Qiang Wang, Xiaoting Fu

 , Xiangzhao Mao, Jiachao Xu, Xin Gao
, Xiangzhao Mao, Jiachao Xu, Xin Gao College of Food Science and Engineering, Ocean University of China, Qingdao 266003, Shandong Province, China
Received: 22 June 2018; Revised: 13 September 2018; Published online: 28 November 2018
Foundation item: Supported by the Grant of Shandong Provincial Key Research and Development Project (2016GSF121034) and by the Grant of Public Science and Technology Research Funds Projects of Ocean (201405040)
Corresponding author: Xiaoting Fu, Tel:+86-532-82032182, Fax:+86-532-82032389, E-mail:xiaotingfu@ouc.edu.cn.
Abstract: [Objective] Marinomonas sp. FW-1 secretes high-activity arylsulfatase. This study was carried out to clone all the possible arylsulfatase in Marinomonas sp. FW-1 and investigate the motif information essential for arylsulfatase.[Methods] We used high-throughput sequencing to obtain the entire genome sequence of Marinomonas sp. FW-1. We carried out gene assembly, gene prediction, functional annotation and Clusters of Orthologous Groups clustering analysis. Thereafter, we cloned arylsulfatase genes in Escherichia coli and analyzed the activity of the recombinant proteins.[Results] The complete genome is of 3964876 bp in length, with an average Guanine and Cytosine content of 44.03%, encoding 3590 protein genes, containing 78 tRNA and 25 rRNA operons. Genome annotation indicated the presence of four possible arylsufatase genes. Among the four candidate genes, three genes contained C-X-P-X-R motif and their recombinant products were found to exhibit arylsulfatase activities.[Conclusion] This study revealed multiple arylsulfatase gene sequences in the complete genome Marinomonas sp. FW-1 strain, which predicted various potential applications of this strain. Moreover, C-X-P-X-R motif was confirmed to be essential for arylsulfatase activity.
Keywords: Marinomonas sp. FW-1complete genome sequencearylsulfatase
一株从红藻中分离出的产芳基硫酸酯酶的海单胞菌FW-1的全基因组测序及结果分析
王嫱, 付晓婷

 , 毛相朝, 许加超, 高昕
, 毛相朝, 许加超, 高昕 中国海洋大学食品科学与工程学院, 山东 青岛 266003
收稿日期:2018-06-22;修改日期:2018-09-13;网络出版日期:2018-11-28
基金项目:山东省重点研发计划(2016GSF121034);国家海洋局国家公益性行业科研专项(201405040)
通信作者:付晓婷, Tel:+86-532-82032182, Fax:+86-532-82032389, E-mail:xiaotingfu@ouc.edu.cn.
摘要:[目的] 海单胞菌Marinomonas sp.FW-1是1株经验证可以获得高活性芳基硫酸酯酶的菌株。为深入研究FW-1菌株产芳基硫酸酯酶机制,进一步筛选高活性的芳基硫酸酯酶基因片段,有必要解析FW-1菌株的全基因组序列信息。[方法] 本研究采用高通量测序技术对FW-1进行全基因组测序,使用相关软件对测序数据进行基因组装、基因预测与功能注释、COG聚类分析等。结合异源表达的方法对其不同基因片段所产生的芳基硫酸酯酶活性进行分析。[结果] 全基因组测序结果表明该基因组大小为3964876 bp,GC含量为44.03%,编码3590个蛋白基因,含有78个tRNA和25个rRNA操纵子。从全基因组测序结果中找到22个可能具有芳基硫酸酯酶活性的基因,对其中4个进一步异源表达后发现FW-1中至少含有的3个具有芳基硫酸酯酶活性的基因,其均含有芳基硫酸酯酶的特异性氨基酸基团C-X-P-X-R基团。[结论] 本研究首次报道了1株含有多个芳基硫酸酯酶基因序列的菌株FW-1的全基因组序列,分析了基因组的基本特征,为芳基硫酸酯酶的进一步应用提供了思路。
关键词:海单胞菌全基因组测序芳基硫酸酯酶
Agar is the polysaccharide extracted from the cell wall of some members of the red algae class Rhodophyta, which is composed of agarose and agaropectin[1]. Agarose is composed of a repeating unit of alternating 1, 4-linked 3, 6-anhydro-α-l- galactosyl residues and 1, 3-linked α-d-galactosyl residues, while the structure of agaropectin additionally contains sulfate groups in C6 position of galactosyl residues and forms l-galactose-6-sulfate[2]. Agarose was widely utilized in biotechnology and pharmaceutical industries, such as the use in electrophoresis and chromatographic resin[3]. Incorporation of sulfate groups in agar usually weakens gel strength due to the avoidance of a cross-linked structure during gelation[4]. There is evidence that the agar quality in terms of gel strength depends on the content of 3, 6-anhydrogalactopyranosyl (3, 6-AG). With increasing demand of agarose, more studies referred to the removal of sulfates for improving the gel strength of agar. The gel strength of the desulfated agarin creased approximately 2-fold compared to control agar[5] and showed a similar separation resolution in agarose gel electrophoresis compared to commercial agarose[3]. The alkali treatment is the traditional method widely used in the agar industry to remove the primary sulfate groups of the galactopyranosyl unit and convert the galatopyranhosyl to 3, 6-AG[6]. However, the alkali treatment has many drawbacks, such as the reduction of polysaccharide yield, the formation of brown color in agar product, and the pollution of environment[7]. Thus, there is a growing need to develop eco-friendly processing technologies for recovery of products from bioresource. Enzymatic processing technology can solve these problems. "Arylsulfatase" which catalyzes the hydrolyseis of arylsulfate ester bonds to the corresponding phenols and inorganic sulfate, it can desulfate agar into agarose[8].
1 Materials and methods 1.1 Materials
1.1.1 Buffer solution: 100 mmol/L Tirs-HCl (pH 7.5), 0.9% NaCl (YongDa, China).
1.1.2 Culture medium (g/L): (1) 2216E solid-agar medium: 2216E (Hope Bio-Technology, Qingdao, China) 37.40, agar (Solarbio, China) 20.00. (2) Seed medium: 2216E 37.40.(3) Fermentation medium: NaCl 25.00, MgCl2·6H2O 4.12, KCl 1.00, CaCl2 0.20, K2HPO4·3H2O 0.13, FeCl2·4H2O 0.014, peptone 5.00, yeast extract 2.00, agar 1.00 (pH 7.5) (peptone and yeast extract both from Hope Bio-Technology, other reagent were from YongDa of China).
1.2 Validation of Marinomonas sp. FW-1 activity and bacterial analysis Samples of seawater, seaweed, and gut of seashells were screened to isolate arylsulfatase producing microorganisms. Enzyme activity was screened as the previously describe method by culturing strains on the 2216E agar medium including 0.1 mL 5 mol/L p-nitrophenyl sulfate (pNPS)[9].
A loop of seed bacteria was picked, diluted with sterilized water, inoculated on the 2216E plate medium and cultured at 27 ℃ for 2 days. The colony morphology was observed, and samples were analyzed by scanning electron microscope in Songshan Hospital of Qingdao.
1.3 Growth and arylsulfatase production curve of Marinomonas sp. FW-1 Marinomonassp. FW-1 was cultured in the seed medium of marine broth 2216 (Difico, Detroit, MI, USA) by shaking at 140 r/min at 28 ℃. The quality of cell growth was estimated by measuring the absorbance of culture broth at 600 nm each 5 h. The Marinomonas sp. FW-1 which was cultured in the seed medium for 12 h was transferred into the fermentation medium with an inoculation concentration of 2% and thereafter incubated by shaking at 140 r/min, 28 ℃. The biomass and enzyme activity were measured each 12 h. Arylsulfatase activity was determined by measuring the amount of formed p-nitrophenl (pNP) at 410 nm. Enzyme activity (U) was defined as the absorbance equivalent of 1 μmol/L pNP produced per minute per milliliter of enzyme solution under the experimental condition.
1.4 Culture of strain FW-1 and extraction of genomic DNA Marinomonassp. FW-1 was cultured in the seed medium by shaking at 140 r/min at 28 ℃. The genomic DNA of Marinomonas sp. FW-1 was extracted using the Puregene Yeast/Bact Kit B (Qiagen, USA). The quantity and quality of the genomic DNA were evaluated by the Agilent 2100 Bioanalyzer (Santa Clara, CA, USA).
1.5 Complete genome analysis of Marinomonas sp. FW-1 The whole genome sequence was conducted with a single-molecule real-time (SMRT) sequencing platform PacBio RS Ⅱ (version 2.3.0, Pacific Biosciences, USA). Totally 914114923 bp of quality-controlled reads were de novo assembled using the RS Hierarchical Genome Assembly Process (HGAP) protocol by SMRT Analysis. Total number of genes, tRNA and rRNA genes were predicted with Prodigal (version 2.6.3), tRNAscan-SE (version 1.3) and Barrnap (version 0.7) programs, respectively. The predicted gene sequences were translated and searched against the National Center for Biotechnology Information (NCBI) nonredundant database, Gene Ontology (GO) database, the Clusters of Orthologous Groups (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases.
1.6 Activity analysis of expressed product The amplified four fragments were ligated with the plasmid of pET-28a (+) and transformed into the E. coli BL21(DE3) cells, respectively. The recombinant strains were cultivated at 37 ℃ until the OD600 reached 0.5-0.8, respectively. It was further incubated at 20 ℃ for 20 h after the addition of isopropyl-β-D-1-thiogalactopyranoside (IPTG with a final concentration of 0.1 mmol/L). After one washing with 0.9% NaCl solution, cells were harvested and ruptured by sonication in pH 7.5 Tris-HCl buffer (100 mmol/L). The cell lysis solution was collected by centrifugation at 10000×g for 20 min at 4 ℃. An aliquot of 2 mL of each supernatant was mixed with 500 μL of pNPS and the arylsulfatase activity was determined.
2 Results 2.1 Analysis of Marinomonassp. FW-1 One arylsulfatase producing microorganism was found to yield yellow circles around the colonies after incubation for 2 days at 28 ℃ (Figure 1-A). The colonies were round-shaped with white color, which showed smooth and wet surface on top of the culture medium. The microorganism was further isolated and characterized to be a Gram- negative bacterium. Its size was (0.7-1.5) μm× (1.8-3.0) μm as analyzed by a scanning electron microscopy at 20 kV (Figure 2). The result of the phylogenetic analysis showed that the strain belonged to the genus of Marinomonas and named as Marinomonassp. FW-1[3].
 |
| Figure 1 Visible enzyme activities of Marinomonas sp. FW-1 at 28 ℃ for 2 days. Marinomonas sp. FW-1 was grown on 2216E solid-agar medium containing pNPS (A) and without pNPS (B). |
| 图选项 |
 |
| Figure 2 Scanning electron micrograph image of Marinomonas sp. FW-1 was observed in 20 kV at 10000× (A) and 20000× (B) magnification, respectively. Its size was (0.7-1.5) μm×(1.8-3.0) μm. |
| 图选项 |
2.2 The growth curve and arylsulfatase production curve of Marinomonas sp. FW-1 The growth curve of Marinomonassp. FW-1 cultured in medium was shown in Figure 3-A. The lag phase was 0-5 h, the logarithmic phase was 5-20 h, the stationary phase was 20-30 h and after 30 h was the decline phase. From the analysis above, we concluded that the whole growth cycle of the bacteria was relatively stable. Arylsulfatase production curve of Marinomonas sp. FW-1 was shown in Figure 3-B. The arylsulfatase production of Marinomonassp. FW-1 increased gradually during the fermentation. When the fermentation time reached 72-84 h, the growth of bacteria was in the decline phase, while the enzyme production reached the highest level of 0.25 U/mL. Thereafter, the arylsulfatase production decreased gradually.
 |
| Figure 3 Growth curve (A) Marinomonas sp. FW-1 was cultured in the seed medium of marine broth 2216E by shaking at 140 r/min at 28 ℃. The quality of cell growth was estimated by measuring the absorbance of culture broth at 600 nm each 5 h. Arylsulfatase production curve (B). The Marinomonas sp. FW-1 was cultured in the seed medium for 12 h were transferred into the fermentation medium (5 g peptone, 2 g yeast extract, 1 g agar, 1 L artificial seawater, pH of 7.5) at 2% inoculums concentration and then shaking at 140 r/min at 28 ℃. |
| 图选项 |
2.3 Complete genome analysis method of Marinomonas sp. FW-1 To understand and improve the application of arylsulfatase, we sequenced the genome of Marinomonas sp. FW-1. The complete genome of Marinomonas sp. FW-1 contained a circular chromosome of 3964876 bp with a G+C content of 44.03% (Figure 4). The total number of genes were 3590 and the coding region accounted for 88.30% of the Marinomonas sp. FW-1 genome. In addition, 103 RNAs including rRNA and tRNA were identified (Table 1). In addition, The COG function classification was shown in Figure 5.
 |
| Figure 4 Circular genome map of Marinomonas sp. FW-1. The outer circles showed the scale marks of the genome. Circles 1 and 2 displayed the protein-coding genes on the forward strand and reverse strand, respectively. Circle 3 displayed the tRNA (black) and rRNA (red) genes on the forward strand. Circle 4 displayed the tRNA (black) and rRNA (red) genes on the reverse strand. Circles 5 and 6 displayed the GC content and skew, respectively. The genome map was made using Circos v0.64 (http://circos.ca/). |
| 图选项 |
Table 1. Genome features of Marinomonas sp. FW-1
| Feature | Value |
| Genome size/bp | 3964876 |
| G+C content/% | 44.03 |
| Total number of genes | 3590 |
| rRNA | 25 |
| tRNA | 78 |
表选项
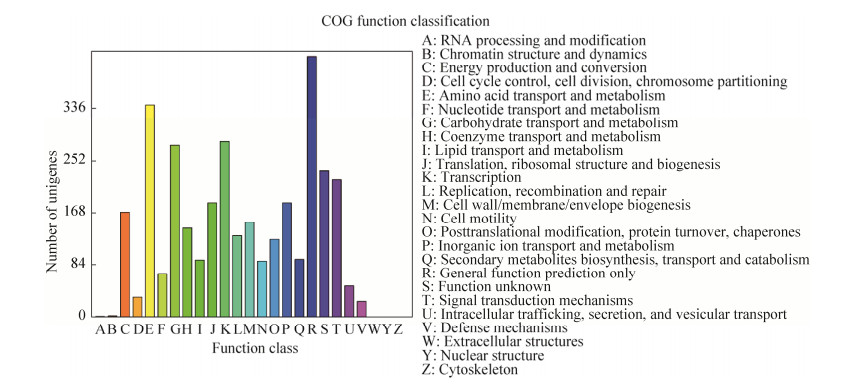 |
| Figure 5 COG function classification of Marinomonas sp. FW-1 genes. |
| 图选项 |
2.4 Activity analysis of expressed product In the genomic sequence of Marinomonas sp. FW-1, two arylsulfatase genes and two sulfatase genes were identified and designated as SulfA, SulfB, ArylsulfA and ArylsulfB, respectively (Table 2). Three genes contained the C-X-P-X-R motif, a characteristic of the cysteine-type sulfatase[9] except the gene SulfA. According to the nucleotide sequences of Marinomonas sp. FW-1 and the fouridentified genes, four pairs of full-length primers were designed and synthesized (Table 3). The color and the absorbance at 410 nm changes of different samples were recorded over time, and the supernatant of Marinomonas sp. FW-1 and E. coli BL21 were used as a positive control and negative control, respectively (Figure 6). The recombinant enzyme from each supernatant of recombinant stairs was designated as SulfA, SulfB, ArylsulfA and ArylsulfB, respectively. ArylsulfB exhibited a very strong arylsulfatase activity because the color changes of the reaction system were observed when the supernatant of the cell lysate of ArylsulfB just contacted with the pNPS. At the reaction time of 5 min, its reaction system showed a deep yellow color and the absorbance at 410 nm was 0.382. The reaction system of FW-1, SulfB, ArylsulfA and ArylsulfB all showed a deep yellow color at the reaction time of 12 h and 24 h. During 24 h, no color changes was observed in supernatant of SulfA and E. coil BL21. To the other three recombinant strains, the overall trend was that the color of the supernatant become deeper and the absorbance at 410 nm increased with time. It was concluded that at least three gene fragments in the whole genome of Marinomonas sp. FW-1 strain assessed for arylsulfatase activity.
Table 2. Predicted arylsulfatase and sulfatase genes in Marinomonas sp. FW-1
| Locus tag | Gene name | Gene product |
| Sulfatase | Anaerobic sulfatase-maturating enzyme homolog AslB | |
| TBCP-2795_1162 | SulfA | Choline-sulfatase |
| TBCP-2795_1775 | SulfB | |
| Arylsulfatase | ||
| TBCP-2795_1161 | ArylsulfA | Arylsulfatase |
| TBCP-2795_2428 | ArylsulfB | Arylsulfatase J |
表选项
Table 3. Sequences of the primers
| Protein name | Primer name | Restriction enzyme | Prime sequence (5′→3′) |
| SulfA | 1-F | BamHⅠ | CGCGGATCCATGAATCAGATTGTAGATCGA |
| 1-R | Hind Ⅲ | CCCAAGCTTTGAATAAAATCTTCCCTTTATCT | |
| SulfB | 2-F | BamHⅠ | CGCGGATCCATGGCAGAAAAGAAACCAAA |
| 2-R | XhoⅠ | CCGCTCGAGCTCTGCTTTAGGGAAACGTGAT | |
| ArylsulfA | 3-F | BamHⅠ | CGCGGATCCATGATTGATCGCTTGCACGA |
| 3-R | SalⅠ | ACGCGTCGACTCGACCACTGTAGGACGCTG | |
| ArylsulfB | 4-F | BamHⅠ | CGCGGATCCATGACAAATATAATCTACAT |
| 4-R | Hind Ⅲ | CCCAAGCTTCCGTTGCGGCATATCCGCCA |
表选项
 |
| Figure 6 Color changes of Marinomonas sp. FW-1, SulfA, SulfB, ArylsulfA, ArylsulfB's supernatant with time. Each image contains two tubes, the left one was a negative control without pNPS, and the right one was the mixture of 2 mL supernatant with 500 μL pNPS, respectively. |
| 图选项 |
3 Discussion Arylsulfatase, is the enzyme that catalyzes the hydrolysis of arylsulfate ester bonds to the corresponding phenols and inorganic sulfate[8]. They are widespread in nature and are found in microorganisms, most animal and human tissues, and plant seeds[9]. A number of arylsulfatases of microbial origin have been characterized. The arylsulfatase producing bacteria including Aspergillus oryzae[10], Klebsiella pneumoniae[11], Marinomoas sp.[3], Pseudoalteromonas carrageenovora[12], Pseudomonas aeruginosa[13], Serratia marcescens[14], Sphingomonas sp.[8] and so on. But the complete genome sequence information of arylsulfatase was rare. Therefore, it is necessary to complete the genome sequencing of arylsulfatase.
There is no clear family classification for arylsulfatase according to the CAZy database (http://www.cazy.org/GH96_characterized.html) and literature [15]. Arylsulfatase genes may have also diverged to an extent that they cannot be readily identified using bioinformatic search tools[15], while the arylsulfatases identified contain a conserved amino acid sequence motif (C/S-X-P-X-R), which is required for enzyme activity[16-17]. In this study, three genes of SulfB, ArylsulfA and ArylsulfB contained the C-X-P-X-R motif and their corresponding proteins exhibited arylsulfatase activity, while the gene of SulfAdidn't contain the motif and its corresponding protein showed no arylsulfatase activity. The result further confirmed that C-X-P-X-R motif was essential for arylsulfatase activity.
In addition to the above mentioned application of arylsulfatase in desulfation agar into agarose, arylsulfatases from different sources have potential applications in many fields of research, industry, and agriculture[17]. Arylsulfatase can release arylphenols like p-cresol and intensify the typical sheep-like flavor in blended cheese[17]. It can degrade endosulfan without producing the toxic metabolite endosulfan sulfate which is an efficient way of remediating contaminated environments[18]. Arylsulfatase activity also could be used as indicator of soil quality[19].
In this study, the complete genome sequence of Marinomonas sp. FW-1 strain was determined and we found in this genome at least three genes with arylsulfatase activity, which provided a valuable reference genome for other species in the genus Marinomonas and gave insights into biological modification of agar for its applications in food, pharmaceutical, industries, and agriculture.
4 Nucleotide sequence accession number The complete genome sequence of Marinomonas sp. FW-1 has been deposited in GenBank under the accession number: CP025987.
References
| [1] | Duckworth M, Yaphe W. The structure of agar: Part Ⅰ. Fractionation of a complex mixture of polysaccharides. Carbohydrate Research, 1971, 16(1): 189-197. DOI:10.1016/S0008-6215(00)86113-3 |
| [2] | Allan GG, Johnson PG, Lai YZ, Sarkanen KV. Marine polymers: Part Ⅰ. A new procedure for the fractionation of agar. Carbohydrate Research, 1971, 17(1): 234-236. DOI:10.1016/S0008-6215(00)81565-7 |
| [3] | Wang XY, Duan DL, Xu J, Gao X, Fu XT. Characterization of a novel alkaline arylsulfatase from Marinomonas sp. FW-1 and its application in the desulfation of red seaweed agar. Journal of Industrial Microbiology & Biotechnology, 2015, 42(10): 1353-1362. |
| [4] | Arnott S, Fulmer A, Scott WE, Dea ICM, Moorhouse R, Rees DA. The agarose double helix and its function in agarose gel structure. Journal of Molecular Biology, 1974, 90(2): 269-284. |
| [5] | Lim JM, Jang YH, Kim HR, Kim YT, Choi TJ, Kim JK, Nam SW. Overexpression of arylsulfatase in E. coli and its application to desulfatation of agar. Journal of Microbiology and Biotechnology, 2004, 14(4): 777-782. |
| [6] | Guiseley KB. The relationship between methoxyl content and gelling temperature of agarose. Carbohydrate Research, 1970, 13(2): 247-256. DOI:10.1016/S0008-6215(00)80831-9 |
| [7] | L?nnerdal B, L??s T. Improved agarose for immunoelectrophoresis. Analytical Biochemistry, 1976, 72(1/2): 527-532. |
| [8] | Kim JH, Byun DS, Godber JS, Choi JS, Choi WC, Kim HR. Purification and characterization of arylsulfatase from Sphingomonas sp. AS6330. Applied Microbiology and Biotechnology, 2004, 63(5): 553-559. DOI:10.1007/s00253-003-1463-8 |
| [9] | Stressler T, Seitl I, Kuhn A, Fischer L. Detection, production, and application of microbial arylsulfatases. Applied Microbiology and Biotechnology, 2016, 100(21): 9053-9067. DOI:10.1007/s00253-016-7838-4 |
| [10] | Benkovic SJ, Vergara EV, Hevey RC. Purification and properties of an arylsulfatase from Aspergillus oryzae. Journal of Biological Chemistry, 1971, 246(16): 4926-4933. |
| [11] | Miech C, Dierks T, Selmer T, Von Figura K, Schmidt B. Arylsulfatase from Klebsiella pneumoniae carries a formylglycine generated from a serine. Journal of Biological Chemistry, 1998, 273(9): 4835-4837. DOI:10.1074/jbc.273.9.4835 |
| [12] | Kim DE, Kim KH, Bae YJ, Lee JH, Jang YH, Nam SW. Purification and characterization of the recombinant arylsulfatase cloned from Pseudoalteromonas carrageenovora. Protein Expression & Purification, 2005, 39(1): 107-115. |
| [13] | Beil S, Kehrli H, James P, Staudenmann W, Cook AM, Leisinger T, Kertesz MA. Purification and characterization of the arylsulfatase synthesized by Pseudomonas aeruginosa PAO during growth in sulfate-free medium and cloning of the arylsulfatase gene (atsA). European Journal of Biochemistry, 1995, 229(2): 385-394. DOI:10.1111/ejb.1995.229.issue-2 |
| [14] | Murooka Y, Yim MH, Harada T. Formation and purification of Serratia marcescens arylsulfatase. Applied and Environmental Microbiology, 1980, 39(4): 812-817. |
| [15] | Ho CL. Phylogeny of algal sequences encoding carbohydrate sulfotransferases, formylglycine-dependent sulfatases, and putative sulfatase modifying factors. Frontiers in Plant Science, 2015, 6: 1057. |
| [16] | Kertesz MA. Riding the sulfur cycle - metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiology Reviews, 2000, 24(2): 135-175. |
| [17] | Stressler T, Leisibach D, Lutz-Wahl S, Kuhn A, Fischer L. Homologous expression and biochemical characterization of the arylsulfatase from Kluyveromyces lactis and its relevance in milk processing. Applied Microbiology and Biotechnology, 2016, 100(12): 5401-5414. DOI:10.1007/s00253-016-7366-2 |
| [18] | Kalyani SS, Sharma J, Singh S, Dureja P, La ta. Enrichment and isolation of endosulfan-degrading microorganism from tropical acid soil. Journal of Environmental Science and Health, Part B: Pesticides, Food Contaminants, and Agricultural Wastes, 2009, 44(7): 663-672. DOI:10.1080/03601230903163665 |
| [19] | Kucharski J, Tomkiel M, Ba?maga M, Borowik A, Wyszkowska J. Enzyme activity and microorganisms diversity in soil contaminated with the boreal 58 WG herbicide. Journal of Environmental Science and Health, Part B: Pesticides, Food Contaminants, and Agricultural Wastes, 2016, 51(7): 446-454. DOI:10.1080/03601234.2016.1159456 |
