 ,*河北农业大学 / 华北作物种质资源研究与利用重点实验室 / 河北省棉花产业协同创新中心, 河北保定 071001
,*河北农业大学 / 华北作物种质资源研究与利用重点实验室 / 河北省棉花产业协同创新中心, 河北保定 071001Genome-wide identification of Laccase gene family in update G. hirsutum L. genome and expression analysis under V. dahliae stress
ZHAO Jing**, LI Xu-Tong**, LIANG Xue-Zhong, WANG Zhi-Cheng, CUI Jing, CHEN Bin, WU Li-Qiang, WANG Xing-Fen, ZHANG Gui-Yin, MA Zhi-Ying, ZHANG Yan ,*Hebei Agricultural University / North China Key Laboratory for Crop Germplasm Resources of Education Ministry / Co-Innovation Center for Cotton Industry of Hebei Province, Baoding 071001, Hebei, China
,*Hebei Agricultural University / North China Key Laboratory for Crop Germplasm Resources of Education Ministry / Co-Innovation Center for Cotton Industry of Hebei Province, Baoding 071001, Hebei, China通讯作者:
第一联系人:
收稿日期:2019-04-2接受日期:2019-06-22网络出版日期:2019-07-13
| 基金资助: |
Received:2019-04-2Accepted:2019-06-22Online:2019-07-13
| Fund supported: |
作者简介 About authors
赵晶,E-mail:824802835@qq.com。

摘要
关键词:
Abstract
Keywords:
PDF (6597KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
赵晶, 李旭彤, 梁学忠, 王志城, 崔静, 陈斌, 吴立强, 王省芬, 张桂寅, 马峙英, 张艳. 陆地棉漆酶基因家族鉴定及在黄萎病菌胁迫下的表达分析 *[J]. 作物学报, 2019, 45(12): 1784-1795. doi:10.3724/SP.J.1006.2019.94053
ZHAO Jing, LI Xu-Tong, LIANG Xue-Zhong, WANG Zhi-Cheng, CUI Jing, CHEN Bin, WU Li-Qiang, WANG Xing-Fen, ZHANG Gui-Yin, MA Zhi-Ying, ZHANG Yan.
棉花(Gossypium spp.)是世界上第一大天然纤维作物, 具有重要的经济价值, 也是食品、饲料和生物燃料的重要来源[1]。在生长过程中, 棉花的品质和产量会受到多种生物和非生物胁迫, 其中危害最大的是由大丽轮枝菌(Verticillium dahliae Kleb.)引起的黄萎病[2]。黄萎病菌寄主范围极其广泛, 其产生的菌核对极端温度变化适应能力强, 可在土壤中存活数十年, 因此防治难度很大, 每年造成巨大经济损失[3,4]。研究表明利用品种的遗传抗性可以有效降低黄萎病带来的危害, 实现棉花的高产、稳产[5]。
细胞壁是植物抵御病原体侵袭的天然屏障, 当病原菌侵染时, 寄主植物细胞在细胞壁、胞间层、细胞质等不同部位产生一系列抗性反应, 可以有效地阻止病原菌的再度侵染和蔓延[6,7,8]。已有研究表明, 在细胞壁的木质化过程中, 从薄壁组织细胞内合成的木质醇单体分泌至胞外经脱氢聚合反应形成木质素[9]。植物细胞壁的木质化能够抵抗病原菌侵入的机械压力、抵抗真菌酶类对细胞壁的降解和阻断病原菌与寄主植物间的物质交流[10]。木质素是构成植物细胞壁的成分之一, 而漆酶又是木质素合成途经中的关键酶之一, 因此漆酶在植物细胞壁形成和抗病过程中的作用值得深入研究。
漆酶属于铜蓝蛋白家族, 是一种结合多个铜离子的糖蛋白, 广泛存在于细菌、真菌和动植物中。目前高等植物中已有部分漆酶基因成员的研究报道, 如短柄草Laccase5对其细胞壁木质化具有重要作用[11]; 亚洲棉漆酶基因GaLAC1导入新疆杨后能够导致转基因植株中总木质素含量的增加, 表明GaLAC1参与了植物木质素的合成[12]; 赵先炎等[13]研究表明, 增强番茄中漆酶基因的活性, 可以提高植株对酚类物质的抗性; 田奇琳等[14]克隆了龙眼DlLac7基因, 并发现其可能参与茉莉酸、水杨酸和脱落酸的逆境胁迫信号转导途径, 调控龙眼多种非生物胁迫应答过程; 黄晨等[15]发现龙井茶树在茉莉酸处理、机械损伤和茶尺蠖取食后, 漆酶基因CsLAC4和CsLAC12的表达量有明显的变化。此外, 在水稻[16]、甜高粱[17]和亚麻[18]中, 漆酶基因家族成员也被相继鉴定出来, 由此可见, 不同植物漆酶基因在生物和非生物胁迫中具有重要功能。
已有关于漆酶基因家族的研究多集中在二倍体植物[16,17,18], 棉花属于异源四倍体, 基因组庞大而复杂, 目前漆酶基因在栽培棉花中的研究报道相对较少, 课题组前期基于SSH文库筛选到GhLAC15, 研究发现其通过防御诱导的木质素的合成增强黄萎病抗性[19]; Hu等[20]超表达棉花GhLAC1基因同时提高了植株对黄萎病菌及棉铃虫的抗性。Balasubramanian等[21,22]以2015年公布的陆地棉TM-1基因组为参考, 通过全基因组鉴定LAC家族成员并明确了不同成员在陆地棉纤维发育中的表达模式。随着测序技术的不断提升和基因组质量的进一步完善, Wang等[23]通过三代基因组获得的最新的陆地棉TM-1基因组较之前版本基因组质量有很大提升, 为全基因组基因家族分析提供更为可靠的参考序列; 另外棉花中漆酶基因家族除目前已报道的两个成员外[19,20], 其他成员是否参与抗黄萎病反应尚不清楚。因此, 本研究参考最新的陆地棉TM-1基因组, 利用生物信息学方法, 提取并鉴定得LAC基因家族成员, 并对各成员的理化性质、基因结构、系统进化和染色体定位等方面进行系统分析, 结合黄萎病胁迫处理后的转录组数据, 明确LAC基因家族成员的表达规律, 为深入解析棉花LAC基因的抗病功能及分子机制奠定基础。
1 材料与方法
1.1 基于陆地棉TM-1三代基因组的漆酶基因家族的鉴定及理化性质分析
本研究利用最新公布的陆地棉TM-1基因组序列为参考, 相关基因组和蛋白质组数据从COTTONGEN (1.2 陆地棉漆酶基因家族的系统进化分析
利用MEGA 7软件中的Clustal W功能对83个陆地棉LAC蛋白序列和17个拟南芥LAC蛋白序列进行序列比对, 然后基于序列比对的结果, 采用邻近法构建系统进化树, 其中Bootstrap值设定为1000。1.3 陆地棉漆酶基因家族的染色体定位、基因结构和保守结构域分析
从陆地棉基因组注释文件中提取LAC基因家族成员的染色体位置信息, 利用MapChart软件2.32绘制LAC基因的染色体定位图, GSDS在线软件(1.4 陆地棉漆酶基因家族在黄萎病胁迫下的表达模式分析
本课题组前期以抗病陆地棉品种农大601为试验材料, 获得在处理组(黄萎病菌胁迫处理后) 2、6、12、24和48 h和对照组(各对应时间点接种水)的根部转录组数据。将陆地棉LAC基因家族各成员的表达量数据进行log2(1+RPKM)处理后, 利用MEV软件, 绘制LAC基因家族各成员的表达热图。1.5 陆地棉漆酶基因家族在黄萎病胁迫下的表达模式分析
根据陆地棉LAC家族基因的cDNA信息, 采用Primer 5.0软件在基因序列5′端、ORF或3′端的特异区域设计引物(表1), 扩增片段约为200 bp。分别以陆地棉农大601根、茎、叶不同组织和接菌后6、12、24、36和48 h的根组织cDNA为模板, 利用qRT-PCR方法检测候选基因的表达情况, 设置每个样本3次重复, 内参基因为GhUBQ14 [25]。qRT-PCR体系为10.0 μL, 其中cDNA 1 μL、2×SYBR预混液5.0 μL、基因特异正向引物(10 μmol L-1) 0.5 μL、反向引物(10 μmol L-1) 0.5 μL、RNase-Free H2O 2.8 μL、ROX校正液0.2 μL。qRT-PCR在ABI7500仪器反应, 反应程序为94℃预变性30 s; 95℃变性5 s, 57℃退火5 s, 72℃延伸34 s, 40个循环。采用2-ΔΔCt方法计算基因的相对表达量[26]。Table 1
表1
表1陆地棉LAC基因家族信息
Table 1
| 基因名称 Gene name | 基因ID Gene ID | 染色体定位 Chromosome location | 基因大小 Gene size (bp) | 蛋白 Protein (aa) | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| GhLAC01 | Ghir_A01G021510 | A01:116737831-116742782 | 4952 | 573 | 胞外Extracellular |
| GhLAC02 | Ghir_A01G021950 | A01:117127088-117132069 | 4982 | 558 | 胞外Extracellular |
| GhLAC03 | Ghir_A02G006480 | A02:10086940-10089681 | 2742 | 580 | 胞外Extracellular |
| GhLAC04 | Ghir_A03G005270 | A03:9138986-9141467 | 2482 | 420 | 胞外Extracellular |
| GhLAC05 | Ghir_A03G005280 | A03:9197235-9204292 | 7058 | 568 | 胞外Extracellular |
| GhLAC06 | Ghir_A03G005300 | A03:9235739-9242786 | 7048 | 571 | 胞外Extracellular |
| GhLAC07 | Ghir_A03G005800 | A03:10413905-10417019 | 3115 | 434 | 胞外Extracellular |
| GhLAC08 | Ghir_A03G007710 | A03:17682089-17684169 | 2081 | 560 | 胞外Extracellular |
| GhLAC09 | Ghir_A04G009430 | A04:71412678-71434702 | 22025 | 531 | 胞外Extracellular |
| GhLAC10 | Ghir_A04G009440 | A04:71412678-71415616 | 2939 | 485 | 胞外Extracellular |
| GhLAC11 | Ghir_A04G009460 | A04:71525265-71528510 | 3246 | 570 | 胞外Extracellular |
| GhLAC12 | Ghir_A04G009470 | A04:71627753-71631136 | 3384 | 576 | 胞外Extracellular |
| GhLAC13 | Ghir_A05G009230 | A05:8476727-8479903 | 3177 | 556 | 胞外Extracellular |
| 基因名称 Gene name | 基因ID Gene ID | 染色体定位 Chromosome location | 基因大小 Gene size (bp) | 蛋白 Protein (aa) | 亚细胞定位 Subcellular localization |
| GhLAC14 | Ghir_A05G010150 | A05:9143475-9146068 | 2594 | 579 | 胞外Extracellular |
| GhLAC15 | Ghir_A05G010190 | A05:9194020-9196191 | 2172 | 574 | 胞外Extracellular |
| GhLAC16 | Ghir_A05G025290 | A05:25890847-25893461 | 2615 | 537 | 胞外Extracellular |
| GhLAC17 | Ghir_A05G025340 | A05:25981068-25989683 | 8616 | 566 | 胞外Extracellular |
| GhLAC18 | Ghir_A05G025350 | A05:26015294-26017921 | 2628 | 563 | 胞外Extracellular |
| GhLAC19 | Ghir_A05G031190 | A05:41692291-41698066 | 5776 | 555 | 胞外Extracellular |
| GhLAC20 | Ghir_A05G031330 | A05:42716069-42718551 | 2483 | 551 | 胞外Extracellular |
| GhLAC21 | Ghir_A06G012170 | A06:67269136-67271306 | 2171 | 558 | 胞外Extracellular |
| GhLAC22 | Ghir_A06G017280 | A06:116079600-116081862 | 2263 | 522 | 胞外Extracellular |
| GhLAC23 | Ghir_A06G017300 | A06:116223123-116225599 | 2477 | 518 | 胞外Extracellular |
| GhLAC24 | Ghir_A06G017320 | A06:116280847-116285027 | 4181 | 562 | 胞外Extracellular |
| GhLAC25 | Ghir_A08G021230 | A08:116971139-116973684 | 2546 | 576 | 胞外Extracellular |
| GhLAC26 | Ghir_A09G016340 | A09:72475738-72478310 | 2573 | 583 | 胞外Extracellular |
| GhLAC27 | Ghir_A10G009410 | A10:18971904-18974558 | 2655 | 562 | 胞外Extracellular |
| GhLAC28 | Ghir_A10G023410 | A10:112560879-112563710 | 2832 | 554 | 胞外Extracellular |
| GhLAC29 | Ghir_A10G024200 | A10:114093313-114095981 | 2669 | 569 | 胞外Extracellular |
| GhLAC30 | Ghir_A11G010610 | A11:9769950-9772299 | 2350 | 570 | 胞外Extracellular |
| GhLAC31 | Ghir_A11G035330 | A11:122965170-122967669 | 2500 | 580 | 胞外Extracellular |
| GhLAC32 | Ghir_A11G035350 | A11:122971778-122974013 | 2236 | 583 | 胞外Extracellular |
| GhLAC33 | Ghir_A11G035490 | A11:123045551-123048240 | 2690 | 556 | 胞外Extracellular |
| GhLAC34 | Ghir_A12G012190 | A12:80351644-80353970 | 2327 | 572 | 胞外Extracellular |
| GhLAC35 | Ghir_A13G001780 | A13:2014792-2017130 | 2339 | 462 | 胞外Extracellular |
| GhLAC36 | Ghir_A13G002160 | A13:2565151-2567561 | 2411 | 553 | 胞外Extracellular |
| GhLAC37 | Ghir_A13G002170 | A13:2580835-2585183 | 4349 | 447 | 胞外Extracellular |
| GhLAC38 | Ghir_A13G002350 | A13:2736016-2738294 | 2279 | 563 | 胞外Extracellular |
| GhLAC39 | Ghir_A13G003100 | A13:3705196-3707259 | 2064 | 537 | 胞外Extracellular |
| GhLAC40 | Ghir_A13G023990 | A13:107564582-107569198 | 4617 | 577 | 胞外Extracellular |
| GhLAC41 | Ghir_D01G023050 | D01:62359392-62363270 | 3879 | 569 | 胞外Extracellular |
| GhLAC42 | Ghir_D01G023480 | D01:62708521-62717375 | 8855 | 558 | 胞外Extracellular |
| GhLAC43 | Ghir_D02G006860 | D02:9508400-9511507 | 3108 | 580 | 胞外Extracellular |
| GhLAC44 | Ghir_D03G010220 | D03:35974903-35978183 | 3281 | 534 | 胞外Extracellular |
| GhLAC45 | Ghir_D03G013060 | D03:43363895-43367056 | 3162 | 556 | 胞外Extracellular |
| GhLAC46 | Ghir_D03G013490 | D03:44287872-44290796 | 2925 | 453 | 胞外Extracellular |
| GhLAC47 | Ghir_D03G013500 | D03:44314668-44316870 | 2203 | 531 | 胞外Extracellular |
| GhLAC48 | Ghir_D03G015740 | D03:48218625-48221114 | 2490 | 576 | 胞外Extracellular |
| GhLAC49 | Ghir_D04G013650 | D04:44922823-44925840 | 3018 | 573 | 胞外Extracellular |
| GhLAC50 | Ghir_D04G013660 | D04:45073981-45077137 | 3157 | 573 | 胞外Extracellular |
| 基因名称 Gene name | 基因ID Gene ID | 染色体定位 Chromosome location | 基因大小 Gene size (bp) | 蛋白 Protein (aa) | 亚细胞定位 Subcellular localization |
| GhLAC51 | Ghir_D04G013670 | D04:45158044-45161359 | 3316 | 592 | 胞外Extracellular |
| GhLAC52 | Ghir_D04G013680 | D04:45181487-45184916 | 3430 | 567 | 胞外Extracellular |
| GhLAC53 | Ghir_D04G013870 | D04:45597220-45599865 | 2646 | 556 | 胞外Extracellular |
| GhLAC54 | Ghir_D05G009240 | D05:7540512-7543586 | 3075 | 556 | 胞外Extracellular |
| GhLAC55 | Ghir_D05G009870 | D05:8212961-8215461 | 2501 | 579 | 胞外Extracellular |
| GhLAC56 | Ghir_D05G025170 | D05:23475679-23484348 | 8670 | 552 | 胞外Extracellular |
| GhLAC57 | Ghir_D05G025180 | D05:23486473-23489452 | 2980 | 531 | 胞外Extracellular |
| GhLAC58 | Ghir_D05G025190 | D05:23514531-23517069 | 2539 | 563 | 胞外Extracellular |
| GhLAC59 | Ghir_D05G025200 | D05:23542441-23557617 | 15177 | 566 | 胞外Extracellular |
| GhLAC60 | Ghir_D05G025220 | D05:23575706-23578427 | 2722 | 562 | 胞外Extracellular |
| GhLAC61 | Ghir_D05G031070 | D05:33904186-33908534 | 4349 | 555 | 胞外Extracellular |
| GhLAC62 | Ghir_D06G012330 | D06:29988284-29990547 | 2264 | 571 | 胞外Extracellular |
| GhLAC63 | Ghir_D06G018210 | D06:59288750-59291253 | 2504 | 568 | 胞外Extracellular |
| GhLAC64 | Ghir_D06G018220 | D06:59357081-59359262 | 2182 | 568 | 胞外Extracellular |
| GhLAC65 | Ghir_D06G018250 | D06:59500342-59502844 | 2503 | 568 | 胞外Extracellular |
| GhLAC66 | Ghir_D08G022010 | D08:63494311-63496810 | 2500 | 576 | 胞外Extracellular |
| GhLAC67 | Ghir_D09G015810 | D09:44179260-44181960 | 2701 | 583 | 胞外Extracellular |
| GhLAC68 | Ghir_D10G009840 | D10:11987141-11989232 | 2092 | 541 | 胞外Extracellular |
| GhLAC69 | Ghir_D10G025960 | D10:66026294-66029136 | 2843 | 554 | 胞外Extracellular |
| GhLAC70 | Ghir_D10G026620 | D10:67161382-67164003 | 2622 | 569 | 胞外Extracellular |
| GhLAC71 | Ghir_D11G010590 | D11:9024231-9027561 | 3331 | 574 | 胞外Extracellular |
| GhLAC72 | Ghir_D11G036190 | D11:72693575-72696286 | 2712 | 570 | 胞外Extracellular |
| GhLAC73 | Ghir_D11G036210 | D11:72700213-72702426 | 2214 | 583 | 胞外Extracellular |
| GhLAC74 | Ghir_D11G036340 | D11:72775350-72777973 | 2624 | 556 | 胞外Extracellular |
| GhLAC75 | Ghir_D12G012430 | D12:41466012-41468206 | 2195 | 569 | 胞外Extracellular |
| GhLAC76 | Ghir_D13G002060 | D13:1759037-1761649 | 2613 | 576 | 胞外Extracellular |
| GhLAC77 | Ghir_D13G002440 | D13:2206264-2213052 | 6789 | 567 | 胞外Extracellular |
| GhLAC78 | Ghir_D13G002640 | D13:2386833-2389243 | 2411 | 563 | 胞外Extracellular |
| GhLAC79 | Ghir_D13G003370 | D13:3233418-3235743 | 2326 | 563 | 胞外Extracellular |
| GhLAC80 | Ghir_D13G003390 | D13:3256191-3258374 | 2184 | 557 | 胞外Extracellular |
| GhLAC81 | Ghir_D13G024730 | D13:62656628-62661237 | 4610 | 435 | 胞外Extracellular |
| GhLAC82 | Ghir_A03G023780 | Scaffold2615:13682-16169 | 2488 | 576 | 胞外Extracellular |
| GhLAC83 | Ghir_A08G026500 | Scaffold2204:49182-53499 | 4318 | 564 | 胞外Extracellular |
新窗口打开|下载CSV
2 结果与分析
2.1 陆地棉LAC基因家族成员的鉴定
通过Hmmsearch搜索和数据库验证, 在陆地棉基因组中共鉴定出83个LAC基因家族成员, 根据其在染色体上的位置信息, 命名为GhirLAC01~ GhirLAC83 (表1), 其中GhirLAC82和GhirLAC83没有定位到染色体上。除A亚组的第7染色体和D亚组的第7染色外, 其他染色体都不均匀地分布着LAC基因(图1)。所有LAC基因所编码蛋白的氨基酸数量在420~583个之间, 差异较小, 且所有基因的亚细胞定位结果都在胞外, 属于分泌蛋白。图1
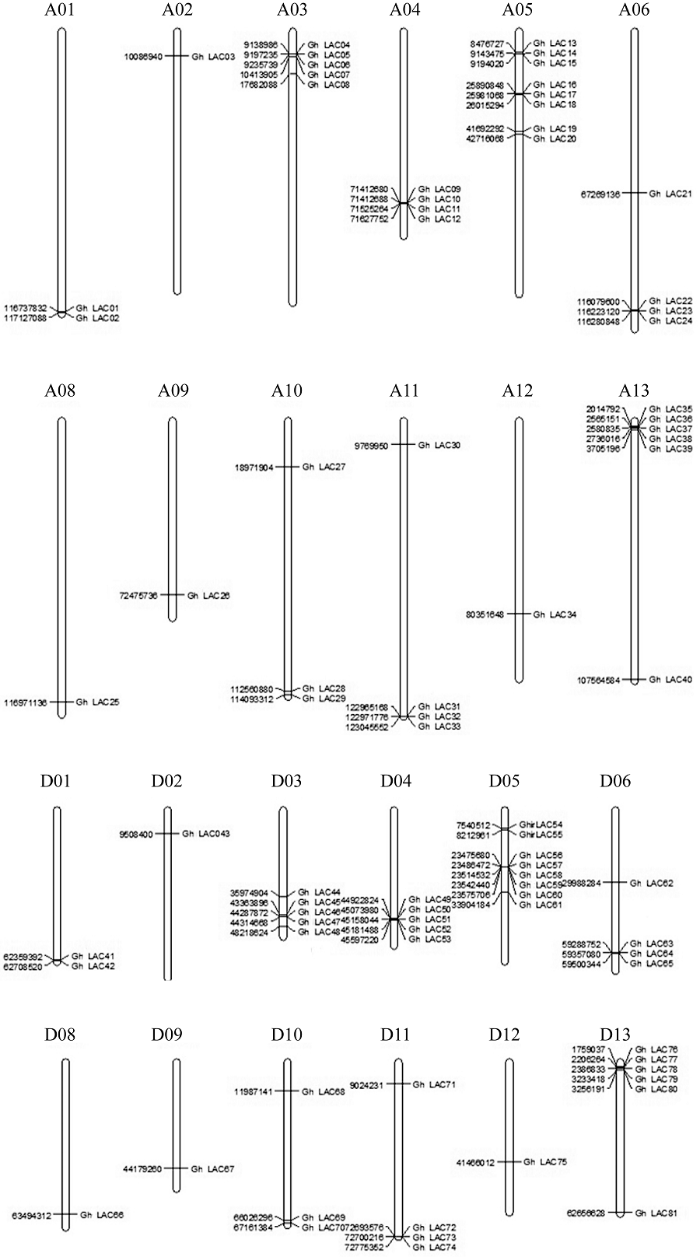 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1陆地棉LAC基因家族成员的染色体定位分析
Fig. 1Chromosome location of LAC family members in G. hirsutum L.
2.2 陆地棉漆酶家族基因系统发育分析
用83个陆地棉GhirLACs蛋白序列和17个拟南芥AtLACs蛋白序列构建系统发育树, 并根据拟南芥漆酶基因的功能[27], 将棉花漆酶分为7组(图2)。第1组包括23个陆地棉漆酶成员和2个拟南芥漆酶成员(AtLAC2和AtKAC17), 第2组包括22个陆地棉漆酶成员和4个拟南芥漆酶成员(AtLAC4、AtLAC10、AtLAC11和AtLAC16), 拟南芥AtKAC17、AtLAC4和AtLAC11在调控木质素单体和细胞壁木质化方面具有重要作用, 属于参与木质素生物合成的单木质素漆酶, 基于该进化树结果, 初步认为棉花第1组与第2组漆酶成员具有类似的功能[27,28,29]。第3组包括13个陆地棉漆酶成员和4个拟南芥漆酶基因(AtLAC3、AtLAC5、AtLAC12和AtLAC13), 其中AtLAC5、AtLAC12和AtLAC13对ABA信号有响应[29]; 第4组包括12个陆地棉漆酶成员和2个拟南芥漆酶基因(AtLAC14和AtLAC15), 可能与干旱、低温和盐胁迫有关[29]; 第5组包括9个陆地棉漆酶成员和3个拟南芥漆酶成员(AtLAC7、AtLAC8和AtLAC9); 第6组只有1个拟南芥漆酶成员AtLAC1; 第7组包括4个陆地棉漆酶成员和1个拟南芥漆酶成员(AtLAC6), 其在生物胁迫下表达会下调[30]。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2陆地棉和拟南芥LACs基因家族进化树分析
Fig. 2Phylogenic tree of LACs family members in Arabidopsis and G. hirsutum L.
2.3 陆地棉漆酶基因家族的结构和保守域分析
对陆地棉LAC基因家族83个成员的基因结构和保守域分析结果如图3所示, 所有LAC基因的外显子个数在4~7个之间变化, 且基本都含有相同的保守结构域(Motif 1、Motif 2、Motif 3、Motif4、Motif5、Motif 9和Motif 10; 图4)。经Pfam数据库验证, Motif 1和Motif 9完全对应于Cu_oxidase_3结构域; Motif 3与Cu_oxidase结构域相对应; Motif2和Motif4则构成了Cu_oxidase_2结构域。某些保守基序只存在于某一亚组中, 如Motif 8只存在于第7亚组中, motif是蛋白质分子中具有特定空间构象和特定功能的结构成分, 是结构域的亚单位, 与特定的功能联系在一起, 推测具有不同motif的漆酶成员在进化和功能上可能存在差异。图3
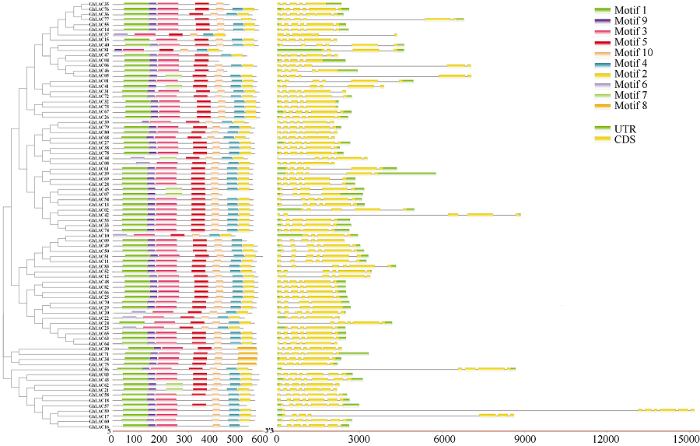 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3陆地棉LAC家族成员基因结构分析
Fig. 3Gene structure analysis of LAC family members in G. hirsutum L.
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4陆地棉LAC基因家族保守基序
Fig. 4Conservative motifs of LAC gene family in G. hirsutum L.
2.4 陆地棉漆酶基因家族在黄萎病胁迫下的表达分析
基于本课题组前期已有的抗病农大601黄萎病菌胁迫下的根转录组数据[31], 将不同处理时间点下的RPKM值进行log2(1+RPKM) 处理后, 构建LAC基因家族成员的表达热图。结果显示, 在83个陆地棉LAC基因中, 有80个基因的表达量在黄萎病胁迫下都发生了显著的变化。80个差异基因在根中的表达模式分为3种类型, 第1类基因呈现出病原菌诱导后表达上调, 包括GhirLAC79、GhirLAC80、GhirLAC43等24个成员。根据该类基因对黄萎病菌的响应速度和程度, 进一步划分为3个亚组, 即快速上调表达基因(I), 包括GhirLAC79、GhirLAC80、GhirLAC06、GhirLAC02和GhirLAC30, 这些成员在接菌后2 h基因表达达到高峰; 较快上调表达基因(II), 包括GhirLAC20、GhirLAC77、GhirLAC15等9个成员, 该亚组基因在接菌后6 h达到表达高峰; 以及第III亚组, 包括GhirLAC01、GhirLAC18、GhirLAC58等10个成员, 受黄萎病菌诱导后这些基因在12~24 h之间达到表达高峰。第2类基因受病原菌胁迫后呈现不同程度下调表达, 包括GhirLAC07、GhirLAC40、GhirLAC45、GhirLAC72等56个成员, 接菌后这些基因的表达明显受到抑制, 暗示这些基因在棉花抗黄萎病反应中起负调控作用; 第3类基因不响应黄萎病菌处理, 推测其不参与棉花抗黄萎病过程, 包括GhirLAC11、GhirLAC27和GhirLAC49 (图5)。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5陆地棉LAC基因在黄萎病胁迫下的表达热图
Fig. 5Expression analysis of LAC genes under V. dahliae stress based on transcriptome data
2.5 陆地棉漆酶基因家族在黄萎病胁迫下的表达分析
结合抗病转录组数据、全长cDNA文库筛选以及功能尚未报道的前提, 本研究进一步挑选出3个分别与拟南芥AtLAC4、AtLAC11和AtLAC12直系同源的新基因, 分别命名为GhLAC4 (GhirLAC02)、GhLAC11 (GhirLAC38)和GhLAC12 (GhirLAC20)。荧光定量PCR结果表明, GhLAC4在接黄萎病后6 h表达量显著提高; GhLAC11受黄萎病菌诱导明显, 接菌后6 h快速响应, 24 h达到表达高峰; GhLAC12的表达在接菌后各时间点均显著上调。这3个基因的荧光定量结果与转录组数据的趋势相吻合, 该结果在进一步明确三个基因的表达规律的同时也反映了转录组数据的可靠性(图6)。组织特异性表达结果显示3个基因在棉花根、茎、叶中均有表达; GhLAC4在茎中表达量最高, GhLAC11和GhLAC12在根中表达量最高(图6)。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6黄萎病菌胁迫下GhLAC4、GhLAC11和GhLAC12基因的表达
Fig. 6Gene relative expression of GhLAC4, GhLAC11, and GhLAC12 under V. dahliae stress
3 讨论
基因组质量的完整性是全基因组水平分析基因家族的基础。随着基因组测序技术的不断深入, 棉花基因组从二倍体棉到四倍体陆地棉不断更新[22,32-33]。基于二代基因组测序组装获得的四倍体陆地棉TM-1的基因组草图为开展全基因组水平分析提供了可能, 但由于棉花属于异源多倍体, 基因组庞大而复杂, 高质量版本的基因组成为提升基因家族分析准确性的必要条件。最新的研究释放了基于超高深度测序、10×Genomics、BioNano光学图谱及Hi-C数据等多技术联合组装的陆地棉TM-1基因组[23], 较以前发表的基因组草图在连续性和完整性上提高了10~20倍, 并对具有高度重复的区域(如着丝粒)进行了成功组装。此外, 新版本基因组预测出72,761。个高度可信的蛋白编码基因, 较旧版本预测的70,478个蛋白编码基因多出了2283个新基因。因此, 利用更新的陆地棉参考基因组分析复杂的基因家族, 将为深入研究该基因家族的功能提供更为准确的信息本研究通过生物信息学方法从高质量的陆地棉TM-1全基因组中鉴定出83个漆酶基因成员, 经过分析发现, 所有基因的外显子个数在4~7之间, 所编码蛋白的氨基酸数量在500个左右, 且基本含有相同的保守基序, 表明基因家族不同成员之间具有较强的保守性。已有研究报道拟南芥AtLAC17、AtLAC4和AtLAC11属于参与木质素生物合成的单木质素漆酶, 在木质素合成方面具有重要功能, 基于本研究棉花漆酶与拟南芥直系同源基因进化树分析, 初步推测分别与拟南芥AtLAC17、AtLAC4和AtLAC11基因聚为一组的棉花漆酶成员可能具有类似的功能[27,28,29]。本研究结果与2016年报道的一篇关于棉花漆酶基因家族的分析结果存在较大差异。本研究基于高质量的TM-1基因组鉴定到83个LAC成员, 除At07和Dt07染色体外, 其他染色体都不均匀地分布着LAC基因, 其中At05和Dt05上LAC基因分布最多, 分别为8个; Balasubramanian等[21]基于旧版本的TM-1基因组草图鉴定到84个LAC成员, 其结果显示在3条染色体At07、Dt07、At08中均没有LAC的分布, LAC成员分布最多的染色体位于At05, 分布着11个LAC基因。这些信息的不匹配性为我们深入研究LAC基因家族带来很大困扰甚至是误导, 因此本研究以目前最高质量的基因组版本系统分析LAC基因家族, 为深入研究LAC重要成员的功能奠定基础。
细胞壁是抵御病原菌入侵的天然屏障, 木质素是细胞壁的重要组成部分, 在植物抗病防御反应中发挥重要功能。漆酶位于木质素合成途径的最后一步, 负责将G、S、H不同木质素单体交联聚合形成多聚物-木质素。自然条件下, 细胞壁作为植物固有的基本组成部分保证植株生长发育, 在此过程中漆酶参与的木质素合成被定义为specific-lignification, 如拟南芥AtLAC17、AtLAC4和AtLAC11[27,28,29]; Chezem等[34]研究表明拟南芥MYB15可以激活防御诱导的木质素的合成, 参与该过程的木质素合成基因受丁香加单包杆菌的诱导表达, 课题组前期的研究也发现GhLAC15 (GhirLAC59)对黄萎病菌迅速响应, 其表达趋势与本研究GhirLAC59在转录组中的表达趋势类似, 超表达GhLAC15基因通过增加植株防御诱导的木质素的合成而提高抗病性[19]。Hu等[20]研究了棉花GhLAC1基因, 发现超表达GhLAC1的植株木质素增加, 从而提高了对黄萎病和棉铃虫的抗性; 抑制GhLAC1基因植株苯丙烷代谢流发生改变, 木质素合成降低而黄酮代谢增强, 同时还激活了植物激素茉莉酸的合成, 从而富集了更多对病虫有害的次生代谢物质。自然条件下, 植物生长面临生物和非生物胁迫等复杂的外界环境, 植物通过能量与物质的重新分配来调控生长与防御的平衡[35]。由此推测, 病菌胁迫条件下, 植物暂停或暂缓生长发育进度, 能量物质向防御反应方向倾斜。从本研究的GhirLAC家族的表达结果来看, 其中属于第二类的各成员受病菌胁迫后, 基因的表达量或快速、或大幅度下调, 推测这些基因在调控生长与防御的平衡方面发挥重要作用。此外, 从第一类基因的表达来看, 该类基因的大部分成员在自然生长状态下处于低或较低表达水平, 受病菌诱导后这些基因在某个时间点达到表达峰值, 随后表达水平逐渐回落, 表明这些基因属于防御诱导的一类抗病漆酶基因, 对病原菌迅速响应, 推测植株一旦防御体系建立, 为了协调防御和植株生长发育, 该类基因的表达随后降低。综上, 本研究基于GhirLAC表达, 认为80个响应黄萎病菌的GhirLAC成员在棉花抗黄萎病反应中具有功能, 但第一类基因可能侧重于增加木质素的合成提高抗病性; 第二类基因可能侧重于改变苯丙烷代谢流来提高抗病性; 二者也可能存在部分交叉。该研究结果为进一步研究GhirLAC的功能提供一定指导。
4 结论
本研究鉴定了陆地棉基因组中的Laccase基因家族, 83个GhirLAC家族成员分布在24条染色体上, 所有GhirLAC蛋白均定位在胞外, GhirLAC家族成员可分为7个亚组。黄萎病菌胁迫下, GhirLAC家族各成员的表达分为3种模式, 其中第1类和第2类GhirLAC基因在棉花抗黄萎病反应中发挥重要功能。GhirLAC02 (GhLAC4)、GhirLAC38 (GhLAC11)和GhirLAC20 (GhLAC12) 3个候选基因均受黄萎病菌诱导表达。本研究为以后深入解析棉花GhirLAC基因的抗病功能及分子机制奠定基础。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1111/j.1365-313X.2011.04491.xURL [本文引用: 1]

P>Cotton is an important cash crop worldwide, and is a significant source of fiber, feed, foodstuff, oil and biofuel products. Considerable effort has been expended to increase sustainable yield and quality through molecular breeding and genetic engineering of new cotton cultivars. Given the recent availability of the whole-genome sequence of cotton, it is necessary to develop molecular tools and resources for large-scale analysis of gene functions at the genome-wide level. We have successfully developed an Agrobacterium-mediated virus-induced gene silencing (VIGS) assay in several cotton cultivars with various genetic backgrounds. The genes of interest were potently and readily silenced within 2 weeks after inoculation at the seedling stage. Importantly, we showed that silencing GhNDR1 and GhMKK2 compromised cotton resistance to the infection by Verticillium dahliae, a fungal pathogen causing Verticillium wilt. Furthermore, we developed a cotton protoplast system for transient gene expression to study gene functions by a gain-of-function approach. The viable protoplasts were isolated from green cotyledons, etiolated cotyledons and true leaves, and responded to a wide range of pathogen elicitors and phytohormones. Remarkably, cotton plants possess conserved, but also distinct, MAP kinase activation with Arabidopsis upon bacterial elicitor flagellin perception. Thus, using gene silencing assays, we have shown that GhNDR1 and GhMKK2 are required for Verticillium resistance in cotton, and have developed high throughput loss-of-function and gain-of-function assays for functional genomic studies in cotton.
DOI:10.1016/j.plantsci.2005.02.008URL [本文引用: 1]
DOI:10.1094/MPMI-03-17-0067-RURL [本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]

本文系统总结了本研究室从1982年起开展棉花黄萎病抗性遗传育种所取得的一些研究结果。 18年4个轮次的研究证明, 棉花黄萎病抗性表现为多个显性单基因的遗传模式, 从而基本 上澄清了国际上长期争论的难题。 通过我国黄萎病病原菌的遗传变异和致病力分化分析, 提出了棉花抗黄萎病育种中鉴别菌株的选择, 抗源的合理利用,
URL [本文引用: 1]

本文系统总结了本研究室从1982年起开展棉花黄萎病抗性遗传育种所取得的一些研究结果。 18年4个轮次的研究证明, 棉花黄萎病抗性表现为多个显性单基因的遗传模式, 从而基本 上澄清了国际上长期争论的难题。 通过我国黄萎病病原菌的遗传变异和致病力分化分析, 提出了棉花抗黄萎病育种中鉴别菌株的选择, 抗源的合理利用,
[本文引用: 1]
DOI:10.1094/Phyto-86-614URL [本文引用: 1]
DOI:10.1111/mpp.2006.7.issue-2URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S0031-9422(96)00595-XURL [本文引用: 1]
DOI:10.1104/pp.114.255489URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.16420/j.issn.0513-353x.2014-1079URL [本文引用: 1]

通过BLAST分析,获得了番茄漆酶(Laccase,LAC)基因相关的15个EST片段,并对这些EST进行了同源比较和序列拼接,得到1个含有小分子RNA miR397识别位点的LAC片段,命名为LeLACmiR397。根据蛋白质结构域推测,LeLACmiR397蛋白存在1个Cu–氧化酶结构域。LeLACmiR397时空表达分析表明,其在番茄根、花、成熟果实和愈伤组织中特异性表达,而在叶中不表达。根据该片段的核苷酸序列信息进行5′-和3′-RACE扩增,得到了番茄LAC基因的全长cDNA(LeLACmiR397,登录号EU503151),其在番茄基因组中对应的基因为Solyc07g049460.2.1。通过在番茄中过表达miR397a基因,发现转基因番茄中LeLACmiR397表达量降低,同时其PPO、POD、SOD含量也有所下降。用丁香酸和芥子酸处理转基因植株幼苗,其根系比非转基因植株更长,抗性增强,表明LeLACmiR397与番茄植株的抗性反应有关。
DOI:10.16420/j.issn.0513-353x.2014-1079URL [本文引用: 1]

通过BLAST分析,获得了番茄漆酶(Laccase,LAC)基因相关的15个EST片段,并对这些EST进行了同源比较和序列拼接,得到1个含有小分子RNA miR397识别位点的LAC片段,命名为LeLACmiR397。根据蛋白质结构域推测,LeLACmiR397蛋白存在1个Cu–氧化酶结构域。LeLACmiR397时空表达分析表明,其在番茄根、花、成熟果实和愈伤组织中特异性表达,而在叶中不表达。根据该片段的核苷酸序列信息进行5′-和3′-RACE扩增,得到了番茄LAC基因的全长cDNA(LeLACmiR397,登录号EU503151),其在番茄基因组中对应的基因为Solyc07g049460.2.1。通过在番茄中过表达miR397a基因,发现转基因番茄中LeLACmiR397表达量降低,同时其PPO、POD、SOD含量也有所下降。用丁香酸和芥子酸处理转基因植株幼苗,其根系比非转基因植株更长,抗性增强,表明LeLACmiR397与番茄植株的抗性反应有关。
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.3390/ijms18010001URL [本文引用: 2]
DOI:10.3389/fpls.2017.00714URL [本文引用: 2]
DOI:10.1186/s12870-017-1072-9URL [本文引用: 2]
DOI:10.1111/mpp.2019.20.issue-3URL [本文引用: 3]
[本文引用: 3]
[本文引用: 2]
DOI:10.1038/nbt.3208URL [本文引用: 2]
DOI:10.1038/s41588-018-0282-xURL [本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1006/meth.2001.1262URL [本文引用: 1]
DOI:10.1105/tpc.110.082792URL [本文引用: 4]

Peroxidases have been shown to be involved in the polymerization of lignin precursors, but it remains unclear whether laccases (EC 1.10.3.2) participate in constitutive lignification. We addressed this issue by studying laccase T-DNA insertion mutants in Arabidopsis thaliana. We identified two genes, LAC4 and LAC17, which are strongly expressed in stems. LAC17 was mainly expressed in the interfascicular fibers, whereas LAC4 was expressed in vascular bundles and interfascicular fibers. We produced two double mutants by crossing the LAC17 (lac17) mutant with two LAC4 mutants (lac4-1 and lac4-2). The single and double mutants grew normally in greenhouse conditions. The single mutants had moderately low lignin levels, whereas the stems of lac4-1 lac17 and lac4-2 lac17 mutants had lignin contents that were 20 and 40% lower than those of the control, respectively. These lower lignin levels resulted in higher saccharification yields. Thioacidolysis revealed that disrupting LAC17 principally affected the deposition of G lignin units in the interfascicular fibers and that complementation of lac17 with LAC17 restored a normal lignin profile. This study provides evidence that both LAC4 and LAC17 contribute to the constitutive lignification of Arabidopsis stems and that LAC17 is involved in the deposition of G lignin units in fibers.
DOI:10.1105/tpc.113.117770URL [本文引用: 3]

The evolution of lignin biosynthesis was critical in the transition of plants from an aquatic to an upright terrestrial lifestyle. Lignin is assembled by oxidative polymerization of two major monomers, coniferyl alcohol and sinapyl alcohol. Although two recently discovered laccases, LAC4 and LAC17, have been shown to play a role in lignin polymerization in Arabidopsis thaliana, disruption of both genes only leads to a relatively small change in lignin content and only under continuous illumination. Simultaneous disruption of LAC11 along with LAC4 and LAC17 causes severe plant growth arrest, narrower root diameter, indehiscent anthers, and vascular development arrest with lack of lignification. Genome-wide transcript analysis revealed that all the putative lignin peroxidase genes are expressed at normal levels or even higher in the laccase triple mutant, suggesting that lignin laccase activity is necessary and nonredundant with peroxidase activity for monolignol polymerization during plant vascular development. Interestingly, even though lignin deposition in roots is almost completely abolished in the lac11 lac4 lac17 triple mutant, the Casparian strip, which is lignified through the activity of peroxidase, is still functional. Phylogenetic analysis revealed that lignin laccase genes have no orthologs in lower plant species, suggesting that the monolignol laccase genes diverged after the evolution of seed plants.
DOI:10.1007/s00425-010-1298-3URL [本文引用: 5]

While laccases, multi-copper glycoprotein oxidases, are often able to catalyze oxidation of a broad range of substrates, such as phenols and amines in vitro, their precise physiological/biochemical roles in higher plants remain largely unclear, e.g., Arabidopsis thaliana contains 17 laccases with only 1 having a known physiological function. To begin to explore their roles in planta, spatial and temporal expression patterns of Arabidopsis laccases were compared and contrasted in different tissues at various development stages using RT-PCR and promoter-GUS fusions. Various cell-specific expressions were noted where specific laccases were uniquely expressed, such as LAC4 in interfascicular fibers and seed coat columella, LAC7 in hydathodes and root hairs, LAC8 in pollen grains and phloem, and LAC15 in seed coat cell walls. Such specific cell-type expression patterns provide new leads and/or strategies into determining their precise physiological/biochemical roles. In addition, there was an apparent redundancy of gene expression patterns for several laccases across a wide variety of tissues, lignified and non-lignified, perhaps indicative of overlapping function(s). Preliminary evidence, based on bioinformatics analyses, suggests that most laccases may also be tightly regulated at both transcriptional (antisense transcripts, histone and DNA methylation) and posttranscriptional (microRNAs) levels of gene expression.
DOI:10.1111/tpj.2009.57.issue-5URL [本文引用: 1]
DOI:10.1186/s12870-018-1565-1URL [本文引用: 1]
DOI:10.1038/ng.2987URL [本文引用: 1]
DOI:10.1038/nbt.3207URL [本文引用: 1]
DOI:10.1105/tpc.16.00954URL [本文引用: 1]
DOI:10.1016/j.molp.2018.10.005URL [本文引用: 1]
