

中国科学院微生物研究所中国科学院微生物生理与代谢工程重点实验室,北京 100101
收稿日期:2020-09-05;接收日期:2021-03-27;网络出版时间:2021-04-29
基金项目:国家重点研发计划(No. 2018YFA0900701),国家自然科学基金(Nos. 31971337, 31961133016, 31971382) 资助
摘要:α-L-鼠李糖苷酶是一个非常重要的工业酶,广泛分布于各种生物中。不同来源的α-L-鼠李糖苷酶具有多样性。细菌来源的α-L-鼠李糖苷酶的最适pH接近中性或偏碱性,而真菌来源的α-L-鼠李糖苷酶的最适pH在酸性范围。除此之外,不同来源的α-L-鼠李糖苷酶在最适温度、热稳定性和底物特异性等方面也不尽相同,酶学性质的差异,决定了其在工业应用时所具有的优势和限制。因此,分析不同来源的α-L-鼠李糖苷酶的酶学性质、阐明其在催化机制和底物特异性等方面的异同点、探究底物的糖苷配体和金属阳离子对酶活性的影响以及L-鼠李糖和葡萄糖对酶的竞争性抑制作用,可以为工业生产中准确选择α-L-鼠李糖苷酶提供参考,进一步推动该酶的工业化应用进程。
关键词:α-L-鼠李糖苷酶酶学性质催化机制酶活
Enzymatic properties of α-L-rhamnosidase and the factors affecting its activity: a review
Xiaochong Zhu, Shuangyan Tang


CAS Key Laboratory of Microbial Physiological and Metabolic Engineering, Institute of Microbiology, Beijing 100101, China
Received: September 5, 2020; Accepted: March 27, 2021; Published: April 29, 2021
Supported by: National Key Research and Development Program of China (No. 2018YFA0900701), National Natural Science Foundation of China (Nos. 31971337, 31961133016, 31971382)
Corresponding author: Shuangyan Tang. Tel: +86-10-64806150; E-mail: tangsy@im.ac.cn.
Abstract: α-L-rhamnosidase is a very important industrial enzyme that is widely distributed in a variety of organisms. α-L-rhamnosidase of different origins show functional diversity. For example, the optimal pH of α-L-rhamnosidase from bacteria is close to neutral or alkaline, while the optimal pH of α-L-rhamnosidase from fungi is in the acidic range. Furthermore, the enzymatic properties of α-L-rhamnosidases of different origins differ in terms of the optimal temperature, the thermal stability, and the substrate specificity, which determine the different applications of these enzymes. In this connection, it is crucial to elucidate the similarities and differences in the catalytic mechanism and substrate specificity of α-L-rhamnosidase of different origins through analyzing its enzymatic properties. Moreover, it is important to explore and understand the effects of aglycon and metal cations on enzyme activity and the competitive inhibition of L-rhamnose and glucose on enzymes. These knowledge can help discover α-L-rhamnosidase of industrial significance and promote its industrial application.
Keywords: α-L-rhamnosidaseenzymatic propertiescatalytic mechanismenzymatic activity
α-L-鼠李糖苷酶可以特异性地水解聚糖或者糖苷类化合物末端的α-1, 2、α-1, 3、α-1, 4、α-1、α-1, 6糖苷键,释放L-鼠李糖,产生新的聚糖或糖苷类化合物[1]。α-L-鼠李糖苷酶在药物、化妆品和食品等领域具有非常重要的应用,因其可以水解果汁和红酒制作过程中产生的天然黄酮类化合物,常被用于果汁脱苦、去除结晶和提升红酒的风味等过程[2]。在医药行业,α-L-鼠李糖苷酶可以将人参皂苷[3]、柴胡皂苷C[4]和薯蓣皂苷[5]等常用中药材中的皂苷类化合物转化成活性成分。另外,水解反应释放的L-鼠李糖可以作为芳香族化合物和黄酮类化合物工业合成的前体物质[6]。
α-L-鼠李糖苷酶广泛存在于动物、植物、真菌和细菌中[7]。CAZy数据库将α-L-鼠李糖苷酶分为3个不同的家族:GH78家族、GH28家族、GH106家族。到目前为止,在被报道的34个不同来源的α-L-鼠李糖苷酶中,有5个α-L-鼠李糖苷酶的晶体结构已被解析,其中4个来自GH78家族[8-11],1个来自GH106家族[12]。
不同来源的α-L-鼠李糖苷酶,甚至相同来源不同类型的α-L-鼠李糖苷酶的酶学特性不尽相同。真菌来源的α-L-鼠李糖苷酶多数偏好偏酸性的环境,细菌来源的α-L-鼠李糖苷酶多数偏好近中性或者偏碱性的环境[13]。嗜热网球菌中的α-L-鼠李糖苷酶具有更好的热稳定性,能耐受较高的反应温度[14],而来自海洋细菌的α-L-鼠李糖苷酶则能在较低的温度下保留较大的活性[15]。除此之外,不同α-L-鼠李糖苷酶对不同底物的亲和性也存在差异。酶的物理化学特性的多样性决定了应用环境的多样性,因此,总结已有的α-L-鼠李糖苷酶的酶学特性,分析不同来源的α-L-鼠李糖苷酶的异同,阐明影响酶活的因素,是进一步推进该酶工业化应用进程的基础。
1 α- L-鼠李糖苷酶的酶学性质酶的适宜温度、适宜pH值和热稳定性是工业生产必须要考虑的因素,也是限制很多酶工业化应用的一个重要因素。反应体系的温度和pH,不仅影响底物和产物的溶解性、稳定性和黏度[7],也决定着整个生产流程的设定和对设备的要求等。而热稳定性强弱决定着酶的利用率、原料损耗和生产成本等[16]。
1.1 最适pH值大多数α-L-鼠李糖苷酶的最适pH值处于酸性或近中性范围(表 1和表 2)[17]。真菌来源的α-L-鼠李糖苷酶多数偏好偏酸性的环境,最适pH值范围为4.0–6.5,有3个偏好碱性的α-L-鼠李糖苷酶:来自白笋顶孢Acrostalagmus luteo albus的α-L-鼠李糖苷酶的最适pH值为8.0,在pH为9.5和11.0的环境中,活性分别保留67%和62%[2];来自串珠镰刀菌Fusarium moniliforme MTCC-2088[18]和黄曲霉Aspergillus flavus MTCC-9606[17]的α-L-鼠李糖苷酶的最适pH值分别为10.5和11.0,是目前报道中pH值最高的两个碱性α-L-鼠李糖苷酶。细菌来源的α-L-鼠李糖苷酶多数偏好近中性或偏碱性的环境[13],最适pH范围为5.0–8.0,有一个偏好酸性的α-L-鼠李糖苷酶:来自乳酸片球菌Pediococcus acidilactici的α-L-鼠李糖苷酶的最适pH值为4.5[19]。
表 1 真菌来源的α-L-鼠李糖苷酶的酶学性质Table 1 The enzymatic property of α-L-rhamnosidases from fungi
| Number | Enzyme | Organism | pHopt | Topt (℃) | Thermal stability | References |
| 1 | Aspergillus flavus MTCC-9606 | 11.0 | 50 | [17] | ||
| 2 | Aspergillus terreus | 6.5 | 40–50 | Stable below 50 ℃ | [23] | |
| 3 | AT-rRha | Aspergillus tubingensis | 4.0 | 50–60 | [13] | |
| 4 | AK-rhaA | Aspergillus kawachii | 4.0 | 50 | Stable | [24] |
| 5 | r-Rha1 | Aspergillus niger | 5.0 | 60 | Stable | [1] |
| 6 | AnRhaE | Aspergillus nidulans | 4.5 | 55 | Stable below 55 ℃ | [25] |
| 7 | RhaA/RhaB | Aspergillus aculeatus | 4.5–5.0 | Stable at 40 ℃ | [22] | |
| 8 | SaRhaA | Streptomyces avermitilis | 6.0 | 50 | Stable below 40 ℃ | [26] |
| 9 | Absidia sp. 39 | 5.0 | 40 | Stable between 30–40 ℃ | [3] | |
| 10 | Absidia sp. 90 | 5.0 | 50 | Stable between 30–40 ℃ | [27] | |
| 11 | Penicillium ulaiense | 5.0 | 60 | Stable | [28] | |
| 12 | Acrostalagmus luteo albus | 8.0 | 55 | Stable below 55 ℃ | [2] | |
| 13 | Curvularia lunata | 4.0 | 50 | Stable between 30–40 ℃ | [29] | |
| 14 | Pichia angusta X349 | 6.0 | 40 | Stable below 30 ℃ | [30] | |
| 15 | Penicillium griseoroseum MTCC-9224 | 6.5 | 57 | Unstable | [31] | |
| 16 | Alternaria alternate SK37.001 | 5.5 | 60 | Stable between 30–50℃ | [32] | |
| 17 | Fusarium moniliforme MTCC-2088 | 10.5 | 50 | Unstable | [18] | |
| 18 | XpoGH78 | Xylaria polymorpha | 6.0–8.0 | 45 | Stable at 40 ℃ | [33] |
表选项
表 2 细菌来源的α-L-鼠李糖苷酶的酶学性质Table 2 The enzymatic property of α-L-rhamnosidases from bacteria
| Number | Enzyme | Organism | pHopt | Topt (℃) | Thermal stability | References |
| 19 | rhmA | Thermophilic bacterium | 7.8 | 60 | Stable | [34] |
| 20 | RamA | Clostridium stercorarium | 7.5 | 60 | Stable | [6] |
| 21 | DtRha | Dictyoglomus thermophilum | 5.0 | 95 | Stable | [14] |
| 22 | RhmA/RhmB | Thermophilic bacterium PRI-1686 | 7.9/5.0–6.9 | 70/70 | Stable at 40℃ | [21] |
| 23 | ramALa | Lactobacillus acidophilus | 6.0 | 37–45 | Unstable | [35] |
| 24 | RhaB1/RhaB2 | Lactobacillus Plantarum NCC245 | 7.0/5.0 | 50/60 | RhaB2 Stable | [36] |
| 25 | Ram/Ram2 | Pediococcus acidilactici | 5.5/4.5 | 50/70 | [19] | |
| 26 | RHA-P | Novosphingobium sp. PP1Y | 6.9 | 37 | [12] | |
| 27 | BtRha | Bacteroides thetaiotaomicron | 6.5 | 60 | Stable | [37] |
| 28 | BbRha | Bifidobacterium breve | 6.5 | 55 | Stable | [38] |
| 29 | rBdRham | Bifidobacterium dentium | 6.0 | 35 | Stable between 30–40℃ | [39] |
| 30 | Fusobacterium K-60 | 5.5–6.5 | [40] | |||
| 31 | Sphingomonas sp. R1 | 8.0 | 50 | Stable below 45 ℃ | [41] | |
| 32 | Pseudomonas paucimobilis FP2001 | 7.8 | 30–50 | Stable below 40 ℃ | [4] | |
| 33 | Pseudoalteromonas sp. 005NJ | 6.0 | 40 | Thermosensitive | [15] | |
| 34 | Bacteroides JY-6 | 7.0 | [42] | |||
| Chloroflexus aurantiacus | 6.0 | 50 | Stable | [43] |
表选项
1.2 最适温度不同α-L-鼠李糖苷酶的最适温度如表 1和表 2所示。大多数真菌来源的α-L-鼠李糖苷酶是嗜热性的酶[20],最适温度为50–60 ℃。细菌来源的不同α-L-鼠李糖苷酶的最适温度差别较大,除了来自嗜热细菌PRI-1686[21]、嗜热网球菌Dictyoglomus thermophilum[14]和P. acidilactici[19]的α-L-鼠李糖苷酶有较高的温度要求外,其他细菌来源的α-L-鼠李糖苷酶的最适温度为30–60 ℃。其中,来自D. thermophilum的α-L-鼠李糖苷酶,其最适温度高达95 ℃[14],是目前报道的适宜温度最高的酶。
1.3 热稳定性α-L-鼠李糖苷酶的热稳定性与蛋白质的糖基化修饰有关[1]。酶的糖基化水平可以在一定程度上被用来衡量其热稳定性的强弱,来源于棘孢曲霉Aspergillus aculeatus的α-L-鼠李糖苷酶RhaA和RhaB,去糖基化后分子质量分别降低24%和15%,在55 ℃孵育4 h后,RhaA和RhaB的活性分别剩余87%和75%,在65 ℃孵育4 h后,二者活性分别剩余60%和55%[22]。与RhaB相比,RhaA的糖基化水平高,热稳定性也较好。除此之外,氢键、疏水相互作用[20]和盐桥[7]等也会影响酶的热稳定性。虽然不同种类的真菌或细菌翻译后修饰机制千差万别,酶蛋白的糖基化水平各不相同,但在一般情况下,嗜热菌株来源的α-L-鼠李糖苷酶能耐受较高的温度,热稳定性较强,如D. thermophilu和构巢曲霉Aspergillus nidulans来源的α-L-鼠李糖苷酶[14]。嗜冷菌株来源的α-L-鼠李糖苷酶在低温下剩余活性较大,但热稳定性弱,多为温敏性糖苷酶,如海洋细菌假交替单胞菌Pseudoalteromonas sp. 005NJ来源的α-L-鼠李糖苷酶[15]。
1.4 底物特异性1.4.1 α- L-鼠李糖苷酶催化机制GH家族的酶可以通过构型翻转(Inverting)机制水解底物中的糖苷键[8],在水解反应中,多数鼠李糖苷酶由酸性氨基酸行使催化作用,例如天冬氨酸和谷氨酸[9]。以鼠李糖苷酶SaRha78A水解糖苷键的过程为例(图 1),底物鼠李糖的O1是易被攻击的位置,鼠李糖苷酶SaRha78A的氨基酸Glu636为该酶的催化质子供体,可以使底物鼠李糖中的O1发生质子化,另外,与底物鼠李糖的O1和鼠李糖苷酶SaRha78A的Glu895有强氢键作用的水分子被氨基酸Glu895去质子化,去质子化的水分子攻击底物中的糖苷键,从而完成水解反应[9]。序列比对和同源建模结果显示,α-L-鼠李糖苷酶的活性位点相对保守[8, 10, 12],在BsRhaB蛋白序列中,与SaRha78A的Glu636和Glu895相对应的保守氨基酸是Glu572和Glu841。Cui等将BsRhaB的Glu572和Glu841突变为谷氨酰胺后酶活明显降低,证明了Glu572和Glu841对该酶催化活性的重要性[8]。同样的,BT0986中的Glu461和Glu593也被推测为典型的催化残基[11]。
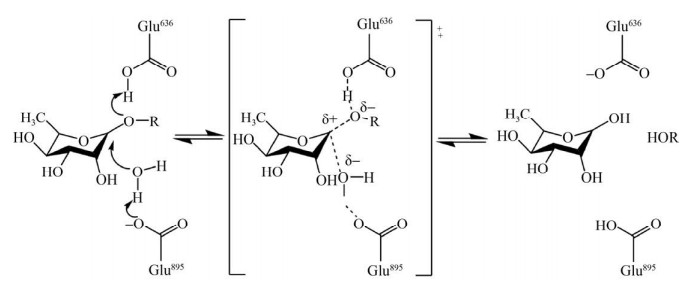 |
| 图 1 α-L-鼠李糖苷酶SaRha78A水解糖苷键的机制[44] Fig. 1 Mechanism of α-L-rhamnosidase SaRha78A in hydrolyzing the glycosidic bond[44]. |
| 图选项 |
1.4.2 α- L-鼠李糖苷酶对糖苷键的选择性不同来源的α-L-鼠李糖苷酶对α-1, 2、α-1, 3、α-1, 4、α-1和α-1, 6五种糖苷键有不同的特异性(表 3)。多数α-L-鼠李糖苷酶既可以水解α-1, 2糖苷键,也可以水解α-1, 6糖苷键,但是,有些酶对α-1, 2糖苷键的亲和性更强,如鞘氨醇单胞菌Sphingomonas sp. R1和白曲霉Aspergillus kawachii来源的鼠李糖苷酶[24-41]。有些酶则对α-1, 6糖苷键的亲和性更强,如齿双歧杆菌Bifidobacterium dentium来源的鼠李糖苷酶[39]。除此之外,RHA-P等6个糖苷酶还可以水解天然黄酮类化合物槲皮甙,如图 2所示,对比芦丁和柚皮素中较为常见的两种糖苷键类型,该化合物中的L-鼠李糖直接连接在糖苷配基上,属于α-1类型糖苷键[12]。还有一些酶的底物特异性较强,只能水解某一种类型的糖苷键,如AT-rRha和FFCDL-39只能特异性水解α-1, 2类型的糖苷键[3-13],相反,RhaB1、RhaB2和FFCDL-90等糖苷酶只能特异性水解α-1, 6类型的糖苷键[27-36]。
表 3 不同来源的α-L-鼠李糖苷酶的底物特异性Table 3 Substrate specificity of α-L-rhamnosidases of different origins
| Number | Enzyme | Organism | Links | Substrates | References |
| 1 | BtRha | Bacteroides thetaiotaomicron | α-1, 2★, α-1, 6 | (1, 3–4, 15) | [37] |
| 2 | AnRhaE | Aspergillus nidulans | α-1, 2+, α-1, 6, α-1, 2★ | (1–2, 4, 10, 15) | [25] |
| 3 | Pichia angusta X349 | α-1, 2+, α-1, 6, α-1 | (1–5) | [30] | |
| 4 | Penicillium ulaiense | α-1, 2, α-1, 6 | (1–3) | [28] | |
| 5 | Acrostalagmus luteo albus | α-1, 2, α-1, 6, α-1 | (1–3, 5) | [2] | |
| 6 | Pseudomonas paucimobilis FP2001 | α-1, 2, α-1, 6, α-1 | (1–6, 16) | [4] | |
| 7 | BbRha | Bifidobacterium breve | α-1, 2, α-1, 6 | (1–4) | [38] |
| 8 | Sphingomonas sp. R1 | α-1, 2+, α-1, 6, α-1 | (1–5) | [41] | |
| 9 | SaRhaA | Streptomyces avermitilis | α-1, 2, α-1, 6 | (1–4, 9) | [26] |
| 10 | Bacteroides JY-6 | α-1, 2+, α-1, 6 | (1–4, 6, 10, 18) | [42] | |
| 11 | RamA | Clostridium stercorarium | α-1, 2, α-1, 6 | (1–3) | [6] |
| 12 | DtRha | Dictyoglomus thermophilum | α-1, 2+, α-1, 6 | (1–2) | [14] |
| 13 | RHA-P | Novosphingobium sp. PP1Y | α-1, 2, α-1, 6, α-1 | (1–5) | [12] |
| 14 | AK-rhaA | Aspergillus kawachii | α-1, 2+, α-1, 6 | (1–3) | [24] |
| 15 | ramALa | Lactobacillus acidophilus | α-1, 2, α-1, 6 | (1–2, 4, 12–13) | [35] |
| 16 | rBdRham | Bifidobacterium dentium | α-1, 2, α-1, 6+ | (1–2, 4, 8, 18) | [39] |
| 17 | RhaA/RhaB | Aspergillus aculeatus | α-1, 2, α-1, 6 | (1–4) | [22] |
| 18 | Fusobacterium K-60 | α-1, 2, α-1, 6, α-1 | (2–5, 18) | [40] | |
| 19 | RhmA | Thermophilic bacterium PRI-1686 | α-1, 2+, α-1, 6 | (1–4) | [34] |
| 20 | RhmB | Thermophilic bacterium PRI-1686 | α-1, 2, α-1, 6 | (1–3) | [34] |
| 21 | AT-rRha | Aspergillus tubingensis | α-1, 2 | (2) | [13] |
| 22 | Absidia sp. 39 | α-1, 2 | (7) | [3] | |
| 23 | RhaB1/RhaB2 | Lactobacillus Plantarum NCC245 | α-1, 6 | (1, 3–4) | [36] |
| 24 | Ram2 | Pediococcus acidilactici | α-1, 6 | (1, 3, 14) | [19] |
| 25 | Absidia sp. 90 | α-1, 6 | (17) | [27] | |
| 26 | r-Rha1 | Aspergillus niger | α-1, 2, α-1, 3, α-1, 4, α-1, 6 | (1–4, 6, 19) | [1, 20] |
| 27 | Pig liver | α-1, 2, α-1, 4 | (11) | [5] | |
| 28 | Penicillium griseoroseum MTCC-9224 | α-1, 6 | (1, 4) | [31] | |
| 29 | Alternaria alternate SK37.001 | α-1, 2, α-1, 6, α-1 | (1–5, 10) | [32] | |
| 30 | Fusarium moniliforme MTCC-2088 | α-1, 2, α-1, 6 | (1–4) | [18] | |
| 31 | Chloroflexus aurantiacus | α-1, 2, α-1, 6+ | (1–4) | [43] | |
| 32 | XpoGH78 | Xylaria polymorpha | α-1, 2 | (1–2) | [33] |
| (1) p-nitrophenyl-a-L-rhamnopyranoside; (2) Naringin; (3) Hesperidin; (4) Rutin; (5) Quercitrin; (6) Saikosaponin C; (7) Ginsenoside Rg2; (8) Ginsenoside Re; (9) Gum Arabic; (10) Neohesperidin; (11) Dioscin; (12) Nicotiflorin; (13) Narirutin; (14) Rutinose; (15) Epimedin C; (16) Proscillaridin A; (17) Gypenoside-5; (18) Poncirin; (19) Myricitrin. The glycosidic bond marked ★ is the glycosidic bond formed between the terminal rhamnose and rhamnose, without ★ means the glycosidic bond is between the terminal rhamnose and glucose, α-1 is the glycosidic bond formed by the terminal rhamnose directly connected to the glycosidic ligand; + indicates that the rhamnosidase has stronger affinity for this type of glycosidic bond than other types of glycosidic bonds. | |||||
表选项
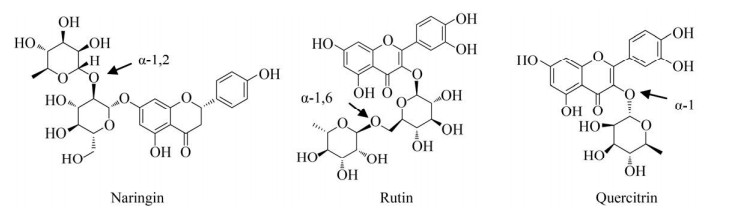 |
| 图 2 黄酮类化合物结构式及所含有的糖苷键类型 Fig. 2 Structures of flavonoids and the types of associated glycosidic bonds. |
| 图选项 |
除了α-1, 2和α-1, 6类型的糖苷键,少动假单胞菌Pseudomonas paucimobilis FP2001和拟杆菌Bacteroides JY-6来源的鼠李糖苷酶还可以水解柴胡皂苷C中的α-1, 4糖苷键[4-42],猪肝中的鼠李糖苷酶可以水解薯蓣皂苷中的α-1, 2和α-1, 4糖苷键[5],芽孢杆菌Bacillus sp. GL1来源的鼠李糖苷酶可以水解结冷胶中的α-1, 3糖苷键[1]。目前,只有来自黑曲霉Aspergillus niger的鼠李糖苷酶可以水解α-1, 2、α-1, 3、α-1, 4和α-1, 6四种类型的糖苷键[38]。值得注意的是,以上所述糖苷键是L-鼠李糖与葡萄糖之间所形成的共价键,Wu等首次证明多形拟杆菌Bacteroides thetaiotaomicron来源的鼠李糖苷酶还可以水解朝藿定C中L-鼠李糖与鼠李糖之间形成的共价键[37],之后来自A. nidulans的鼠李糖苷酶也被发现具有水解该类糖苷键的活性[25]。
虽然α-L-鼠李糖苷酶的活性位点相对保守,催化机制类似,但是α-L-鼠李糖苷酶的底物特异性却存在差异[12]。遗憾的是,由于目前关于α-L-鼠李糖苷酶结晶的研究较少,还没有研究者针对底物特异性差异给出合理的解释。所以,总结已有α-L-鼠李糖苷酶的糖苷键作用类型,不仅有利于该酶的工业化应用,还可以为鼠李糖苷酶底物特异性原理研究提供证据。
2 影响酶活性的因素2.1 糖苷配体对α-L-鼠李糖苷酶底物特异性的影响α-L-鼠李糖苷酶水解天然黄酮类化合物末端的L-鼠李糖时,底物的糖苷配体会影响酶的特异性。糖苷配体的影响主要是通过产生氢键或空间位阻[21]来实现。Guillotin等利用Doking模型,揭示了鼠李糖苷酶DtRha对柚皮苷的亲和性高的原因,是柚皮苷的糖苷配体与DtRha的活性中心形成氢键,使催化复合体更加稳定[14]。嗜热细菌PRI-1686[21]、A. aculeatus[22]和短双歧杆菌Bifidobacterium breve[38]的鼠李糖苷酶对同为α-1, 6糖苷键的芦丁和橘皮苷亲和性不同,是因为在柚皮苷中糖侧链与C-1位置相连,在芦丁中糖侧链与C-3位置相连,不同的位置造成不同的空间位阻,从而影响酶对底物的催化活性[22]。
2.2 阳离子和有机试剂对α-L-鼠李糖苷酶活性的影响微生物代谢或体外酶反应过程中,阳离子的参与会对酶的活性产生影响。Mensitieri等通过对比已有的5个α-L-鼠李糖苷酶晶体结构,揭示了Ca2+对α-L-鼠李糖苷酶催化过程的重要作用[12]。如图 3所示,在SaRha78A鼠李糖苷酶中,Ca2+结合在离催化结构域很近的一个独立结构域上,当底物与酶的活性位点结合时,Ca2+可以与底物鼠李糖的O3和O4形成配位键,从而促进反应进行[9]。被金属螯合剂EDTA抑制的酶活,也可以通过添加Ca2+完全恢复。Miake等[4]和Hashimoto等[41]的实验结果也证明了Ca2+对糖苷酶活性的积极作用。与之相反,Hg2+和硫基试剂会影响酶的硫基基团,从而抑制一些α-L-鼠李糖苷酶的活性[30],当1 mmol/L Hg2+存在时,BtRha和RamA等糖苷酶的活性明显降低甚至被完全抑制[6, 37]。除了受阳离子影响外,一些α-L-鼠李糖苷酶的活性会被对氯甲基苯乙烯(1-(chloromethyl)-4- ethenylbenzene,PCMS) 抑制,这些酶的底物特异性可能与半胱氨酸残基有关,因为PCMS可以和半胱氨酸形成共价键,从而影响酶与底物结合[40]。
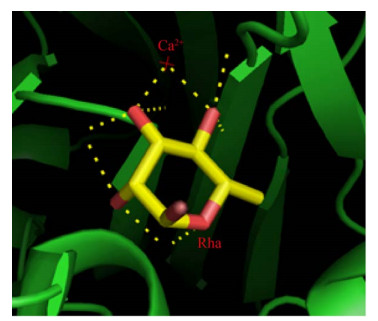 |
| 图 3 α-L-鼠李糖苷酶SaRha78A与L -鼠李糖结合的三维结构图 Fig. 3 The crystal structure of α-L-rhamnosidase SaRha78A complex bound with L-rhamnose. Stereo view of the sugar-binding pocket in domain D (PDB code 3W5N). |
| 图选项 |
2.3 L -鼠李糖和葡萄糖对α-L-鼠李糖苷酶的竞争性抑制作用L-鼠李糖和葡萄糖可以作为竞争性抑制剂对很多α-L-鼠李糖苷酶的活性产生抑制作用[2],且抑制效果具有剂量依赖性[45]。在文献记载的34个不同来源的α-L-鼠李糖苷酶中,接近一半的鼠李糖苷酶被报道了末端产物L-鼠李糖对酶活性的抑制作用,其中来自P. angusta X349和L. plantarum NCC245的鼠李糖苷酶的活性被L-鼠李糖轻微抑制[30, 36],来自Pseudoalteromonas sp. 005NJ、P. acidilactici和梭杆菌Fusobacterium K-60等多个鼠李糖苷酶的活性被L-鼠李糖严重抑制[15, 19, 40]。虽然葡萄糖与鼠李糖的结构类似,但葡萄糖与L-鼠李糖的抑制效果不是完全一致,如被L-鼠李糖严重抑制活性的r-Rha1和005NJ鼠李糖苷酶,在高浓度葡萄糖存在下,仍能保留70%左右的活性[1, 15],除了P. acidilactici的Ram2被葡萄糖严重抑制活性外[19],P. angusta X349和嗜热细菌PRI-1686等多个鼠李糖苷酶的活性被葡萄糖轻微抑制[21, 30]。总体来说,葡萄糖对鼠李糖苷酶活性的抑制效果不如L-鼠李糖的抑制效果。值得一提的是来自宾曲霉Aspergillus tubingensis的鼠李糖苷酶AT-rRha的活性不会被L-鼠李糖和葡萄糖影响[13],这个特点非常有利于工业生产。
3 总结与展望α-L-鼠李糖苷酶是非常重要的工业酶,在食品和制药领域具有广泛的应用[2]。近年来,越来越多的****围绕不同来源的α-L-鼠李糖苷酶的酶学特性和底物特异性进行了研究,展示了鼠李糖苷酶的多样性。本论文总结了不同来源的α-L-鼠李糖苷酶的最适pH、最适反应温度、热稳定性和底物特异性,概括了α-L-鼠李糖苷酶的催化机制和影响鼠李糖苷酶的因素,为选择适合不同应用环境的鼠李糖苷酶提供线索,有利于α-L-鼠李糖苷酶的工业应用。
L-鼠李糖的存在会对菌株产生碳代谢压力,上调细胞内α-L-鼠李糖苷酶的合成,但L-鼠李糖又会对鼠李糖苷酶的活性产生抑制作用,这个矛盾的关系可以通过异源表达的方式解决,异源表达能消除碳源代谢压力,也可以满足工业上大量生产α-L-鼠李糖苷酶的需求[1]。但是异源表达系统可能缺乏必要的翻译后修饰机制,如蛋白糖基化,因此通过异源表达合成的α-L-鼠李糖苷酶的热稳定性是否会受到影响有待进一步研究。
参考文献
| [1] | Li LJ, Yu Y, Zhang X, et al. Expression and biochemical characterization of recombinant α-L-rhamnosidase r-Rha1 from Aspergillus niger JMU-TS528. Int J Biol Macromol, 2016, 85: 391-399. DOI:10.1016/j.ijbiomac.2015.12.093 |
| [2] | Rojas NL, Voget CE, Hours RA, et al. Purification and characterization of a novel alkaline α-L-rhamnosidase produced by Acrostalagmus luteoalbus. J Ind Microbiol Biotechnol, 2011, 38(9): 1515-1522. DOI:10.1007/s10295-010-0938-8 |
| [3] | Yu HS, Gong JM, Zhang CZ, et al. Purification and characterization of ginsenoside-a-L-rhamnosidase. Chem Pharm Bull, 2002, 50(2): 175-178. DOI:10.1248/cpb.50.175 |
| [4] | Miake F, Satho T, Takesue H, et al. Purification and characterization of intracellular α-L-rhamnosidase from Pseudomonas paucimobilis FP2001. Arch Microbiol, 2000, 173(1): 65-70. DOI:10.1007/s002030050009 |
| [5] | Qian S, Yu HS, Zhang CZ, et al. Purification and characterization of dioscin-α-L-rhamnosidase from Pig liver. Chem Pharm Bull, 2005, 53(8): 911-914. DOI:10.1248/cpb.53.911 |
| [6] | Zverlov VV, Hertel C, Bronnenmeier K, et al. The thermostable α-L-rhamnosidase RamA of Clostridium stercorarium: biochemical characterization and primary structure of a bacterial α-L-rhamnoside hydrolase, a new type of inverting glycoside hydrolase. Mol Microbiol, 2000, 35(1): 173-179. DOI:10.1046/j.1365-2958.2000.01691.x |
| [7] | Ge L, Li DD, Wu T, et al. B-factor-saturation mutagenesis as a strategy to increase the thermostability of α-L-rhamnosidase from Aspergillus terreus. J Biotechnol, 2018, 275: 17-23. DOI:10.1016/j.jbiotec.2018.03.013 |
| [8] | Cui ZL, Maruyama Y, Mikami B, et al. Crystal structure of glycoside hydrolase family 78 alpha-L-rhamnosidase from Bacillus sp. GL1. J Mol Biol, 2007, 374(2): 384-398. DOI:10.1016/j.jmb.2007.09.003 |
| [9] | Fujimoto Z, Jackson A, Michikawa M, et al. The structure of a Streptomyces avermitilis α-L-rhamnosidase reveals a novel carbohydrate- binding module CBM67 within the six-domain arrangement. J Biol Chem, 2013, 288(17): 12376-12385. DOI:10.1074/jbc.M113.460097 |
| [10] | O'Neill EC, Stevenson CEM, Paterson MJ, et al. Crystal structure of a novel two domain GH78 family α-rhamnosidase from Klebsiella oxytoca with rhamnose bound. Proteins, 2015, 83(9): 1742-1749. DOI:10.1002/prot.24807 |
| [11] | Ndeh D, Rogowski A, Cartmell A, et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature, 2017, 544(7648): 65-70. DOI:10.1038/nature21725 |
| [12] | Mensitieri F, De Lise F, Strazzulli A, et al. Structural and functional insights into RHA-P, a bacterial GH106 α-L-rhamnosidase from Novosphingobium sp. PP1Y. Arch Biochem Biophys, 2018, 648: 1-11. DOI:10.1016/j.abb.2018.04.013 |
| [13] | Li LJ, Gong JY, Wang S, et al. Heterologous expression and characterization of a new clade of Aspergillus α-L-rhamnosidase suitable for citrus juice processing. J Agric Food Chem, 2019, 67(10): 2926-2935. DOI:10.1021/acs.jafc.8b06932 |
| [14] | Guillotin L, Kim H, Traore Y, et al. Biochemical characterization of the α-L-rhamnosidase DtRha from Dictyoglomus thermophilum: application to the selective derhamnosylation of natural flavonoids. ACS Omega, 2019, 4(1): 1916-1922. DOI:10.1021/acsomega.8b03186 |
| [15] | Orrillo AG, Ledesma P, Delgado OD, et al. Cold-active α-L-rhamnosidase from psychrotolerant bacteria isolated from a sub-antarctic ecosystem. Enzyme Microb Technol, 2007, 40(2): 236-241. DOI:10.1016/j.enzmictec.2006.04.002 |
| [16] | Kaur A, Singh S, Singh RS, et al. Hydrolysis of citrus peel naringin by recombinant α-L-rhamnosidase from Clostridium stercorarium. J Chem Technol Biotechnol, 2010, 85(10): 1419-1422. DOI:10.1002/jctb.2433 |
| [17] | Yadav V, Yadav S, Yadava S, et al. α-L-rhamnosidase from Aspergillus flavus MTCC-9606 isolated from lemon fruit peel. Int J Food Sci Technol, 2011, 46(2): 350-357. DOI:10.1111/j.1365-2621.2010.02498.x |
| [18] | Kumar D, Yadav S, Yadava S, et al. An alkali tolerant α-L-rhamnosidase from Fusarium moniliforme MTCC-2088 used in de-rhamnosylation of natural glycosides. Bioorg Chem, 2019, 84: 24-31. DOI:10.1016/j.bioorg.2018.11.027 |
| [19] | Michlmayr H, Brandes W, Eder R, et al. Characterization of two distinct glycosyl hydrolase family 78 α-L-rhamnosidases from Pediococcus acidilactici. Appl Environ Microbiol, 2011, 77(18): 6524-6530. DOI:10.1128/AEM.05317-11 |
| [20] | Li LJ, Wu ZY, Yu Y, et al. Development and characterization of an α-L-rhamnosidase mutant with improved thermostability and a higher efficiency for debittering orange juice. Food Chem, 2018, 245: 1070-1078. DOI:10.1016/j.foodchem.2017.11.064 |
| [21] | Birgisson H, Hreggvidsson GO, Fridjónsson OH, et al. Two new thermostable α-L-rhamnosidases from a novel thermophilic bacterium. Enzyme Micro Technol, 2004, 34(6): 561-571. DOI:10.1016/j.enzmictec.2003.12.012 |
| [22] | Manzanares P, Van Den Broeck HC, De Graaff LH, et al. Purification and characterization of two different alpha-L-rhamnosidases, RhaA and RhaB, from Aspergillus aculeatus. Appl Environ Microbiol, 2001, 67(5): 2230-2234. DOI:10.1128/AEM.67.5.2230-2234.2001 |
| [23] | Custodio MVG, Otamendi FP, Vidal DR, et al. Production and characterization of an Aspergillus terreus α-L-rhamnosidase of oenological interest. Z Lebensm Unters Forch, 1996, 203(6): 522-527. DOI:10.1007/BF01193157 |
| [24] | Koseki T, Mese Y, Nishibori N, et al. Characterization of an α-L-rhamnosidase from Aspergillus kawachii and its gene. Appl Microbiol Biotechnol, 2008, 80(6): 1007-1013. DOI:10.1007/s00253-008-1599-7 |
| [25] | Lyu YB, Zeng WZ, Du GC, et al. Efficient bioconversion of epimedin C to icariin by a glycosidase from Aspergillus nidulans. Bioresour Technol, 2019, 289: 121612. DOI:10.1016/j.biortech.2019.121612 |
| [26] | Ichinose H, Fujimoto Z, Kaneko S. Characterization of an α-L-rhamnosidase from Streptomyces avermitilis. Biosci Biotechnol Biochem, 2013, 77(1): 213-216. DOI:10.1271/bbb.120735 |
| [27] | Yu HS, Liu H, Zhang CZ, et al. Purification and characterization of gypenoside-α-L-rhamnosidase hydrolyzing gypenoside-5 into ginsenoside Rd. Proc Biochem, 2004, 39(7): 861-867. DOI:10.1016/S0032-9592(03)00196-1 |
| [28] | Rajal VB, Cid AG, Ellenrieder G, et al. Production, partial purification and characterization of α-L-rhamnosidase from Penicillium ulaiense. World J Microbiol Biotechnol, 2009, 25(6): 1025-1033. DOI:10.1007/s11274-009-9979-4 |
| [29] | Feng B, Hu W, Ma BP, et al. Purification, characterization, and substrate specificity of a glucoamylase with steroidal saponin-rhamnosidase activity from Curvularia lunata. Appl Microbiol Biotechnol, 2007, 76(6): 1329-1338. DOI:10.1007/s00253-007-1117-3 |
| [30] | Yanai T, Sato M. Purification and characterization of an α-L-rhamnosidase from Pichia angusta X349. Biosci Biotechnol Biochem, 2000, 64(10): 2179-2185. DOI:10.1271/bbb.64.2179 |
| [31] | Yadav S, Yadava S, Yadav KDS. α-L-rhamnosidase selective for rutin to isoquercitrin transformation from Penicillium griseoroseum MTCC-9224. Bioorg Chem, 2017, 70: 222-228. DOI:10.1016/j.bioorg.2017.01.002 |
| [32] | Zhang T, Yuan WB, Li ML, et al. Purification and characterization of an intracellular α-L-rhamnosidase from a newly isolated strain, Alternaria alternata SK37.001. Food Chem, 2018, 269: 63-69. DOI:10.1016/j.foodchem.2018.06.134 |
| [33] | Nghi D H, Bittner B, Kellner H, et al. The wood rot ascomycete Xylaria polymorpha produces a novel GH78 glycoside hydrolase that exhibits α-L-rhamnosidase and feruloyl esterase activities and releases hydroxycinnamic acids from lignocelluloses. Appl Environ Microbiol, 2012, 78(14): 4893-4901. DOI:10.1128/AEM.07588-11 |
| [34] | Birgisson H, Wheat JO, Hreggvidsson GO, et al. Immobilization of a recombinant Escherichia coli producing a thermostable α-L-rhamnosidase: creation of a bioreactor for hydrolyses of naringin. Enzyme Microb Technol, 2007, 40(5): 1181-1187. DOI:10.1016/j.enzmictec.2006.08.026 |
| [35] | Beekwilder J, Marcozzi D, Vecchi S, et al. Characterization of rhamnosidases from Lactobacillus plantarum and Lactobacillus acidophilus. Appl Environ Microbiol, 2009, 75(11): 3447-3454. DOI:10.1128/AEM.02675-08 |
| [36] | ávila M, Jaquet M, Moine D, et al. Physiological and biochemical characterization of the two α-L-rhamnosidases of Lactobacillus plantarum NCC245. Microbiology, 2009, 155(Pt 8): 2739-2749. |
| [37] | Wu T, Pei JJ, Ge L, et al. Characterization of a α-L-rhamnosidase from Bacteroides thetaiotaomicron with high catalytic efficiency of epimedin C. Bioorg Chem, 2018, 81: 461-467. DOI:10.1016/j.bioorg.2018.08.004 |
| [38] | Zhang R, Zhang BL, Xie T, et al. Biotransformation of rutin to isoquercitrin using recombinant α-L-rhamnosidase from Bifidobacterium breve. Biotechnol Lett, 2015, 37(6): 1257-1264. DOI:10.1007/s10529-015-1792-6 |
| [39] | Bang SH, Hyun YJ, Shim J, et al. Metabolism of rutin and poncirin by human intestinal microbiota and cloning of their metabolizing α-L-rhamnosidase from Bifidobacterium dentium. J Microbiol Biotechnol, 2015, 25(1): 18-25. DOI:10.4014/jmb.1404.04060 |
| [40] | Park SY, Kim JH, Kim DH. Purification and characterization of quercitrin-hydrolyzing α-L-rhamnosidase from Fusobacterium K-60, a human intestinal bacterium. J Microbiol Biotechnol, 2005, 15(3): 519-524. |
| [41] | Hashimoto W, Murata K. alpha-L-rhamnosidase of Sphingomonas sp. R1 producing an unusual exopolysaccharide of sphingan. Biosci Biotechnol Biochem, 1998, 62(6): 1068-1074. DOI:10.1271/bbb.62.1068 |
| [42] | Jang IS, Kim DH. Purification and characterization of α-L-rhamnosidase from Bacteroides JY-6, a human intestinal bacterium. Biol Pharm Bull, 1996, 19(12): 1546-1549. DOI:10.1248/bpb.19.1546 |
| [43] | Shin KC, Seo MJ, Oh DK, et al. Cloning and characterization of α-L-rhamnosidase from Chloroflexus aurantiacus and its application in the production of isoquercitrin from rutin. Biotechnol Lett, 2019, 41(3): 419-426. DOI:10.1007/s10529-019-02648-8 |
| [44] | David LZ, Stephen GW. Glycosidase mechanisms: anatomy of a finely tuned catalyst. Acc Chem Res, 2000, 33(1): 11-18. DOI:10.1021/ar970172 |
| [45] | Mazzaferro LS, Orrillo GA, Ledesma P, et al. Dose-dependent significance of monosaccharides on intracellular α-L-rhamnosidase activity from Pseudoalteromonas sp. Biotechnol Lett, 2008, 30(12): 2147-2150. DOI:10.1007/s10529-008-9810-6 |
