 ,1,2,3,*
,1,2,3,*Genome-wide association analysis and candidate genes prediction of waterlogging-responding traits in Brassica napus L.
LI Yang-Yang1,2,3,**, JING Rong-Rong1,3,**, LYU Rong-Rong1,2,3, SHI Peng-Cheng1,2,3, LI Xin1,2,3, WANG Qin1,2,3, WU Dan1,2,3, ZHOU Qing-Yuan1,3, LI Jia-Na1,2,3, TANG Zhang-Lin ,1,2,3,*
,1,2,3,*通讯作者:
第一联系人:
收稿日期:2019-02-25接受日期:2019-06-24网络出版日期:2019-07-16
| 基金资助: |
Received:2019-02-25Accepted:2019-06-24Online:2019-07-16
| Fund supported: |
作者简介 About authors
李阳阳,E-mail:liyangyangswu@163.com;。
荆蓉蓉,E-mail:m15525440862@163.com。

摘要
关键词:
Abstract
Keywords:
PDF (9249KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李阳阳, 荆蓉蓉, 吕蓉蓉, 石鹏程, 李欣, 王芹, 吴丹, 周清元, 李加纳, 唐章林. 甘蓝型油菜湿害胁迫响应性状的全基因组关联分析及候选基因预测[J]. 作物学报, 2019, 45(12): 1806-1821. doi:10.3724/SP.J.1006.2019.94027
LI Yang-Yang, JING Rong-Rong, LYU Rong-Rong, SHI Peng-Cheng, LI Xin, WANG Qin, WU Dan, ZHOU Qing-Yuan, LI Jia-Na, TANG Zhang-Lin.
水分是影响作物生长发育的重要非生物因子, 水分过多或过少均会对作物产生不同程度的危害。湿害是由于土壤水分超过田间最大持水量并长期处于饱和状态, 对作物生长发育产生的危害[1], 主要是对作物产生次生胁迫[2], 导致作物根系缺氧, 出现生理失水, 严重时会使根系腐烂发臭[3]。湿害可造成细胞质膜过氧化, 离子渗漏量增加, 无机营养吸收在植株体内重新分布, 丙二醛含量升高, 总糖量和淀粉酶活性降低, 保护酶系统受损, 严重时会使蛋白质分解、原生质结构破坏而致死[4]。湿害会使植株变矮, 生长缓慢, 生育期延长, 叶片由下至上逐渐黄化, 绿叶面积减少, 与光合作用相关的酶活性降低, 光合作用减弱, 光合产物减少且运输减慢, 从而影响植株生长发育和作物产量、品质[5,6,7,8]。
油菜是我国重要的油料作物之一, 也是我国食用植物油的主要来源, 其种植面积和总产量均占全球的30%左右[9]。长江流域是我国冬油菜主要种植区域, 其种植面积和产量均占全国90%以上[10]。该区域秋季多阴雨且实行水旱轮作制度, 土壤黏重、透气性差、排水困难、地下水位高[11], 是油菜湿涝灾害的多发地和重灾区。
作物的耐湿能力取决于形态结构和生理代谢上对缺氧的适应能力。作物对湿害胁迫的响应极其复杂, 不仅表现出长期适应性[12], 在细胞结构、组织和形态, 甚至线粒体、内质网和核糖体等亚细胞结构方面也表现出适应反应[13], 同时还通过基因表达调控调节自身的内环境和代谢途径[14]。油菜的不同类型、品种或品系的耐湿性不同, 张学昆等[15]研究甘蓝型油菜耐湿性发现, 缺氧胁迫下, 耐湿品种的活性氧清除系统能降低膜脂过氧化程度, 同时显著增加渗透调节的有机物含量, 提高对抗缺氧胁迫的能力。目前, 针对油菜, 特别是甘蓝型油菜耐湿性的研究较少, 且主要采用缺氧萌发种子或其他水淹处理, 通过测定幼苗相关性状筛选耐湿资源[11,16-18]。在遗传研究方面, 丛野等[19]发现, 甘蓝型油菜耐湿性的遗传受2对完全显性的主基因+加性-显性多基因控制。李真[4]利用150个甘蓝型油菜DH系, 通过183个SSR标记和157个AFLP标记在对照和湿害胁迫下共检测到45个QTL, 其中在2个环境下同时检测到9个, 与耐湿系数相关的有11个。
本研究调查湿害胁迫响应相关性状, 构建耐湿系数, 基于60K Illumina Infinium SNP芯片基因型数据进行全基因组关联分析, 并预测耐湿候选基因, 旨在为利用分子标记辅助选择培育耐湿甘蓝型油菜新品种和揭示甘蓝型油菜湿害响应分子机制研究奠定基础。
1 材料与方法
1.1 试验材料及处理
将248份甘蓝型油菜品种(系)播种于重庆市油菜工程技术研究中心歇马基地旱棚的盆钵(直径25 cm, 高30 cm)中, 当幼苗长至四叶一心时, 选取幼苗长势均匀、植株大小一致的盆钵进行正常灌溉(well watering, WW)和湿害胁迫(waterlogging, WL)处理, WW组土壤含水量维持在20%~23%之间, WL组维持土壤最大持水量(30%左右)且土壤表面无积水, 采用浙江托普仪器有限公司的TZS-1K土壤水分测定仪测定土壤含水量。持续处理4周后调查幼苗的绿叶数(green leaf number, GLN)、地上部鲜重(shoot fresh weight, SFW)和干重(shoot dry weight, SDW)、地下部鲜重(root fresh weight, RFW)和干重(root dry weight, RDW)以及叶片过氧化物酶(peroxidase, POD)活性、可溶性蛋白(soluble protein, Protein)含量和丙二醛(malondialdehyde, MDA)含量, 并计算各性状的耐湿系数(waterlogging resistance index, WLRI): 耐湿系数 = WL性状表型值/WW性状表型值。1.2 性状调查和测定
1.2.1 GLN调查 心叶完全平展视为1片叶, 绿色面积超过叶片面积1/2的视为绿叶。1.2.2 SFW、SDW、RFW和RDW测定 随机选取每材料WL和WW各3株, 称其地上部鲜重(g), 将根部洗净擦干后称其地下部鲜重(g)。然后于110℃杀青30 min, 75℃烘至恒重, 称其地上部干重(g)和地下部干重(g)。
1.2.3 叶片POD活性和Protein、MDA含量测定
取倒数第二、三片叶的混合样品, 采用南京建成生物工程研究所提供的试剂盒(A084-3)测定POD活性(U g-1 FW), 参照邹琦[20]所用考马斯亮蓝-G250染色法测定Protein含量(mg g-1 FW), 参照李合生[21]所用硫代巴比妥酸法测定MDA含量(μmol g-1 FW)。各材料每个处理3个生物学重复。
1.3 基因型分析
采用油菜60K Illumina Infinium SNP芯片(包含52,157个SNP标记)进行基因型分析, 按照Qu等[22]的方法删除位置不确定的标记以及缺失率大于20%和最小等位基因频率小于5%的标记, 保留32,839个SNP标记用于后续分析。1.4 连锁不平衡分析
使用Tassel 5.2.1[23]基于32,839个SNP标记对各染色体进行连锁不平衡分析, r2的衰减阈值定为0.2。1.5 群体结构分析
使用STRUCTURE 2.3.4[24]进行群体结构分析, K值依次取1~10, 每个K值运算5次, 蒙特卡罗迭代(Markov Chain Monte Carlo, MCMC)和模拟参数迭代(length of bum-in period)值均为100,000。1.6 亲缘关系分析
使用Tassel 5.2.1进行亲缘关系分析, 小于0的亲缘关系值取为0[25]。1.7 关联分析、单倍型分析和候选基因预测
利用Tassel 5.2.1及6种模型(na?ve, Q, PCA, K, K+Q, K+PCA)对8个性状的WLRI进行关联分析, 根据各模型检测到的P值与期望P值的偏离程度选择最佳模型, 利用MATLAB 2015b绘制QQ图(Quantile-quantile Plot)和最佳模型Manhattan图。显著关联SNP标记的阈值为3.045×10-5(1/32,839)。对各关联SNP不同基因型材料的耐湿系数进行t测验, 将不同SNP基因型间具有显著(P<0.05)或极显著(P<0.01)差异的标记称为显著性标记。利用haploView[26]对显著性标记所在LD区间的SNP进行单倍型分析, 参照Qu等[22]设定参数。根据已公布的甘蓝型油菜“Darmor-Bzh”基因组信息(http://www. genoscope.cns.fr/brassicanapus/)获取显著性标记所在的Block区间的基因及序列, 通过BLAST与拟南芥基因进行同源性比对, 根据拟南芥基因注释(https://www.arabidopsis.org/)预测甘蓝型油菜耐湿响应相关候选基因的功能。
2 结果与分析
2.1 表型数据分析
8个性状的方差分析结果表明, 每个性状在水分间、材料间以及水分×材料互作间均存在极显著(P<0.01)差异(表1)。由图1可知, 绝大多数材料GLN、SDW、SFW和Protein的耐湿系数小于1, MDA、POD的耐湿系数略大于1; 部分材料RFW和RDW的耐湿系数小于1, 但大多数材料RFW和RDW的耐湿系数接近于1。由此可见, 湿害会降低油菜幼苗的绿叶数、地上部鲜重、干重和叶片的可溶性蛋白含量, 会使叶片的丙二醛含量、POD活性升高, 地下部干重和地下部鲜重的变化在多数材料中表现为降低, 在有些材料中表现为升高, 但变化幅度较小。图1
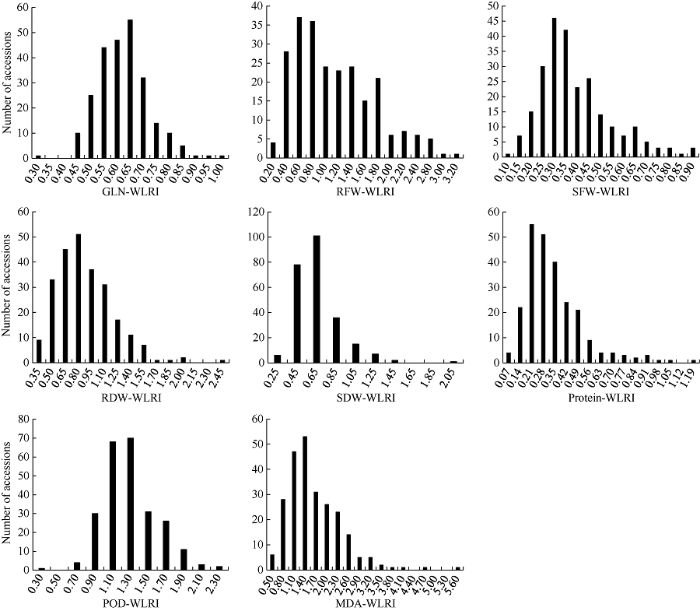 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1甘蓝型油菜幼苗湿害胁迫响应性状耐湿系数频数分布图
缩写同
Fig. 1Frequency distribution for WLRI (waterlogging resistance index) of each waterlogging-responding trait at seedling stage in B. napus
Abbreviations are the same as those given in
Table 1
表1
表1甘蓝型油菜幼苗湿害胁迫响应性状的方差分析
Table 1
| 变异来源 Source of variation | 自由度 df | F-value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| GLN | SFW | SDW | RFW | RDW | POD | MDA | Protein | ||
| 水分间Water | 1 | 4840.51** | 29,719.19** | 6028.69** | 67.37** | 1113.40** | 1167.63** | 2314.55** | 17,856.96** |
| 材料间Variety | 245 | 7.89** | 38.19** | 17.30** | 11.25** | 13.21** | 12.81** | 34.72** | 20.95** |
| 水分×材料Water × Variety | 245 | 1.95** | 20.63** | 8.97** | 8.08** | 7.62** | 13.27** | 31.70** | 17.80** |
| 误差Error | 984 | ||||||||
新窗口打开|下载CSV
2.2 亲缘关系和群体结构
由图2可知, 91%的材料间的亲缘系数小于0.2, 其中亲缘系数为0的材料占56%。由此可见, 试验材料间的亲缘关系较弱。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2248份甘蓝型油菜品种(系)的亲缘系数频率分布图
Fig. 2Frequency distribution of relative kinship coefficient in 248 B. napus accessions
由群体结构分析结果(图3)可知, 当K=2时, ΔK有最大值, 试验材料可分为2个亚群(P1和P2), 如图4所示。将Q矩阵的临界值设为0.7, 其中16个材料属于P1亚群, 大多数为来源于丹麦、加拿大和中国的春性甘蓝型油菜; 168个材料属于P2亚群, 大多数为来源于中国的冬性或半冬性甘蓝型油菜。64个材料位于P1亚群与P2亚群之间, 大多数为来源于中国的半冬性甘蓝型油菜。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图32个连续K值所对应的后验概率的变化速率(ΔK)
Fig. 3Estimation of ΔK based on change rate of ln P(D) between successive K value
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4248份甘蓝型油菜品种(系)群体结构分布图
Fig. 4Population structure of 248 B. napus accessions
2.3 LD分析
连锁不平衡分析LD (r2)平均值与遗传距离(Mb)之间的变化如图5所示, 当r2阈值设为0.2时, 估算的各染色体衰减距离如表2所示。由图5和表2可知, A亚基因组的衰减速率较C亚基因组快。图5
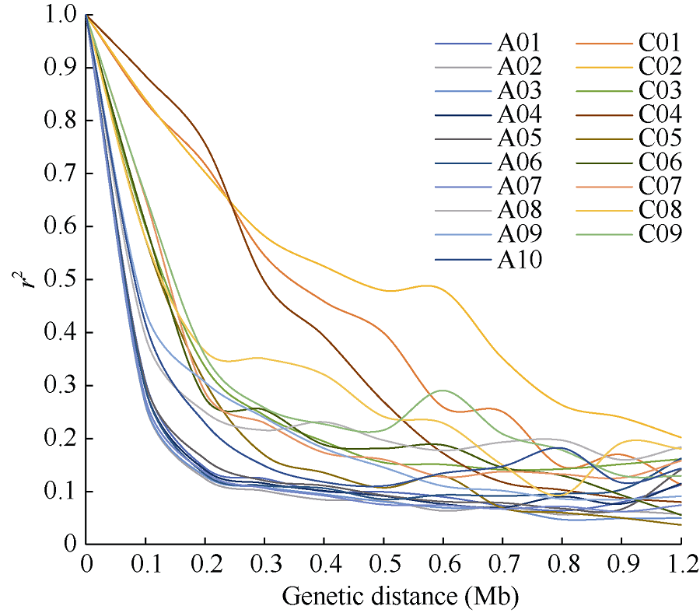 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5甘蓝型油菜染色体连锁不平衡衰减曲线
Fig. 5LD decay of chromosomes in B. napus
Table 2
表2
表2甘蓝型油菜各染色体的衰减距离
Table 2
| 染色体Chromosome | 衰减距离 LD decay (kb) | 染色体Chromosome | 衰减距离 LD decay (kb) |
|---|---|---|---|
| A01 | 180 | C01 | 750 |
| A02 | 180 | C02 | 1200 |
| A03 | 180 | C03 | 400 |
| A04 | 180 | C04 | 590 |
| A05 | 180 | C05 | 290 |
| A06 | 180 | C06 | 400 |
| A07 | 180 | C07 | 380 |
| A08 | 500 | C08 | 620 |
| A09 | 380 | C09 | 710 |
| A10 | 210 |
新窗口打开|下载CSV
2.4 关联分析
为了较好地控制关联分析的假阳性和假阴性, 利用32,839个有效SNP位点, 基于na?ve、Q、PCA、K、K+Q和K+PCA 六种模型对湿害胁迫响应的8个性状进行关联分析, 结果如图6、图7和表3所示。根据各性状的WLRI, 基于6种模型的-lg(P)观察值与期望值的接近程度, 选择最佳模型。最终, SDW和RFW的最佳模型为Q模型, SFW和GLN的最佳模型为PCA模型, RDW的最佳模型为K+Q模型, POD、MDA和Protein的最佳模型为K+PCA模型。图6
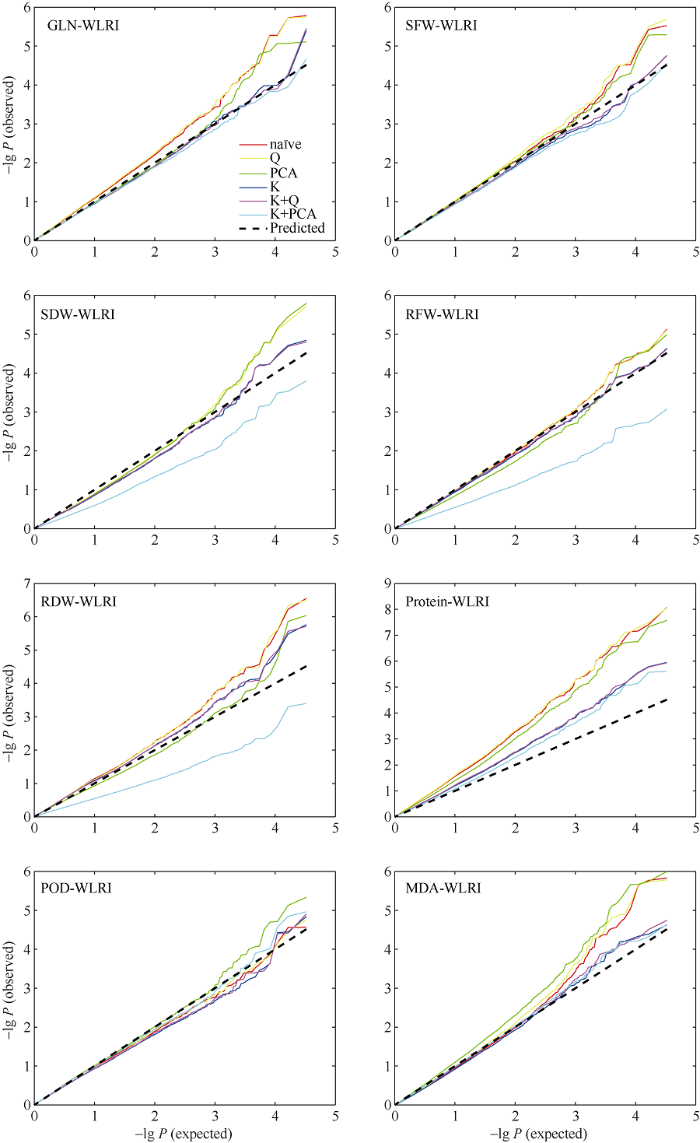 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6甘蓝型油菜湿害胁迫响应性状耐湿系数基于6种模型的关联分析QQ图
Fig. 6QQ plots of GWAS for WLRI of waterlogging-responding traits based on six models in B. napus
图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7甘蓝型油菜湿害胁迫响应性状耐湿系数基于最佳模型关联分析的Manhattan图
Fig. 7Manhattan plots of GWAS for WLRI of waterlogging-responding traits based on the best model in B. napus
Table 3
表3
表3甘蓝型油菜湿害胁迫响应性状WLRI显著关联的SNP标记
Table 3
| 性状 Trait | 标记 Marker | 染色体 Chromosome | 位置 Position | P值 P-value | 贡献率 R2 (%) |
|---|---|---|---|---|---|
| GLN | Bn-scaff_23954_1-p1045541 | C03 | 10,880,567 | 1.61E-05 | 9.82 |
| Bn-scaff_20901_1-p1110262 | C05 | 2,905,509 | 5.17E-06 | 10.69 | |
| Bn-A03-p11122273 | C07 | 41,947,271 | 5.09E-06 | 10.70 | |
| 性状 Trait | 标记 Marker | 染色体 Chromosome | 位置 Position | P值 P-value | 贡献率 R2 (%) |
| SFW | Bn-A02-p13803435 | A02 | 10,432,008 | 1.36E-05 | 9.71 |
| Bn-A04-p6975646a | A04 | 8,262,555 | 8.53E-06 | 10.06 | |
| Bn-A04-p6978163b | A04 | 8,265,085 | 8.53E-06 | 10.06 | |
| Bn-A04-p6980459 | A04 | 8,267,518 | 1.64E-05 | 9.57 | |
| Bn-A10-p15740896c | A10 | 15,809,264 | 1.46E-05 | 9.65 | |
| Bn-scaff_16531_1-p428890d | C04 | 21,584,887 | 7.76E-06 | 10.13 | |
| SDW | Bn-A04-p6975646a | A04 | 8,262,555 | 1.71E-05 | 9.74 |
| Bn-A04-p6978163b | A04 | 8,265,085 | 1.71E-05 | 9.74 | |
| Bn-A10-p15740896c | A10 | 15,809,264 | 1.94E-06 | 11.40 | |
| Bn-scaff_21884_1-p644186 | C01 | 37,469,955 | 7.66E-06 | 10.36 | |
| Bn-scaff_16531_1-p428890d | C04 | 21,584,887 | 4.67E-06 | 10.73 | |
| Bn-scaff_17919_1-p94976 | C09 | 34,508,780 | 2.82E-05 | 8.28 | |
| RFW | Bn-scaff_18062_1-p214669 | C04 | 31,128,751 | 2.45E-05 | 9.47 |
| Bn-scaff_21124_1-p301252 | C05 | 37,592,772 | 7.89E-06 | 10.35 | |
| RDW | Bn-A08-p15326882 | A08 | 12,894,573 | 2.74E-06 | 12.39 |
| Bn-A09-p20263302 | A09 | 17,272,800 | 1.95E-06 | 12.71 | |
| Bn-scaff_18826_1-p776957 | C05 | 36,457,927 | 1.02E-05 | 11.14 | |
| Bn-scaff_17888_1-p170198 | C09 | 15,826,668 | 1.74E-05 | 10.64 | |
| Protein | Bn-A01-p6538362 | A01 | 6,002,545 | 2.06E-05 | 10.54 |
| Bn-A02-p12822311 | A02 | 9,572,851 | 8.39E-06 | 11.47 | |
| Bn-A03-p7079712 | A03 | 6,365,301 | 2.87E-05 | 11.61 | |
| Bn-A03-p20100490 | A03 | 18967298 | 1.57E-05 | 10.83 | |
| Bn-A07-p20887568 | A07 | 22,470,590 | 7.18E-06 | 11.63 | |
| Bn-A09-p21935602 | A09 | 19,325,386 | 2.67E-06 | 12.65 | |
| Bn-scaff_20675_1-p32364 | C01 | 9,755,072 | 2.08E-05 | 11.96 | |
| Bn-scaff_21188_1-p1142 | C02 | 17,317,511 | 8.30E-06 | 12.95 | |
| Bn-scaff_17067_1-p509745 | C02 | 27,930,748 | 2.18E-05 | 11.91 | |
| Bn-scaff_15585_1-p1047449 | C04 | 44,459,141 | 2.45E-06 | 12.74 | |
| POD | Bn-A01-p637586 | A01 | 228,343 | 4.62E-06 | 9.61 |
| Bn-A03-p12250401 | A03 | 11,367,398 | 1.93E-05 | 9.62 | |
| Bn-A09-p30788157 | A09 | 28,540,524 | 7.44E-06 | 10.35 | |
| Bn-A10-p2246688 | A10 | 1,588,516 | 2.00E-05 | 9.59 | |
| MDA | Bn-A06-p3088460 | C05 | 3,221,158 | 2.31E-05 | 9.38 |
新窗口打开|下载CSV
对各性状WLRI共筛选到36个显著相关的SNP位点, 分布在A01、A02、A03、A04、A07、A08、A09、A10、C01、C02、C03、C04、C05、C07、C09等15条染色体上, 可解释的表型变异为8.28%~ 12.95% (表3)。对SFW-WLRI与SDW-WLRI检测到4个相同的显著关联SNP位点(Bn-A04-p6975646、Bn-A04-p6978163、Bn-A10-p15740896和Bn-scaff_ 16531_1-p428890), 与SFW-WLRI显著关联的Bn- A04-p6980459、Bn-A04-p6975646和Bn-A04-p6978163位于同一LD区间, 与GLN-WLRI显著关联的Bn- scaff_20901_1-p1110262和Bn-A06-p3088460位于同一LD区间。
t测验结果显示, 17个SNP标记(后续称为显著性SNP标记)所关联的WLRI在不同的SNP基因型间具有显著(P<0.05)或极显著(P<0.01)的差异(表4)。
Table 4
表4
表4甘蓝型油菜湿害胁迫响应性状显著关联SNP标记不同基因型的耐湿系数
Table 4
| 性状 Trait | SNP标记 SNP marker | 基因型 Loci | 耐湿系数 WLRI | 方差 Variance |
|---|---|---|---|---|
| GLN | Bn-scaff_23954_1-p1045541 | (GG)5 | 0.793 Aa | 0.005 |
| (TG)25 | 0.627 Bb | 0.012 | ||
| (TT)168 | 0.589 Bb | 0.008 | ||
| Bn-scaff_20901_1-p1110262 | (AA)50 | 0.656 Aa | 0.013 | |
| (CC)155 | 0.593 Bb | 0.007 | ||
| (AC)20 | 0.570 Bb | 0.009 | ||
| Bn-A03-p11122273 | (CC)25 | 0.681 Aa | 0.010 | |
| (TC)5 | 0.647 ABab | 0.007 | ||
| (TT)202 | 0.586 Bb | 0.008 | ||
| 性状 Trait | SNP标记 SNP marker | 基因型 Loci | 耐湿系数 WLRI | 方差 Variance |
| SFW | Bn-A02-p13803435 | (TT)210 | 0.361 Aa | 0.022 |
| (TC)12 | 0.347 ABab | 0.016 | ||
| (CC)16 | 0.296 Bb | 0.006 | ||
| Bn-A04-p6975646 | (AA)12 | 0.560 Aa | 0.036 | |
| (CC)220 | 0.355 Bb | 0.020 | ||
| (AC)8 | 0.273 Bb | 0.017 | ||
| Bn-A04-p6978163 | (TT)12 | 0.560 Aa | 0.036 | |
| (GG)222 | 0.356 Bb | 0.021 | ||
| (TG)8 | 0.273 Bb | 0.017 | ||
| Bn-A04-p6980459 | (GG)13 | 0.570 Aa | 0.034 | |
| AA)218 | 0.355 Bb | 0.021 | ||
| Bn-A10-p15740896 | (TT)14 | 0.546 Aa | 0.039 | |
| (TC)3 | 0.435 ABab | 0.026 | ||
| (CC)216 | 0.357 Bb | 0.021 | ||
| Bn-scaff_16531_1-p428890 | (CC)4 | 0.767 Aa | 0.022 | |
| (TC)17 | 0.380 Bb | 0.014 | ||
| (TT)200 | 0.358 Bb | 0.023 | ||
| SDW | Bn-A04-p6975646 | (AA)12 | 0.862 Aa | 0.178 |
| (CC)220 | 0.548 ABb | 0.046 | ||
| (AC)8 | 0.416 Bc | 0.016 | ||
| Bn-A04-p6978163 | (TT)12 | 0.862 Aa | 0.178 | |
| (GG)222 | 0.550 ABb | 0.047 | ||
| (TG)8 | 0.416 Bc | 0.016 | ||
| Bn-A10-p15740896 | TT)14 | 0.865 Aa | 0.098 | |
| (TC)3 | 0.756 ABab | 0.170 | ||
| (CC)216 | 0.544 Bb | 0.048 | ||
| Bn-scaff_21884_1-p644186 | (GG)4 | 1.125 Aa | 0.094 | |
| (AA)174 | 0.566 Bb | 0.053 | ||
| (AG)25 | 0.512 Bb | 0.050 | ||
| Bn-scaff_16531_1-p428890 | (CC)4 | 1.145 Aa | 0.138 | |
| (TC)17 | 0.618 Bb | 0.061 | ||
| (TT)200 | 0.545 Bb | 0.049 | ||
| Bn-scaff_17919_1-p94976 | (AA)213 | 0.559 Aa | 0.051 | |
| (AC)23 | 0.455 Bb | 0.020 | ||
| RFW | Bn-scaff_18062_1-p214669 | (AC)11 | 2.538 Aa | 2.529 |
| (CC)67 | 1.222 Ab | 0.347 | ||
| (AA)143 | 1.291 Ab | 0.850 | ||
| Bn-scaff_21124_1-p301252 | (TT)208 | 1.363 Aa | 0.969 | |
| (CC)28 | 1.317 Aa | 0.697 | ||
| (TC)6 | 0.988 Aa | 0.132 | ||
| RDW | Bn-A08-p15326882 | (AA)215 | 1.464 Aa | 0.365 |
| (AG)27 | 1.321 Aa | 0.241 | ||
| Bn-A09-p20263302 | (TT)198 | 1.471 Aa | 0.349 | |
| (TA)19 | 1.414 Aab | 0.412 | ||
| 性状 Trait | SNP标记 SNP marker | 基因型 Loci | 耐湿系数 WLRI | 方差 Variance |
| (AA)24 | 1.195 Ab | 0.217 | ||
| Bn-scaff_18826_1-p776957 | (TC)26 | 1.546 Aa | 0.318 | |
| (CC)69 | 1.469 Aa | 0.334 | ||
| (TT)136 | 1.367 Aa | 0.317 | ||
| Bn-scaff_17888_1-p170198 | (AC)30 | 1.534 Aa | 0.321 | |
| (CC)77 | 1.505 Aa | 0.423 | ||
| (AA)124 | 1.343 Aa | 0.232 | ||
| Protein | Bn-A01-p6538362 | (TT)117 | 0.293 Aa | 0.025 |
| (TG)108 | 0.291 Aa | 0.023 | ||
| Bn-A02-p12822311 | (AA)174 | 0.302 Aa | 0.028 | |
| (AG)67 | 0.302 Aa | 0.024 | ||
| Bn-A03-p7079712 | (AG)7 | 0.681 Aa | 0.122 | |
| (AA)33 | 0.312 Ab | 0.034 | ||
| (GG)197 | 0.295 Ab | 0.024 | ||
| Bn-A03-p20100490 | (TC)50 | 0.318 Aa | 0.029 | |
| (CC)189 | 0.296 Aa | 0.025 | ||
| Bn-A07-p20887568 | (TC)75 | 0.299 Aa | 0.023 | |
| (TT)162 | 0.298 Aa | 0.027 | ||
| Bn-A09-p21935602 | (GG)217 | 0.314 Aa | 0.031 | |
| (AG)26 | 0.230 Bb | 0.008 | ||
| Bn-scaff_20675_1-p32364 | (GG)9 | 0.358 Aa | 0.039 | |
| (TG)14 | 0.305 Aa | 0.028 | ||
| (TT)208 | 0.291 Aa | 0.023 | ||
| Bn-scaff_21188_1-p1142 | (GG)3 | 0.350 Aa | 0.046 | |
| (TT)193 | 0.293 Aa | 0.022 | ||
| (TG)33 | 0.282 Aa | 0.033 | ||
| Bn-scaff_17067_1-p509745 | (TG)16 | 0.330 Aa | 0.054 | |
| (TT)33 | 0.313 Aa | 0.029 | ||
| (GG)191 | 0.297 Aa | 0.024 | ||
| Bn-scaff_15585_1-p1047449 | (GG)211 | 0.301 Aa | 0.026 | |
| (AG)28 | 0.286 Aa | 0.030 | ||
| POD | Bn-A01-p637586 | (AA)212 | 1.195 Aa | 0.089 |
| (AG)28 | 1.118 Aa | 0.066 | ||
| Bn-A03-p12250401 | (AA)40 | 1.188 Aa | 0.086 | |
| (GG)162 | 1.186 Aa | 0.076 | ||
| (AG)34 | 1.150 Aa | 0.121 | ||
| Bn-A09-p30788157 | (TT)225 | 1.201 Aa | 0.084 | |
| (CC)13 | 0.980 Bb | 0.059 | ||
| Bn-A10-p2246688 | (CC)34 | 1.282 Aa | 0.115 | |
| (AA)182 | 1.168 Aa | 0.081 | ||
| (AC)20 | 1.146 Aa | 0.092 | ||
| MDA | Bn-A06-p3088460 | (GG)195 | 1.513 Aa | 0.489 |
| (AG)47 | 1.294 Ab | 0.403 |
新窗口打开|下载CSV
2.5 单倍型分析
对17个显著性标记所在LD区间的SNP进行单倍型分析表明, 显著性标记Bn-A02-p13803435、Bn-A09- p20263302、Bn-A09-p21935602、Bn-A10- p15740896、Bn-scaff_16531_1-p428890、Bn-scaff_ 17919_1-p94976、Bn-A06-p3088460和Bn-scaff_18062_1-p214669与至少1个SNP标记位于同一Block, 显著性标记Bn- A04-p6975646、Bn-A04-p6978163和Bn-A04- p6980459与其他7个SNP位于同一个72 kb跨度的Block, Bn-A03-p7079712、Bn-A03-p11122273、Bn-scaff_21884_1-p644186、Bn-A09-p30788157、Bn-scaff_23954_1-p1045541和Bn-scaff_20901_1- p1110262未与其他SNP形成Block (图8)。由于在后续分析中未筛选到候选基因, 因此Bn-A03- p11122273、Bn-scaff_17919_1-p94976和Bn-scaff_ 21884_1-p644186未在图8中列出。图8
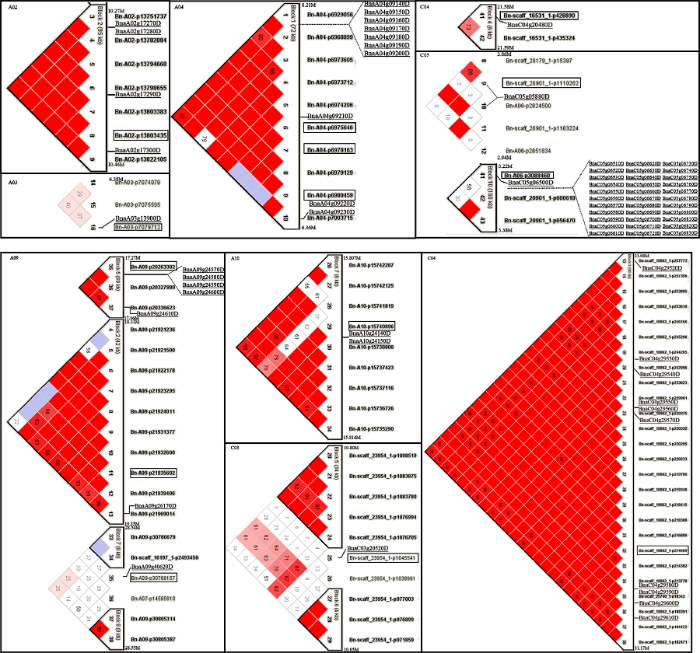 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8显著性标记所在的Blocks及候选基因
显著性标记用黑色边框标示, 候选基因用下画线标示。
Fig. 8Blocks including significant SNP markers and candidate genes
Significant markers are marked with black border and the candidate genes are underlined.
2.6 候选基因预测
显著性标记所在Block区间共包含71个候选基因(图8和表5), 其中BnaA04g09210D、BnaA09g 24580D、BnaC04g29550D、BnaC04g29560D、Bna C05g06560D、BnaC05g06730D和BnaC05g06770D共7个未Blast到同源的拟南芥基因, 其余64个均可通过同源的拟南芥基因注释预测其功能, 它们主要编码转录因子(如BnaA02g17270D, transcription initiation factor IIF; BnaA09g24610D, bZIP68; Bna A10g24150D, myb-like HTH transcriptional regulator family protein), 转运蛋白(如BnaA04g09160D, SUC2), 液泡蛋白质分拣蛋白[如BnaC05g05880D, vacuolar protein sorting 41 (VPS41)], 生长抑制子(如BnaC04g 29580D, ING2), DNA/RNA结合蛋白[如BnaC05g 06750D、BnaC05g06760D和BnaC05g 06810D, basic helix-loop-helix (bHLH) DNA-binding superfamily protein; BnaC05g06790D, RNA-binding (RRM/RBD/ RNP motifs) family protein], 酶类(如BnaC05g06690D, protein phosphatase 2C family protein; BnaA04g09140D, UDP-glucose 6-dehydrogenase family protein; Bna A04g09150D, HXXXD-type acyl-transferase family protein; BnaA04g09230D, saccharopine dehydrogenase; BnaA09g24570D, alpha/beta-Hydrolases superfamily protein), 激素响应蛋白[如BnaA02g17300D, SAUR- like auxin-responsive protein family; BnaA09g26170D, auxin response factor 8 (ARF8); BnaA10g24140D, arginine decarboxylase 2 (ADC2); BnaC05g06520D, nuclear factor Y (NF-YC9)], 氧化胁迫、渗透胁迫、盐胁迫或水分剥夺响应蛋白[如BnaC05g06500D, EARLY RESPONSE TO DEHYDRATION 6 (ERD6); BnaC05g 06530D, NPK1-related protein kinase 1 (NP1); BnaC 04g20480D, TIR-NBS-LRR; BnaC05g06780D, calreticulin 1b (CRT1b)]等。BnaC04g29570D和BnaC05g 06590D所编码的蛋白功能未知。Table 5
表5
表5甘蓝型油菜湿害胁迫响应性状耐湿系数候选基因相关信息
Table 5
| 油菜基因 B. napus gene | 染色体 Chr. | 物理位置 Position (bp) | 拟南芥基因 AT gene | 功能 Function |
|---|---|---|---|---|
| BnaA02g17270D | A02 | 10,385,018-10,386,352 | AT1G75510 | Transcription initiation factor IIF |
| BnaA02g17280D | A02 | 10,389,281-10,390,641 | AT1G75520 | SHI-related sequence 5 (SRS5) |
| BnaA02g17290D | A02 | 10,428,468-10,429,854 | AT1G75540 | Salt tolerance homolog 2 (STH2) |
| BnaA02g17300D | A02 | 10,461,565-10,461,997 | AT1G75580 | SAUR-like auxin-responsive protein family |
| BnaA03g13900D | A03 | 6,364,664-6,366,026 | AT2G30360 | SOS3-interacting protein 4 (SIP4) |
| BnaA04g09140D | A04 | 8,212,219-8,213,486 | AT5G39320 | UDP-glucose 6-dehydrogenase family protein |
| BnaA04g09150D | A04 | 8,215,808-8,217,193 | AT5G39050 | HXXXD-type acyl-transferase family protein |
| BnaA04g09160D | A04 | 8,231,509-8,231,884 | AT1G22710 | ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 2 (SUC2) |
| BnaA04g09170D | A04 | 8,231,950-8,233,460 | AT5G39340 | Histidine-containing phosphotransmitter 3 (AHP3) |
| BnaA04g09180D | A04 | 8,233,647-8,235,665 | AT5G39350 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| BnaA04g09190D | A04 | 8,235,944-8,237,287 | AT5G39360 | EID1-like 2 (EDL2) |
| BnaA04g09200D | A04 | 8,238,128-8,241,036 | AT5G39380 | Plant calmodulin-binding protein-related |
| BnaA04g09210D | A04 | 8,261,813-8,262,055 | — | |
| BnaA04g09220D | A04 | 8,280,136-8,282,045 | AT5G39400 | PTEN1 |
| BnaA04g09230D | A04 | 8,282,166-8,284,176 | AT5G39410 | Saccharopine dehydrogenase |
| BnaA09g24570D | A09 | 17,276,994-17,280,062 | AT1G32190 | Alpha/beta-Hydrolases superfamily protein |
| BnaA09g24580D | A09 | 17,282,321-17,283,676 | — | |
| BnaA09g24590D | A09 | 17,303,368-17,305,678 | AT1G46984 | F-box family protein |
| BnaA09g24600D | A09 | 17,320,367-17,322,665 | AT1G46984 | F-box family protein |
| BnaA09g24610D | A09 | 17,360,901-17,363,794 | AT1G32150 | Basic region/leucine zipper transcription factor 68 (bZIP68) |
| BnaA09g26170D | A09 | 19,351,820-19,355,843 | AT5G37020 | Auxin response factor 8 (ARF8) |
| BnaA09g40620D | A09 | 28,538,438-28,542,878 | AT2G26260 | 3 beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 (3BETAHSD/D2) |
| BnaA10g24140D | A10 | 15,809,477-15,810,280 | AT4G34710 | Arginine decarboxylase 2 (ADC2) |
| BnaA10g24150D | A10 | 15,810,902-15,813,379 | AT5G06800 | Myb-like HTH transcriptional regulator family protein |
| BnaC03g20520D | C03 | 10,879,272-10,881,026 | AT2G37110 | PLAC8 family protein |
| BnaC04g20480D | C04 | 21,584,060-21,585,266 | AT1G63740 | Disease resistance protein (TIR-NBS-LRR class) family |
| BnaC04g29520D | C04 | 31,077,933-31,080,151 | AT4G12790 | P-loop containing nucleoside triphosphate hydrolases superfamily protein |
| BnaC04g29530D | C04 | 31,085,683-31,088,293 | AT4G12740 | HhH-GPD base excision DNA repair family protein |
| 油菜基因 B. napus gene | 染色体 Chr. | 物理位置 Position (bp) | 拟南芥基因 AT gene | 功能 Function |
| BnaC04g29540D | C04 | 31,096,099-31,098,880 | AT4G12650 | Endomembrane protein 70 protein family |
| BnaC04g29550D | C04 | 31,113,638-31,113,859 | — | |
| BnaC04g29560D | C04 | 31,116,424-31,116,591 | — | |
| BnaC04g29570D | C04 | 31,121,991-31,124,968 | AT4G12680 | Unknown protein |
| BnaC04g29580D | C04 | 31,130,755-31,131,747 | AT1G54390 | INHIBITOR OF GROWTH 2 (ING2) |
| BnaC04g29590D | C04 | 31,133,548-31,136,282 | AT5G35380 | Protein kinase protein with adenine nucleotide alpha hydrolases-like domain |
| BnaC04g29600D | C04 | 31,158,769-31,162,504 | AT5G35430 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| BnaC04g29610D | C04 | 31,170,772-31,172,233 | AT5G35520 | MINICHROMOSOME INSTABILITY 12 (MIS12)-LIKE (MIS12) |
| BnaC05g05880D | C05 | 2,904,698-2,910,332 | AT1G08190 | Vacuolar protein sorting 41 (VPS41) |
| BnaC05g06500D | C05 | 3,218,324-3,221,856 | AT1G08930 | EARLY RESPONSE TO DEHYDRATION 6 (ERD6) |
| BnaC05g06510D | C05 | 3,222,332-3,224,004 | AT1G08940 | Phosphoglycerate mutase family protein |
| BnaC05g06520D | C05 | 3,224,198-3,228,539 | AT1G08970 | Nuclear factor Y |
| BnaC05g06530D | C05 | 3,228,942-3,233,245 | AT1G09000 | NPK1-related protein kinase 1 (NP1) |
| BnaC05g06540D | C05 | 3,233,325-3,237,550 | AT1G09010 | Glycoside hydrolase family 2 protein |
| BnaC05g06550D | C05 | 3,239,409-3,244,162 | AT1G09020 | Homolog of yeast sucrose nonfermenting 4 (SNF4) |
| BnaC05g06560D | C05 | 3,246,008-3,247,080 | — | |
| BnaC05g06570D | C05 | 3,248,229-3,248,627 | AT1G09030 | Nuclear factor Y |
| BnaC05g06580D | C05 | 3,258,505-3,259,753 | AT2G19610 | RING/U-box superfamily protein |
| BnaC05g06590D | C05 | 3,261,801-3,265,778 | AT1G09050 | Unknown protein |
| BnaC05g06600D | C05 | 3,266,250-3,266,895 | AT1G09060 | Zinc finger protein |
| BnaC05g06610D | C05 | 3,267,487-3,268,443 | AT1G09070 | Soybean gene regulated by cold-2 (SRC2) |
| BnaC05g06620D | C05 | 3,269,846-3,271,741 | AT1G09130 | ATP-dependent caseinolytic (Clp) protease/crotonase family protein |
| BnaC05g06630D | C05 | 3,275,956-3,279,260 | AT1G09140 | SERINE-ARGININE PROTEIN 30 (ATSRP30) |
| BnaC05g06640D | C05 | 3,279,332-3,280,873 | AT1G09150 | Containing protein |
| BnaC05g06650D | C05 | 3,281,248-3,282,283 | AT1G09155 | Phloem protein 2-B15 (PP2-B15) |
| BnaC05g06660D | C05 | 3,298,672-3,298,972 | AT1G09155 | Phloem protein 2-B15 (PP2-B15) |
| BnaC05g06670D | C05 | 3,299,052-3,303,054 | AT1G09155 | Phloem protein 2-B15 (PP2-B15) |
| BnaC05g06680D | C05 | 3,303,126-3,306,459 | AT2G43445 | F-box and associated interaction domains-containing protein |
| BnaC05g06690D | C05 | 3,308,135-3,309,772 | AT1G09160 | Protein phosphatase 2C family protein |
| BnaC05g06700D | C05 | 3,311,000-3,312,454 | AT1G09190 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| BnaC05g06710D | C05 | 3,312,506-3,313,708 | AT1G56330 | Secretion-associated RAS 1B (SAR1B) |
| BnaC05g06720D | C05 | 3,316,670-3,319,893 | AT5G08020 | RPA70-kDa subunit B (RPA70B) |
| BnaC05g06730D | C05 | 3,321,779-3,322,096 | — | |
| BnaC05g06740D | C05 | 3,324,873-3,325,493 | AT5G09390 | CD2-binding protein-related |
| BnaC05g06750D | C05 | 3,329,829-3,330,444 | AT1G09250 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| BnaC05g06760D | C05 | 3,330,666-3,331,280 | AT1G09250 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| BnaC05g06770D | C05 | 3,349,687-3,350,292 | — | |
| BnaC05g06780D | C05 | 3,356,126-3,359,392 | AT1G09210 | Calreticulin 1b (CRT1b) |
| BnaC05g06790D | C05 | 3,360,115-3,363,017 | AT1G09230 | RNA-binding (RRM/RBD/RNP motifs) family protein |
| BnaC05g06800D | C05 | 3,370,500-3,371,711 | AT1G09240 | Nicotianamine synthase 3 (NAS3) |
| BnaC05g06810D | C05 | 3,372,700-3,373,315 | AT1G09250 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| BnaC05g06820D | C05 | 3,378,631-3,379,149 | AT1G09260 | Chaperone DnaJ-domain superfamily protein |
| BnaC05g06830D | C05 | 3,379,950-3,383,047 | AT1G09270 | Importin alpha isoform 4 (IMPA-4) |
新窗口打开|下载CSV
3 讨论
张学昆等[15]研究发现, 油菜苗期湿害处理使植株变得矮小, 叶片发黄, 茎秆纤细, 可溶性糖和丙二醛含量升高, SOD酶活性增强, 而POD酶活性在耐湿材料中显著提高, 在湿害敏感材料中显著降低。李浩杰等[27]认为, 湿害胁迫抑制了油菜生长, 使得根冠比、根干重下降。李真等[11]对油菜DH群体苗期的耐湿性鉴定发现, 湿害胁迫后各家系的根干重、地上部干重都较正常灌溉组下降。涂玉琴等[28]研究发现甘蓝型油菜幼苗的绿叶数在湿害胁迫后明显减少, 黄叶数增加, 地上部鲜重和干重显著低于对照。本研究结果显示, 湿害胁迫后, 甘蓝型油菜幼苗绿叶数减少, 地上部鲜重、干重均降低, POD酶活性和MDA含量升高, 这与前人研究结果基本一致。陈龙等[29]认为植物在遭受逆境胁迫时, 一些原有的蛋白质合成受到抑制, 逆境蛋白被诱发合成以提高植物的耐胁迫能力。湿害胁迫对油菜幼苗可溶性蛋白含量的影响, 前人的研究结果不尽一致。范其新[30]研究发现湿害胁迫前后油菜幼苗的可溶性蛋白含量变化不大。张学昆等[15]发现湿害处理使油菜幼苗可溶性蛋白含量升高。在薛远超[31]研究中, 湿害胁迫下油菜苗期可溶性蛋白含量耐湿系数为0.68~1.44, 说明在有的材料中可溶性蛋白含量升高, 而有的材料降低。陈娟妮等[18]对长江流域主要甘蓝型油菜品种苗期耐湿性鉴定发现, 湿害使可溶性蛋白有不同程度的降低, 本研究的结果与此一致。湿害胁迫下, 可溶性蛋白含量的变化可能与试验材料的耐湿性强弱、湿害胁迫程度和时间有关。本研究的湿害胁迫时间为4周, 可能由于多数材料可溶性蛋白的分解加剧, 合成受阻, 导致可溶性蛋白含量有所降低。
李真等[11]和涂玉琴等[28]发现, 湿害胁迫后油菜幼苗地下部鲜重和干重均显著低于对照, 而薛远超[31]对油菜苗期进行淹水处理(保持水面高于土面3 cm), 发现不同材料地下部干重耐湿系数为0.24~3.91, 平均为1.03, 说明大多数材料在受到湿害胁迫后地下部干重会增加。本研究的结果与此一致, 其可能原因是本研究的湿害胁迫处理土壤表面无积水, 对部分耐湿性强的材料根系生长抑制较小, 且受到湿害胁迫后形成了部分不定根, 使根的总量超过了正常灌溉的对照。李真等[11]也发现油菜DH群体受到淹水处理后部分耐湿性强的株系植株下胚轴接近水表面处形成了许多不定根。
本研究对8个湿害胁迫响应性状的耐湿系数进行全基因组关联分析, 共检测到36个显著相关的SNP位点, 可解释8.28%~12.95%的表型变异, 其中17个显著性标记所关联的耐湿系数在不同的SNP基因型间具有显著(P<0.05)或极显著(P<0.01)的差异, 而其余19个差异不显著, 原因可能是关联分析所采用的阈值(1/32,839)对于某些性状偏高, 导致出现假阳性的关联位点。17个显著性标记所在的Blocks覆盖了71个候选基因, 其中64个可通过同源的拟南芥基因注释预测其功能。有研究表明, ARF8为根生长发育调控所必须, 参与生长素激活的信号通路[32], 精氨酸脱羧酶ADC2响应ABA、JA、冷胁迫、渗透胁迫、氧化胁迫、盐胁迫、机械损伤等[33], TIR- NBS-LRR蛋白具有应激响应和信号传导功能[34], ERD6响应ABA、盐胁迫和水分剥夺[35], NF-YC9参与脱落酸激活和赤霉素介导的信号通路[36], NPK1- related protein kinase 1 (NP1)响应氧化胁迫[37], CRT1b响应氧化胁迫和盐胁迫[38,39]。它们对应的甘蓝型油菜同源基因在本研究中都被预测到。许多研究发现, miRNA和可变剪接在湿害胁迫响应过程中具有重要作用, Zou等[40]对甘蓝型油菜苗期湿害胁迫下根部转录组分析检测到了一系列“DNA/RNA binding”基因。本研究检测到了3个DNA binding候选基因和1个RNA binding候选基因。根据这些候选基因注释的功能可以推测, 甘蓝型油菜可能通过miRNA或可变剪接调控、转录因子调控、生长调节、激素响应及一系列酶类和功能蛋白等响应湿害胁迫, 同时也可能存在与其他非生物胁迫和应激响应相关的基因参与湿害胁迫响应过程。本研究筛选的与甘蓝型油菜湿害胁迫响应性状显著关联的分子标记和候选基因, 为进一步进行基因功能验证、阐释湿害胁迫响应机制奠定了基础, 将有助于耐湿品种的培育。
4 结论
湿害胁迫较正常灌溉, 油菜幼苗绿叶数减少, 地上部干重和鲜重以及叶片可溶性蛋白含量降低, 叶片POD酶活性和MDA含量略有升高。地下部干重和鲜重在多数材料中降低, 而在有些材料中升高, 变化幅度均较小。共检测到与耐湿系数显著关联的SNP标记36个。17个所关联的耐湿系数在基因型间具有显著(P<0.05)或极显著(P<0.01)差异, 其所在的Blocks共覆盖71个候选基因, 其中64个Blast到同源拟南芥基因, 主要编码转录因子、转运蛋白、生长抑制子、蛋白磷酸酶、DNA/RNA结合蛋白、激素响应蛋白以及氧化胁迫、渗透胁迫、盐胁迫或水分剥夺响应蛋白等。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]

采用人工模拟淹水的试验方法,研究了五个不同生育时期根际土壤淹水对6个冬小麦品种光合作用和经济产量的影响。结果表明,不同生育时期根际土壤淹水对产量影响的相对受害率(RIR)差异及耐湿性不同品种产量的RIR差异达极显著水平(方差分析结果分别为F=111.68**,υ1=4,υ2=20,F0.01=4.43;F=18.51**,υ1=5,υ2=20,F0.0
URL [本文引用: 1]

采用人工模拟淹水的试验方法,研究了五个不同生育时期根际土壤淹水对6个冬小麦品种光合作用和经济产量的影响。结果表明,不同生育时期根际土壤淹水对产量影响的相对受害率(RIR)差异及耐湿性不同品种产量的RIR差异达极显著水平(方差分析结果分别为F=111.68**,υ1=4,υ2=20,F0.01=4.43;F=18.51**,υ1=5,υ2=20,F0.0
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 5]

【目的】评价甘蓝型油菜双单倍体(doubled haploid,DH)群体苗期耐湿性,筛选极端耐湿性DH系。【方法】采用盆栽试验,以株高(PH)、根长(RL)、地上部干重(SDW)、根干重(RDW)、根冠比(R/S)、总干重(TDW)等6个性状的耐湿系数作为耐湿性评价指标,对甘蓝型油菜DH群体的118个株系及其亲本进行苗期耐湿性评价。【结果】(1)与对照相比,湿害严重抑制了甘蓝型油菜苗期的生长,6个评价指标均表现出显著差异,其中根干重的变异系数最大;(2)在对照和湿害条件下,群体各株系各性状均表现出超亲连续分离,大部分呈正态分布,湿害条件下的分离更为明显;(3)相关性分析表明,地上部干重、根干重和总干重耐湿系数可作为甘蓝型油菜苗期耐湿性的主要评价指标。【结论】本研究综合应用地上部干重耐湿系数、地上部干重耐湿系数和总干重耐湿系数3个评价指标,筛选出005、007、040为候选的极端耐湿性基因型,086、110、119等为候选的极端不耐湿基因型,为耐湿相关研究及育种提供新材料。
URL [本文引用: 5]

【目的】评价甘蓝型油菜双单倍体(doubled haploid,DH)群体苗期耐湿性,筛选极端耐湿性DH系。【方法】采用盆栽试验,以株高(PH)、根长(RL)、地上部干重(SDW)、根干重(RDW)、根冠比(R/S)、总干重(TDW)等6个性状的耐湿系数作为耐湿性评价指标,对甘蓝型油菜DH群体的118个株系及其亲本进行苗期耐湿性评价。【结果】(1)与对照相比,湿害严重抑制了甘蓝型油菜苗期的生长,6个评价指标均表现出显著差异,其中根干重的变异系数最大;(2)在对照和湿害条件下,群体各株系各性状均表现出超亲连续分离,大部分呈正态分布,湿害条件下的分离更为明显;(3)相关性分析表明,地上部干重、根干重和总干重耐湿系数可作为甘蓝型油菜苗期耐湿性的主要评价指标。【结论】本研究综合应用地上部干重耐湿系数、地上部干重耐湿系数和总干重耐湿系数3个评价指标,筛选出005、007、040为候选的极端耐湿性基因型,086、110、119等为候选的极端不耐湿基因型,为耐湿相关研究及育种提供新材料。
DOI:10.1126/sciadv.1501302URLPMID:27652334 [本文引用: 1]

Many forest ecosystems have experienced recent declines in productivity; however, in some alpine regions, tree growth and forest expansion are increasing at marked rates. Dendrochronological analyses at the upper limit of alpine forests in the Tibetan Plateau show a steady increase in tree growth since the early 1900s, which intensified during the 1930s and 1960s, and have reached unprecedented levels since 1760. This recent growth acceleration was observed in small/young and large/old trees and coincided with the establishment of trees outside the forest range, reflecting a connection between the physiological performance of dominant species and shifts in forest distribution. Measurements of stable isotopes (carbon, oxygen, and nitrogen) in tree rings indicate that tree growth has been stimulated by the synergistic effect of rising atmospheric CO2 and a warming-induced increase in water and nutrient availability from thawing permafrost. These findings illustrate the importance of considering soil-plant-atmosphere interactions to understand current and anticipate future changes in productivity and distribution of forest ecosystems.
DOI:10.1111/nph.1987.106.issue-3URL [本文引用: 1]
[本文引用: 1]
URL [本文引用: 3]

【目的】试图阐明不同耐湿基因型甘蓝型油菜苗期对缺氧胁迫的生理差异响应。【方法】试验采用不同耐湿基因型的甘蓝型油菜材料,对发芽种子和幼苗(3叶期)缺氧处理,测定种子发芽性状和幼苗可溶性糖、脯氨酸、相对电导率、丙二醛(MDA)、过氧化物酶(POD)活性、过氧化氢酶(CAT)活性、超氧化物歧化酶(SOD)活性等生理指标。【结果】表明耐湿与不耐湿基因型相比具有显著的差异。缺氧处理后耐湿材料发芽种子具有较高的相对发芽率、相对苗长、根长、苗重和活力指数,而相对电导率较低。3叶期幼苗缺氧处理后,耐湿材料可溶性糖和脯氨酸含量提高幅度显著高于不耐湿材料,MDA和相对电导率则显著较低;耐湿基因型POD活性显著提高,CAT活性显著降低,SOD酶活性有较大幅度提高并显著高于不耐湿材料;不耐湿基因型则相反,POD活性显著降低,CAT活性显著提高,SOD酶活性幅度提高较小。【结论】缺氧胁迫下,甘蓝型油菜耐湿基因型能显著增加渗透调节有机物含量,同时启动活性氧清除系统,减少膜脂化程度,提高抵御缺氧胁迫。
URL [本文引用: 3]

【目的】试图阐明不同耐湿基因型甘蓝型油菜苗期对缺氧胁迫的生理差异响应。【方法】试验采用不同耐湿基因型的甘蓝型油菜材料,对发芽种子和幼苗(3叶期)缺氧处理,测定种子发芽性状和幼苗可溶性糖、脯氨酸、相对电导率、丙二醛(MDA)、过氧化物酶(POD)活性、过氧化氢酶(CAT)活性、超氧化物歧化酶(SOD)活性等生理指标。【结果】表明耐湿与不耐湿基因型相比具有显著的差异。缺氧处理后耐湿材料发芽种子具有较高的相对发芽率、相对苗长、根长、苗重和活力指数,而相对电导率较低。3叶期幼苗缺氧处理后,耐湿材料可溶性糖和脯氨酸含量提高幅度显著高于不耐湿材料,MDA和相对电导率则显著较低;耐湿基因型POD活性显著提高,CAT活性显著降低,SOD酶活性有较大幅度提高并显著高于不耐湿材料;不耐湿基因型则相反,POD活性显著降低,CAT活性显著提高,SOD酶活性幅度提高较小。【结论】缺氧胁迫下,甘蓝型油菜耐湿基因型能显著增加渗透调节有机物含量,同时启动活性氧清除系统,减少膜脂化程度,提高抵御缺氧胁迫。
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 2]

以32 个长江流域主要甘蓝型油菜品种为材料, 在模拟田间湿害环境下, 研究了甘蓝型油菜不同种质资源耐湿性及其适宜的耐湿性指标。结果表明, 湿害使甘蓝型油菜脯氨酸含量显著升高, 根系活力、叶绿素含量、蛋白质含量、根干重、总干重不同程度降低。甘蓝型油菜耐湿性受基因型控制, 遗传差异较大, 其中耐湿性强的品种有6 个(“中油821”、“黔油18 号、“中双11 号”、“09L553×L559”、“中双9 号”及“SWU7”), 占总品种数的18.75%; 耐湿性差的品种有3 个(“先油杂2 号”、“川油58”、“川油20”), 占总品种数的9.38%; 其他属于中等耐湿品种。相关分析表明, 不同品种各性状湿害指数与综合湿害指数相关性均达显著或极显著水平, 各性状湿害指数之间相关性大部分达显著或极显著水平, 其中根系活力、总干重和脯氨酸含量3 项指标对湿害综合指数贡献率达79.41%, 可作为鉴定甘蓝型油菜幼苗耐湿性的指标。
URL [本文引用: 2]

以32 个长江流域主要甘蓝型油菜品种为材料, 在模拟田间湿害环境下, 研究了甘蓝型油菜不同种质资源耐湿性及其适宜的耐湿性指标。结果表明, 湿害使甘蓝型油菜脯氨酸含量显著升高, 根系活力、叶绿素含量、蛋白质含量、根干重、总干重不同程度降低。甘蓝型油菜耐湿性受基因型控制, 遗传差异较大, 其中耐湿性强的品种有6 个(“中油821”、“黔油18 号、“中双11 号”、“09L553×L559”、“中双9 号”及“SWU7”), 占总品种数的18.75%; 耐湿性差的品种有3 个(“先油杂2 号”、“川油58”、“川油20”), 占总品种数的9.38%; 其他属于中等耐湿品种。相关分析表明, 不同品种各性状湿害指数与综合湿害指数相关性均达显著或极显著水平, 各性状湿害指数之间相关性大部分达显著或极显著水平, 其中根系活力、总干重和脯氨酸含量3 项指标对湿害综合指数贡献率达79.41%, 可作为鉴定甘蓝型油菜幼苗耐湿性的指标。
DOI:10.3724/SP.J.1006.2009.01462URL [本文引用: 1]

油菜湿害是我国特有的自然灾害,对油菜种子发芽、出苗和幼苗生长造成严重影响,导致单产显著下降。为研究油菜种子发芽耐湿性的遗传规律,本文利用耐湿性遗传差异较大的2个甘蓝型油菜纯系中双9号和GH01的杂交后代衍生的世代家系群体构建油菜耐湿性遗传体系,并以主基因+多基因家系世代联合分析方法对油菜耐湿性的遗传规律进行分析。结果表明,中双9号×GH01组合的耐湿性的遗传受2对完全显性主基因+加性-显性多基因控制,表现为耐湿对不耐湿完全显性。该组合的第1对主基因加性效应与显性效应相等,为0.0696;第2对主基因的加性效应与显性效应相等,为0.0530。多基因加性效应为0.3275,显性效应为负值([h]= –0.2137)。主基因存在显性效应,该组合的耐湿性存在杂种优势,多基因显性效应为负,多基因显性效应使F1代耐湿性降低。F2家系的主基因遗传力为73.57%,表现出较高的遗传力,建议育种工作者对耐湿性在早期选择效率较高。
DOI:10.3724/SP.J.1006.2009.01462URL [本文引用: 1]

油菜湿害是我国特有的自然灾害,对油菜种子发芽、出苗和幼苗生长造成严重影响,导致单产显著下降。为研究油菜种子发芽耐湿性的遗传规律,本文利用耐湿性遗传差异较大的2个甘蓝型油菜纯系中双9号和GH01的杂交后代衍生的世代家系群体构建油菜耐湿性遗传体系,并以主基因+多基因家系世代联合分析方法对油菜耐湿性的遗传规律进行分析。结果表明,中双9号×GH01组合的耐湿性的遗传受2对完全显性主基因+加性-显性多基因控制,表现为耐湿对不耐湿完全显性。该组合的第1对主基因加性效应与显性效应相等,为0.0696;第2对主基因的加性效应与显性效应相等,为0.0530。多基因加性效应为0.3275,显性效应为负值([h]= –0.2137)。主基因存在显性效应,该组合的耐湿性存在杂种优势,多基因显性效应为负,多基因显性效应使F1代耐湿性降低。F2家系的主基因遗传力为73.57%,表现出较高的遗传力,建议育种工作者对耐湿性在早期选择效率较高。
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:28292259 [本文引用: 2]

B. napus (oilseed) is an important source of edible vegetable oil, and its nutritional and economic value is determined by its fatty acid composition and content.
DOI:10.1093/bioinformatics/btm308URLPMID:17586829 [本文引用: 1]

Association analyses that exploit the natural diversity of a genome to map at very high resolutions are becoming increasingly important. In most studies, however, researchers must contend with the confounding effects of both population and family structure. TASSEL (Trait Analysis by aSSociation, Evolution and Linkage) implements general linear model and mixed linear model approaches for controlling population and family structure. For result interpretation, the program allows for linkage disequilibrium statistics to be calculated and visualized graphically. Database browsing and data importation is facilitated by integrated middleware. Other features include analyzing insertions/deletions, calculating diversity statistics, integration of phenotypic and genotypic data, imputing missing data and calculating principal components.
DOI:10.1111/j.1365-294X.2005.02553.xURLPMID:15969739 [本文引用: 1]

The identification of genetically homogeneous groups of individuals is a long standing issue in population genetics. A recent Bayesian algorithm implemented in the software STRUCTURE allows the identification of such groups. However, the ability of this algorithm to detect the true number of clusters (K) in a sample of individuals when patterns of dispersal among populations are not homogeneous has not been tested. The goal of this study is to carry out such tests, using various dispersal scenarios from data generated with an individual-based model. We found that in most cases the estimated 'log probability of data' does not provide a correct estimation of the number of clusters, K. However, using an ad hoc statistic DeltaK based on the rate of change in the log probability of data between successive K values, we found that STRUCTURE accurately detects the uppermost hierarchical level of structure for the scenarios we tested. As might be expected, the results are sensitive to the type of genetic marker used (AFLP vs. microsatellite), the number of loci scored, the number of populations sampled, and the number of individuals typed in each sample.
DOI:10.1038/ng1702URLPMID:16380716 [本文引用: 1]

As population structure can result in spurious associations, it has constrained the use of association studies in human and plant genetics. Association mapping, however, holds great promise if true signals of functional association can be separated from the vast number of false signals generated by population structure. We have developed a unified mixed-model approach to account for multiple levels of relatedness simultaneously as detected by random genetic markers. We applied this new approach to two samples: a family-based sample of 14 human families, for quantitative gene expression dissection, and a sample of 277 diverse maize inbred lines with complex familial relationships and population structure, for quantitative trait dissection. Our method demonstrates improved control of both type I and type II error rates over other methods. As this new method crosses the boundary between family-based and structured association samples, it provides a powerful complement to currently available methods for association mapping.
DOI:10.1093/bioinformatics/bth457URLPMID:15297300 [本文引用: 1]

Research over the last few years has revealed significant haplotype structure in the human genome. The characterization of these patterns, particularly in the context of medical genetic association studies, is becoming a routine research activity. Haploview is a software package that provides computation of linkage disequilibrium statistics and population haplotype patterns from primary genotype data in a visually appealing and interactive interface.
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.1104/pp.15.00265URLPMID:25941314 [本文引用: 1]

Legume root nodules convert atmospheric nitrogen gas into ammonium through symbiosis with a prokaryotic microsymbiont broadly called rhizobia. Auxin signaling is required for determinant nodule development; however, the molecular mechanism of auxin-mediated nodule formation remains largely unknown. Here, we show in soybean (Glycine max) that the microRNA miR167 acts as a positive regulator of lateral root organs, namely nodules and lateral roots. miR167c expression was up-regulated in the vasculature, pericycle, and cortex of soybean roots following inoculation with Bradyrhizobium japonicum strain USDA110 (the microsymbiont). It was found to positively regulate nodule numbers directly by repressing the target genes GmARF8a and GmARF8b (homologous genes of Arabidopsis [Arabidopsis thaliana] AtARF8 that encode auxin response factors). Moreover, the expression of miR167 and its targets was up- and down-regulated by auxin, respectively. The miR167-GmARF8 module also positively regulated nodulation efficiency under low microsymbiont density, a condition often associated with environmental stress. The regulatory role of miR167 on nodule initiation was dependent on the Nod factor receptor GmNFR1α, and it acts upstream of the nodulation-associated genes nodule inception, nodulation signaling pathway1, early nodulin40-1, NF-YA1 (previously known as HAEM activator protein2-1), and NF-YA2. miR167 also promoted lateral root numbers. Collectively, our findings establish a key role for the miR167-GmARF8 module in auxin-mediated nodule and lateral root formation in soybean.
DOI:10.1016/j.plaphy.2015.04.005URLPMID:25914135 [本文引用: 1]

Plant stress induced by high salinity has leading to an important reduction in crop yields. Due to their tropical origin, citrus fruits are highly sensitive to salts. Rootstocks are the root system of fruit trees, regulating ion uptake and transport to the canopy. Therefore, increasing their salt tolerance could improve the salt tolerance of the fruit tree. For this, we genetically-transformed an important rootstock for lemon, Citrus macrophylla W, to constitutively express the CBF3/DREB1A gene from Arabidopsis, a well-studied salinity tolerance transcription factor. Transgenic lines showed normal size, with no dwarfism. Under salt stress, some transgenic lines showed greater growth, similar accumulation of chloride and sodium in the leaves and better stomatal conductance, in comparison to wild-type plants. Quantitative real-time analyses showed a similar expression of several CBF3/DREB1A target genes, such as COR15A, LEA 4/5, INV, SIP1, P5CS, GOLS, ADC2 and LKR/SDH, in transgenic lines and wild type plants, with the exception of INV that shows increased expression in line 4C15. Under salt stress, all measured transcript increased in both wild type and transgenics lines, with the exception of INV. Altogether, these results suggest a higher salt tolerance of transgenic C.?macrophylla plants induced by the overexpression of AtCBF3/DREB1A.
DOI:10.1111/j.1365-313X.2006.02688.xURLPMID:16623885 [本文引用: 1]

In a cross between the two resistant accessions Col-0 and Ler-0, a 15:1 segregation was found in F2, suggesting the presence of unlinked resistance loci to Leptosphaeria maculans. One hundred Col-4 x Ler-0, and 50 Ler-2 x Cvi-1 recombinant inbred lines, and seven susceptible Ler-0 x Ws-0 F2 progenies were examined to identify the two loci. Resistance in Col-4, Ws-0 and Cvi-1 (RLM1) was mapped to the marker m305 on chromosome 1. Col-4 x Ler-0 and Ler-2 x Cvi-1 mapping populations located RLM2(Ler) on the same arm of chromosome 4. A tight physical location of RLM2 was established through near-isogenic lines. This region was found to correspond to an ancient duplication event between the RLM1 and RLM2 loci. Two independent T-DNA mutants in a TIR-NB-LRR R gene (At1g64070) displayed susceptibility, and L. maculans susceptible mutant phenotypes were confirmed to be allelic for rlm1 in F1 after crosses with susceptible rlm1(Ler)rlm2(Col) plants. Complementation of rlm1(Ler)rlm2(Col) with the genomic Col-0 sequence of At1g64070 conferred resistance. In addition, two T-DNA mutants in a neighbouring homologous TIR-NB-LRR gene (At1g63880) displayed moderate susceptibility to L. maculans. Sequence analysis revealed that At1g64070 was truncated by a premature stop codon, and that At1g63880 was absent in Ler-0. RNA interference confirmed that Ler-0 resistance is dependent on genes structurally related to RLM1. Camalexin was identified as a quantitative co-dominant resistance factor of Col-0 origin, but independent of RLM1. RLM1/RLM2 resistance was, however, found to require RAR1 and partially HSP90.1.
DOI:10.1074/jbc.M109.054288URLPMID:19901034 [本文引用: 1]

Sugars play indispensable roles in biological reactions and are distributed into various tissues or organelles via transporters in plants. Under abiotic stress conditions, plants accumulate sugars as a means to increase stress tolerance. Here, we report an abiotic stress-inducible transporter for monosaccharides from Arabidopsis thaliana that is termed ESL1 (ERD six-like 1). Expression of ESL1 was induced under drought and high salinity conditions and with exogenous application of abscisic acid. Promoter analyses using beta-glucuronidase and green fluorescent protein reporters revealed that ESL1 is mainly expressed in pericycle and xylem parenchyma cells. The fluorescence of ESL1-green fluorescent protein-fused protein was detected at tonoplast in transgenic Arabidopsis plants and tobacco BY-2 cells. Furthermore, alanine-scanning mutagenesis revealed that an N-terminal LXXXLL motif in ESL1 was essential for its localization at the tonoplast. Transgenic BY-2 cells expressing mutated ESL1, which was localized at the plasma membrane, showed an uptake ability for monosaccharides. Moreover, the value of K(m) for glucose uptake activity of mutated ESL1 in the transgenic BY-2 cells was extraordinarily high, and the transport activity was independent from a proton gradient. These results indicate that ESL1 is a low affinity facilitated diffusion transporter. Finally, we detected that vacuolar invertase activity was increased under abiotic stress conditions, and the expression patterns of vacuolar invertase genes were similar to that of ESL1. Under abiotic stress conditions, ESL1 might function coordinately with the vacuolar invertase to regulate osmotic pressure by affecting the accumulation of sugar in plant cells.
DOI:10.1007/s11103-017-0661-1URLPMID:28924726 [本文引用: 1]

Nuclear factor Y (NF-Y) family proteins are involved in many developmental processes and responses to environmental cues in plants, but whether and how they regulate phytohormone abscisic acid (ABA) signaling need further studies. In the present study, we showed that over-expression of the NF-YC9 gene confers ABA hypersensitivity in both the early seedling growth and stomatal response, while down-regulation of NF-YC9 does not affect ABA response in these processes. We also showed that over-expression of the NF-YC9 gene confers salt and osmotic hypersensitivity in early seedling growth, which is likely to be directly associated with the ABA hypersensitivity. Further, we observed that NF-YC9 physically interacts with the ABA-responsive bZIP transcription factor ABA-INSENSITIVE5 (ABI5), and facilitates the function of ABI5 to bind and activate the promoter of a target gene EM6. Additionally, NF-YC9 up-regulates expression of the ABI5 gene in response to ABA. These findings show that NF-YC9 may be involved in ABA signaling as a positive regulator and likely functions redundantly together with other NF-YC members, and support the model that the NF-YC9 mediates ABA signaling via targeting to and aiding the ABA-responsive transcription factors such as ABI5.
DOI:10.1073/pnas.97.6.2940URLPMID:10717008 [本文引用: 1]

Despite the recognition of H(2)O(2) as a central signaling molecule in stress and wounding responses, pathogen defense, and regulation of cell cycle and cell death, little is known about how the H(2)O(2) signal is perceived and transduced in plant cells. We report here that H(2)O(2) is a potent activator of mitogen-activated protein kinases (MAPKs) in Arabidopsis leaf cells. Using epitope tagging and a protoplast transient expression assay, we show that H(2)O(2) can activate a specific Arabidopsis mitogen-activated protein kinase kinase kinase, ANP1, which initiates a phosphorylation cascade involving two stress MAPKs, AtMPK3 and AtMPK6. Constitutively active ANP1 mimics the H(2)O(2) effect and initiates the MAPK cascade that induces specific stress-responsive genes, but it blocks the action of auxin, a plant mitogen and growth hormone. The latter observation provides a molecular link between oxidative stress and auxin signal transduction. Finally, we show that transgenic tobacco plants that express a constitutively active tobacco ANP1 orthologue, NPK1, display enhanced tolerance to multiple environmental stress conditions without activating previously described drought, cold, and abscisic acid signaling pathways. Thus, manipulation of key regulators of an oxidative stress signaling pathway, such as ANP1/NPK1, provides a strategy for engineering multiple stress tolerance that may greatly benefit agriculture.
DOI:10.1007/s00299-013-1497-zURL [本文引用: 1]

The calreticulin triple knockout mutant shows growth defects in response to abiotic stress.
The endoplasmic reticulum (ER) is an essential organelle that is responsible for the folding and maturation of proteins. During ER stress, unfolded protein aggregates accumulate in the cell, leading to the unfolded protein response (UPR). The UPR up-regulates the expression of ER-stress-responsive genes encoding calreticulin (CRT), an ER-localized Ca2+-binding protein. To understand the function of plant CRTs, we generated a triple knockout mutant, t123, which lacks CRT1, CRT2 and CRT3 and examined the roles of calreticulins in abiotic stress tolerance. A triple knockout mutant increased sensitivity to water stress which implies that calreticulins are involved in the Arabidopsis response to water stress. We identified that the cyclophilin AtCYP21-2, which is located in the ER, was specifically enhanced in the t123 mutants. Seed germination of the atcyp21-1 mutant was retarded by water stress. Taken together, these results suggest that regulatory proteins that serve to protect plants from water stress are folded properly in part with the help of calreticulins. The AtCYP21-2 may also participate in this protein-folding process in association with calreticulins.
DOI:10.1016/j.bbrc.2011.01.082URLPMID:21277284 [本文引用: 1]

VIGG is a putative endoplasmic reticulum (ER) resident protein induced by virus infection and ER stress, and is correlated with fruit quality in grapevine. The present study was undertaken to determine the biological function of VIGG in grapevine. Experiments using fluorescent protein-VIGG fusion protein demonstrated that VIGG is localized in ER and the ER targeting sequence is in the N-terminus. The overexpression of VIGG in Arabidopsis plant led to growth retardation. The rosette leaves of VIGG-overexpressing plants were smaller than those of the control plants and rolled at 42days after seeding. VIGG-overexpressing plants revealed robustness to ER stress as well as the low expression of ER stress marker proteins, such as the luminal binding proteins. These characteristics of VIGG-overexpressing plants were supported by a microarray experiment that demonstrated the disruption of genes related to ER stress response and flowering, as well as cation mobility, in the plants. Finally, cation homeostasis in the plants was disturbed by the overexpression of VIGG. Taken together, these results suggest that VIGG may disturb cation homeostasis in plant, which is correlated with the robustness to ER stress and growth retardation.
DOI:10.3390/ijms14022637URLPMID:23358252 [本文引用: 1]

Although rapeseed (Brassica napus L.) is known to be affected by waterlogging, the genetic basis of waterlogging tolerance by rapeseed is largely unknown. In this study, the transcriptome under 0 h and 12 h of waterlogging was assayed in the roots of ZS9, a tolerant variety, using digital gene expression (DGE). A total of 4432 differentially expressed genes were identified, indicating that the response to waterlogging in rapeseed is complicated. The assignments of the annotated genes based on GO (Gene Ontology) revealed there were more genes induced under waterlogging in &quot;oxidation reduction&quot;, &quot;secondary metabolism&quot;, &quot;transcription regulation&quot;, and &quot;translation regulation&quot;; suggesting these four pathways are enhanced under waterlogging. Analysis of the 200 most highly expressed genes illustrated that 144 under normal conditions were down-regulated by waterlogging, while up to 191 under waterlogging were those induced in response to stress. The expression of genes involved under waterlogging is mediated by multiple levels of transcriptional, post-transcriptional, translational and post-translational regulation, including phosphorylation and protein degradation; in particular, protein degradation might be involved in the negative regulation in response to this stress. Our results provide new insight into the response to waterlogging and will help to identify important candidate genes.
