Jianhua Tong, Gibson Kamau Gicharu, Xun Hu, Xiaojing Fan, Tao Zhuo, Huasong Zou


Fujian University Key Laboratory for Plant-Microbe Interaction, College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian Province, China
Received: 1 November 2018; Revised: 20 January 2019; Published online: 13 March 2019
Foundation item: Supported by the National Natural Science Foundation of China (31671988, 31872919) and by the Guiding Project from Fujian Province (2016N0006)
Corresponding author: Huasong Zou, Tel: +86-591-83789246; E-mail: hszou@fafu.edu.cn.
Abstract: [Objective] Ralstonia solanacearum is the causative agent of a devastating bacterial wilt disease in solanaceous plants. The purpose of this work was to identify genes involved in the pathogenesis of R. solanacearum.[Methods] We used a Tn5-based mutagenesis strategy to generate random insertion mutants that were then assayed for biofilm formation, cell motility and pathogenicity. Thermal asymmetric interlaced PCR (TAIL-PCR) was performed to identify the Tn5 insertion site of mutants with altered phenotypes.[Results] A total of 400 mutants were generated in the model strain GMI1000. Two mutants were found to be unable to form biofilms and had reduced swimming and swarming motility on soft agar media. When inoculated into tomato plants, both mutants failed to cause wilting disease symptoms. Both mutants carried a Tn5 insertion within the NADH dehydrogenase subunit F gene (nuoF), and the insertion sites were at 103 bp and 225 bp from the translational start site. A complemented strain expressing nuoF under the control of the ripAY promoter fully restored the wild-type phenotype.[Conclusion] The NADH dehydrogenase complex is the first enzyme in the respiratory electron transport chain in microbes. Our data here indicated that NADH dehydrogenase plays a role in biofilm formation, cell motility and pathogenicity in R. solanacearum.
Keywords: Ralstonia solanacearumNADH dehydrogenasepathogenicitynuoFmotility
茄科作物青枯菌NADH脱氢酶F亚基在细胞运动和致病性中的功能研究
童建华, Gibson Kamau Gicharu, 户勋, 范晓静, 卓涛, 邹华松


福建农林大学植物保护学院, 植物与微生物相互作用福建省高校重点实验室, 福建 福州 350002
收稿日期:2018-11-01;修改日期:2019-01-20;网络出版日期:2019-03-13
基金项目:国家自然科学基金(31671988,31872919);福建省科技厅引导项目(2016N0006)
通信作者:邹华松, Tel:+86-591-83789246; E-mail:hszou@fafu.edu.cn.
摘要:[目的] 劳尔氏菌(Ralstonia solanacearum)在茄科作物上引起严重的细菌性青枯病,本研究旨在发掘青枯劳尔氏菌与致病相关的基因。[方法] 利用Tn5转座子构建随机插入突变体,分析生物膜形成、细胞运动和致病性;对有表型变化的突变体,运用TAIL-PCR方法鉴定Tn5插入位点,确定所突变的基因。[结果] 以模式菌株GMI000为出发菌,总共获得了400个突变体,其中2个突变体不能形成生物膜,在软琼脂平板上的运动能力下降;接种感病番茄植物,这2个突变体都不能引起萎焉症状。TAIL-PCR结果显示,2个突变体的Tn5插入位点都在NADH脱氢酶F亚基(nuoF)中,距离翻译起始位点分别为103-bp和225-bp。ripAY基因启动子推动的nuoF基因互补载体,完全恢复了2个突变体的表型。[结论] NADH脱氢酶复合物是微生物呼吸电子传递链中的第一步催化酶。我们的结果表明,NADH脱氢酶复合物对R.solanacearum生物膜形成、细胞运动和致病性也有重要作用。
关键词:青枯菌NADH脱氢酶致病性nuoF运动
The bacterial respiratory complex Ⅰ enzyme is the main entry point for electrons from NADH into the respiratory chains of mitochondria and bacteria[1]. Type Ⅰ NADH dehydrogenase represents a minimal form of this enzyme complex. NADH dehydrogenase is small, relatively simple, and hence used as a model to identify and characterize the mechanisms by which cells regulate the synthesis and assembly of large respiratory complexes[2]. The complex Ⅰ enzyme family has been well characterized in Paracoccus denitrificans, and a model of the subunit structure and function has been inferred[3]. The subunit consists of 14 proteins referred to collectively as NADH:ubiquinone oxidoreductase (Nuo)[4]. The proteins are organized into two groups: integral membrane subunits (NuoA, NuoH, and NuoJ–N) and peripheral subunits (NuoB–G and NuoL). NuoE, NuoF and NuoG assemble a NADH dehydrogenase module that oxidizes NADH along with other known redox cofactors[5].
Bacterial respiratory complex Ⅰ contributes to a range of growth and metabolic processes and thus has evolved to accommodate substantially different energetic and metabolic needs[6]. Studies conducted in Escherichia coli have shown that cells lacking complex Ⅰ have a competitive disadvantage in stationary-phase[7], as microbes use complex Ⅰ to support anaerobic fumarate respiration during facultative anaerobic growth[8]. In Rhodobacter capsulatus, complex Ⅰ is required for phototrophic growth, driving NADH synthesis from quinol using the proton motive force. In Campylobacter jejuni, complex Ⅰ does not require NADH but uses flavodoxin, which is an electron donor[9]. Many studies have shed light on the structure and mechanism of action of bacterial complex Ⅰ[10], but only a few studies have addressed the biological roles of the enzyme.
R. solanacearum is a soil-borne, rod-shaped, Gram-negative bacterium belonging to the β-subdivision of the proteobacteria. This bacterium causes a devastating wilt disease in tropical and subtropical agriculture, and thus is ranked as the second most important bacterial plant pathogen[11]. Biofilm formation and motility are two factors that contribute to R. solanacearum virulence. A mature biofilm structure is usually developed on the host after invasion into the leaf intercellular space[12]. Mutation of the lectin encoding gene lecM results in a significant decrease in biofilm formation and a loss of virulence on tomato plants. The aerotaxis and extracellular nuclease proteins are both required for normal biofilm formation in vitro. A non-motile flagellin (fliC) mutant had reduced virulence on unwounded tomato plants[13]. When bacterial cell density is high in the xylem vessels after invasion, the quorum-sensing signal 3-hydroxy palmitic acid methyl ester (3-OH PAME) accumulates extracellularly and subsequently promotes the production of virulence factors and loss of motility[14].
In this work, we aimed to discover new genes involved in the pathogenicity of R. solanacearum, the causative agent of bacterial wilt disease. Through a Tn5 random mutagenesis strategy, two mutants with an inactivated nuoF gene were identified and had altered biofilm formation, cell motility, and pathogenicity. Our results demonstrated that the function of NADH dehydrogenase is not restricted to NADH oxidation in respiration, and it is also required for other biological processes facilitating the pathogenesis of R. solanacearum.
1 Materials and methods 1.1 Bacterial strains and plasmids The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown at 37 ℃ in Luria-Bertani medium. R. solanacearum strain GMI1000 and its mutants were grown at 28 ℃ on nutrient agar (NA) or in nutrient broth (NB)[15]. NYGB media containing 0.3% and 0.5% agar were used for swimming and swarming assays, respectively[16]. Antibiotics were used at the following concentrations: kanamycin (50 mg/L), ampicillin (100 mg/L), spectinomycin (20 mg/L), gentamycin (10 mg/L), and polymyxin B (20 mg/L).
Table 1. Bacterial strains and plasmids used in this study
| Strains or plasmids | Relevant characteristics | Source |
| Strains | ||
| ??Ralstonia solanacearum | ||
| ??GMI1000 | Wild-type, phylotype Ⅰ, biovar 3, race 1 | [17] |
| ??M1 | Kmr, a mutant with Tn5 insertion at 103 bp site in nuoF gene derived from GMI1000 | This study |
| ??M2 | Kmr, a mutant with Tn5 insertion at 225 bp site in nuoF gene derived from GMI1000 | This study |
| ??CM1 | KmrGmr, M1 carrying pBnuoF | This study |
| ??CM2 | KmrGmr, M2 carrying pBnuoF | This study |
| ??Escherichia coli | ||
| ??DH5α | F'Φ80dlacZDM15D(lacZYA-argF)U169 endA1 deoR recA1 hsdR17(rK2 mK+) phoA supE44 l2 thi-l gyrA96 relA1 | Invitrogen |
| Plasmids | ||
| ??pUTKm | Ampr Kmr, delivery plasmid for Tn5, R6Kreplion | [18] |
| ??pRK2073 | Spr, helper plasmid supplying IncP tra functions | [19] |
| ??pBBR1MCS-5 | Gmr, 5.1-kb broad-host range plasmid, lacZ | [20] |
| ??pBnuoF | Gmr, the 1296-bp nuoF gene under control of RSp1022 promoter in pBBR1MCS-5 | This study |
表选项
1.2 Construction of Tn5 insertion mutants The transposon plasmid pUTKm was introduced into wild-type R. solanacearum strain GMI1000 by triparental mating[19]. The recipient strain GMI1000, helper strain pRK2073 and donor strain carrying pUTKm were cultured to an OD600 of 1.0. From each cell suspension, 300 μL was collected into 1.5 mL tubes and centrifuged at 5000 r/min for 10 min. After discarding the supernatant, the cell pellets were suspended in 1 mL of distilled water and centrifuged again at 5000 r/min for 10 min. The cells were then suspended in 50 μL of distilled water and transferred onto NA plates lacking antibiotics. After incubation at 28 ℃ for 24 h, the cells were spread on NA plates supplemented with both polymyxin and kanamycin. GMI1000 cells carrying the Tn5 transposon insertion were isolated after incubation at 28 ℃ for 72 h. Individual colonies were transferred onto NA plates containing polymyxin and kanamycin for further analysis.
1.3 Identification of mutants defective in biofilm formation To test for biofilm formation, mutant cells were cultured in 4 mL of NB broth. All cultures were adjusted to an OD600 of 1.0 and then 3 mL of the cell suspension was incubated in sterilized 5-mL glass bottles at 28 ℃ without shaking for one week. The planktonic cells were gently removed from the bottles and the bottles were then washed twice with distilled water. Bacterial adhesion to the bottle walls was assessed after staining with 1% crystal violet for 15 min at room temperature. Excess stain was removed by washing with distilled water. Wild type GMI1000 inoculated into NB broth without antibiotics was used as a positive control. Each experiment was repeated three times.
1.4 Swimming and swarming ability Swarming motility was assayed on NYGB media solidified with 0.5% agar, while swimming motility was assays on NYGB containing 0.3% agar[16]. Cells were allowed to grow overnight in NB broth containing kanamycin and the OD600 was adjusted to 0.5. For each strain, 1 μL of each cell suspension was placed on the surface of the NYGB media. Cell motility was determined from the diameter of each colony at 3 d post inoculation. The experiments were repeated four times. Statistical analyses were performed using the Student's t-test.
1.5 Pathogenicity test Tomato plants (Lycopersicon esculentum Mill. cv. 'Hongyangli'), which were susceptible to R. solanacearum, were used to assess pathogenicity by the stem inoculation method. For each strain, eight 4-week-old tomato plants were used for pathogenicity analysis. The plants were wounded with hypodermic needles at the junction of the third leaf from the top. The cultured wild-type and mutant cells were prepared at an OD600=0.5. Then, 20 μL of each cell suspension was placed on the wound. Bacterial wilt progress was rated daily according to the standard disease index. The percent severity index was calculated using a previously described method[15]. The mean disease index for each treatment for each day was analyzed by analysis of variance (ANOVA) at the 95% level. All experiments were repeated three times.
1.6 Identification of Tn5 insertion sites To identify the transposon insertion sites, bacterial genomic DNA was isolated using a TIANamp Genomic DNA Kit (Tiangen, Beijing, China). One set of nested primers specific to the end of the Tn5 transposon (SP1, 5′-GCCTGGTATGAGT CAGCAACA-3′; SP2, 5′-GGCAGACCTCAGCGCT ATTC-3′; SP3, 5′-ATCACGACTGTGCTGGTCATT A-3′) and one arbitrary degenerate primer (5′- NGTCGA[G/C][A/T]GA[A/T/C/G]A[A/T]GAA-3′) were used to perform TAIL-PCR. The parameters for the three-step TAIL-PCR were adopted from a previously described protocol[21]. TAIL-PCR products were electrophoresed, extracted and purified from a gel, and the cloned into pMD19-T simple vector (TaKaRa, Dalian, China) for confirmation via sequencing. DNA and protein sequences were analyzed by BLASTn/BLASTx in GenBank (http://www.ncbi.nlm.nih.gov/BLAST).
1.7 Construction of a nuoF complemented strains To complement the nuoF mutation, a 487 bp DNA fragment containing the promoter region of the RipAY gene was PCR amplified from wild-type genomic DNA and inserted into plasmid pBBR1MCS-5 at the Kpn Ⅰ and XhoⅠ sites (using primers 5′-TTCGGTACCGTCCACAGATGTCGCT TCAC-3′ and 5′-TTCCTCGAGATTCTTTCCATGG CT-3′). The 1296-bp nuoF gene was amplified and ligated into plasmid pBBR1MCS-5 at the EcoR Ⅰ and BamH Ⅰ sites (using primers 5′-TCGAATTCAT GACCTCCCTGCACGATC-3′ and 5′-TCGGATCCT TAGATGTAGGTGGGCACCAT-3′). The resultant recombinant pBnuoF construct was transformed into mutants to produce the complemented strains.
2 Results 2.1 Isolation of mutants defective in biofilm formation To identify genes required for biofilm formation, the transposon pUTKm was introduced into wild-type GMI1000 by triparental mating. Through 10 rounds of triparental mating, approximately 400 mutants were derived. The random insertion mutants were then assayed for adhesion to glass bottle walls in order to assess their ability to form biofilms. When grown in NB media, the two isolated mutant strains aggregated at the bottom of the culture tubes leaving a clear column of media above. This indicated that the isolated mutants were unable to move about freely within the liquid media (Figure 1). A wild-type culture maintained under the same conditions continued to increase in turbidity, indicating the cells remained motile. The two mutant strains, hereafter referred to as M1 and M2, were completely defective for biofilm formation (Figure 1).
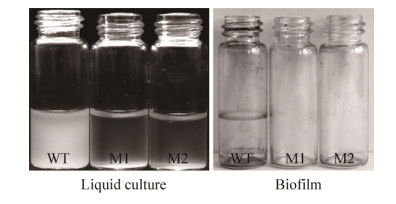 |
| Figure 1 Liquid cultures and biofilm formation of mutants M1 and M2. |
| 图选项 |
2.2 Identification of insertion sites within the nuoF gene The insertion sites in the mutants were identified using TAIL-PCR. For each mutant, one specific PCR band was produced after three-step TAIL-PCR. Nucleotide sequences and BLASTn results indicated that both mutants harbored Tn5 insertions in the nuoF gene (RSc2057), which is a 1290-bp gene that encodes the NADH dehydrogenase I chain F subunit. The two identified mutants were named M1 and M2, and carried the Tn5 insertion at 103 and 225 bp from the translational start site, respectively (Figure 2). In R. solanacearum GMI1000, the nuo cluster is composed of 14 genes located from 2221198 bp to 2235935 bp in the bacterial genome, and has a genetic organization similar to that found in E. coli[10].
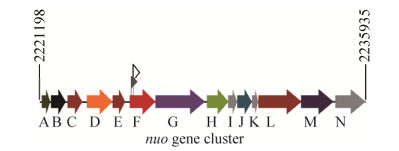 |
| Figure 2 Genetic organization of the 14 nuo genes in the nuo operon of R. solanacearum. The gray and blank flags indicate the Tn5 insertion sites in M1 and M2, respectively. |
| 图选项 |
For complementation studies, the promoter region of ripAY (RSp1022) and the entire coding region of nuoF were PCR amplified and cloned into the plasmid pBBR1MCS-5 to generate the construct pBnuoF (Table 1). The recombinant plasmid was transformed into both M1 and M2 mutants to produce complemented strains, hereafter referred to as CM1 and CM2, respectively. Both complemented strains were tested for their growth in NB media to assess motility (i.e., turbid media versus aggregation at the bottom of the vessel). The wild-type phenotypes were restored in each of the complemented mutants. In addition, biofilm formation was fully restored in CM1 and CM2 with biofilm rings observed on the glass bottle walls (Figure 3). For the wild-type strain, bacteria formed a biofilm ring on the walls of their container at the air/liquid interphase and remained turbid in liquid media. These results indicate that the nuoF insertion mutations have abrogated biofilm formation, which is an important for infection by these bacteria.
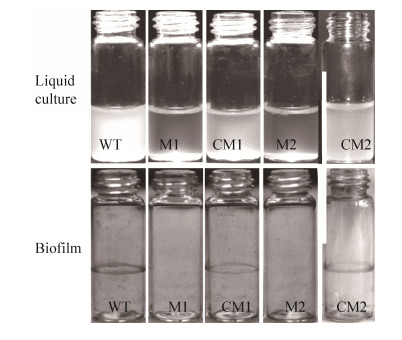 |
| Figure 3 Comparison of biofilm formation between wild-type, mutant and complemented mutant strains. |
| 图选项 |
2.3 NuoF mutants have reduced cell motility on soft agar plates Cell motility was tested on NYGB medium after all cultures were adjusted to an OD600 of 0.5. On 0.5% agar plates, the diameter of the wild-type colonies reached 1.3 cm, while the mutant M1 and M2 strains reached diameters of 0.7 cm and 0.5 cm, representing 46% and 62% reductions in swarming motility, respectively (Figure 4). On 0.3% agar plates, the diameter of the wild-type colonies reached 1.5 cm, while the diameters of M1 and M2 colonies were 0.9 cm and 0.7 cm, representing reductions of about 40% and 53% in swimming ability, respectively (Figure 4). After complementing both mutants, swarming and swimming phenotypes were restored. Insertions in nuoF significantly reduced cell swarming and swimming abilities.
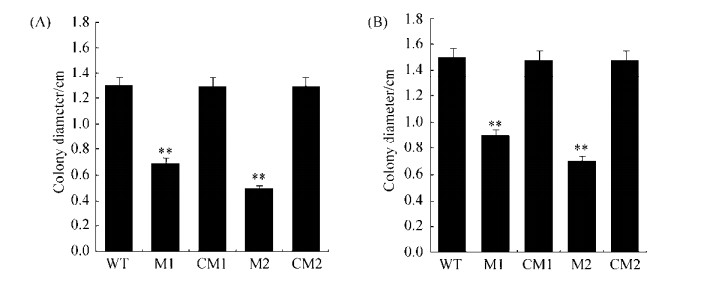 |
| Figure 4 Cell motility assay of nuoF mutants. The asterisks indicate significant differences to wild type according to Student's t-tests. **:P < 0.01. A: swarming; B: swimming. |
| 图选项 |
2.4 NuoF is essential for bacterial pathogenicity in tomato plants Because the mutants exhibited a loss of biofilm formation, an important virulence factor, we wanted to explore the effect of these mutations on virulence in plants. Disease progression was investigated after application of the strains using the stem inoculation method. At 3 d post inoculation, plants inoculated with wild-type R. solanacearum started showing signs of wilt and by the tenth day had totally collapsed. In contrast, plants inoculated with the mutant strains did not show any signs of wilting apart from small lesions at the point of inoculation (Figure 5). Following inoculation with the complemented mutant strains, the pathogenicity phenotype was restored to wild type levels (Figure 5). These results demonstrate that NADH dehydrogenase plays a crucial role in the development of bacterial wilt disease.
 |
| Figure 5 The pathogenicity of two nuoF mutants in tomato plants. The inoculum was introduced into the wound at the junction of the third leaf from the top and the stem. For each strain in every repeat, wild type, mutants and complemented strains were tested on eight 4-week-old tomato plants. Disease symptoms of representative plants were photographed at 6 d post inoculation. The disease index was recorded every day post inoculation until day 10. The asterisks denote statistical significance in comparison with the wild type. *: P < 0.01. |
| 图选项 |
3 Discussion Genome-wide mutant libraries are collections of non-isogenic mutants in essential genes and are powerful tools for investigating the genetics of organisms. One of the simplest methods to generate mutant libraries is through random transposon mutagenesis[17]. This technique has been used to construct genome-wide mutant libraries for many pathogenic bacteria, such as Pseudomonas aeruginosa[22] and Xanthomonas citri subsp. citri[23]. We used this method to generate 400 R. solanacearum mutants. Even though the number of mutants did not guarantee saturation of the GMI1000 genome, two nouF mutants were identified to be deficient in cell motilty and pathogenicity in this study.
The role of cell motility in the virulence of phytopathogens is not well understood, as studies in various bacteria have shown conflicting results. A deficiency in motility led to significantly reduced virulence in Pseudomonas syringae pv. glycinea, Erwinia amylovora and P. phaseolicola[24-26]. In Agrobacterium tumefaciens, non-motile mutants were found to be fully virulent when inoculated by direct immersion; however, they remained avirulent when inoculated by planting wounded seedlings into the soil infested with the bacteria[27]. Flagella-driven cell motility is required for full virulence of R. solanacearum when inoculated onto unwounded plants through soil drench methods[13]. This suggests that flagella-driven cell motility promotes movement to the root surface by soil-borne R. solanacearum. In this study, the nuoF mutants M1 and M2 harbored different Tn5 insertion sites, but they both exhibited reduced cell motility and a complete loss of pathogenicity. It seems that disruption of nuoF gene affects not only flagella-driven cell motility, but also other virulence factors required for R. solanacearum infection.
R. solanacearum infects healthy plants through plant roots, wounds, or natural openings. It enters the intercellular spaces of the root cortex, moves through the vascular parenchyma and xylem vessels and finally spreads into the upper parts of the plant[11]. After invasion into host vasculature, R. solanacearum produces an acidic high molecular mass extracellular polysaccharide, which is a major virulence factor[11]. A recent study has indicated that R. solanacearum is able to produce mature mushroom-type biofilms in vitro on the surface of tomato cells after invasion into the intercellular spaces[12]. This is further supported by the finding that within the xylem vessels, R. solanacearum uses DNases to modulate biofilm structure for systemic spread and virulence[12]. Even though several methods have been successfully developed to study biofilm formation under in vitro culture conditions, direct evidence is required from interaction of the bacteria with host plants. Here, the nuoF mutants derived from GMI1000 failed to produce biofilms under lab culturing conditions during our experiments. Further work is required to determine whether the mutants can produce biofilms within xylem vessels.
Respiratory complex Ⅰ provides the proton motive force that is essential for energy-consuming processes[1]. Because R. solanacearum is a xylem-specific pathogen, its movement within the xylem is particularly important for wilt symptom development[11]. We speculate that the biological energy derived from NADH dehydrogenase is essential for R. solanacearum to colonize hosts. This may be the reason that nuoF mutants failed to cause typical wilt symptoms in tomato plants. Generally, the nuo operon is comprised of fourteen genes that provide the minimal structural framework of the proton-pump Nuo[4]. Except for the nuoF mutants, we did not obtain any other mutants in the nuo locus in this study. Further studies are needed to elucidate the roles of the other nuo genes of NADH dehydrogenase, which will provide more substantive evidence on the biological function of NADH dehydrogenase in R. solanacearum.
In conclusion, two nuoF mutants derived from R. solanacearum GMI1000 were obtained by Tn5-based mutagenesis in this study. The insertional inactivation of nuoF led to a reduction in cell motility and a complete loss of pathogenicity. Our results highlight the necessity of NADH dehydrogenase for the pathogenicity by R. solanacearum.
References
| [1] | Yagi T, Matsuno-Yagi A. The proton-translocating NADH-quinone oxidoreductase in the respiratory chain: the secret unlocked. Biochemistry, 2003, 42(8): 2266-2274. DOI:10.1021/bi027158b |
| [2] | Hirst J. Mitochondrial complex Ⅰ. Annual Review of Biochemistry, 2013, 82: 551-575. DOI:10.1146/annurev-biochem-070511-103700 |
| [3] | Yagi T. Purification and characterization of NADH dehydrogenase complex from Paracoccus denitrificans. Archives of Biochemistry and Biophysics, 1986, 250(2): 302-311. DOI:10.1016/0003-9861(86)90731-9 |
| [4] | Falk-Krzesinski HJ, Wolfe AJ. Genetic analysis of the nuo locus, which encodes the proton-translocating NADH dehydrogenase in Escherichia coli. Journal of Bacteriology, 1998, 180(5): 1174-1184. |
| [5] | Calhoun MW, Gennis RB. Demonstration of separate genetic loci encoding distinct membrane-bound respiratory NADH dehydrogenases in Escherichia coli. Journal of Bacteriology, 1993, 175(10): 3013-3019. DOI:10.1128/jb.175.10.3013-3019.1993 |
| [6] | Friedrich T, Scheide D. The respiratory complex Ⅰ of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Letters, 2000, 479(1/2): 1-5. |
| [7] | Zambrano MM, Kolter R. Escherichia coli mutants lacking NADH dehydrogenase Ⅰ have a competitive disadvantage in stationary phase. Journal of Bacteriology, 1993, 175(17): 5642-5647. DOI:10.1128/jb.175.17.5642-5647.1993 |
| [8] | Tran QH, Bongaerts J, Vlad D, Unden G. Requirement for the proton-pumping NADH dehydrogenase Ⅰ of Escherichia coli in respiration of NADH to fumarate and its bioenergetic implications. European Journal of Biochemistry, 1997, 244(1): 155-160. DOI:10.1111/j.1432-1033.1997.00155.x |
| [9] | Weerakoon DR, Olson JW. The Campylobacter jejuni NADH: ubiquinone oxidoreductase (complex Ⅰ) utilizes flavodoxin rather than NADH. Journal of Bacteriology, 2008, 190(3): 915-925. DOI:10.1128/JB.01647-07 |
| [10] | Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex Ⅰ. Nature, 2013, 494(7438): 443-448. DOI:10.1038/nature11871 |
| [11] | Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. Top 10 plant pathogenic bacteria in molecular plant pathology. Molecular Plant Pathology, 2012, 13(6): 614-629. DOI:10.1111/j.1364-3703.2012.00804.x |
| [12] | Mori Y, Inoue K, Ikeda K, Nakayashiki H, Higashimoto C, Ohnishi K, Kiba A, Hikichi Y. The vascular plant-pathogenic bacterium Ralstonia solanacearum produces biofilms required for its virulence on the surfaces of tomato cells adjacent to intercellular spaces. Molecular Plant Pathology, 2016, 17(6): 890-902. DOI:10.1111/mpp.12335 |
| [13] | Tans-Kersten J, Huang HY, Allen C. Ralstonia solanacearum needs motility for invasive virulence on tomato. Journal of Bacteriology, 2001, 183(12): 3597-3605. DOI:10.1128/JB.183.12.3597-3605.2001 |
| [14] | Li P, Yin WF, Yan JL, Chen YF, Fu SN, Song SH, Zhou JN, Lyu MF, Deng YY, Zhang LH. Modulation of inter-kingdom communication by PhcBSR quorum sensing system in Ralstonia solanacearum phylotype Ⅰ strain GMI1000. Frontiers in Microbiology, 2017, 8: 1172. DOI:10.3389/fmicb.2017.01172 |
| [15] | Sun DL, Zhuo T, Hu X, Fan XJ, Zou HS. Identification of a Pseudomonas putida as biocontrol agent for tomato bacterial wilt disease. Biological Control, 2017, 114: 45-50. DOI:10.1016/j.biocontrol.2017.07.015 |
| [16] | Zhuo T, Rou W, Song X, Guo J, Fan XJ, Kamau GG, Zou HS. Molecular study on the carAB operon reveals that carB gene is required for swimming and biofilm formation in Xanthomonas citri subsp. citri. BMC Microbiology, 2015, 15: 225. DOI:10.1186/s12866-015-0555-9 |
| [17] | Boucher CA, Barberis PA, Trigalet AP, Demery DA. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. Journal of General Microbiology, 1985, 131(9): 2449-2457. |
| [18] | Herrero M, de Lorenzo V, Timmis KN. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. Journal of Bacteriology, 1990, 172(11): 6557-6567. DOI:10.1128/jb.172.11.6557-6567.1990 |
| [19] | Better M, Helinski DR. Isolation and characterization of the recA gene of Rhizobium meliloti. Journal of Bacteriology, 1983, 155(1): 311-316. |
| [20] | Kovach ME, Phillips RW, Elzer PH, Roop Ⅱ RM, Peterson KM. pBBR1MCS: a broad-host-range cloning vector. Biotechniques, 1994, 16(5): 800-802. |
| [21] | Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics, 1995, 25(3): 674-681. DOI:10.1016/0888-7543(95)80010-J |
| [22] | Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Liu C R, Guenthner D, Bovee D, Olson MV, Manoil C. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(24): 14339-14344. DOI:10.1073/pnas.2036282100 |
| [23] | Song X, Guo J, Ma WX, Ji ZY, Zou LF, Chen GY, Zou HS. Identification of seven novel virulence genes from Xanthomonas citri subsp. citri by Tn5-based random mutagenesis. Journal of Microbiology, 2015, 53(5): 330-336. DOI:10.1007/s12275-015-4589-3 |
| [24] | Panopoulos NJ, Schroth MN. Role of flagellar motility in the invasion of bean leaves by Pseudomonas phaseolicola. Phytopathology, 1974, 64(11): 1389-1397. DOI:10.1094/Phyto-64-1389 |
| [25] | Bayot RG, Ries SM. Role of motility in apple blossom infection by Erwinia amylovora and studies of fire blight control with attractant and repellent compounds. Phytopathology, 1986, 76(4): 441-445. DOI:10.1094/Phyto-76-441 |
| [26] | Hatterman DR, Ries SM. Motility of Pseudomonas syringae pv. glycinea and its role in infection. Phytopathology, 1989, 79(3): 284-289. DOI:10.1094/Phyto-79-284 |
| [27] | Hawes MC, Smith LY. Requirement for chemotaxis in pathogenicity of Agrobacterium tumefaciens on roots of soil-grown pea plants. Journal of Bacteriology, 1989, 171(10): 5668-5671. DOI:10.1128/jb.171.10.5668-5671.1989 |
