Cell cycle),Term17:ECM受体互作(ECM-receptor interaction),Term18:间隙连接(Gap junction)
Fig. 5KEGG function classification of mRNA differentially expressed genes of GY and JD between 21 ed and 27 ed
两个品种富集到5个共有KEGG通路(表8),其中,Term2(apla00020,Citrate cycle (TCA cycle))、Term6(apla00190,Oxidative phosphorylation)、Term9(apla01200,Carbon metabolism)由上调基因所富集,主要与能量代谢有关;Term12(apla03030,DNA replication)、Term15(apla04110,Cell cycle)由下调基因所富集,主要与DNA复制和细胞周期有关。每个共有KEGG通路下的共有基因数量见表8。
Table 8
表8
表8高邮鸭、金定鸭21胚龄和27胚龄之间显著富集的共有KEGG通路
Table 8
| KEGG通路 KEGG pathway | KEGG描述 KEGG description | 标记 Mark | 共有基因数 Number of shared genes | 上/下调 Up or down |
|---|---|---|---|---|
| apla00020 | 三羧酸循环(TCA循环) Citrate cycle (TCA cycle) | Term2 | 17 | 上调Up |
| apla00190 | 氧化磷酸化 Oxidative phosphorylation | Term6 | 64 | 上调Up |
| apla01200 | 碳的代谢 Carbon metabolism | Term9 | 34 | 上调Up |
| apla03030 | DNA复制 DNA replication | Term12 | 16 | 下调Down |
| apla04110 | 细胞周期 Cell cycle | Term15 | 36 | 下调Down |
新窗口打开|下载CSV
2.9 GO、KEGG联合分析
对共有GO条目的基因和共有KEGG通路的基因取交集,筛选出16个上调基因和15个下调基因(表9)。上调基因大多为辅酶Q相关基因、ATP酶合成相关基因、细胞色素C相关基因,下调基因大多为微型染色体维持蛋白(MCM)相关基因、复制因子C(RFC)相关基因。Table 9
表9
表9GO与KEGG联合分析后的共有基因
Table 9
| 基因名 Gene name | 基因描述 Gene description | Ensembl No. | 上/下调 Up or down | 基因名 Gene name | 基因描述 Gene description | Ensembl No. | 上/下调 Up or down | |
|---|---|---|---|---|---|---|---|---|
| ATP5F1 | ATP合成酶:H+转运线粒体Fo复合物亚基5F1 ATP synthase, H+ transporting, mitochondrial Fo complex subunit 5F1 | ENSAPLG00000013428 | 上调 Up | ANAPC2 | (细胞分裂)后期启动复合物亚基2 Anaphase promoting complex subunit 2 | ENSAPLG00000001836 | 下调Down | |
| ATP5H | ATP合成酶:H+转运线粒体Fo复合物亚基5H ATP synthase, H+ transporting, mitochondrial Fo complex subunit 5H | ENSAPLG00000006404 | 上调 Up | DNA2 | DNA复制解螺旋/核酸酶2 DNA replication helicase/nuclease 2 | ENSAPLG00000015576 | 下调Down | |
| ATP5J | ATP合成酶:H+转运线粒体Fo复合物亚基5J ATP synthase, H+ transporting, mitochondrial Fo complex subunit 5J | ENSAPLG00000009600 | 上调 Up | FEN1 | Flap结构特异性核酸内切酶1 Flap structure-specific endonuclease 1 | ENSAPLG00000001725 | 下调Down | |
| COX5A | 细胞色素c氧化酶亚基5A Cytochrome c oxidase subunit 5A | ENSAPLG00000014886 | 上调 Up | MAD1L1 | MAD1有丝分裂阻滞缺陷类似物1 MAD1 mitotic arrest deficient like 1 | ENSAPLG00000015788 | 下调Down | |
| COX7A2L | 细胞色素c氧化酶亚基类7A2 Cytochrome c oxidase subunit 7A2 like | ENSAPLG00000012898 | 上调 Up | MCM2 | 微型染色体维持复合物组分2 Minichromosome maintenance complex component 2 | ENSAPLG00000004520 | 下调Down | |
| LOC101800937 | 线粒体中细胞色素c氧化酶亚基7B Cytochrome c oxidase subunit 7B, mitochondrial | ENSAPLG00000007286 | 上调 Up | MCM3 | 微型染色体维持复合物组分3 Minichromosome maintenance complex component 3 | ENSAPLG00000013058 | 下调Down | |
| NDUFA5 | NADH:泛醌氧化还原酶亚基A5 NADH: Ubiquinone oxidoreductase subunit A5 | ENSAPLG00000014770 | 上调 Up | MCM4 | 微型染色体维持复合物组分4 Minichromosome maintenance complex component 4 | ENSAPLG00000003491 | 下调 Down | |
| NDUFA7 | NADH:泛醌氧化还原酶亚基A7 NADH: Ubiquinone oxidoreductase subunit A7 | ENSAPLG00000011378 | 上调 Up | MCM5 | 微型染色体维持复合物组分5 Minichromosome maintenance complex component 5 | ENSAPLG00000007931 | 下调 Down | |
| NDUFB1 | NADH:泛醌氧化还原酶亚基B1 NADH: Ubiquinone oxidoreductase subunit B1 | ENSAPLG00000006000 | 上调 Up | ORC5 | 起点识别复合物亚基5 Origin recognition complex subunit 5 | ENSAPLG00000002905 | 下调 Down | |
| NDUFB3 | NADH:泛醌氧化还原酶亚基B3 NADH: Ubiquinone oxidoreductase subunit B3 | ENSAPLG00000015508 | 上调 Up | POLE | DNA聚合酶ε催化亚基 DNA polymerase epsilon, catalytic subunit | ENSAPLG00000009950 | 下调 Down | |
| NDUFB4 | NADH:泛醌氧化还原酶亚基B4 NADH: Ubiquinone oxidoreductase subunit B4 | ENSAPLG00000007537 | 上调 Up | RB1 | RB转录共阻遏因子1 RB transcriptional corepressor 1 | ENSAPLG00000007734 | 下调 Down | |
| NDUFB8 | NADH:泛醌氧化还原酶亚基B8 NADH: Ubiquinone oxidoreductase subunit B8 | ENSAPLG00000015152 | 上调 Up | RFC1 | 复制因子C亚基1 Replication factor C subunit 1 | ENSAPLG00000010594 | 下调 Down | |
| NDUFC2 | NADH:泛醌氧化还原酶亚基C2 NADH: Ubiquinone oxidoreductase subunit C2 | ENSAPLG00000006009 | 上调 Up | RFC2 | 复制因子C亚基2 Replication factor C subunit 2 | ENSAPLG00000009389 | 下调 Down | |
| NDUFS6 | NADH:泛醌氧化还原酶亚基S6 NADH: Ubiquinone oxidoreductase subunit S6 | ENSAPLG00000014333 | 上调 Up | TFDP1 | 转录因子Dp-1 Transcription factor Dp-1 | ENSAPLG00000014438 | 下调 Down | |
| SDHD | 琥珀酸盐脱氢酶复合物亚基D Succinate dehydrogenase complex subunit D | ENSAPLG00000005936 | 上调 Up | YWHAZ | 酪氨酸3-单氧酶/色氨酸5-单氧酶活化蛋白ζ Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | ENSAPLG00000002635 | 下调 Down | |
| UQCRB | 泛醌-细胞色素c还原酶结合蛋白 Ubiquinol-cytochrome c reductase binding protein | ENSAPLG00000012746 | 上调 Up |
新窗口打开|下载CSV
3 讨论
肌肉发育是一个持续不断累积的过程,本课题组前期对高邮鸭、金定鸭的研究[8,13]以及其他课题组的多个试验均报道鸡[14]、鸭[6,7]胚胎发育中后期,胸肌生长迟缓,甚至出现萎缩现象。火鸡胚胎后期胸肌横截面积减小、肌肉萎缩的同时,肌肉卫星细胞的有丝分裂活性也有降低[5]。为了解这一现象背后的分子机制,本试验以高邮鸭、金定鸭21胚龄、27胚龄胚胎胸肌为试验素材,通过RNA-seq比较品种之间以及21胚龄、27胚龄两个时间点之间的mRNA差异表达基因。挑选部分显著差异表达基因进行qRT-PCR验证,结果表明RNA-seq结果与荧光定量PCR结果相似,高通量测序具有较高的可信度。RNA-seq水平相关性检查发现,同一时间点、品种内的3个生物学重复之间的表达模式相似度较高,能够保证后续差异基因的可靠分析。同一时间点不同品种之间的相关性高于同一品种不同时间点之间的相关性。高邮鸭21胚龄、27胚龄的差异基因和金定鸭21胚龄、27胚龄的差异基因数量远多于21胚龄金定鸭、高邮鸭之间的差异基因数量和27胚龄金定鸭、高邮鸭之间的差异基因数量。对高邮鸭、金定鸭两个时间点的基因按照其RPKM值进行层次聚类分析,21胚龄高邮鸭、金定鸭聚为一类,27胚龄高邮鸭、金定鸭聚为一类。以上结果均表明,在21胚龄到27胚龄的发育过程中,高邮鸭与金定鸭的胸肌表达模式具有相似性,在胸肌发育过程中时间因素的影响要大于品种之间遗传因素的影响。
在GO和KEGG富集分析中,上调基因富集到的GO条目、KEGG通路主要与线粒体能量代谢有关;下调基因富集到的GO条目、KEGG通路主要与细胞周期、DNA复制有关。筛选出GO条目和KEGG通路中的共有基因,发现与能量代谢有关的上调基因主要为辅酶Q相关基因、ATP酶合成相关基因以及细胞色素c相关基因,与DNA复制和细胞周期有关的下调基因主要为微型染色体维持蛋白(MCM)相关基因、复制因子C(RFC)相关基因。辅酶Q对体内呼吸链中的质子移位及电子传递起重要作用,是细胞代谢和细胞呼吸的激活剂[15,16]。细胞色素c也是呼吸链中可流动的递氢体或递电子体。上调基因中,辅酶Q相关基因(NDUF*等)分别参与了NADH还原酶1α和1β等复合物的编码合成;ATP合成酶相关基因(ATP5F1、ATP5H和ATP5J)参与了ATP合成酶复合物的编码合成。21胚龄到27胚龄阶段,鸭胸肌组织中这些基因的上调,增强了能量的转化水平和能量代谢频率。真核细胞DNA的复制和有丝分裂需要微型染色体维持蛋白(MCM,Minichromosome maintenance protein complex)[17,18,19]和复制因子C[20,21,22]的参与。21胚龄到27胚龄鸭胸肌DNA复制和细胞周期相关基因下调, 说明在此阶段胸肌细胞的增殖速度开始减缓。细胞周期阻滞是后续细胞融合和分化的必要条件,细胞的融合、分化也将使组织器官获取相应的功能和合适的尺寸[23]。
动物机体肌肉的形成可分为多个步骤。在胚胎期,成肌细胞增殖、分化形成多核的肌管,肌管再分化形成肌纤维,最后组装成肌肉组织[2,24-25]。肌肉发育过程需要有许多调控因子的参与,其中IGF1、MUSTN1、MyoD1、MyoG和MSTN等起着重要作用,因此本试验筛选这5个基因进行详细分析。IGF1是由肝脏主要分泌的调控生长的一种生长因子,近来的研究表明,IGF1也可在肌肉组织局部表达,以旁分泌/自分泌的形式,通过IGF1-Akt/PKB途径调节骨骼肌生长[26],还可通过IGF1-CaN-NFATc3途径促进成肌细胞分化和肌纤维类型的转换[27]。MUSTN1基因在肌纤维分化、融合过程中具有关键作用,并调节下游靶基因MyoD1和MyoG[28]。北京鸭上的研究发现,MUSTN1 mRNA表达水平与鸭胸腿肌的相对生长率相关[29]。MyoD1和MyoG为MRFs(生肌调控因子)的两个重要成员。在动物胚胎期MyoD1可以诱导肌祖细胞向生肌细胞系转变[30,31],体外试验也表明,MyoD1可将其他类型细胞转变为成肌细胞[32,33,34]。MyoG在成肌细胞脱离细胞周期并从增殖过程转变到分化过程中起着重要作用,但不影响成肌细胞的增殖[35,36,37]。MSTN是一种重要的负调控骨骼肌生长的关键基因。在MSTN功能缺失的牛[38,39,40]、鼠[41,42,43],其肌肉量显著增加。本研究发现,在21胚龄到27胚龄胸肌发育阻滞的同时,MUSTN1显著上调(高邮鸭和金定鸭基因表达水平分别提高6.5倍和4.7倍),表明MUSTN1在鸭胸肌发育中起着重要作用。MyoD1与MUSTN1的mRNA表达模式相似,21胚龄到27胚龄也表现显著上调,而IGF1和MyoG均显著下调,MSTN均没有显著变化。本试验前期研究中对21胚龄、27胚龄高邮鸭、金定鸭胸肌肌肉生长相关基因进行了qRT-PCR定量分析,发现两品种21胚龄到27胚龄期间IGF1均显著下调,MSTN均略微上调,MSTN表达在21胚龄和27胚龄两个时间点均没有品种差异[13]。在对北京鸭的研究中,GU等发现21胚龄到27胚龄阶段,MyoG表达水平持续下降,而MSTN表达水平持续上升[7]。除了MSTN基因,IGF1、MyoG和MyoD1结果都与本试验RNA-seq结果相似。MyoD1的上调将促使更多的肌祖细胞向成肌细胞转化,这为出雏后肌纤维的快速生长提供了更多的材料来源。细胞周期与DNA复制相关基因mRNA表达水平显著下降,这可能导致肌肉卫星细胞的有丝分裂活性降低;另外IGF1、MyoG的下调,最终使得鸭胚在接近出雏时胸肌发育阻滞。
4 结论
本试验利用高通量测序技术对高邮鸭、金定鸭胚胎中后期胸肌组织的转录组进行了测序分析,结果表明在此阶段胸肌组织中与能量代谢相关的基因显著上调,与DNA复制、细胞周期相关的基因显著下调,一些肌肉发育相关的关键基因,如IGF1、MyoG显著下调,最终导致胸肌肌肉细胞增殖减缓,胸肌发育阻滞。该发现将为下一步深入探索鸭胸肌发育阻滞机制奠定基础。(责任编辑 林鉴非)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.3382/ps.0420283URL [本文引用: 1]
DOI:10.1051/rnd:2002035URL [本文引用: 3]
[本文引用: 1]
DOI:10.2527/jas1973.372536xURLPMID:4748487 [本文引用: 1]

Swatland HJ.
DOI:10.3923/ijps.2005.138.142URL [本文引用: 2]
DOI:10.1113/expphysiol.2011.01083.xURLPMID:22787243 [本文引用: 2]

Unlike the mammalian fetus, whose growth is supported by the sustained provision of maternal nutrients, poultry embryos undergo development in a relatively closed space, and the yolk sac serves as the sole nutrient supply for embryonic development throughout the whole incubation period. To increase our understanding of the muscle developmental patterns in the final stage of incubation and early days posthatching, we used late-term duck embryos and newly hatched ducklings as animal models. Pectoralis muscle samples were collected at 22 days (22E) of incubation, 25 days (25E) of incubation, hatching and day 7 posthatching. The pectoralis muscle mass, muscle fibre bundles and myofibre cross-sectional area showed a marked reduction from 22E to hatching, but they increased dramatically by day 7 posthatching. The mRNA expression of Atrogin-1, a key mediator of the ubiquitin system responsible for protein degradation, increased dramatically with the age of late-term duck embryos, but it decreased by day 7 and reached a very low level. The extent of mRNA expression of FoxO1, one of the transcription factors of the Atrogin-1 gene, exhibited a transient increase at 25E and then decreased from hatching to day 7. The phosphorylated p70 ribosomal protein S6 kinase 1 (S6K1)/S6K1 ratio exhibited a dramatic reduction from 22E to hatching (P < 0.05) and then increased by day 7. The results of the present study indicated that there was a developmental transition of pectoralis muscle from atrophy in late-term duck embryos to hypertrophy in neonates.
DOI:10.4238/2013.December.13.6URLPMID:24391014 [本文引用: 4]

To confirm the entire developmental process and transition point of embryonic Pekin duck pectoral muscle, and to investigate the association between pectoral muscle development and their regulating genes, anatomical and morphological analyses of embryonic Pekin duck skeletal muscles were performed, and the expression patterns of its regulating genes were investigated. The anatomical analysis revealed that body weight increased with age. while increases in pectoral muscle weight nearly ceased after the embryo was 20 days of hatching (E20). The developmental morphological characteristics of Pekin duck pectoral muscle at the embryonic stage showed that E20 was the transition point (from proliferation to fusion) of Pekin duck pectoral muscle. The expression patterns of MRF4. MyoG, and MSTN indicated that E19 or E20 was the fastest point of pectoral muscle development and the crucial transition for Pekin duck pectoral muscle development during the embryonic stage. Together. these findings imply that E20 is the crucial transition point (from proliferation to fusion) of Pekin duck pectoral muscle and that there is no muscle fiber hypertrophy after E20. Results of this study provide further understanding of the developmental process and transition point of Pekin duck pectoral muscle during the embryo stage.
DOI:10.3864/j.issn.0578-1752.2016.02.016URL [本文引用: 2]

【目的】选择生长速度不同的高邮鸭和金定鸭为试验模型,比较2个不同品种鸭胚胎期和出雏早期骨骼肌中胰岛素样生长因子I(insulin-like growth factor I,IGF-I)、肌肉生长抑素A(myostatin A,MSTN-A)mRNA的表达规律及其与胸浅肌和腓肠肌(简称为胸腿肌)发育模式的相关性。【方法】在鸭13、17、21、25、27胚龄和出雏后7日龄时,记录体重、胸浅肌(PM)和腓肠肌外侧头(LM)的重量,采用实时荧光定量PCR方法研究骨骼肌中IGF-I和MSTN mRNA的表达规律。【结果】本试验证明鸭早期发育过程中,体重和骨骼肌重的变化呈现极显著的品种和时间特异性;鸭胚胎期胸/腿肌的IGF-I和MSTN-A mRNA的表达与体重、胸/腿肌重均呈极显著的负相关,各自与胸/腿体指数则呈反式的相关性(正/负),均在13胚龄有一个表达高峰,IGF-I和MSTN-A mRNA表达之间存在极显著的正相关;鸭PM中IGF-I mRNA表达转折点的出现时间与胸肌绝对生长和相对生长变化趋势是吻合的,而LM中IGF-I mRNA的表达与腿肌生长和肌纤维特性变化均不相符;鸭胚胎期MSTN-A mRNA的表达趋势与骨骼肌生长和肌纤维数量形成高峰同步;胸腿肌中IGF-I/MSTN-A mRNA表达比值的变化趋势和肌纤维特性变化吻合,PM中的IGF-I/MSTN-A mRNA表达比值的差异点和PM重量出现品种差异的时间点一致。【结论】在胚胎发育中后期和出雏后早期,鸭的胸腿肌呈现异步发育模式,骨骼肌中IGF-I和MSTN mRNA表达的相对水平参与骨骼肌生长速率的调节。
DOI:10.3864/j.issn.0578-1752.2016.02.016URL [本文引用: 2]

【目的】选择生长速度不同的高邮鸭和金定鸭为试验模型,比较2个不同品种鸭胚胎期和出雏早期骨骼肌中胰岛素样生长因子I(insulin-like growth factor I,IGF-I)、肌肉生长抑素A(myostatin A,MSTN-A)mRNA的表达规律及其与胸浅肌和腓肠肌(简称为胸腿肌)发育模式的相关性。【方法】在鸭13、17、21、25、27胚龄和出雏后7日龄时,记录体重、胸浅肌(PM)和腓肠肌外侧头(LM)的重量,采用实时荧光定量PCR方法研究骨骼肌中IGF-I和MSTN mRNA的表达规律。【结果】本试验证明鸭早期发育过程中,体重和骨骼肌重的变化呈现极显著的品种和时间特异性;鸭胚胎期胸/腿肌的IGF-I和MSTN-A mRNA的表达与体重、胸/腿肌重均呈极显著的负相关,各自与胸/腿体指数则呈反式的相关性(正/负),均在13胚龄有一个表达高峰,IGF-I和MSTN-A mRNA表达之间存在极显著的正相关;鸭PM中IGF-I mRNA表达转折点的出现时间与胸肌绝对生长和相对生长变化趋势是吻合的,而LM中IGF-I mRNA的表达与腿肌生长和肌纤维特性变化均不相符;鸭胚胎期MSTN-A mRNA的表达趋势与骨骼肌生长和肌纤维数量形成高峰同步;胸腿肌中IGF-I/MSTN-A mRNA表达比值的变化趋势和肌纤维特性变化吻合,PM中的IGF-I/MSTN-A mRNA表达比值的差异点和PM重量出现品种差异的时间点一致。【结论】在胚胎发育中后期和出雏后早期,鸭的胸腿肌呈现异步发育模式,骨骼肌中IGF-I和MSTN mRNA表达的相对水平参与骨骼肌生长速率的调节。
DOI:10.1038/ng.2657URLPMID:4003391 [本文引用: 1]

The duck (Anas platyrhynchos) is one of the principal natural hosts of influenza A viruses. We present the duck genome sequence and perform deep transcriptome analyses to investigate immune-related genes. Our data indicate that the duck possesses a contractive immune gene repertoire, as in chicken and zebra finch, and this repertoire has been shaped through lineage-specific duplications. We identify genes that are responsive to influenza A viruses using the lung transcriptomes of control ducks and ones that were infected with either a highly pathogenic (A/duck/Hubei/49/05) or a weakly pathogenic (A/goose/Hubei/65/05) H5N1 virus. Further, we show how the duck defense mechanisms against influenza infection have been optimized through the diversification of its -defensin and butyrophilin-like repertoires. These analyses, in combination with the genomic and transcriptomic data, provide a resource for characterizing the interaction between host and influenza viruses.
DOI:10.1093/bioinformatics/btp120URLPMID:19289445 [本文引用: 1]

Abstract MOTIVATION: A new protocol for sequencing the messenger RNA in a cell, known as RNA-Seq, generates millions of short sequence fragments in a single run. These fragments, or 'reads', can be used to measure levels of gene expression and to identify novel splice variants of genes. However, current software for aligning RNA-Seq data to a genome relies on known splice junctions and cannot identify novel ones. TopHat is an efficient read-mapping algorithm designed to align reads from an RNA-Seq experiment to a reference genome without relying on known splice sites. RESULTS: We mapped the RNA-Seq reads from a recent mammalian RNA-Seq experiment and recovered more than 72% of the splice junctions reported by the annotation-based software from that study, along with nearly 20,000 previously unreported junctions. The TopHat pipeline is much faster than previous systems, mapping nearly 2.2 million reads per CPU hour, which is sufficient to process an entire RNA-Seq experiment in less than a day on a standard desktop computer. We describe several challenges unique to ab initio splice site discovery from RNA-Seq reads that will require further algorithm development. AVAILABILITY: TopHat is free, open-source software available from http://tophat.cbcb.umd.edu. SUPPLEMENTARY INFORMATION: Supplementary data are available at Bioinformatics online.
DOI:10.2307/2346101URL [本文引用: 1]

The common approach to the multiplicity problem calls for controlling the familywise error rate (FWER). This approach, though, has faults, and we point out a few. A different approach to problems of multiple significance testing is presented. It calls for controlling the expected proportion of falsely rejected hypotheses-the false discovery rate. This error rate is equivalent to the FWER when all hypotheses are true but is smaller otherwise. Therefore, in problems where the control of the false discovery rate rather than that of the FWER is desired, there is potential for a gain in power. A simple sequential Bonferroni-type procedure is proved to control the false discovery rate for independent test statistics, and a simulation study shows that the gain in power is substantial. The use of the new procedure and the appropriateness of the criterion are illustrated with examples.
DOI:10.1186/gb-2010-11-2-r14URLPMID:20132535 [本文引用: 1]

GOseq is a method for GO analysis of RNA-seq data that takes into account the length bias inherent in RNA-seq We present GOseq, an application for performing Gene Ontology (GO) analysis on RNA-seq data. GO analysis is widely used to reduce complexity and highlight biological processes in genome-wide expression studies, but standard methods give biased results on RNA-seq data due to over-detection of differential expression for long and highly expressed transcripts. Application of GOseq to a prostate cancer data set shows that GOseq dramatically changes the results, highlighting categories more consistent with the known biology.
DOI:10.1016/j.gene.2015.01.009URLPMID:25577952 [本文引用: 2]

61The ontogeny of early growth of two different duck breeds was compared.61The pattern of HPGA-related genes mRNA expression of two duck breeds was compared.61Breed specificity in duck early growth was correlated with HPGA genes expression.61Species variance was found regarding HPGA-related gene expression of bird embryo.61Hepatic IGF-1 may be involved in mediating a genetic effect on ducks early growth.
DOI:10.1016/S1096-6374(02)00136-3URLPMID:12550077 [本文引用: 1]

Insulin-like growth factor-I (IGF-I) and myostatin (MSTN) are paracrine regulators of muscle growth. The present study was conducted to relate their expression with muscle fibre development in chickens selected for high breast meat yield and their controls. Both mRNA levels were measured by real-time RT-PCR in the Pectoralis major (PM) muscle between 14 days in ovo and 6 weeks post-hatch and in the Sartorius (SART) muscle between 2 and 6 weeks. The data show that PM growth was slow during in ovo development and rapid in the early post-hatch period. Chickens from the selected genotype exhibited significantly higher breast muscle yields from 2 to 6 weeks of age, and muscle fibre hypertrophy. In the PM, IGF-I and MSTN mRNA levels decreased markedly around hatch, while the IGF-I/MSTN ratio increased, suggesting that it could contribute to the explosive growth observed in the early post-hatch period. Between 4 and 6 weeks of age in selected chickens, IGF-I mRNA levels were significantly higher (=0.04) with a similar trend in MSTN mRNA levels (=0.07) in the PM muscle but not in the SART muscle. Our results support the hypothesis that the relative levels of IGF-I and MSTN mRNA may participate to set muscle growth rate along development, while other factors are required to explain differences between genotypes.
DOI:10.1038/35046114URLPMID:11117751 [本文引用: 1]

Uncoupling proteins (UCPs) are thought to be intricately controlled uncouplers that are responsible for the futile dissipation of mitochondrial chemiosmotic gradients, producing heat rather than ATP. They occur in many animal and plant cells and form a subfamily of the mitochondrial carrier family. Physiological uncoupling of oxidative phosphorylation must be strongly regulated to avoid deterioration of the energy supply and cell death, which is caused by toxic uncouplers. However, an H+ transporting uncoupling function is well established only for UCP1 from brown adipose tissue, and the regulation of UCP1 by fatty acids, nucleotides and pH remains controversial. The failure of UCP1 expressed in Escherichia coli inclusion bodies to carry out fatty-acid-dependent H+ transport activity inclusion bodies made us seek a native UCP cofactor. Here we report the identification of coenzyme Q (ubiquinone) as such a cofactor. On addition of CoQ10 to reconstituted UCP1 from inclusion bodies, fatty-acid-dependent H+ transport reached the same rate as with native UCP1. The H+ transport was highly sensitive to purine nucleotides, and activated only by oxidized but not reduced CoQ. H+ transport of native UCP1 correlated with the endogenous CoQ content.
DOI:10.1016/j.bbamem.2003.11.012URLPMID:14757233 [本文引用: 1]

Coenzyme Q (CoQ) is present in all cells and membranes and in addition to be a member of the mitochondrial respiratory chain it has also several other functions of great importance for the cellular metabolism. This review summarizes the findings available to day concerning CoQ distribution, biosynthesis, regulatory modifications and its participation in cellular metabolism. There are a number of indications that this lipid is not always functioning by its direct presence at the site of action but also using e.g. receptor expression modifications, signal transduction mechanisms and action through its metabolites. The biosynthesis of CoQ is studied in great detail in bacteria and yeast but only to a limited extent in animal tissues and therefore the informations available is restricted. However, it is known that the CoQ is compartmentalized in the cell with multiple sites of biosynthesis, breakdown and regulation which is the basis of functional specialization. Some regulatory mechanisms concerning amount and biosynthesis are established and nuclear transcription factors are partly identified in this process. Using appropriate ligands of nuclear receptors the biosynthetic rate can be increased in experimental system which raises the possibility of drug-induced upregulation of the lipid in deficiency. During aging and pathophysiological conditions the tissue concentration of CoQ is modified which influences cellular functions. In this case the extent of disturbances is dependent on the localizaion and the modified distribution of the lipid at cellular and membrane levels.
DOI:10.1046/j.1365-2443.2002.00544.xURLPMID:12059957 [本文引用: 1]

To maintain genome integrity in eukaryotes, DNA must be duplicated precisely once before cell division occurs. A process called replication licensing ensures that chromosomes are replicated only once per cell cycle. Its control has been uncovered by the discovery of the CDKs (cyclin dependent kinases) as master regulators of the cell cycle and the initiator proteins of DNA replication, such as the Origin Recognition Complex (ORC), Cdc6/18, Cdt1 and the MCM complex. At the end of mitosis, the MCM complex is loaded on to chromatin with the aid of ORC, Cdc6/18 and Cdt1, and chromatin becomes licensed for replication. CDKs, together with the Cdc7 kinase, trigger the initiation of replication, recruiting the DNA replicating enzymes on sites of replication. The activated MCM complex appears to play a key role in the DNA unwinding step, acting as a replicating helicase and moves along with the replication fork, at the same time bringing the origins to the unlicensed state. The cycling of CDK activity in the cell cycle separates the two states of replication origins, the licensed state in G1-phase and the unlicensed state for the rest of the cell cycle. Only when CDK drops at the completion of mitosis, is the restriction on licensing relieved and a new round of replication is allowed. Such a CDK-regulated licensing control is conserved from yeast to higher eukaryotes, and ensures that DNA replication takes place only once in a cycle. Xenopus laevis and mammalian cells have an additional system to control licensing. Geminin, whose degradation at the end of mitosis is essential for a new round of licensing, has been shown to bind Cdt1 and negatively regulate it, providing a new insight into the regulation of DNA replication in higher eukaryotes. <p>
DOI:10.1016/S0167-4781(98)00033-5URL [本文引用: 1]
DOI:10.1146/annurev.biochem.68.1.649URL [本文引用: 1]
DOI:10.1093/nar/26.17.3877URL [本文引用: 1]
DOI:10.1074/jbc.272.15.10065URLPMID:9092550 [本文引用: 1]

Replication factor C (RFC) and proliferating cell nuclear antigen (PCNA) are processivity factors for eukaryotic DNA polymerases delta and epsilon. RFC binds to a DNA primer end and loads PCNA onto DNA in an ATP-dependent reaction. The five RFC subunits p140, p40, p38, p37, and p36, all of which are required to form the active RFC complex, share regions of high homology including the defined RFC boxes II-VIII. RFC boxes III and V constitute a putative ATP binding site, whereas the function of the other conserved boxes is unknown. To study the individual subunits in the RFC complex and the role of the RFC boxes, deletion mutations were created in all subunits. Sequences close to the C terminus of each of the small subunits are required for formation of the five subunit complex. A N-terminal region of the small subunits, containing the RFC homology box II, plays a critical role in the function of these subunits, deletion of which reduces but does not abolish RFC activity in loading PCNA onto DNA and in supporting an RFC-dependent replication reaction. The N termini of p37 and p40, although highly homologous, are not interchangeable, suggesting unique functions for the individual subunits.
DOI:10.1073/pnas.96.5.1869URLPMID:10051561 [本文引用: 1]

Proliferating cell nuclear antigen (PCNA) is a processivity factor required for DNA polymerase δ (or ε)-catalyzed DNA synthesis. When loaded onto primed DNA templates by replication factor C (RFC), PCNA acts to tether the polymerase to DNA, resulting in processive DNA chain elongation. In this report, we describe the identification of two separate peptide regions of human PCNA spanning amino acids 36-55 and 196-215 that bind RFC by using the surface plasmon resonance technique. Site-directed mutagenesis of residues within these regions in human PCNA identified two specific sites that affected the biological activity of PCNA. Replacement of the aspartate 41 residue by an alanine, serine, or asparagine significantly impaired the ability of PCNA to (i) support the RFC/PCNA-dependent polymerase δ -catalyzed elongation of a singly primed DNA template; (ii) stimulate RFC-catalyzed DNA-dependent hydrolysis of ATP; (iii) be loaded onto DNA by RFC; and (iv) activate RFC-independent polymerase δ -catalyzed synthesis of poly dT. Introduction of an alanine at position 210 in place of an arginine also reduced the efficiency of PCNA in supporting RFC-dependent polymerase δ -catalyzed elongation of a singly primed DNA template. However, this mutation did not significantly alter the ability of PCNA to stimulate DNA polymerase δ in the absence of RFC but substantially lowered the efficiency of RFC-catalyzed reactions. These results are in keeping with a model in which surface exposed region of PCNA interact with RFC and the subsequent loading of PCNA onto DNA orients the elongation complex in a manner essential for processive DNA synthesis.
DOI:10.1016/j.mod.2016.03.004URLPMID:27039019 [本文引用: 1]

61The Drosophila Erect wing is required for development of the indirect flight muscles.61Erect wing null in myoblasts leads to mispatterned and delayed differentiation of IFM.61Erect wing governs the myoblast fusion events by regulating the expression of Cyclin E.
DOI:10.1080/00071668.2011.590795URLPMID:21919569 [本文引用: 1]

1. The objective of the research was to investigate the molecular evolutionary relationships between the duck myogenic determination factors (MYOD) gene family members and their roles in muscle development. 2. The four members of the duck MYOD gene family were cloned using RT-PCR, and their relative mRNA expression during duck muscle development was measured using qRT-PCR. 3. The results showed that MyoD and Myf5 clustered together, as did MyoG and MRF4 based on their complete amino acid sequence and the basic helix-loop-helix domain. Results of the evolutionary level analysis were consistent with that of the differential expression patterns during duck breast muscle development. As determined by qRT-PCR, MyoD and Myf5 were highly expressed in 22-day embryos, while MyoG and MRF4 expression was high in 14-day embryos. 4. We conclude that the entire MYOD gene family in the duck originated from a common ancestral gene and evolved after two duplication events. The roles of the MYOD gene family members in duck muscle development are similar to those in mammals.
DOI:10.1046/j.1469-7580.2003.00139.xURLPMID:12587921 [本文引用: 1]

During embryogenesis, skeletal muscle forms in the vertebrate limb from progenitor cells originating in the somites. These cells delaminate from the hypaxial edge of the dorsal part of the somite, the dermomyotome, and migrate into the limb bud, where they proliferate, express myogenic determination factors and subsequently differentiate into skeletal muscle. A number of regulatory factors involved in these different steps have been identified. These include Pax3 with its target c-met, Lbx1 and Mox2 as well as the myogenic determination factors Myf5 and MyoD and factors required for differentiation such as Myogenin, Mrf4 and Mef2 isoforms. Mutants for genes such as Lbx1 and Mox2 , expressed uniformly in limb muscle progenitors, reveal unexpected differences between fore and hind limb muscles, also indicated by the differential expression of Tbx genes. As development proceeds, a secondary wave of myogenesis takes place, and, postnatally, satellite cells become located under the basal lamina of adult muscle fibres. Satellite cells are thought to be the progenitor cells for adult muscle regeneration, during which similar genes to those which regulate myogenesis in the embryo also play a role. In particular, Pax3 as well as its orthologue Pax7 are important. The origin of secondary/fetal myoblasts and of adult satellite cells is unclear, as is the relation of the latter to so-called SP or stem cell populations, or indeed to potential mesangioblast progenitors, present in blood vessels. The oligoclonal origin of postnatal muscles points to a small number of founder cells, whether or not these have additional origins to the progenitor cells of the somite which form the first skeletal muscles, as discussed here for the embryonic limb.
DOI:10.1186/2044-5040-1-4URLPMID:21798082 [本文引用: 1]

A highly conserved signaling pathway involving insulin-like growth factor 1 (IGF1), and a cascade of intracellular components that mediate its effects, plays a major role in the regulation of skeletal muscle growth. A central component in this cascade is the kinase Akt, also called protein kinase B (PKB), which controls both protein synthesis, via the kinases mammalian target of rapamycin (mTOR) and glycogen synthase kinase 3?? (GSK3??), and protein degradation, via the transcription factors of the FoxO family. In this paper, we review the composition and function of this pathway in skeletal muscle fibers, focusing on evidence obtained in vivo by transgenic and knockout models and by muscle transient transfection experiments. Although this pathway is essential for muscle growth during development and regeneration, its role in adult muscle response to mechanical load is less clear. A full understanding of the operation of this pathway could help to design molecularly targeted therapeutics aimed at preventing muscle wasting, which occurs in a variety of pathologic contexts and in the course of aging.
DOI:10.1128/MCB.20.17.6600-6611.2000URLPMID:86143 [本文引用: 1]

The differentiation and maturation of skeletal muscle cells into functional fibers is coordinated largely by inductive signals which act through discrete intracellular signal transduction pathways. Recently, the calcium-activated phosphatase calcineurin (PP2B) and the family of transcription factors known as NFAT have been implicated in the regulation of myocyte hypertrophy and fiber type specificity. Here we present an analysis of the intracellular mechanisms which underlie myocyte differentiation and fiber type specificity due to an insulinlike growth factor 1 (IGF-1)-calcineurin-NFAT signal transduction pathway. We demonstrate that calcineurin enzymatic activity is transiently increased during the initiation of myogenic differentiation in cultured C2C12 cells and that this increase is associated with NFATc3 nuclear translocation. Adenovirus-mediated gene transfer of an activated calcineurin protein (AdCnA) potentiates C2C12 and Sol8 myocyte differentiation, while adenovirus-mediated gene transfer of noncompetitive calcineurin-inhibitory peptides (cain or DeltaAKAP79) attenuates differentiation. AdCnA infection was also sufficient to rescue myocyte differentiation in an IGF-depleted myoblast cell line. Using 10T1/2 cells, we demonstrate that MyoD-directed myogenesis is dramatically enhanced by either calcineurin or NFATc3 cotransfection, while a calcineurin inhibitory peptide (cain) blocks differentiation. Enhanced myogenic differentiation directed by calcineurin, but not NFATc3, preferentially specifies slow myosin heavy-chain expression, while enhanced differentiation through mitogen-activated protein kinase kinase 6 (MKK6) promotes fast myosin heavy-chain expression. These data indicate that a signaling pathway involving IGF-calcineurin-NFATc3 enhances myogenic differentiation whereas calcineurin acts through other factors to promote the slow fiber type program.
DOI:10.1152/ajpcell.00553.2009URLPMID:20130207 [本文引用: 1]

Mustn1 (Mustang, musculoskeletal temporally activated novel gene) was originally identified in fracture callus tissue, but its greatest expression is detected in skeletal muscle. Thus, we conducted experiments to investigate the expression and function of Mustn1 during myogenesis. Temporally, quantitative real-time PCR analysis of muscle samples from embryonic day 17 to 12 mo of age reveals that Mustn1 mRNA expression is greatest at 3 mo of age and beyond, consistent with the expression pattern of Myod. In situ hybridization shows abundant Mustn1 expression in somites and developing skeletal muscles, while in adult muscle, Mustn1 is localized to some peripherally located nuclei. Using RNA interference (RNAi), we investigated the function of Mustn1 in C2C12 myoblasts. Though silencing Mustn1 mRNA had no effect on myoblast proliferation, it did significantly impair myoblast differentiation, preventing myofusion. Specifically, when placed in low-serum medium for up to 6 days, Mustn1-silenced myoblasts elongated poorly and were mononucleated. In contrast, control RNAi-treated and parental myoblasts presented as large, multinucleated myotubes. Further supporting the morphological observations, immunocytochemistry of Mustn1-silenced cells demonstrated significant reductions in myogenin (Myog) and myosin heavy chain (Myhc) expression at 4 and 6 days of differentiation as compared with control and parental cells. The decreases in Myog and Myhc protein expression in Mustn1-silenced cells were associated with robust ( approximately 3-fold or greater) decreases in the expression of Myod and desmin (Des), as well as the myofusion markers calpain 1 (Capn1), caveolin 3 (Cav3), and cadherin 15 (M-cadherin; Cadh15). Overall, we demonstrate that Mustn1 is an essential regulator of myogenic differentiation and myofusion, and our findings implicate Myod and Myog as its downstream targets.
DOI:10.4238/2015.May.4.2URLPMID:25966217 [本文引用: 1]

Musculoskeletal embryonic nuclear protein 1 (MUSTN1) gene is involved in myogenic fusion and differentiation in rats. We previously showed the differential expression of MUSTN1 in week (W) 2 and W6 breast muscles of Pekin ducks. In this study, we further investigated its molecular characteristics and expression profiles in different tissues at W7 and in breast and leg muscles at W1, W3, W5, W7, and W9. The relationship between muscle development and muscle fiber areas was also investigated. A 358-bp cDNA sequence was obtained. The coding sequence of duck MUSTN1 cDNA encoded a 78-amino acid sequence, which showed high similarity with those of other species (96% similarity with zebra finch and 94% with chicken). In addition, a 6435-bp genomic DNA sequence of MUSTN1 was obtained. In total, 231 transcription factor-binding sites were found in the promoter region, and many of these transcription factors were involved in the regulation of muscle development. MUSTN1 expression in breast muscle increased from W1 to W5 and then decreased at W9. In leg muscle, the expression increased from W1 to W3 and then decreased. The relative growth rates of breast and leg muscle fibers reached their peaks at W3-W5 and W1-W3, respectively. Since the greatest relative growth rates appeared at the highest expression levels of the MUSTN1 gene, it was thought to play roles in duck muscle development. Our findings would be helpful in understanding the molecular characteristics and functions of the MUSTN1 gene in breast muscle development of ducks.
DOI:10.1126/science.1315077URLPMID:1315077 [本文引用: 1]

The molecular basis of skeletal muscle lineage determination was investigated by analyzing DNA control elements that regulate the myogenic determination gene myoD. A distal enhancer was identified that positively regulates expression of the human myoD gene. The myoD enhancer and promoter were active in myogenic and several nonmyogenic cell lines. In transgenic mouse embryos, however, the myoD enhancer and promoter together directed expression of a lacZ transgene specifically to the skeletal muscle lineage. These data suggest that during development myoD is regulated by mechanisms that restrict accessibility of myoD control elements to positive trans-acting factors.
[本文引用: 1]
DOI:10.1073/pnas.87.20.7988URLPMID:2172969 [本文引用: 1]

Shortly after their birth, postmitotic mononucleated myoblasts in myotomes, limb buds, and conventional muscle cultures elongate and assemble a cohort of myofibrillar proteins into definitively striated myofibrils. MyoD induces a number of immortalized and/or transformed nonmuscle cells to express desmin and several myofibrillar proteins and to fuse into myosacs. We now report that MyoD converts normal dermal fibroblasts, chondroblasts, gizzard smooth muscle, and pigmented retinal epithelial cells into elongated postmitotic mononucleated striated myoblasts. The sarcomeric localization of antibodies to desmin, -actinin, titin, troponin-I, -actin, myosin heavy chain, and myomesin in these converted myoblasts are indistinguishable from in vivo and in vitro normal myoblasts. Converted myoblasts fuse into typical anisodiametric multinucleated myotubes that often contract spontaneously. Conversion and subsequent expression of the skeletal myogenic program are autonomous events, occurring in four nonmuscle microenvironments consisting of different combinations of foreign extracellular matrix molecules. Early events associated with conversion by MyoD involve (i) withdrawal from the cell cycle, (ii) down-regulation of the subverted cell's ongoing differentiation program, and (iii) initiation of desmin synthesis in presumptive myoblasts and dramatic redistribution of microtubules and desmin intermediate filaments in postmitotic myoblasts.
DOI:10.1016/0092-8674(87)90585-XURLPMID:3690668 [本文引用: 1]

5-azacytidine treatment of mouse C3H10T1 2 embryonic fibroblasts converts them to myoblasts at a frequency suggesting alteration of one or only a few closely linked regulatory loci. Assuming such loci to be differentially expressed as poly(A) + RNA in proliferating myoblasts, we prepared proliferating myoblast-specific, subtracted cDNA probes to screen a myocyte cDNA library. Based on a number of criteria, three cDNAs were selected and characterized. We show that expression of one of these cDNAs transfected into C3H10T1 2 fibroblasts, where it is not normally expressed, is sufficient to convert them to stable myoblasts. Myogenesis also occurs, but to a lesser extent, when this cDNA is expressed in a number of other cell lines. The major open reading frame encoded by this cDNA contains a short protein segment similar to a sequence present in the myc protein family.
DOI:10.1073/pnas.86.14.5434URLPMID:2748593 [本文引用: 1]

MyoD is a master regulatory gene for myogenesis. Under the control of a retroviral long terminal repeat, MyoD was expressed in a variety of differentiated cell types by using either a DNA transfection vector or a retrovirus. Expression of muscle-specific proteins was observed in chicken, human, and rat primary fibroblasts and in differentiated melanoma, neuroblastoma, liver, and adipocyte lines. The ability of MyoD to activate muscle genes in a variety of differentiated cell lines suggests that no additional tissue-specific factors other than MyoD are needed to activate the downstream program for terminal muscle differentiation or that, if such factors exist, they are themselves activated by MyoD expression.
DOI:10.1038/364501a0URL [本文引用: 1]
DOI:10.1038/364532a0URL [本文引用: 1]
[本文引用: 1]
DOI:10.1101/gr.7.9.910URL [本文引用: 1]
DOI:10.1073/pnas.94.23.12457URLPMID:9356471 [本文引用: 1]

Myostatin (GDF-8) is a member of the transforming growth factor 尾 superfamily of secreted growth and differentiation factors that is essential for proper regulation of skeletal muscle mass in mice. Here we report the myostatin sequences of nine other vertebrate species and the identification of mutations in the coding sequence of bovine myostatin in two breeds of double-muscled cattle, Belgian Blue and Piedmontese, which are known to have an increase in muscle mass relative to conventional cattle. The Belgian Blue myostatin sequence contains an 11-nucleotide deletion in the third exon which causes a frameshift that eliminates virtually all of the mature, active region of the molecule. The Piedmontese myostatin sequence contains a missense mutation in exon 3, resulting in a substitution of tyrosine for an invariant cysteine in the mature region of the protein. The similarity in phenotypes of double-muscled cattle and myostatin null mice suggests that myostatin performs the same biological function in these two species and is a potentially useful target for genetic manipulation in other farm animals.
DOI:10.1038/ng0997-71URL [本文引用: 1]
DOI:10.1038/387083a0URL [本文引用: 1]
DOI:10.1073/pnas.1525795113URLPMID:26858428 [本文引用: 1]

Sarcopenia, or skeletal muscle atrophy, is a debilitating comorbidity of many physiological and pathophysiological processes, including normal aging. There are no approved therapies for sarcopenia ...
DOI:10.1016/S0006-291X(02)02953-4URLPMID:12559968 [本文引用: 1]

A human therapeutic that specifically modulates skeletal muscle growth would potentially provide a benefit for a variety of conditions including sarcopenia, cachexia, and muscular dystrophy. Myostatin, a member of the TGF-尾 family of growth factors, is a known negative regulator of muscle mass, as mice lacking the myostatin gene have increased muscle mass. Thus, an inhibitor of myostatin may be useful therapeutically as an anabolic agent for muscle. However, since myostatin is expressed in both developing and adult muscles, it is not clear whether it regulates muscle mass during development or in adults. In order to test the hypothesis that myostatin regulates muscle mass in adults, we generated an inhibitory antibody to myostatin and administered it to adult mice. Here we show that mice treated pharmacologically with an antibody to myostatin have increased skeletal muscle mass and increased grip strength. These data show for the first time that myostatin acts postnatally as a negative regulator of skeletal muscle growth and suggest that myostatin inhibitors could provide a therapeutic benefit in diseases for which muscle mass is limiting.
