李紫云#, 贺艳艳#, 王娟, 王明钰, 徐海


山东大学微生物技术国家重点实验室, 山东 青岛 266237
收稿日期:2020-06-01;修回日期:2020-07-27;网络出版日期:2020-08-05
基金项目:国家重点研究发展计划(2017YFD0400301);国家自然科学基金(31770042, 31770043);山东省重点研究发展计划(2016GSF121040, 2018GSF118008);山东大学基础研究基金(2017JC028, 2018JC013, 2018JC027);山东大学微生物技术开放项目基金国家重点实验室(M2017-07, M2018-07);济南文化产业发展基金
*通信作者:徐海, Tel/Fax: +86-532-58630856;E-mail: haixu@sdu.edu.cn.
#共同第一作者。
摘要:[目的] 本文对山东某屠宰场的肉食鸡内脏中的大肠杆菌进行了β-内酰胺类抗生素的耐药性监测和分析。[方法] 从屠宰场的肉食鸡中获取内脏样品,处理并筛选得到对β-内酰胺类耐药的大肠杆菌。通过抑菌圈法对细菌耐药性进行分析。提取细菌DNA,进行系统发育亚型分析。检测并鉴定菌株中β-内酰胺酶基因和整合子的结构,并进行了接合转移实验。[结果] 检测到对3种及以上的β-内酰胺类药物同时具有耐药性的大肠杆菌占总菌的80%以上。编码A类β-内酰胺酶的blaTEM和blaCTX-M耐药基因存在率较高,分别为86.7%和81%,但仅blaCTX-M与β-内酰胺类药物的耐药性呈现显著相关性。B1和D2亚型的大肠杆菌中β-内酰胺耐药基因的检出率较高,并且显著增强了大肠杆菌对β-内酰胺类药物的耐药性,而A0和A1亚型菌株对β-内酰胺类药物较敏感。尽管整合子在大肠杆菌中普遍存在,但它们伴随β-内酰胺酶基因转移到受体菌的比例很低,说明二者之间的相关性较低。[结论] 本文结果显示肉鸡源大肠杆菌对β-内酰胺类药物的耐药程度较高,多重耐药情况普遍存在。本文明确了大肠杆菌中β-内酰胺类抗生素的耐药性与系统发育之间的联系,为肉鸡源大肠杆菌中β-内酰胺耐药性的流行监测提供参考依据。
关键词:超广谱β-内酰胺酶大肠杆菌整合子多重耐药性肉食鸡
Detection of extended spectrum β-lactamase producing E. coli from broiler chickens
Ziyun Li#, Yanyan He#, Juan Wang, Mingyu Wang, Hai Xu


State Key Laboratory of Microbial Technology, Shandong University, Qingdao 266237, Shandong Province, China
Received: 1 June 2020; Revised: 27 July 2020; Published online: 5 August 2020
*Corresponding author: Hai Xu, Tel/Fax: +86-532-58630856; E-mail: haixu@sdu.edu.cn.
Foundation item: Supported by the National Key Research and Development Program of China (2017YFD0400301), by the National Natural Science Foundation of China (31770042, 31770043), by the Shandong Province Key Research and Development Program (2016GSF121040, 2018GSF118008), by the Fundamental Research Funds of Shandong University (2017JC028, 2018JC013, 2018JC027), by the State Key Laboratory of Microbial Technology Open Project Funds, Shandong University (M2017-07, M2018-07) and by the Jinan Cultural Industry Development Fund
#These authors contributed equally to this work.
Abstract: [Objective] We detected and analyzed β-lactam resistance in E. coli in broiler chickens in Shandong province. [Methods] The viscera samples were obtained from broiler chickens in the slaughterhouse. We screened β-lactam resistant E. coli, used the disk diffusion assay to determine antimicrobial-resistance, extracted bacterial DNA, conducted phylogenetic and biochemical assays, identified β-lactam-resistance genes and the structure of integrons and performed conjugation assays. [Results] More than 80% isolates were resistant to 3 or more β-lactams. The blaTEM and blaCTX-M resistance genes encoding class A β-lactamases have a higher rate, 86.7% and 81%, respectively, but only blaCTX-M and β-lactam resistance showed a significant correlation. The rate of β-lactam resistance genes in B1 and D2 subtypes of E. coli was higher, and the resistance was significantly enhanced, while the A0 and A1 subtypes were more sensitive. Although integrons were commonly detected in E. coli, their correlation with β-lactamase genes is low. [Conclusion] The results show that broiler-derived E. coli has highly resistant to β-lactams, and multi-drug resistance is widespread. The relationship between the resistance of β-lactams and the phylogeny in E. coli is clarified, and it provides a reference for the epidemiological surveillance of β-lactam resistance in E. coli from broilers.
Keywords: extended spectrum β-lactamaseE. coliintegronmultidrug-resistancebroiler chicken
抗生素作为人类抵抗细菌感染的主要武器,在医疗业和畜牧养殖业中被广泛使用。与医疗部门相比,畜牧养殖业使用的抗生素数量更多,据统计,在2013年,中国使用抗生素的总量为162000 t,其中人类处方使用量占48% (77760 t),而畜牧业使用量占52% (84240 t)[1]。在抗生素压力下,细菌适应性进化导致抗生素耐药性的产生[2],监测并了解食源性细菌中的抗生素耐药性状况是检测食品安全和探索抗生素耐药性解决方案的首要任务。
人类首次发现的青霉素属于β-内酰胺类抗生素,直至现在医疗上最常使用的处方药还是以β-内酰胺类抗生素为主,例如头孢菌素和碳青霉烯类药物。β-内酰胺类抗生素的广泛使用导致了细菌对β-内酰胺类抗生素产生耐药性[1],最常见的机制是产生可以水解抗生素的β-内酰胺酶[3],根据氨基酸序列同源性将这些酶分为A, B, C和D四类[3]。其中,A类最常见,重要基因包括blaTEM和blaCTX-M等;B类酶的活性需要锌的参与;C类有可以水解头孢菌素的AmpC β-内酰胺酶,重要基因有blaCMY;D类酶能够水解苯唑西林,如blaOXA基因,而这些耐药基因可能会参与β-内酰胺类抗生素耐药性的传播。耐药性的传播机制涉及多种可移动遗传元件,如质粒,转座子,以及整合子[4]。整合子能够将耐药基因整合到染色体或质粒中,进一步通过遗传或水平转移进行传播[4],根据整合酶基因的序列相似性,整合子又可以被分为五类,其中1, 2和3类整合子已经得到了广泛的研究[5]。
大肠杆菌是重要的人畜共患病的条件致病菌,也是产超广谱β-内酰胺酶的主要菌株之一,它作为一种胃肠道病原菌,可以引起各种疾病,例如肠胃炎,泌尿道感染和新生儿脑膜炎等。食物中大肠杆菌含量超标会造成食物中毒事件[6],这会导致大规模召回食物,更导致消费者患病。而且,(O157:H7)作为大肠杆菌中最致命的血清型之一,会引起出血性腹泻,肾衰竭,甚至导致儿童,老人和免疫力低下的人死亡[7]。据美国卫生部在2013年的统计显示,大约有90例患者由于感染碳青霉烯耐药的大肠杆菌而死亡,而感染产超广谱β-内酰胺酶的大肠杆菌导致死亡的病例约为600例[8],因此在2013年,美国疾病控制与预防中心(the US Centers for Disease Control and Prevention,CDC)就将产超广谱β-内酰胺酶和对碳青霉烯类耐药的肠杆菌科(Enterobacteriaceae)细菌确定为紧急的公共卫生威胁[8]。由此可见,大肠杆菌对β-内酰胺类抗生素产生耐药性可能会对临床治疗产生严重的阻碍。
本文从山东某屠宰场的肉食鸡内脏中鉴定并分离了大肠杆菌,分析讨论了分离菌株的β-内酰胺类抗生素耐药性,阐明了β-内酰胺酶基因在肉食鸡中的流行和传播,并进一步研究了β-内酰胺基因与可移动遗传元件的相互关系。这些信息将有助于进一步了解食源性病原体的耐药性,从而增强食品安全性并减少耐药性的传播。
1 材料和方法 1.1 样品采集和大肠杆菌的分离鉴定 从山东某屠宰场的89只鸡的内脏各部位按照之前的方法进行取样[9],然后将处理后的样品接种到分别补充有阿米卡星(AMK,32 μg/mL),阿莫西林-克拉维酸钾(AMC,32/16 μg/mL),环丙沙星(CIP,4 μg/mL),磺胺间甲氧嘧啶(SXT,4/76 μg/mL),强力霉素(DC,16 μg/mL)和舒巴坦-头孢哌酮(SBT/CPZ,60/64 μg/mL)的6种曙红亚甲基蓝琼脂平板上,最终筛选得到341株大肠杆菌。接下来,按照细菌DNA提取试剂盒(GK1072,Generay Biotech)的说明,提取细菌DNA。基于重复元素序列的聚合酶链反应(REP-PCR)鉴定并分析去除重复的菌株[10],最终筛选并得到158株非重复的耐药大肠杆菌:在AMC板、CIP板、AMK板、DC板、CPZ板、SXT板上分别为28、27、35、29、18、21株。
1.2 药敏试验 为了分析筛选出的耐药大肠杆菌对常见的抗生素的耐药情况,我们按照CLSI (Clinical and Laboratory Standards Institute,M100-S25)的指南[11],使用大肠杆菌ATCC 25922作为质量控制菌株;使用以下抗生素:头孢他啶(CAZ)、氨苄西林(AMP)、头孢噻肟(CTX)、头孢吡肟(CPE)、加替沙星(GAT)、萘啶酸(NAL)、CIP、磷霉素、(FOS)、四环素(TET)、甲氧苄啶(TMP)、氯霉素(CHL)、卡那霉素(KAN)、链霉素(STR)、AMC、头孢西丁(CFX)、亚胺培南(IMP)和美罗培南(MEM);使用Müller-Hinton琼脂(MHA)平板进行K-B纸片扩散试验,测量抑菌圈并计算是否耐药。在本实验中,每个样品对每种抗生素的耐药性测试均设置3个平行,并且定义对3种以上的抗生素耐药的菌株被称为多重耐药菌。
1.3 DNA序列分析及β-内酰胺酶活性分析 按照先前报道的方法和引物[12-16],PCR扩增β-内酰胺酶基因和整合酶基因片段,然后进行测序分析。整合子基因盒阵列的鉴定使用先前报道的限制性片段长度多态性方法(PCR-RFLP)[17]。接下来根据先前报道的碘量法[18]测定了β-内酰胺酶的活性。
1.4 系统发育亚型分组 根据已报道的方法[19],使用PCR确定了chuA, yjaA和TSPE4.C2的存在,对大肠杆菌进行了亚型分组。依次根据chuA/ yjaA/ TSPE4.C2的有无分为A0 (-/-/-);A1 (-/yjaA/-);B1 (-/-/ TSPE4.C2);B21 (chuA/yjaA/-);B22 (chuA/yjaA/ TSPE4.C2);D1 (chuA/-/-)和D2 (chuA/-/ TSPE4.C2)[20-21]。
1.5 接合转移实验 接合转移实验使用有固体支持面的接合测定法[9],选择对叠氮化钠(NaN3)具有抗性的大肠杆菌J53作为受体菌。挑选在含有AMP (100 μg/mL)和NaN3 (100 μg/mL)两种抗生素的平板上生长的接合转移子,提取DNA并进行β-内酰胺酶和整合酶基因的检测。
1.6 统计学分析 使用双尾皮尔森相关性分析法计算β-内酰胺酶基因与β-内酰胺抗性之间的相关性;使用超几何统计学分布分析抗生素耐药性和不同系统发育亚组中β-内酰胺耐药性基因的存在关系。一般认为概率P < 0.05时,结果具有统计学意义。
2 结果和分析 2.1 大肠杆菌的系统发育亚型以及耐药性分析 158株非重复的大肠杆菌分离株的系统发育亚型如下:A0 24株(15.19%),A1 30株(18.99%),B1 44株(27.85%),B21 4株(2.53%),B22 10株(6.33%),D1 21株(13.29%)和D2 25株(15.82%) (表 1)。分析这些分离株对14种抗生素的敏感水平,结果发现除了对CFP和FOS耐药的分离株所占比例较低,分别为13.29%和26.58%,对其余12种抗生素的耐药的分离株所占比例均大于50% (图 1-A)。158株分离株中,99.37%为多重耐药菌,并且大多数对多达10-13种抗生素具有耐药性(图 1-B)。测试了分离株对5种β-内酰胺类抗生素的敏感水平,发现对3种及以上的β-内酰胺类抗生素耐药的菌株超过80%,有且仅有4株大肠杆菌对这5种抗生素均表现为敏感(图 1-C)。这表明,山东省的肉鸡源大肠杆菌对β-内酰胺类抗生素的耐药程度较高,多重耐药情况普遍存在;大肠杆菌系统遗传亚型中的B1和D2亚型与抗生素耐药显著相关,而A0和A1亚型与抗生素耐药无显著相关(表 2),大肠杆菌的B1和D2亚型与3种头孢菌素CAZ,CTX和CPE的耐药存在显著相关性。
表 1. 大肠杆菌分离株中β-内酰胺耐药基因及其分型和整合子及其基因盒序列的分析 Table 1. Prevalence of β-lactam resistance genes and integrons in isolated E. coli strains
| Strains | Subtypes | blaCTX-M | blaTEM | blaCMY | Class 2 integron gene cassette | Class 1 integron gene cassette | |
| Types | Subtypes | ||||||
| JN16 | D1 | CTX-M-9 | CTX-M-65 | TEM | dfrA17-aadA5 | ||
| JN18 | A1 | CTX-M-9 | CTX-M-65 | dfrA17-aadA5 | |||
| JN22 | A1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN23 | A0 | TEM | Empty | ||||
| JN26 | D1 | CTX-M-9 | CTX-M-65 | TEM | Empty | ||
| JN27 | A1 | CTX-M-1 | CTX-M-55 | TEM | |||
| JN28 | B1 | CTX-M-1 | CTX-M-55 | TEM | dfrA12-orfF-aadA2 | ||
| JN29 | D2 | CTX-M-1 | CTX-M-55 | TEM | catB3-arr3 | ||
| JN58 | A1 | TEM | |||||
| JN61 | A0 | CTX-M-9 | CTX-M-65 | TEM | |||
| JN63 | B22 | CTX-M-1 | CTX-M-55 | TEM | catB3-arr3 | ||
| JN73 | B21 | CTX-M-9 | CTX-M-65 | TEM | dfrA1-sat2-aadA1 | dfrA17-aadA5 | |
| JN104 | A0 | CTX-M-9 | CTX-M-14 | TEM | Empty | ||
| JN106 | D1 | CTX-M-1 | CTX-M-55 | TEM | dfrA12-orfF-aadA2 | ||
| JN109 | A1 | CTX-M-1 | CTX-M-55 | TEM | dfrA17-aadA5 | ||
| JN122 | A1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN127 | D2 | CTX-M-9 | CTX-M-27 | TEM | dfrA1-sat2-aadA1 | ||
| JN130 | B1 | CTX-M-9 | CTX-M-27 | TEM | Empty | ||
| JN131 | B22 | ||||||
| JN134 | A1 | TEM | |||||
| JN138 | B1 | CTX-M-1 | CTX-M-55 | dfrA1-aadA1 | |||
| JN143 | D2 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN144 | A0 | Empty | |||||
| JN149 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN155 | D2 | CTX-M-9 | CTX-M-14 | TEM | Empty | ||
| JN163 | D2 | CTX-M-1 | CTX-M-55 | TEM | dfrA17-aadA5 | ||
| JN166 | A1 | CTX-M-1 | CTX-M-55 | TEM | dfrA17-aadA5 | ||
| JN170 | B1 | CTX-M-1 | CTX-M-55 | ||||
| JN183 | D2 | CTX-M-9 | CTX-M-14 | TEM | |||
| JN184 | B1 | TEM | Empty | ||||
| JN186 | D1 | CTX-M-1 | CTX-M-55 | TEM | dfrA1-sat2-aadA1 | dfrA17-aadA5 | |
| JN193 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN199 | D2 | CTX-M-9 | CTX-M-27 | TEM | |||
| JN202 | D1 | CTX-M-1 | CTX-M-55 | TEM | |||
| JN203 | D1 | CTX-M-1 | CTX-M-55 | TEM | dfrA12-orfF-aadA2 | ||
| JN205 | D1 | CTX-M-9 | CTX-M-65 | TEM | dfrA17-aadA5 | ||
| JN209 | B1 | CTX-M-9 | CTX-M-14 | TEM | dfrA1-sat2-aadA1 | aacA4-cmlA1 | |
| JN210 | B1 | CTX-M-1 | CTX-M-55 | TEM | dfrA1-sat2-aadA1 | Empty | |
| JN215 | B1 | CTX-M-1 | CTX-M-55 | TEM | |||
| JN220 | B1 | CTX-M-9 | CTX-M-65 | TEM | dfrA1-sat2-aadA1 | dfrA17-aadA5 | |
| JN221 | B1 | CTX-M-1 | CTX-M-55 | TEM | |||
| JN231 | B1 | CTX-M-1 | CTX-M-55 | dfrA1-aadA1 | |||
| JN236 | A1 | CTX-M-9 | CTX-M-14 | CMY-2 | Empty | ||
| JN239 | D1 | CTX-M-1 | CTX-M-55 | CMY-2 | Empty | ||
| JN241 | D1 | CTX-M-9 | CTX-M-65 | Empty | |||
| JN248 | A0 | TEM | |||||
| JN261 | A1 | CTX-M-9 | CTX-M-65 | TEM | dfrA17-aadA5 | ||
| JN262 | D1 | CTX-M-9 | CTX-M-65 | TEM | Empty | ||
| JN264 | A0 | CTX-M-9 | CTX-M-65 | TEM | Empty | ||
| JN268 | A0 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN272 | B1 | CTX-M-9 | CTX-M-65 | TEM | Empty | Empty | |
| JN273 | A0 | CTX-M-1 | CTX-M-55 | TEM | |||
| JN274 | A1 | dfrA12-orfF-aadA2 | |||||
| JN276 | A0 | TEM | Empty | ||||
| JN287 | D1 | CTX-M-9 | CTX-M-65 | TEM | |||
| JN288 | D1 | CTX-M-1 | CTX-M-55 | CMY-2 | Empty | ||
| JN292 | A1 | CTX-M-9 | CTX-M-14 | Empty | |||
| JN293 | D2 | CTX-M-9 | CTX-M-27 | TEM | dfrA12-orfF-aadA2 | ||
| JN294 | B1 | CTX-M-9 | CTX-M-14 | TEM | dfrA17-aadA5 | ||
| JN295 | A1 | TEM | Empty | ||||
| JN297 | D2 | CTX-M-1 | CTX-M-123 | TEM | Empty | ||
| JN307 | B1 | CTX-M-1 | CTX-M-55 | TEM | |||
| JN308 | A1 | CTX-M-9 | CTX-M-65 | TEM | dfrA12-orfF-aadA2 | ||
| JN310 | A0 | CTX-M-9 | CTX-M-65 | TEM | Empty | ||
| JN311 | B1 | CTX-M-9 | CTX-M-14 | TEM | dfrA17-aadA5 | ||
| JN326 | A1 | TEM | dfrA17-aadA5 | ||||
| JN331 | B1 | CTX-M-9 | CTX-M-14 | TEM | dfrA17-aadA5 | ||
| JN336 | D2 | CTX-M-1 | CTX-M-55 | TEM | dfrA17-aadA5 | ||
| JN338 | D2 | CTX-M-9 | CTX-M-65 | TEM | dfrA17-aadA5 | ||
| JN339 | B1 | CTX-M-9 | CTX-M-14 | TEM | dfrA12-orfF-aadA2 | ||
| JN340 | A1 | TEM | dfrA12-orfF-aadA2 | ||||
| JN341 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN343 | D1 | CTX-M-9 | CTX-M-65 | TEM | dfrA1-sat2-aadA1 | dfrA12-orfF-aadA2 | |
| JN344 | A0 | CTX-M-9 | CTX-M-65 | TEM | Empty | ||
| JN345 | A0 | CTX-M-1 | CTX-M-123 | Empty | |||
| JN349 | D2 | CTX-M-1 | CTX-M-123 | TEM | Empty | ||
| JN351 | A1 | TEM | dfrA1-aadA1 | ||||
| JN353 | A0 | CTX-M-9 | CTX-M-65 | TEM | Empty | ||
| JN355 | B22 | CTX-M-9 | CTX-M-65 | Empty | |||
| JN357 | A1 | TEM | Empty | ||||
| JN358 | A1 | CTX-M-9 | CTX-M-65 | TEM | dfrA17-aadA5 | ||
| JN359 | A1 | TEM | dfrA1-sat2-aadA1 | dfrA1-aadA1 | |||
| JN362 | B1 | CTX-M-9 | CTX-M-65 | TEM | |||
| JN363 | A0 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN365 | B1 | CTX-M-9 | CTX-M-65 | TEM | Empty | ||
| JN368 | B1 | CTX-M-9 | CTX-M-14 | TEM | dfrA17-aadA5 | ||
| JN371 | A1 | TEM | Empty | ||||
| JN374 | A1 | CTX-M-9 | CTX-M-14 | Empty | |||
| JN376 | A1 | CTX-M-9 | CTX-M-14 | Empty | |||
| JN379 | B1 | CTX-M-9 | CTX-M-65 | TEM | dfrA17-aadA5 | ||
| JN381 | A0 | CTX-M-1 | CTX-M-55 | TEM | dfrA12-orfF-aadA2 | ||
| JN382 | A0 | CTX-M-9 | CTX-M-27 | TEM | dfrA1-sat2-aadA1 | dfrA12-orfF-aadA2 | |
| JN388 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN389 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN398 | B22 | CTX-M-1 | CTX-M-55 | TEM | dfrA17-aadA5 | ||
| JN399 | B1 | CTX-M-1 | CTX-M-55 | TEM | dfrA12-orfF-aadA2 | ||
| JN407 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN408 | D2 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN418 | D2 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN420 | B22 | CTX-M-1 | CTX-M-55 | TEM | catB3-arr3 | ||
| JN421 | B22 | CTX-M-1 | CTX-M-55 | TEM | dfrA12-orfF-aadA2 | ||
| JN430 | B1 | CTX-M-1 | CTX-M-55 | TEM | dfrA12-orfF-aadA2 | ||
| JN431 | A0 | CTX-M-1 | CTX-M-55 | TEM | dfrA12-orfF-aadA2 | ||
| JN432 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN436 | B1 | CTX-M-9 | CTX-M-14 | ||||
| JN437 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN439 | D1 | CTX-M-9 | CTX-M-14 | TEM | dfrA17-aadA5 + aacA4-cmlA1 | ||
| JN440 | A1 | CTX-M-1 | CTX-M-123 | TEM | dfrA12-orfF-aadA2 | ||
| JN441 | B21 | CTX-M-1 | CTX-M-55 | dfrA1-sat2-aadA1 | Empty | ||
| JN443 | D1 | CTX-M-9 | CTX-M-65 | TEM | dfrA12-orfF-aadA2 | ||
| JN445 | A1 | TEM | Empty | ||||
| JN446 | B22 | CTX-M-9 | CTX-M-27 | dfrA1-aadA1 | |||
| JN447 | D1 | CTX-M-1 | CTX-M-55 | dfrA1-sat2-aadA1 | Empty | ||
| JN452 | D2 | CTX-M-9 | CTX-M-65 | TEM | dfrA1-sat2-aadA1 | Empty | |
| JN455 | B1 | CTX-M-1 | CTX-M-55 | TEM | dfrA1-sat2-aadA1 | Empty | |
| JN460 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN461 | A1 | TEM | Empty | ||||
| JN463 | D1 | CTX-M-1 | CTX-M-55 | TEM | |||
| JN470 | D2 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN472 | D2 | CTX-M-9 | CTX-M-65 | TEM | dfrA17-aadA5 | ||
| JN474 | D2 | CTX-M-1 | CTX-M-55 | TEM | Empty | Empty | |
| JN475 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | Empty | |
| JN478 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN484 | B1 | CTX-M-9 | CTX-M-65 | TEM | Empty | dfrA17-aadA5 | |
| JN486 | B1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN495 | A0 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN499 | D1 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN500 | B22 | CTX-M-9 | CTX-M-27 | dfrA1-aadA1 | |||
| JN501 | A0 | CTX-M-1 | CTX-M-55 | TEM | dfrA17-aadA5 | ||
| JN507 | A1 | CTX-M-9 | CTX-M-65 | TEM | dfrA1-sat2-aadA1 | dfrA12-orfF-aadA2 | |
| JN511 | B1 | CTX-M-1 | CTX-M-55 | TEM | dfrA1-sat2-aadA1 | Empty | |
| JN514 | B21 | CTX-M-1 | CTX-M-55 | TEM | CMY-2 | dfrA12-orfF-aadA2 | |
| JN525 | D2 | CTX-M-9 | CTX-M-14 | TEM | dfrA12-orfF-aadA2 | ||
| JN566 | B22 | CTX-M-1 | CTX-M-55 | TEM | dfrA17-aadA5 | ||
| JN567 | A0 | CTX-M-9 | CTX-M-65 | TEM | Empty | ||
| JN573 | D1 | CTX-M-1 | CTX-M-123 | TEM | Empty | ||
| JN583 | B1 | CTX-M-9 | CTX-M-65 | TEM | dfrA17-aadA5 | ||
| JN592 | A1 | CTX-M-9 | CTX-M-65 | dfrA17-aadA5 | |||
| JN593 | A0 | CTX-M-9 | CTX-M-14 | TEM | dfrA17-aadA5 | ||
| JN597 | A0 | CTX-M-1 | CTX-M-55 | dfrA1-sat2-aadA1 | Empty | ||
| JN598 | B22 | CTX-M-1 | CTX-M-123 | TEM | dfrA12-orfF-aadA2 | ||
| JN599 | A1 | dfrA17-aadA5 | |||||
| JN600 | D1 | dfrA1-sat2-aadA1 | Empty | ||||
| JN601 | B21 | CTX-M-1 | CTX-M-55 | dfrA1-sat2-aadA1 | Empty | ||
| JN602 | B1 | CTX-M-1 | CTX-M-55 | Empty | |||
| JN603 | D2 | CTX-M-1 | CTX-M-55 | ||||
| JN604 | A1 | CTX-M-9 | CTX-M-65 | TEM | Empty | ||
| JN606 | D1 | CTX-M-1 | CTX-M-123 | TEM | dfrA1-sat2-aadA1 | Empty | |
| JN608 | A0 | CTX-M-9 | CTX-M-65 | TEM | Empty | ||
| JN610 | B1 | CTX-M-1 | CTX-M-55 | TEM | dfrA1-sat2-aadA1 | Empty | |
| JN615 | B1 | CTX-M-1 | CTX-M-55 | TEM | dfrA1-sat2-aadA1 | dfrA17-aadA5 | |
| JN616 | D2 | CTX-M-1 | CTX-M-55 | TEM | dfrA1-sat2-aadA1 | Empty | |
| JN617 | D2 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN631 | D2 | CTX-M-1 | CTX-M-55 | TEM | Empty | ||
| JN633 | B1 | CTX-M-9 | CTX-M-14 | ||||
| JN641 | A0 | Empty | |||||
| JN660 | D2 | CTX-M-1 | CTX-M-55 | TEM | dfrA1-sat2-aadA1 | dfrA12-orfF-aadA2 | |
| JN662 | D2 | CTX-M-9 | CTX-M-65 | TEM | CMY-2 | dfrA1-aadA1 | |
表选项
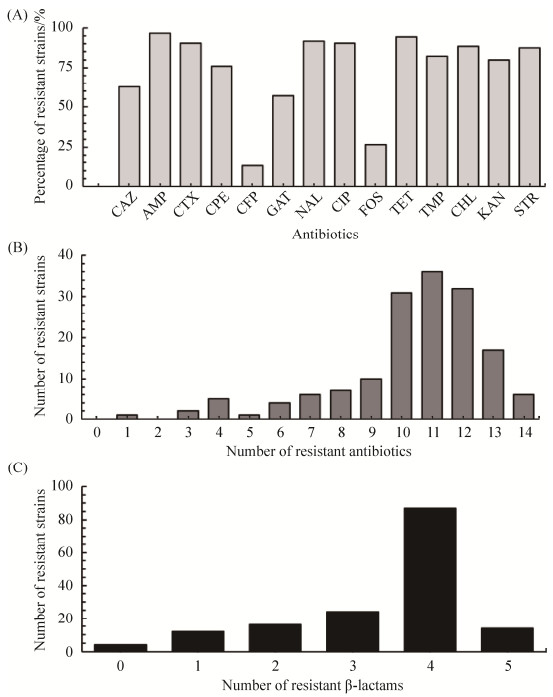 |
| 图 1 大肠杆菌耐药分离株的抗生素耐药分析 Figure 1 Resistance analysis of isolated drug-resistant E. coli to antibiotics. A: The percentage of E. coli strains to different antibiotics; B: The number of strains resistant to different numbers of antibiotics; C: The number of strains resistant to different numbers of β-lactams. |
| 图选项 |
表 2. 大肠杆菌系统发育亚型与抗生素耐药性菌株和敏感性菌株之间相关性的统计分析 Table 2. Statistical analysis of the correlation between E. coli phylogenetic subgroups and resistance/sensitivity to antibiotics
| Sub-types | CAZ | AMP | CTX | CPE | NAL | CIP | STR | FOS | TET | TMP | CHL | KAN | CFP | GAT | Overall |
| A0 pR pS | (11) 1.0 0.054 | (22) 1.0 0.2 | (20) 0.9 0.2 | (15) 1.0 0.09 | (21) 0.9 0.3 | (21) 0.9 0.4 | (21) 0.6 0.6 | (4) 0.9 0.2 | (22) 0.9 0.4 | (15) 1.0 0.01 | (22) 0.5 0.8 | (17) 0.9 0.2 | (4) 0.4 0.8 | (16) 0.2 0.9 | (24) 1.0 0.01 |
| A1 pR pS | (10) 1.0 3.0×10-4 | (27) 1.0 0.03 | (21) 1.0 5.6×10 –4 | (15) 1.0 5.0×10 –4 | (26) 0.9 0.2 | (27) 0.7 0.5 | (20) 1.0 7.3×10 –4 | (5) 0.9 0.1 | (27) 0.9 0.2 | (27) 0.2 0.9 | (22) 1.0 0.008 | (18) 1.0 0.005 | (1) 1.0 0.06 | (16) 0.7 0.4 | (30) 1.0 2.7×10 –9 |
| B1 pR pS | (34) 0.01 1.0 | (44) 0.1 1.0 | (43) 0.04 0.8 | (39) 0.01 1.0 | (43) 0.08 1.0 | (42) 0.2 0.9 | (40) 0.3 0.9 | (8) 0.9 0.1 | (43) 0.2 1.0 | (41) 0.02 1.0 | (42) 0.07 1.0 | (40) 0.02 0.1 | (9) 0.09 1.0 | (24) 0.7 0.4 | (44) 5.1×10 –5 1.0 |
| B21 pR pS | (3) 0.5 0.9 | (4) 0.9 1.0 | (4) 0.7 1.0 | (4) 0.3 1.0 | (3) 1.0 0.3 | (4) 0.7 1.0 | (4) 0.6 1.0 | (1) 0.7 0.7 | (4) 0.8 1.0 | (3) 0.9 0.5 | (4) 0.6 1.0 | (4) 0.4 1.0 | (1) 0.41 0.9 | (2) 0.8 0.6 | (4) 0.2 0.9 |
| B22 pR pS | (8) 0.2 0.9 | (9) 1.0 0.3 | (10) 0.4 1.0 | (7) 0.8 0.5 | (10) 0.4 1.0 | (9) 0.8 0.6 | (9) 0.6 0.8 | (3) 0.5 0.8 | (10) 0.5 1.0 | (8) 0.7 0.6 | (7) 1.0 0.09 | (8) 0.7 0.6 | (2) 0.4 0.9 | (4) 0.9 0.2 | (10) 0.5 0.6 |
| D1 pR pS | (12) 0.8 0.4 | (21) 0.4 1.0 | (20) 0.4 0.7 | (16) 0.6 0.6 | (18) 0.9 0.2 | (20) 0.4 0.9 | (19) 0.5 0.8 | (10) 0.02 1.0 | (19) 0.9 0.3 | (18) 0.5 0.8 | (20) 0.3 0.9 | (17) 0.6 0.7 | (2) 0.8 0.4 | (17) 0.01 1.0 | (21) 0.06 1.0 |
| D2 pR pS | (21) 0.01 1.0 | (25) 0.4 1.0 | (25) 0.07 1.0 | (24) 0.006 0.7 | (24) 0.4 0.9 | (21) 1.0 0.2 | (25) 0.02 1.0 | (10) 0.07 1.0 | (24) 0.6 0.2 | (18) 0.9 0.1 | (23) 0.4 0.8 | (22) 0.2 0.9 | (2) 0.9 0.3 | (11) 0.9 0.1 | (25) 0.02 1.0 |
| pR and pS: p-values for correlation of antibiotic resistance and susceptibility, respectively. Significant correlation is marked with bold numbers, p-values smaller than 0.05 are considered statistically significant. | |||||||||||||||
表选项
2.2 β-内酰胺类耐药基因的检测及分型 对3种常见的β-内酰胺耐药基因blaTEM、blaCTX-M和blaCMY进行检测,分离株中blaCTX-M和blaTEM耐药基因的存在最普遍,存在率分别是86.7%和81% (图 2)。我们对存在率最高的blaCTX-M家族进行了分型,其中有77株菌属于blaCTX-M-1家族,有60株菌属于blaCTX-M-9家族(图 2)。blaCTX-M-1家族中,blaCTX-M-55和blaCTX-M-123的存在率分别为90.91%和9.09%;blaCTX-M-9家族中,blaCTX-M-65、blaCTX-M-14和blaCTX-M-27家族的存在率分别为58.33%、30%和11.67% (图 2)。仅在5个分离株中发现了编码AmpC型β-内酰胺酶的blaCMY基因,并且均为blaCMY-2家族。分析发现blaCMY-2与blaCTX-M两个基因在同一个分离株中有共存的现象,但是blaCMY-2与β-内酰胺类抗生素的耐药性之间没有相关性(表 3)。尽管先前的研究显示blaTEM作为最流行的β-内酰胺耐药基因之一,是环境中最普遍的β-内酰胺酶基因[22],但是进一步对耐药表型和基因型的相关性分析,发现β-内酰胺类抗生素的耐药性与blaCTX-M基因有显著的相关性,而与blaTEM基因没有显著相关性(表 3)。与其他亚型相比,blaCTX-M基因普遍存在于大肠杆菌的B1和D2亚型中(P值分别为0.0046和0.0167) (表 2),与之呼应的是大肠杆菌的B1和D2这两个亚型与β-内酰胺类抗生素的耐药性存在显著的相关性。这一发现表明在肉鸡源大肠杆菌中,blaCTX-M基因在表达β-内酰胺耐药性中起着更加重要的作用。
 |
| 图 2 β-内酰胺耐药基因的流行性分析图 Figure 2 Prevalence of β-lactam resistance genes. TEM: blaTEM; CMY: blaCMY; CTX-M: blaCTX-M; CTX-M-9: blaCTX-M-9; CTX-M-1: blaCTX-M-1; CTX-M-14: blaCTX-M-14; CTX-M-27: blaCTX-M-27; CTX-M-55: blaCTX-M-55; CTX-M-65: blaCTX-M-65; CTX-M-123: blaCTX-M-123. |
| 图选项 |
表 3. β-内酰胺耐药表现型与β-内酰胺耐药基因存在的相关性 Table 3. Correlation between β-lactam resistance and the prevalence of β-lactam resistance genes
| β-lactam resistance and resistance genes | blaCTX-M prevalence | blaTEM prevalence | blaCMY-2 prevalence | Number of β-lactam resistance genes |
| Correlation | 0.607 (n=158) | 0.107 (n=158) | 0.001 (n=158) | 0.448 (n=158) |
| P-value | 2.64×10–17 (n=158) | 0.181 (n=158) | 0.988 (n=158) | 3.68×10–9 (n=158) |
表选项
2.3 大肠杆菌中整合子的检测及鉴定 整合子作为重要的可移动遗传元件,可以参与耐药基因的整合剪切和转移。对分离株中1、2和3整合子的调查显示,携带1类整合子的分离株有135株(85.44%),其中有72株(52.94%)的整合子中没有基因盒阵列(表 1)。携带2类整合子的分离株有29株(18.35%),其中23株(79.31%)的整合子中含有通用基因盒阵列(dfrA1-sat2-aadA1),其余6株(20.69%)的整合子中没有基因盒阵列,然而3类整合子在所有样品中均未检测到(表 1)。肉鸡源大肠杆菌中普遍存在整合子,这表明整合子在耐药性的传播中可能发挥作用。
2.4 β- 内酰胺耐药基因的接合转移与整合子相关性较低 研究肉食鸡源的大肠杆菌中β-内酰胺类耐药基因以及整合子基因与接合转移的相互关系,发现除了blaCMY-2基因以外,blaTEM和blaCTX-M基因均可以通过接合被转移至受体菌中。产生β-内酰胺酶活性的分离株占所有分离株的55.28%,然而仅有大约30%的β-内酰胺类耐药基因可以通过接合转移进行传播,我们推测这是因为β-内酰胺酶基因中的其中1个基因发生转移后,就能表达出足够的β-内酰胺酶。尽管整合子在肉鸡源大肠杆菌中存在率很高,但相关性分析后发现β-内酰胺类的耐药基因与整合酶基因之间的相关性较弱,分离株的整合酶基因中,伴随着β-内酰胺耐药基因同时被转移进受体菌中的整合酶基因比例不足10% (表 4),这表明β-内酰胺类耐药基因的接合转移对整合子的依赖程度较低,这也可以解释为什么分离株的整合子基因盒阵列中缺少β-内酰胺酶基因。
表 4. β-内酰胺类耐药基因和整合酶基因的接合转移比率 Table 4. Ratio of conjugation of β-lactam resistance genes and integrase genes
| Number of strains | intI1 | intI2 | blaCTX-M | blaTEM | blaCMY-2 |
| Gene-harboring strains | 135 | 29 | 137 | 128 | 5 |
| Successful conjugation count | 5 | 3 | 42 | 45 | 0 |
| Ratio of conjugation/% | 3.70 | 10.34 | 30.66 | 35.16 | 0.00 |
表选项
3 讨论 农业部门中抗生素的大范围和大规模使用导致耐药性食品病原菌的数量不断增加,这对人类健康造成威胁,这个问题在肉类产品中表现的更加严重,因为:(1)在畜牧业中,抗生素一般会直接注射或者喂食给动物,所以肉类产品中出现耐药性的机会比蔬菜和水果中高得多。(2)在食用肉类产品或者鸡蛋时,因为不同的饮食习惯或者为了获得更好的口感,不完全煮熟就食用的情况经常存在。然而肉类中经常会含有细菌、真菌和病毒[23],鸡蛋内容物中也普遍携带大肠杆菌和沙门氏菌等致病菌[24],不完全加热就食用经常会引起胃肠道发生细菌感染,而抗生素被广泛的应用到治疗细菌感染中,这可能会促进耐药基因在胃肠道中传播。β-内酰胺类药物常用于治疗大肠杆菌引起的感染,也是畜牧业中常见的饲料添加剂[25]。因此这项研究主要涉及肉食鸡内脏中大肠杆菌的β-内酰胺耐药性:(1)普遍性和耐药原因。(2)抗生素耐药性与系统发育之间的联系;(3)β-内酰胺类耐药的表现型与耐药基因以及整合酶基因之间的相互关系。
研究发现,山东省肉鸡源大肠杆菌对β-内酰胺类药物的耐药程度较高,多重耐药情况普遍存在,这表明肉鸡源大肠杆菌绝大多数都是对β-内酰胺耐药的多重耐药菌,并且blaCTX-M和blaTEM两种β-内酰胺酶基因在肉食鸡中流行性高。这一结论与其他省份的研究相吻合,证明肉鸡源大肠杆菌在不同地区都普遍存在耐药性较高的情况[26]。基因水平转移向来被认为是细菌间耐药性传播的最有效方法,而整合子作为可移动的遗传元件一直与抗生素耐药性的传播有关,这项工作分离出的耐药大肠杆菌中整合子的高流行率证实了这一点。然而与β-内酰胺酶基因较高的转移率相比,整合子的转移比率较低,表明β-内酰胺耐药基因的接合转移对整合子的依赖性较低,这与整合子的基因盒阵列中缺乏β-内酰胺酶基因是一致的。此外肉鸡源大肠杆菌的B1和D2亚型与抗生素耐药显著相关,而A0和A1亚型与抗生素耐药无显著相关,这是令人担忧的,因为大肠杆菌的高致病率血清型绝大部分属于D亚型[19]。
肉鸡源大肠杆菌中的β-内酰胺耐药性已经逐渐成为一个全民关注的问题,一旦饮食不当,致使耐药菌进入人体内并引发感染,将导致抗生素治疗效果降低,甚至会威胁到免疫力较低的人群的生命。目前人类无法做到消除耐药基因,因此不仅应该呼吁大家尽可能的避免食用未煮熟的肉类蛋类食品,还要注意卫生避免感染携带耐药基因的耐药菌,此外在抗生素和消炎药的使用上,一定要严格遵从医嘱,避免不当的药物压力促使耐药基因快速传播。
References
| [1] | Zhang QQ, Ying GG, Pan CG, Liu YS, Zhao JL. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environmental Science & Technology, 2015, 49(11): 6772-6782. |
| [2] | Bonomo RA. β-lactamases: a focus on current challenges. Cold Spring Harbor Perspectives in Medicine, 2017, 7(1): a025239. DOI:10.1101/cshperspect.a025239 |
| [3] | Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Molecular Microbiology, 1995, 15(4): 593-600. |
| [4] | Mazel D. Integrons: agents of bacterial evolution. Nature Reviews Microbiology, 2006, 4(8): 608-620. DOI:10.1038/nrmicro1462 |
| [5] | FDA (United States Food and Drug Administration). FDA investigated multistate outbreak of E. coli O157 infections linked to alfalfa sprouts from Jack and the Green Sprouts. (2016-03-25). http://www.fda.gov/food/recallsoutbreaksemergencies/outbreaks/ucm487651.htm. |
| [6] | Saeedi P, Yazdanparast M, Behzadi E, Salmanian AH, Mousavi SL, Nazarian S, Amani J. A review on strategies for decreasing E. coli O157:H7 risk in animals. Microbial Pathogenesis, 2017, 103: 186-195. DOI:10.1016/j.micpath.2017.01.001 |
| [7] | U.S. Department of Health and Human Services. Antibiotic resistance threats in the United States, 2013. Atlanta, 2013. |
| [8] | Wu H, Xia SB, Bu FY, Qi J, Liu YQ, Xu H. Identification of integrons and phylogenetic groups of drug-resistant Escherichia coli from broiler carcasses in China. International Journal of Food Microbiology, 2015, 211: 51-56. DOI:10.1016/j.ijfoodmicro.2015.07.004 |
| [9] | Xia RR, Ren Y, Guo XH, Xu H. Molecular diversity of class 2 integrons in antibiotic-resistant gram-negative bacteria found in wastewater environments in China. Ecotoxicology, 2013, 22: 402-414. DOI:10.1007/s10646-012-1034-9 |
| [10] | Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 25th Informational Supplement. CLSI Document M100-S25. Wayne PA: Clinical and Laboratory Standards Institute, 2015. |
| [11] | Achour NB, Mercuri PS, Power P, Belhadj C, Moussa MB, Galleni M, Belhadj O. First detection of CTX-M-28 in a Tunisian hospital from a cefotaxime-resistant Klebsiella pneumoniae strain. Pathologie Biologie, 2009, 57(5): 343-348. DOI:10.1016/j.patbio.2008.07.016 |
| [12] | Fujita S, Yosizaki K, Ogushi T, Uechi K, Takemori Y, Senda Y. Rapid identification of gram-negative bacteria with and without CTX-M extended-spectrum β-lactamase from positive blood culture bottles by PCR followed by microchip gel electrophoresis. Journal of Clinical Microbiology, 2011, 49(4): 1483-1488. DOI:10.1128/JCM.01976-10 |
| [13] | Kurpiel PM, Hanson ND. Association of IS5 with divergent tandem blaCMY-2 genes in clinical isolates of Escherichia coli. Journal of Antimicrobial Chemotherapy, 2011, 66(8): 1734-1738. DOI:10.1093/jac/dkr212 |
| [14] | Schwaber MJ, Navon-Venezia S, Chmelnitsky I, Leavitt A, Schwartz D, Carmeli Y. Utility of the VITEK 2 advanced expert system for identification of extended-spectrum β-lactamase production in Enterobacter spp.. Journal of Clinical Microbiology, 2006, 44(1): 241-243. DOI:10.1128/JCM.44.1.241-243.2006 |
| [15] | Xu H, Davies J, Miao V. Molecular characterization of class 3 integrons from Delftia spp.. Journal of Bacteriology, 2007, 189(17): 6276-6283. DOI:10.1128/JB.00348-07 |
| [16] | Guo X, Xia R, Han N, Xu H. Genetic diversity analyses of class 1 integrons and their associated antimicrobial resistance genes in Enterobacteriaceae strains recovered from aquatic habitats in China. Letters in Applied Microbiology, 2011, 52(6): 667-675. DOI:10.1111/j.1472-765X.2011.03059.x |
| [17] | Livermore DM, Brown DFJ. Detection of β-lactamase-mediated resistance. Journal of Antimicrobial Chemotherapy, 2001, 48(S1): 59-64. |
| [18] | Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Applied and Environmental Microbiology, 2000, 66(10): 4555-4558. DOI:10.1128/AEM.66.10.4555-4558.2000 |
| [19] | Han N, Sheng DH, Xu H. Role of Escherichia coli strain subgroups, integrons, and integron-associated gene cassettes in dissemination of antimicrobial resistance in aquatic environments of Jinan, China. Water Science and Technology, 2012, 66(11): 2385-2392. DOI:10.2166/wst.2012.473 |
| [20] | Figueira V, Serra E, Manaia CM. Differential patterns of antimicrobial resistance in population subsets of Escherichia coli isolated from waste- and surface waters. Science of the Total Environment, 2011, 409(6): 1017-1023. DOI:10.1016/j.scitotenv.2010.12.011 |
| [21] | Yan L, Liu D, Wang XH, Wang YK, Zhang B, Wang MY, Xu H. Bacterial plasmid-mediated quinolone resistance genes in aquatic environments in China. Scientific Reports, 2017, 7: 40610. DOI:10.1038/srep40610 |
| [22] | Zhen ZY, Hu XJ, Xu LY, Wang YL, Niu XC. Research progress on analytical methods used in meat microbial diversity. Chinese Journal of Bioprocess Engineering, 2020, 18(3): 381-385. (in Chinese) 甄宗圆, 胡雪洁, 徐留艳, 王艺霖, 牛玺程. 肉类微生物多样性分析方法的研究进展. 生物加工过程, 2020, 18(3): 381-385. DOI:10.3969/j.issn.1672-3678.2020.03.017 |
| [23] | Sun DD, Qin YC, Li JG. Analysis and control technology for egg production quality and safety. China Poultry, 2015, 37(5): 45-49. (in Chinese) 孙丹丹, 秦玉昌, 李军国. 鸡蛋生产质量安全问题分析及控制研究进展. 中国家禽, 2015, 37(5): 45-49. |
| [24] | Hu GZ, Hu H, Liu BG, Yuan L, Liu JH, Pan YS, Wu H, Chen YX. Characterization of blaCTX-M gene in one Klebsiella pneumoniae isolate from sick chickens in China. Journal of Integrative Agriculture, 2012, 11(10): 1714-1720. DOI:10.1016/S2095-3119(12)60175-0 |
| [25] | Chen X, Zhao XF, Che J, Zhang YF, Yuan M, Bai XM, Li J. Antibiotic resistance characteristics of 527 Escherichia coli isolates from broilers. Disease Surveillance, 2020, 35(1): 46-51. (in Chinese) 陈霞, 赵晓菲, 车洁, 张云飞, 袁敏, 白雪梅, 李娟. 527株肉鸡源大肠埃希菌的药物敏感性特征研究. 疾病监测, 2020, 35(1): 46-51. |
