
 , 周国华1,2
, 周国华1,21 中国药科大学生命科学与技术学院,江苏 南京 210009
2 南京军区南京总医院药理科,江苏 南京 210002
基金项目:国家科技重大专项 (No. 2013ZX10004103),江苏省临床科技专项 (No. BL2012061),国家自然科学基金 (Nos. 31300704, 21475151, 21405176, 21275161),中国博士后基金特别资助项目基金 (Nos. 2013T60938, 2014T71011),江苏省六大人才高峰项目 (No. 2015-WSN-085) 资助
摘要: Flap核酸内切酶1 (Flap endonuclease 1, FEN1) 是一种能催化核酸侵入反应的核酸内切酶,可应用于信号放大检测方法,但该酶详细的表达纯化工艺尚无报道,并且活性难以准确测定,限制了其应用。通过合成嗜热古球菌Archaeoglobus fulgidus来源的FEN1基因序列,构建了pET24a(+)-FEN1-His重组质粒,并通过优化表达条件,得到了FEN1最优表达条件为:37 ℃、200 r/min振荡培养8 h后,加入诱导剂IPTG至终浓度为0.05 mmol/L,再于37 ℃、200 r/min诱导表达11 h,最终经镍亲和层析成功纯化得到了分子量约为38 kDa的重组FEN1。同时建立了基于荧光标记探针的FEN1活性测定方法,准确测定了重组FEN1的活性,为建立基于该酶的核酸检测方法提供了可靠的酶活力依据。最终将重组FEN1用于实时荧光PCR偶联高特异核酸侵入信号扩增法检测了乙醛脱氢酶2基因 (aldh2) 的基因型,得到了准确的分型结果,表明重组FEN1能用于基因多态性的分型检测中,为发展基于核酸侵入反应的核酸检测方法提供了可靠的工具酶。
关键词: flap核酸内切酶1 信号放大 活性测定
Expression and activity assay of recombinant flap endonuclease
Sheng Nan1, Ma Yinjiao1, Wang Jianping1, Zou Bingjie2

 , Zhou Guohua1,2
, Zhou Guohua1,2 1 School of Life Science and Technology, China Pharmaceutical University, Nanjing 210009, Jiangsu, China;
2 Department of Pharmacology, Nanjing General Hospital, Nanjing 210002, Jiangsu, China
Received: February 23, 2016; Accepted: May 16, 2016
Supported by:National Key Science & Technology Special Project (No. 2013ZX10004103), Jiangsu Province’s Clinical Science & Technology Special Project (No. BL2012061), National Natural Science Foundation of China (Nos. 31300704, 21475151, 21405176, 21275161), China Postdoctoral Science Special Foundation (Nos. 2013T60938, 2014T71011), Six Talent Peaks Project in Jiangsu Province (No. 2015-WSN-085)
Corresponding authors:Guohua Zhou. Tel/Fax: +86-25-80860195; Fax: +86-25-80860196; E-mail: ghzhou@nju.edu.cn
Abstract: Flap endonuclease 1 (FEN1) is an endonuclease that catalyzes invasive reaction. It can be used in signal-amplification reaction-based nucleic acid assay. However, the application of FEN1 is hampered due to the lack of detailed protocols to express and purify the enzyme, and to quantify the enzyme activity. In this paper, the DNA fragment coding the gene of FEN1 from Archaeoglobus fulgidus was synthesized, and inserted into the plasmid of pET24a(+) to express recombinant FEN1 with His-tag. After optimizing the expression, detailed expression protocol of FEN1 was obtained by culturing the recombinant E. coli at 37 ℃with 200 r/min of shaking for 8 h, followed by inducing with 0.05 mmol/L IPTG at 37 ℃ for 11 h. The purified recombinant FEN1 with the molecular mass of 38 kDa was obtained by Ni-affinity chromatography. Moreover, we developed a accurate quantification method with fluorescence-labelled probes. Finally, the recombinant FEN1 was used in real-time PCR coupled with high specific invader assay for aldh2 gene genotyping to obtain the correct typing results, indicating that the recombinant FEN1 can be used in gene polymorphism detection. We provide a reliable enzyme for developing invasive reaction-based nucleic acid assay.
Key words: flap endonuclease 1 signal amplification activity assay
Flap核酸内切酶1 (Flap endonuclease 1,FEN1) 存在于多种真核生物及古细菌中,它能够识别核酸双链中上游片段3′端侵入到下游5′区域形成的单个碱基重叠结构 (“核酸侵入”结构) 并切割5′翘起的flap片段,在DNA的复制、重组和修复过程中发挥着重要作用[1-2]。利用FEN1的识别与切割活性,可针对待测核酸序列设计一对探针,使其与待测模板杂交后形成“核酸侵入”结构,引发核酸侵入反应 (图 1),切割产生flap片段,当反应温度接近下游探针与模板的熔解温度 (Tm) 时,会形成探针杂交与flap切割的动态循环,产生flap片段的累积,形成信号放大。该反应将待测模板转化为与模板序列无关的flap信号分子,避免了模板扩增引发的产物交叉污染的风险,非常适用于核酸检测[3-4]。因此,利用核酸侵入反应开发了很多核酸信号扩增检测方法,如级联侵入信号放大技术 (Invader assay)[5]、偶联PCR侵入信号放大技术 (Invader plus)[6-8]、偶联滚环扩增侵入信号放大技术 (Invader RCA assay)[9]、偶联切刻内切酶侵入信号放大技术 (Cascade enzymatic signal amplification,CESA)[10]、偶联链置换延伸侵入信号放大技术 (Invasive reaction assisted strand- displacement signal amplification,IRASA)[11]及偶联纳米金探针可视化核酸检测技术 (Invasive reaction with nicking endonuclease-assisted nanoparticles amplification,IR-NEANA)[12]等,并在单核苷酸多态性 (Single nucleotide polymorphism,SNP) 分型检测[13-15]、突变检 测[16-19]、病原体分型检测[20-22]及RNA检测[23-24]等方面有着广泛应用。但FEN1表达纯化工艺未见详细报道,并且该酶的活性测定方法尚未建立,常规的电泳分析蛋白方法[25-26]只能根据酶切片段的有无以及条带的深浅对酶活性进行半定量,不能准确定量其活性,导致在应用该酶时难以优化用量。因此,本文研究了FEN1表达纯化的方法,通过合成古细菌Archaeoglobus fulgidus来源的FEN1基因序列,构建了pET24a(+)-FEN1-His重组质粒,成功表达了重组Afu FEN1,利用镍亲和层析纯化获得最终分子量约为38 kDa的重组酶。
 |
| 图 1 FEN1作用于核酸侵入反应的原理[10] Figure 1 Schematic illustration of FEN1 function in the invasive reaction[10]. When a complete complementary upstream-probe (UP) and a partial complementary downstream-probe (DP) hybridize with the target nucleic acid (Target),forming a tripartite structure with one-base overlapping structure at the 3′ end of the UP,FEN1 recognizes this overlapping structure and cleaves DP into two parts,then the unmatched part (Flap) dissociates from DP. Since the reaction temperature is close to the melting temperature of DP,DP hybridizes to the target with UP and assemble the tripartite structure,FEN1 cleaves DP continuously and forms a circulation,dissociated flaps finally accumulate and lead to signal amplification. |
| 图选项 |
为了克服FEN1无法准确定量测定活性的难题,本文利用荧光标记探针建立了FEN1活性测定方法,根据国际酶组织的酶活力单位定义,准确测定重组酶的活力单位,为基于该酶的其他核酸检测方法提供了酶活依据,便于核酸侵入反应的条件优化,最终成功将表达的酶应用于基于实时荧光PCR偶联核酸侵入反应[27]的SNP检测中。
1 材料与方法1.1 材料1.1.1 质粒与菌株表达载体pET24a(+)、宿主菌大肠杆菌BL21(DE3) 均为本实验室保存。
1.1.2 主要仪器与试剂PTC-225 PCR扩增仪 (美国MJ Research公司);Power PAC1000高压电源电泳仪/槽 (美国BIO-RAD公司);ChemilmagerTm凝胶电泳成像仪 (美国Alpha Innotech公司);THZ-03M2R小型冷冻空气浴恒温摇床 (上海申能博彩生物科技有限公司);Gene SpeIII微量核酸蛋白测定仪 (日本Naka Intruments公司);PolyScience 9105低温水浴箱 (美国PolyScience公司);电热恒温培养箱 (沈阳利港净化设备有限公司);UVPLM-26E高效2UV透照仪 (美国Upland公司);21R贝克曼台式高速冷冻离心机 (美国Beckman公司) 等。
限制性内切酶XbaⅠ、XhoⅠ、Taq DNA聚合酶、T4 DNA连接酶、λ-EcoT14Ⅰ digest DNA Marker、TaKaRa Agarose Gel DNA Purification Kit Ver. 2.0等购自宝生物工程 (大连) 有限公司;硅胶膜PCR产物纯化试剂盒购自上海赛百胜基因技术有限公司;IPTG、Lysozyme等购自Amresco公司;酵母提取物、胰蛋白胨等购自Oxoid公司;Wizard? Genomic DNA Purification Kit购自美国Promega公司;其他试剂均为国产分析纯;修饰探针由大连宝生物公司合成制备并经HPLC纯化;未修饰的寡核苷酸片段为英潍捷基 (上海) 贸易有限公司合成制备并经PAGE纯化,所有核酸片段序列见表 1。
表 1 核酸序列信息Table 1 Oligonucleotides sequences used in this study
| Primers/probes | Sequences (5′-3′) |
| UPa | ATGTCACTTCCCCTTGGTTCTCTCC |
| DPa | AGCAGGACGGGATCTGGCCTGGTGC |
| Targeta | CTATTGCACCAGGCCAGATGAGAGAACCAAGGGGAAGTGACAT |
| Flapb | ACGGACGCGGAGT |
| Fc | GGGTCAACTGCTATGATGTGTTTGG |
| Rc | GCCCCCAGCAGGTCCC |
| Invaderc | GGGCTGCAGGCATACACTT |
| WPc | CGCGCCGAGGGAAGTGAAAACTGTGAGTG-PO3 |
| MPc | ACGGACGCGGAGAAAGTGAAAACTGTGAGTG-PO3 |
| Hairpin1c | VIC-TCTT (BHQ1) AGCCGGTTTTCCGGCTAAGACCTCGGCGCG-C6-NH2 |
| Hairpin2b,c | FAM-TCTT (BHQ1) AGCCGGTTTTCCGGCTAAGACTCCGCGTCCGT-C6-NH2 |
| a: sequences of probes and the target in the preliminary activity verification system; b: sequences of probes in the activity assay system; c: sequences of oligonucleotides used in SNP genotyping. | |
表选项
1.2 方法1.2.1 工程菌的构建与筛选根据NCBI获得A. fulgidus来源的FEN1基因序列 (Gene ID: 1483479),由上海捷瑞生物工程有限公司合成并插入质粒pET24a(+),于大肠杆菌XL10-Gold保存,命名为pET24a(+)-FEN1- His,将重组质粒转化入大肠杆菌BL21(DE3),送上海华大公司测序,筛选得到阳性重组菌命名为BL21(DE3)-FEN1。
1.2.2 诱导时间的优化将重组菌BL21(DE3)-FEN1接种于5 mL的LB (Kan+,30 μg/mL) 液体培养基中,37 ℃、200 r/min振摇过夜,次日按1 %的接种量接种到100 mL的LB (Kan+,30 μg/mL)中,37 ℃、200 r/min振摇培养8 h时加入IPTG (终浓度0.1 mmol/L) ,37 ℃、200 r/min分别诱导0、3、6、11和16 h后进行SDS-PAGE (12%)分析。
1.2.3 诱导剂IPTG浓度的优化调整诱导剂IPTG的用量,使终浓度分别为0、0.05、0.1、0.5和1 mmol/L,37 ℃、200 r/min诱导表达11 h后对诱导表达产物进行SDS-PAGE (12%)分析。
1.2.4 诱导时机的选择调整诱导剂IPTG的加入时机,分别在1、4、8、10和12 h时加入IPTG (终浓度0.05 mmol/L),37 ℃、200 r/min均诱导表达11 h后进行SDS- PAGE (12 %)分析。
1.2.5 重组FEN1的分离纯化及活性初步验证将重组菌按优化后的条件培养,收集诱导表达的菌体,超声破菌并离心,由于重组蛋白含有组氨酸标签,故采用镍亲和层析系统进行纯化,低浓度咪唑 (60 mmol/L) 除杂蛋白,高浓度咪唑 (300 mmol/L) 洗脱靶蛋白,收集洗脱液得到重组FEN1,SDS-PAGE分析后超滤除盐处理 (采用截留分子量为10 kDa的超滤膜),保存于FEN1储存缓冲液中。
应用侵入信号放大反应初步测定重组FEN1的活性。配制反应体系,包含1×反应缓冲液,2.25 mmol/L MgCl2,0.01 μmol/L上游探针,0.1 μmol/L下游探针和71 ng FEN1,分别加入终浓度为0.1、0.01和0.005 μmol/L靶核酸。加入FEN1前,反应管先置于95 ℃孵育5 min,待反应温度降至63 ℃再加入FEN1,63 ℃孵育 2 h。对产物进行尿素变性聚丙烯酰胺凝胶电泳分析。
1.2.6 重组FEN1酶切反应信号的监测配制30 μL的酶切反应体系:1×缓冲液 (10 mmol/L MOPS,50 mmol/L KCl,7.5 mmol/L MgCl2,pH 7.5),0.33 μmol/L flap探针,0.33 μmol/L FAM标记的荧光共振转移发卡探针,5 μL重组FEN1,蒸馏水补齐体系。实时荧光程序设置为94 ℃ 5 s;63 ℃ 10 s,采集荧光信号,99个循环。
1.2.7 FAM荧光标准曲线的绘制配制30 μL反应体系:1×缓冲液 (10 mmol/L MOPS,50 mmol/L KCl,7.5 mmol/L MgCl2,pH 7.5),加入5 μL不同浓度带有FAM荧光基团标记的模板,FAM加入浓度以2.5μmol/L为起始对半稀释6个梯度,蒸馏水补齐体系,按酶切反应程序采集荧光信号。
1.2.8 实时荧光PCR偶联invader高特异性核酸侵入反应实时荧光PCR偶联invader高特异性核酸侵入反应体系的配制和程序设置参考文献[27]。
2 结果与分析2.1 重组FEN1诱导表达条件的优化在成功构建筛选到阳性重组菌后,为了获得较高的重组FEN1蛋白产率,分别对诱导时间、IPTG浓度和诱导时机3个影响因素进行了优化,考察结果如图 2所示。诱导时间的变化影响了蛋白的表达 (图 2A),SDS-PAGE电泳图进一步用BandScan软件分析重组FEN1在菌体总蛋白中的百分比 (图 2B),结果表明,随着诱导时间的增加,目的蛋白表达量也有所增加,诱导表达11 h后再延长诱导时间表达量变化不大,因此选择11 h作为最佳诱导时间。而不同浓度IPTG诱导表达的产物经SDS-PAGE (图 2C) 和BandScan软件分析 (图 2D) 均表明IPTG浓度为0.05 mmol/L时目的蛋白表达量最高,因此选择0.05 mmol/L IPTG作为最佳诱导剂浓度。
外源蛋白的表达会影响工程菌的生长,因此在已确立的诱导时间和诱导剂浓度条件下,考察了诱导时机对重组蛋白表达的影响,重组菌过夜活化,分别在培养不同的时间后进行诱导表达,产物经SDS-PAGE (图 2E) 和BandScan软件分析 (图 2F),结果表明当重组菌培养时间增至8 h时,目的蛋白的比例大大提高,进一步增加培养时间,目的蛋白增幅不大。从节约操作时间的角度考虑,选择培养8 h作为IPTG的最佳诱导时机。
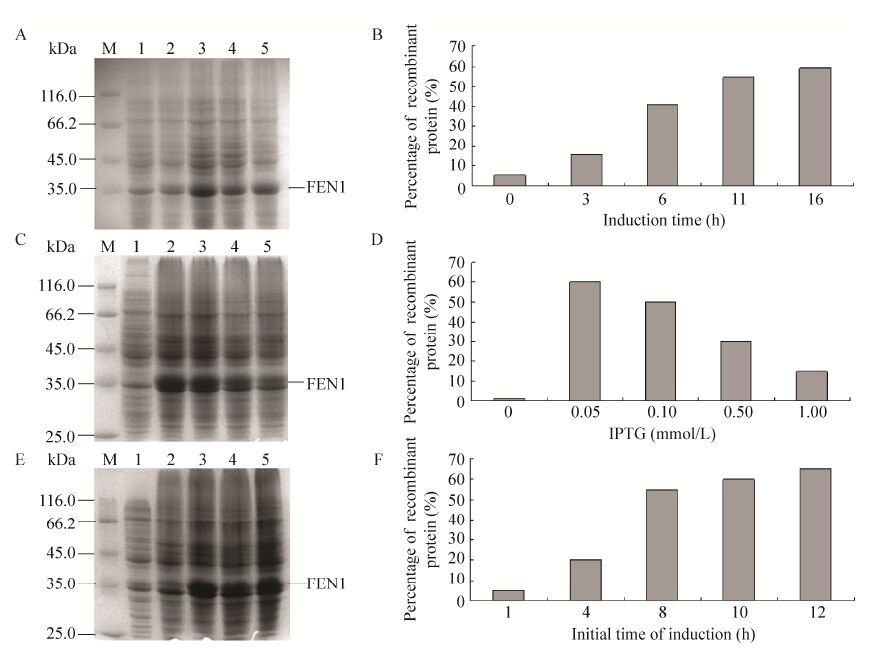 |
| 图 2 诱导表达条件优化结果图 Figure 2 Optimization of expression conditions. (A) The electrophoregram of the supernatant of the lysate of recombinant E. coli after inducing 0,3,6,11,and 16 h (Lane 1-5). (C) The electrophoregram of the supernatant of the lysate of recombinant E. coli induced with 0,0.05,0.1,0.5,and 1 mmol/L IPTG (Lane 1-5). (E) The electrophoregram of the supernatant of the lysate of recombinant E. coli with the initial time of induction of 1,4,8,10 and 12 h (Lane 1-5). (B) (D) (F) The percentage of FEN1 in the supernatant of the lysate by analysing the bands of electrophoregrams with BandScan software. M: protein marker. |
| 图选项 |
2.2 重组FEN1的分离纯化及活性初步验证由于重组菌BL21(DE3)-FEN1表达的重组FEN1融合有His-Tag,所以可经镍亲和层析纯化。采用低浓度咪唑 (60 mmol/L) 缓冲液洗涤除去非特异性结合的蛋白,再用高浓度咪唑 (300 mmol/L) 缓冲液洗脱特异性吸附的靶蛋白,收集流出液进行SDS-PAGE (图 3),电泳结果显示有清晰的目的条带且无杂蛋白,表明纯化后的重组FEN1纯度可达电泳纯,基本满足生物催化和酶学研究的要求。
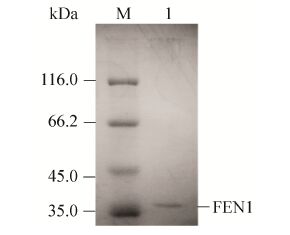 |
| 图 3 SDS-PAGE分析纯化后的重组FEN1 Figure 3 SDS-PAGE analysis of the purified recombinant FEN1. M: protein marker; 1: purified recombinant FEN1. |
| 图选项 |
为了获知纯化后的重组FEN1是否具有活性,设计了能形成核酸侵入结构的靶序列 (Target)、上游探针 (UP) 和下游探针 (DP),有活性的FEN1识别侵入结构后会发生酶切反应,下游探针5′端的flap片段被切割,形成游离的flap片段,切割产物通过电泳鉴别。结果如图 4所示,由泳道1可见,在FEN1的作用下,下游片段基本都被切割成片段长度更短的切割产物,证明所表达的FEN1具有催化侵入信号放大反应的活性。泳道1、2、3显示,随着靶序列在反应体系中终浓度的降低,下游片段的切割产物也逐渐减少,进一步说明表达的重组FEN1可应用于侵入信号放大反应,并可对靶序列进行定量检测。但是,电泳法难以准确定量重组FEN1的活性,故需重新建立FEN1的活性测定方法以准确获知其酶切能力。
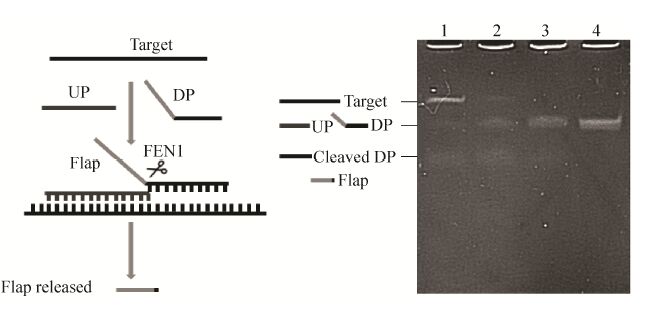 |
| 图 4 核酸侵入反应产物的尿素变性聚丙烯酰胺凝胶电泳分析 Figure 4 Urea denaturing PAGE analysis of the products of invasive reaction. Lane 1-4: invasive reaction with 0.1,0.01,0.005,and 0 μmol/L targets respectively. |
| 图选项 |
2.3 重组FEN1活性测定方法的建立文献报道[1]的FEN1活性主要依靠条带的深浅判定酶活性的强弱,该方法难以准确测定酶活性。为了便于表达的重组FEN1的进一步应用,我们建立了基于荧光探针的FEN1活性测定方法。参照国际酶学会规定,我们定义重组FEN1的活力单位为在63 ℃条件下、1 min内能切割产生1 μmol/L底物的酶量为一个酶活力单位,即1 U。为了能够测定FEN1切割产生的flap量,将形成侵入结构的下游探针5′端进行荧光标记 (图 5A),并在间隔3个碱基位置标记一淬灭基团来使下游探针不发荧光,当FEN1切割下游探针的flap时,荧光基团与淬灭基团分离,产生荧光,通过实时测定荧光强度来定量切割产生的flap片段的量,从而计算FEN1的活力。
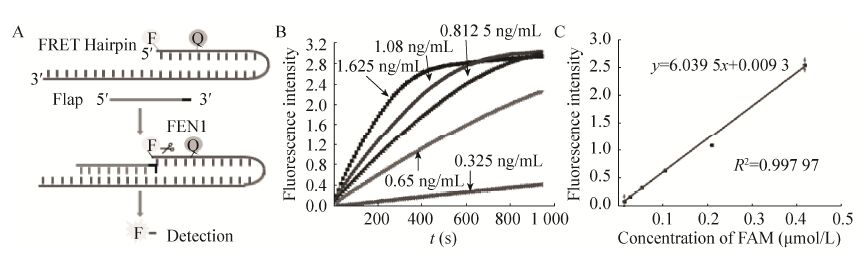 |
| 图 5 荧光标记探针法测定FEN1活性 Figure 5 Measuring the activity of FEN1 with fluorescence-lablelled probes. (A) The principle of FEN1 activity determination. (B) The curves of real-time fluorescence with varies concentrations of FEN1. (C) The standard curve of FAM signals with the concentrations of FAM labelled probe. FRET hairpin: fluorescence resonance energy transfer hairpin probe. |
| 图选项 |
根据米氏方程 (1),米氏常数与酶的种类有关,反应过程中反应速度只与底物浓度有关,当底物浓度[S]远远大于米氏常数Km时,反应速度满足方程 (2),此时反应以最大反应速度进行,酶结合的底物数目决定了反应速度,因此,此时反应速度与酶浓度成正比。为了准确测定单位时间内FEN1切割产生的flap量,首先需要优化得到测活使用的FEN1量,保证在测活时间内底物远远过量,反应呈恒速进行。
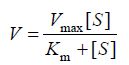 | (1) |
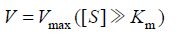 | (2) |
为了确定测活时的最佳酶量,将表达纯化获得的FEN1稀释后按1.625、1.08、0.812 5、0.65和0.325 ng/mL加入酶切反应体系,10 s读一次荧光值观察底物转化量,作出荧光信号值与时间的关系图 (图 5B),结果表明1 000 s反应时间内,FEN1的加入浓度为0.65 ng/mL和0.325 ng/mL时,荧光信号值保持线性增长,此时反应处于恒速阶段,能准确测定产生的flap片段的量,但0.325 ng/mL时荧光信号值较低易造成误差,因此0.65 ng/mL的FEN1浓度为最适反应浓度。向反应体系中分别加入仅有FAM荧光标记的标准探针,以2.5 μmol/L为起始浓度 (终浓度0.416 7 μmol/L) 对半稀释6个梯度绘制荧光标准曲线,测定荧光信号值与FAM探针浓度的关系,如图 5C所示。通过荧光强度与探针浓度的标准曲线,可计算得到本次表达纯化的重组FEN1活性为1 634.44 U/μL,紫外测定重组酶浓度为6.5 mg/mL,因此比活为 2.514 5×105 U/mg。
2.4 重组FEN1在SNP位点分型检测中的应用为了验证表达的FEN1的实用性,将重组FEN1用于本实验室建立的实时荧光PCR偶联高特异性核酸侵入反应法对酒精代谢相关aldh2基因SNP位点 (rs671,G>A) 进行分型检测[27],结果显示3种基因型均能得到良好的分型扩增曲线,与焦磷酸测序法对比显示,实时荧光PCR偶联高特异性核酸侵入反应法的分型检测结果与测序结果完全一致,表明所表达的经酶活标定的FEN1可用于SNP分型检测。
3 结论本文成功构建了A. fulgidus来源的FEN1工程菌,通过优化诱导表达条件,成功表达并纯化了重组FEN1;经电泳初步验证,所表达的重组酶具有切割活性;为了准确测定FEN1的活性,设计了基于荧光标记探针的FEN1酶活测定方法,并测定了自表达的重组FEN1活性,为进一步应用该酶发展新型核酸检测技术提供了准确测定酶活的方法,便于检测体系中酶量的优化;最终将重组FEN1应用于实时荧光PCR偶联高特异性核酸侵入反应法检测SNP位点,获得了很好的分型结果,表明重组FEN1能够用于核酸侵入反应的SNP检测中,为进一步发展基于核酸侵入反应的检测新方法提供了良好的工具酶。
参考文献
| [1] | Kaiser MW, Lyamicheva N, Ma W, et al. A comparison of eubacterial and archaeal structure-specific 5′-exonucleases.J Biol Chem,1999, 274(30): 21387–21394.DOI: 10.1074/jbc.274.30.21387 |
| [2] | Feng M, Patel D, Dervan JJ, et al. Roles of divalent metal ions in flap endonuclease-substrate interactions.Nat Struct Mol Biol,2004, 11(5): 450–456.DOI: 10.1038/nsmb754 |
| [3] | Oh H, Smith CL. Evolving methods for single nucleotide polymorphism detection: factor V leiden mutation detection.J Clin Lab Anal,2011, 25(4): 259–288.DOI: 10.1002/jcla.v25.4 |
| [4] | Ginocchio CC, Barth D, Zhang F. Comparison of the third wave invader human papillomavirus (HPV) assay and the digene HPV hybrid capture 2 assay for detection of high-risk HPV DNA.J Clin Microbiol,2008, 46(5): 1641–1646.DOI: 10.1128/JCM.01824-07 |
| [5] | Hall JG, Eis PS, Law SM, et al. Sensitive detection of DNA polymorphisms by the serial invasive signal amplification reaction.Proc Natl Acad Sci USA,2000, 97(15): 8272–8277.DOI: 10.1073/pnas.140225597 |
| [6] | Takanashi M, Ito S, Kaneto H, et al. Development and clinical application of an InvaderPlus assay for the detection of genital mycoplasmas.J Infect Chemother,2015, 21(7): 157–164. |
| [7] | Tang YW, Allawi HT, DeLeon-Carnes M, et al. Detection and differentiation of wild-type and vaccine mutant varicella-zoster viruses using an Invader Plus method.J Clin Virol,2007, 40(2): 129–134.DOI: 10.1016/j.jcv.2007.07.007 |
| [8] | Kan T, Hashimoto S, Kawabe N, et al. The clinical features of patients with a Y93H variant of hepatitis C virus detected by a PCR invader assay.J Gastroenterol,2016, 51(1): 63–70.DOI: 10.1007/s00535-015-1080-1 |
| [9] | Nie B, Shortreed MR, Smith LM. Scoring single-nucleotide polymorphisms at the single-molecule level by counting individual DNA cleavage events on surfaces.Anal Chem,2005, 77(20): 6594–6600.DOI: 10.1021/ac051025p |
| [10] | Zou BJ, Ma YJ, Wu HP, et al. Ultrasensitive DNA detection by cascade enzymatic signal amplification based on Afu flap endonuclease coupled with nicking endonuclease.Angew Chem Int Ed,2011, 50(32): 7395–7398.DOI: 10.1002/anie.v50.32 |
| [11] | Zou BJ, Song QX, Wang JP, et al. Invasive reaction assisted strand-displacement signal amplification for sensitive DNA detection.Chem Commun (Camb),2014, 50(89): 13722–13724.DOI: 10.1039/C4CC06079B |
| [12] | Zou BJ, Cao XM, Wu HP, et al. Sensitive and specific colorimetric DNA detection by invasive reaction coupled with nicking endonuclease- assisted nanoparticles amplification.Biosens Bioelectron,2015, 66: 50–54.DOI: 10.1016/j.bios.2014.10.077 |
| [13] | Kani S, Tanaka Y, Matsuura K, et al. Development of new IL28B genotyping method using invader plus assay.Microbiol Immunol,2012, 56(5): 318–323.DOI: 10.1111/mim.2012.56.issue-5 |
| [14] | Oler AT, Attie AD. A rapid, microplate SNP genotype assay for the leptinob allele.J Lipid Res,2008, 49(5): 1126–1129.DOI: 10.1194/jlr.D800002-JLR200 |
| [15] | Boonyarit H, Mahasirimongkol S, Chavalvechakul N, et al. Development of a SNP set for human identification: a set with high powers of discrimination which yields high genetic information from naturally degraded DNA samples in the Thai population.Forensic Sci Int Genet,2014, 11(4): 166–173. |
| [16] | Tadokoro K, Kobayashi M, Suzuki F, et al. Comparative quantitative analysis of hepatitis C mutations at amino acids 70 and 91 in the core region by the Q-Invader assay.J Virol Methods,2013, 189(1): 221–227.DOI: 10.1016/j.jviromet.2012.10.011 |
| [17] | Tadokoro K, Suzuki F, Kobayashi M, et al. Rapid detection of drug-resistant mutations in hepatitis B virus by the PCR-Invader assay.J Virol Methods,2011, 171(1): 67–73.DOI: 10.1016/j.jviromet.2010.10.001 |
| [18] | Fujikura J, Nakao K, Sone M, et al. Induced pluripotent stem cells generated from diabetic patients with mitochondrial DNA A3243G mutation.Diabetologia,2012, 55(6): 1689–1698.DOI: 10.1007/s00125-012-2508-2 |
| [19] | Mabuchi H, Nohara A, Noguchi T, et al. Genotypic and phenotypic features in homozygous familial hypercholesterolemia caused by proprotein convertase subtilisin/kexin type 9 (PCSK9) gain-of-function mutation.Atherosclerosis,2014, 236(1): 54–61.DOI: 10.1016/j.atherosclerosis.2014.06.005 |
| [20] | Wong AK, Chan RC, Nichols WS, et al. Invader human papillomavirus (HPV) type 16 and 18 assays as adjuncts to HPV screening of cervical papanicolaou smears with atypical squamous cells of undetermined significance.Cancer,2009, 115(4): 823–832.DOI: 10.1002/cncr.v115:4 |
| [21] | Wong DK, Yuen MF, Yuan H, et al. Quantitation of covalently closed circular hepatitis B virus DNA in chronic hepatitis B patients.Hepatology,2004, 40(3): 727–737.DOI: 10.1002/(ISSN)1527-3350 |
| [22] | Hjertner B, Meehan B, McKillen J, et al. Adaptation of an Invader assay for the detection of African swine fever virus DNA.J Virol Methods,2005, 124(1/2): 1–10. |
| [23] | Allawi HT, Dahlberg JE, Olson S, et al. Quantitation of microRNAs using a modified Invader assay.RNA,2004, 10(7): 1153–1161.DOI: 10.1261/rna.5250604 |
| [24] | Tian H, Cao L, Tan Y, et al. Multiplex mRNA assay using electrophoretic tags for high- throughput gene expression analysis.Nucl Acids Res,2004, 32(16): 219–231. |
| [25] | Wang JP, Zou BJ, Chen ZY, et al. Characterization of recombinant single-stranded DNA-binding protein from Escherichia coli and its application in accurate pyrosequencing.Chin J Biotech,2011, 27(10): 1513–1520.(in Chinese). 王建平, 邹秉杰, 陈之遥, 等. 重组大肠杆菌单链结合蛋白性质表征及其在提高焦磷酸测序准确性中的应用.生物工程学报, 2011, 27(10): 1513-1520. |
| [26] | Xu S, Zou BJ, Wang JP, et al. Expression of thermostable recombiant Luciola lateralis luciferase and development of heat-stable pyrosequencing system.Chin J Biotech,2012, 28(6): 763–771.(in Chinese). 徐澍, 邹秉杰, 王建平, 等. 耐热荧光素酶的表达及热稳定焦磷酸测序体系的建立.生物工程学报, 2012, 28(6): 763-771. |
| [27] | Zheng ML, Qi XM, Tong H, et al. Detection of single nucleotide polymorphism genotyping by real-time polymerase chain reaction coupled with high specific invader assay in single tube.Chin J Anal Chem,2015, 43(7): 1001–1008.(in Chinese). 郑梦琳, 齐谢敏, 童欢, 等. 实时荧光PCR偶联高特异性核酸侵入反应对SNP实现单管低成本无污染检测.分析化学, 2015, 43(7): 1001-1008.DOI:10.1016/S1872-2040(15)60838-6 |
