贺振1,2, 李文凤3, 李世访2


1.扬州大学园艺与植物保护学院, 江苏 扬州 225009;
2.中国农业科学院植物保护研究所, 植物病虫害生物学国家重点实验室, 北京 100193;
3.云南省农业科学院甘蔗研究所, 云南省甘蔗遗传改良重点实验室, 云南 开远 661699
收稿日期:2016-03-27;修回日期:2016-06-27;网络出版日期:2016-07-19
基金项目:国家自然科学基金(31601604);植物病虫害生物学国家重点实验室开放课题(SKLOF201518);扬州大学科技创新培育基金(2015CXJ043)
*通信作者:李世访, Tel:+86-10-62890875;E-mail:sfli@ippcaas.cn
摘要: [目的]利用NIa基因,阐明甘蔗线条花叶病毒(Sugarcane streak mosaic virus,SCSMV)的种系发生关系,为预测SCSMV流行变异趋势及科学防控提供理论依据。[方法]从云南蔗区和国家甘蔗种质资源圃采集感病样品,RT-PCR扩增获得SCSMV NIa基因序列后,使用Splits Tree、RDP、PhyML、DnaSP等软件分析SCSMV中国分离物的系统发生、选择压力及基因流动等特征。[结果]共获得23条SCSMV NIa基因序列。这些序列间未发生重组,云南蔗区的部分序列形成1个新簇,且云南蔗区与国家甘蔗种质资源圃之间的基因交流不显著。此外,选择压力分析表明,NIa基因受很强的负选择压力作用。[结论]与P1、HC-Pro和CP等基因类似,SCSMV在NIa基因上也包含5个簇;SCSMV云南分离物具有较高的遗传多样性和清晰的地理相关性。
关键词: 甘蔗线条花叶病毒 NIa基因 系统发生
A novel phylogenetic lineage clustered by NIa gene of Sugarcane streak mosaic virus Yunnan isolates
He Zhen1,2, Li Wenfeng3, Li Shifang2


1.School of Horticulture and Plant Protection, Yangzhou University, Yangzhou 225009, Jiangsu Province, China;
2.State Key Laboratory of Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China;
3.Yunnan Key Laboratory of Genetic Improvement of Sugarcane, Sugarcane Research Institute, Yunnan Academy of Agricultural Sciences, Kaiyuan 661699, Yunnan Province, China
Received 27 March 2016; Revised 27 June 2016; Published online 19 July 2016
*Corresponding author: Shifang Li, Tel:+86-10-62890875;E-mail:sfli@ippcaas.cn
Supported by the National Natural Science Foundation of China (31601604), by the State Key Laboratory for Biology of Plant Diseases and Insect Pests (SKLOF201518) and by the Yangzhou University Science and Technology Innovation Fund (2015CXJ043)
Abstract: [Objective]We assessed the phylogenetic relationship of Sugarcane streak mosaic virus (SCSMV) according to NIa sequences, to infer the prevalence and variation of SCSMV and to prevent and control this virus.[Methods]Leaf samples with mosaic symptom were collected from sugarcane-growing areas in Yunnan province and the Chinese national nursery of sugarcane germplasm resources (NNSGR). NIa sequences of SCSMV were determined by RT-PCR, and analyzed by Splits Tree, RDP, PhyML and DnaSP softwares, in aspect of phylogenetic, selection, and gene flow.[Results]We obtained 23 NIa sequences; clear recombination site was not found in NIa; a novel cluster formed by SCSMV Yunnan isolates determined here was found; strong purifying selection was found in NIa of SCSMV; and the gene flow of SCSMV subpopulations between sugarcane-growing areas in Yunnan province and the NNSGR was not frequent.[Conclusion]Similar with P1, HC-Pro and CP genes, SCSMV isolates could be divided into five clusters. NIa of SCSMV Yunnan isolates showed high genetic diversity and clear geographical distribution.
Key words: Sugarcane streak mosaic virus NIa phylogenetic
甘蔗线条花叶病毒(Sugarcane streak mosaic virus, SCSMV)属于马铃薯Y病毒科(Potyviridae)禾本科病毒属(Poacevirus)。该病毒首先在甘蔗花叶病株上发现,自然条件下能够侵染甘蔗、高粱和一些禾本科杂草[1-4]。SCSMV侵染甘蔗引起长短不一的线条形花叶症状,发生普遍时能够引起甘蔗减产20%左右,目前在印度、印度尼西亚、泰国等南亚、东南亚国家广泛发生,已成为该地区甘蔗花叶病的主要病原[1, 4]。2008-2011年,我们在云南省甘蔗产区首次发现SCSMV,后续调查发现,SCSMV在云南省甘蔗主产区发生越来越普遍,部分地区呈现流行爆发趋势[2, 5-6]。因此,需要采取有效防控措施,控制SCSMV的进一步扩散和蔓延。
明确SCSMV种群的遗传多样性及分子进化可以为其防控提供指导。研究发现[7-8]SCSMV印度分离物的CP基因和HC-Pro基因具有较高的遗传多样性。我国分离的SCSMV云南分离物与印度分离物是2个独立遗传的种群,两者之间基因交换的频率很低[6]。我们的前期研究发现[6],依据P1、HC-Pro和CP基因,SCSMV包含5个系统发育簇,而NIa基因只含有4簇,可能是由于NIa基因分离物的数目和多样性水平较低造成。本研究对采自云南省沅江、弥勒、红河、新平、常宁和国家甘蔗种质资源圃内的16个甘蔗花叶病样品的NIa基因进行克隆测序,基于NIa基因分析SCSMV的重组、系统发生、选择压力、种群结构以及基因流动,从而确定NIa基因的分子进化特征。研究结果能够为制定合理的SCSMV病毒病害防控策略提供依据。
1 材料和方法 1.1 病毒分离物 2008-2011年,从中国云南省沅江、弥勒、红河、新平、常宁等县市和国家甘蔗种质资源圃内(源自日本、印度尼西亚样品)采集具有典型花叶症状的甘蔗样品。新鲜叶片样品经RT-PCR法检测鉴定,将SCSMV阳性样品经冷冻干燥后,-80 ℃保存备用。本研究选取其中不同地区具有代表性的16个样品,详细信息见表 1。
表 1. 本研究中的SCSMV样品采集表 Table 1. The Sugarcane streak mosaic virus isolates collected in this study
| Isolate | Origin | Collection time | Cultivar | Symptom |
| W23 | Japan* | 2010.8 | Japan 1 | Mosaic |
| W24 | Japan* | 2010.8 | Japan 6 | Mosaic |
| W32 | Indonesia* | 2010.8 | Ti20-0 | Mosaic |
| M55 | Changning,China (常宁) | 2008.6 | Q170 | Mosaic |
| M61 | Yuanjiang,China (沅江) | 2008.11 | Unknown | Mosaic |
| M62 | Xinping,China (新平) | 2009.7 | Yunyin10 | Mosaic |
| M85 | Honghe,China (红河) | 2010.8 | Yue79-177 | Mosaic |
| M86 | Mile,China (弥勒) | 2010.8 | Yun99-91 | Mosaic |
| M112 | Yuanjiang,China (沅江) | 2011.6 | ROC22 | Mosaic |
| M116 | Yuanjiang,China (沅江) | 2011.6 | Yun03-258 | Mosaic |
| M117 | Yuanjiang,China (沅江) | 2011.6 | Yun98-136 | Mosaic |
| M118 | Yuanjiang,China (沅江) | 2011.6 | De03-83 | Mosaic |
| M119 | Yuanjiang,China (沅江) | 2011.6 | Yunyin58 | Mosaic |
| M121 | Yuanjiang,China (沅江) | 2011.6 | Yunyin58 | Mosaic |
| M124 | Yuanjiang,China (沅江) | 2011.6 | SP81-3250 | Mosaic |
| M126 | Yuanjiang,China (沅江) | 2011.6 | Yun07-912 | Mosaic |
| *: sugarcane samples stored in the Chinese national nursery of sugarcane germplasm resources (NNSGR). | ||||
表选项
1.2 克隆测序 利用RNA prep pure Plant Kit (天根)提取甘蔗叶片总RNA,方法参照试剂盒说明书。取2 μL总RNA用反向引物SCSMNIa-R (5′-CAAGTGCTC AACTCTTCGT-3′)进行反转录获得cDNA,反应体系参照MLV反转录试剂盒(Promega)说明书。然后使用引物SCSMNIa-F (5′-ATTGGGATGAT GGAAAACAG-3′)和SCSMNIa-R扩增NIa基因。PCR反应体系为:cDNA 2 μL,引物SCSMNIa-F (10 μmol/L)和SCSMNIa-R (10 μmol/L)各2 μL,10×PCR缓冲液5 μL,dNTPs (2.5 mmol/L) 4 μL,Long Taq DNA polymerase (5 U/μL) 0.5 μL,加ddH2O至50 μL。PCR反应条件为:94 ℃ 5 min;94 ℃ 30 s,55 ℃ 30 s,72 ℃ 1 min,共30个循环;72 ℃ 10 min,4 ℃保存。PCR产物经过凝胶纯化后克隆至pMD18-T载体(TaKaRa)上,并转化至大肠杆菌(Escherichia coli) DH5α感受态细胞中。经菌落PCR鉴定获得阳性重组质粒,筛选3-6个阳性克隆送至北京六合华大基因科技股份有限公司测序。测序结果通过峰图及序列比对分析排除由PCR扩增引起的突变。
1.3 重组分析 以TriMV (NC012779)作为比对分析的外组(outgroup)[9]。将GenBank中已登录的SCSMV NIa基因序列与本研究获得的序列一起进行分析。首先,利用CLUSTAL X2[10]和TRANSALIGN(由Georg Weiller教授惠赠)进行核苷酸序列和氨基酸序列的比对,得到能够正确编译出氨基酸的核酸序列。然后,使用Datamonkey (http://www.da tamonkey.org/)中GARD和RDP 4.0软件包[11]中的RDP[12]、GENECONV[13]、BOOTSCAN[14]、MAXCHI[15]、CHIMAERA[16]、3SEQ[17]和SISCAN[18] 7个软件进行重组检测。各检测方法的参数采用默认值,Bonferroni校正的P-value为0.05和0.01,当某分离物有至少3个方法的检测结果P < 1.0×10-6时,支持该分离物为重组体[19-20]。最后,删除分析中的外组序列,去除TriMV对SCSMV序列造成的gap影响,直接检测确认SCSMV在NIa基因区的重组位点。
1.4 系统发生与多样性分析 利用最大似然法(Maximum-likelihood, ML)、邻接法(Neighbor-joining, NJ)及邻接网法(Neighbor-net, NN)进行系统发生分析,所用软件分别为PhyML 3.0[21]、MEGA 6.0[22]和Splits Tree 4.11.3[23]。在ML法分析中,通过jModeltest 0.1.1[24]软件确定NIa基因序列的最适核苷酸替代模型为GTR+I+Г4。在ML和NJ法分析中,支长皆用自举法(bootstrap)进行1000模拟复制计算检验。系统发育树由Tree View[25]展示。
使用DnaSP 5.0[26]计算不同种群分离物的核苷酸多样性(nucleotide diversity)和单体型多样性(haplotype diversity)。SCSMV NIa基因的核苷酸间的多样性分布通过SDT 1.0[27]中的Clustal W法计算获得。
1.5 选择压力分析 基于ML法计算NIa基因的dN/dS值(非同义突变和同义突变之间的比值),预测其承受的选择压力。首先,利用Datamonkey中SLAC (single-like lihood ancestor counting)、FEL (fixed-effects likelihood)和REL (random-effects likelihood)检测NIa基因不同位置密码子的选择压力;其次,应用MEGA 6.0[22]中的Pamilo-Bianchi-Li method[28]计算不同系统发育分支(组)的选择压力。当dN/dS < 1时,表示该组分离物处于纯化或负向选择压力下;当dN/dS=1时,表示该组分离物处于中性选择压力中;当dN/dS > 1时,表示该组分离物受正向选择或多样化选择作用。
1.6 基因漂移分析 种群间的基因交流用Fst和Nm值来衡量。Fst的绝对值在0-1之间,当Fst的绝对值小于统计阈值0.33时,说明两个地区之间存在频繁的基因交流,反之,则交流的频率很低[6, 29-31]。若Nm < 1时,种群间很容易发生遗传漂变,故遗传漂变是促使群体发生遗传分化的主要原因;若Nm > 1说明地缘关系较近或者种群间存在可以发生基因交流的渠道[31]。
种群之间的遗传差异用Ks*,Z和Snn[32]衡量。Ks*由不同序列的数量决定,与序列的来源无关;Z值是秩统计量(rank statistic),代表Z1和Z2的加权和;Snn是指近邻统计(nearest-neighbor statistic),适用于样品采集来源于2个或者2个以上的地区,目的是计算在相同地理学空间上近邻序列出现的频率。如果Z和Ks*统计值很小,P < 0.05,则拒绝无遗传差异的假设。上述统计由DnaSP 5.0[26]完成。
2 结果和分析 2.1 SCSMV NIa基因的序列特征与重组分析 在云南省不同蔗区和国家甘蔗种质资源圃内采集样品,经检测后,从中选取16个具有典型花叶症状的SCSMV阳性样品,对SCSMV的NIa基因进行克隆测序。共获得23条可用序列(登录号:KU314373-KU314395)。NIa基因的序列长度为714 nt。
将获得的23条序列与从GenBank中下载的43条SCSMV NIa基因序列进行重组分析。结果(图 1)发现,除了SCSMV-TPT分离物,不同分离物间没有明显的网状交叉。表明不同分离物间未发生重组。此外,GARD和RDP 4.0重组分析也未发现重组位点。
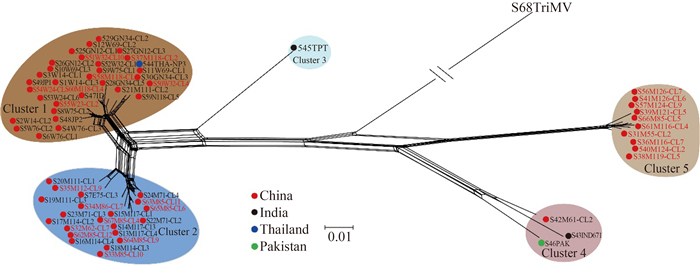 |
| 图 1. SCSMV NIa基因的网状树 Figure 1. Phylogenetic analysis of the NIa gene sequences of Sugarcane streak mosaic virus (SCSMV) using neighbor-net method. The SCSMV sequences with red colour were determined in this study. |
| 图选项 |
2.2 系统发育树重建 在NN法构建的网状树中,SCSMV分成5簇(图 1)。中国分离物集中在第1、2和5簇。值得指出的是,部分云南分离物形成1个新簇——第5簇(Cluster 5,图 1)。这不同于之前的报道[6]。此外,ML法和NJ法生成的系统发育树具有相似的拓扑结构(ML树如图 2所示)。可分为3组:第Ⅲ、Ⅴ和第Ⅰ+Ⅳ组[11],其中第Ⅴ组又可以分为3个亚组(Ⅴ-1、Ⅴ-2和Ⅴ-3)。由图 2可知,中国SCSMV分离物分布于整个系统树,而印度分离物则集中于第Ⅴ和Ⅰ+Ⅳ组。
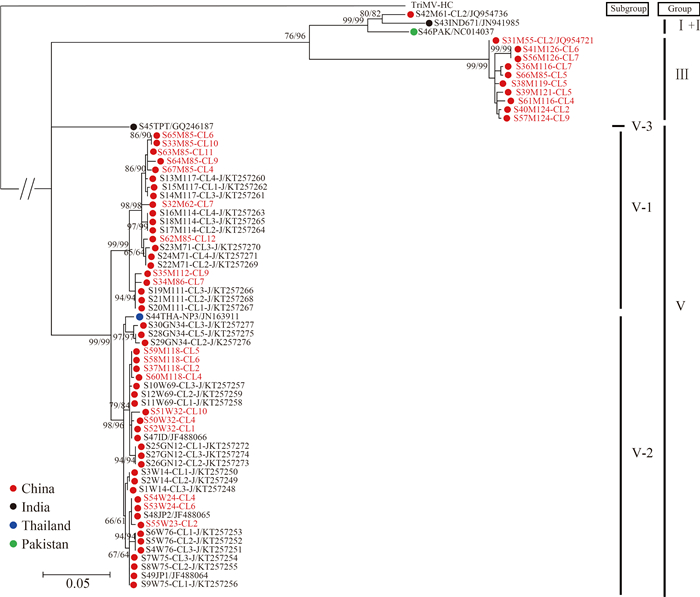 |
| 图 2. 利用ML法对SCSMV NIa基因的系统发生分析 Figure 2. Phylogenetic analysis of the NIa sequences of Sugarcane streak mosaic virus isolates using ML method. The SCSMV sequences with red colour were determined in this study. |
| 图选项 |
基于ML系统树分析发现,NIa基因处于强烈的负选择压力作用下(dN/dS值低至0.014),Datamonkey检测结果显示,NIa基因中未发现正向选择作用位点。
2.3 多样性分析 图 3中统计了SCSMV不同分离物在NIa基因上的核苷酸多样性分布。与系统发生分析结果相一致,SCSMV序列核酸相似性差异可以显著的分为3个组,不同组间NIa基因相似性高于80% (图 3)。在系统发育所分的3个组中,第Ⅲ和第Ⅴ组分别具有较高的单体型多样性(0.978±0.054;0.980±0.007)和较低的核苷酸多样性(0.01147±0.00126;0.02661±0.00259)(表 2)。相对于SCSMV印度分离物,SCSMV中国分离物间遗传相似性程度更高(表 3)。
 |
| 图 3. SCSMV NIa基因序列一致率分布图 Figure 3. The distribution of pairwise identity scores of the NIa sequences of Sugarcane streak mosaic virus isolates. |
| 图选项 |
表 2. 甘蔗线条花叶病毒NIa基因的多样性分析 Table 2. Nucleotide diversity of Sugarcane streak mosaic virus NIa gene
| Group | n | Haplotype diversity (H) | Nucleotide diversity (Πa) |
| Ⅲ | 10 | 0.978±0.054 | 0.01147±0.00126 |
| Ⅰ+Ⅳ | 3 | 1.000±0.272 | 0.05169±0.01451 |
| Ⅴ | 54 | 0.980±0.007 | 0.02661±0.00259 |
| China | 63 | 0.985±0.006 | 0.07439±0.01084 |
| India | 2 | 1.000±0.500 | 0.18841±0.09420 |
| a: nucleotide diversity was estimated by the average pairwise difference between sequences in a sample, based on all sites. | |||
表选项
表 3. 甘蔗线条花叶病毒NIa基因的基因漂移和遗传差异分析 Table 3. Gene flow and genetic differentiation of the NIa of Sugarcane streak mosaic virus
| Region[the number of sequences] | Parametera | ||||
| Ks (P-valueb) | Z (P-value) | Snn (P-value) | Fst | Nm | |
| China[n=63] vs. India[n=2] | 3.28388 (0.0190 *) | 1009.58807 (0.0320 *) | 0.95385 (0.2680) | 0.07208 | 3.22 |
| NN[n=27] vs. India[n=2] | 1.89135 (0.0000 ***) | 175.00000 (0.0040 **) | 0.93103 (0.0550) | 0.25235 | 0.74 |
| YN[n=36] vs. India[n=2] | 3.51550 (0.0010 **) | 341.96667 (0.0780) | 0.92105 (0.2720) | 0.03656 | 6.59 |
| YN[n=36] vs. NN[n=27] | 2.82730 (0.0000 ***) | 749.68054 (0.0000 ***) | 1.00000 (0.0000 ***) | 0.35185 | 0.46 |
| a, Ks* and Z are the sequence-based statistics considered by Hudson (2000). Snn is the nearest-neighbor statistic. Fst is the interpopulation component of genetic variation of the standardized variance in allele frequencies across populations. An absolute value of F+ < 0.33 suggests frequent gene flow. N is the population size of each subpopulation. m is the migration fraction per generation. b, P < 0.05 was considered as the criterion for rejecting the null hypothesis that there is no genetic differentiation between two subpopulations. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001. | |||||
表选项
2.4 基因漂移分析 通过DnaSP 5.0中Ks*、Z和Snn 3个统计量分析SCSMV不同种群间的遗传差异。结果发现,SCSMV在不同组间的遗传差异显著,中国不同地区亚种群间的遗传差异也十分明显(表 3)。基因流动分析中,SCSMV在云南蔗区和资源圃形成的种群间的Fst为0.352,大于0.33(表 3),表明SCSMV分离物在云南蔗区和资源圃间的基因流动频率较低。SCSMV不同种群间Nm < 1(表 3),说明SCSMV种群受到遗传漂变的影响。
3 讨论 SCSMV作为甘蔗花叶病病原之一,目前在印度、印度尼西亚和中国等蔗区的发生分布越来越广泛,并有在东南亚诸国广泛流行发生的趋势[4, 6, 8, 33]。目前,针对该病毒的研究主要集中于遗传进化领域。相较于P1、HC-Pro和CP基因,Potyviridae基因组中NIa基因的保守性较高,重组发生的频率较低[6, 19, 29-30, 34]。本研究测定了采自云南蔗区和国家甘蔗种质资源圃内的23个SCSMV NIa基因序列,经研究未发现其具有重组位点,这进一步支持先前的研究结果。
He等报道发现,SCSMV依据P1、HC-Pro和CP构建的网状树包含5簇,而NIa基因只含有4簇,原因可能是由于NIa基因系统发生分析中SCSMV分离物数量不足,无法完整显现其多样性分布[6]。本研究发现由云南分离物形成的1个新簇——第5簇,从而证明NIa基因与P1、HC-Pro和CP基因在进化上的一致性。
系统发生显示,依据NIa基因SCSMV包含5个分组(亚组),该结果与网状树形成的5簇相一致,具有清晰的地理特异性。SCSMV云南蔗区分离物分布在除V-3亚组外的所有分支,具有高度的遗传多样性。SCSMV资源圃分离物主要集中在V-2亚组,与蔗区分离物间有比较明显的遗传差异。结合SCSMV在云南蔗区与资源圃间的遗传差异和较低频率的基因交流,表明SCSMV在中国可能有2个不同的起源。
本研究确定了SCSMV云南分离物在NIa基因上的多样性、聚类分布和地理特异性特征,明确了SCSMV在云南蔗区和资源圃内具有不同的种群分布,为设计合理的甘蔗花叶病病害防治策略提供了理论依据。
参考文献
| [1] | Hema M, Sreenivasulu P, Savithri HS. Taxonomic position of Sugarcane streak mosaic virus in the family Potyviridae.Archives of Virology, 2002, 147(10): 1997–2007DOI:10.1007/s00705-002-0851-1. |
| [2] | Li WF, He Z, Li SF, Huang YK, Zhang ZX, Jiang DM, Wang XY, Luo ZM. Molecular characterization of a new strain of Sugarcane streak mosaic virus (SCSMV).Archives of Virology, 2011, 156(11): 2101–2104DOI:10.1007/s00705-011-1090-0. |
| [3] | Xu DL, Zhou GH, Xie YJ, Mock R, Li R. Complete nucleotide sequence and taxonomy of Sugarcane streak mosaic virus, member of a novel genus in the family Potyviridae.Virus Genes, 2010, 40(3): 432–439DOI:10.1007/s11262-010-0457-8. |
| [4] | Putra LK, Kristini A, Achadian EM, Damayanti TA. Sugarcane streak mosaic virus in Indonesia:distribution, characterisation, yield losses and management approaches.Sugar Tech, 2014, 16(4): 392–399DOI:10.1007/s12355-013-0279-9. |
| [5] | He Z, Li WF, Yasaka R, Huang YK, Zhang ZX, Ohshima K, Li SF. Molecular variability of Sugarcane streak mosaic virus in China based on an analysis of the P1 and CP protein coding regions.Archives of Virology, 2014, 159(5): 1149–1154DOI:10.1007/s00705-013-1854-9. |
| [6] | He Z, Yasaka R, Li WF, Li SF, Ohshima K. Genetic structure of populations of Sugarcane streak mosaic virus in China:comparison with the populations in India.Virus Research, 2016, 211: 103–116DOI:10.1016/j.virusres.2015.09.020. |
| [7] | Viswanathan R, Balamuralikrishnan M, Karuppaiah R. Characterization and genetic diversity of Sugarcane streak mosaic virus causing mosaic in sugarcane.Virus Genes, 2008, 36(3): 553–564DOI:10.1007/s11262-008-0228-y. |
| [8] | Bagyalakshmi K, Parameswari B, Chinnaraja C, Karuppaiah R, Ganesh Kumar V, Viswanathan R. Genetic variability and potential recombination events in the HC-Pro gene of Sugarcane streak mosaic virus.Archives of Virology, 2012, 157(7): 1371–1375DOI:10.1007/s00705-012-1297-8. |
| [9] | Fellers JP, Seifers D, Ryba-White M, Martin TJ. The complete genome sequence of Triticum mosaic virus, a new wheat-infecting virus of the High Plains.Archives of Virology, 2009, 154(9): 1511–1515DOI:10.1007/s00705-009-0462-1. |
| [10] | Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0.Bioinformatics, 2007, 23(21): 2947–2948DOI:10.1093/bioinformatics/btm404. |
| [11] | Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3:a flexible and fast computer program for analyzing recombination.Bioinformatics, 2010, 26(19): 2462–2463DOI:10.1093/bioinformatics/btq467. |
| [12] | Martin D, Rybicki E. RDP:detection of recombination amongst aligned sequences.Bioinformatics, 2000, 16(6): 562–563DOI:10.1093/bioinformatics/16.6.562. |
| [13] | Sawyer SA. GENECONV:a computer package for the statistical detection of gene conversion.St.Louis: Department of Mathematics, Washington University, 1999. |
| [14] | Salminen MO, Carr JK, Burke DS, McCutchan FE. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning.AIDS Research and Human Retroviruses, 1995, 11(11): 1423–1425DOI:10.1089/aid.1995.11.1423. |
| [15] | Smith JM. Analyzing the mosaic structure of genes.Journal of Molecular Evolution, 1992, 34(2): 126–129. |
| [16] | Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences:computer simulations.Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(24): 13757–13762DOI:10.1073/pnas.241370698. |
| [17] | Boni MF, Posada D, Feldman MW. An exact nonparametric method for inferring mosaic structure in sequence triplets.Genetics, 2007, 176(2): 1035–1047. |
| [18] | Gibbs MJ, Armstrong JS, Gibbs AJ. Sister-scanning:a monte carlo procedure for assessing signals in recombinant sequences.Bioinformatics, 2000, 16(7): 573–582DOI:10.1093/bioinformatics/16.7.573. |
| [19] | Ohshima K, Yamaguchi Y, Hirota R, Hamamoto T, Tomimura K, Tan ZY, Sano T, Azuhata F, Walsh JA, Fletcher J, Chen JS, Gera A, Gibbs A. Molecular evolution of Turnip mosaic virus:evidence of host adaptation, genetic recombination and geographical spread.Journal of General Virology, 2002, 83(6): 1511–1521DOI:10.1099/0022-1317-83-6-1511. |
| [20] | Tomitaka Y, Ohshima K. A phylogeographical study of the Turnip mosaic virus population in East Asia reveals an 'emergent' lineage in Japan.Molecular Ecology, 2006, 15(14): 4437–4457DOI:10.1111/mec.2006.15.issue-14. |
| [21] | Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies:assessing the performance of PhyML 3.0.Systematic Biology, 2010, 59(3): 307–321DOI:10.1093/sysbio/syq010. |
| [22] | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6:molecular evolutionary genetics analysis version 6.0.Molecular Biology and Evolution, 2013, 30(12): 2725–2729DOI:10.1093/molbev/mst197. |
| [23] | Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies.Molecular Biology and Evolution, 2006, 23(2): 254–267. |
| [24] | Posada D. jModelTest:phylogenetic model averaging.Molecular Biology and Evolution, 2008, 25(7): 1253–1256DOI:10.1093/molbev/msn083. |
| [25] | Page RDM. Tree view:an application to display phylogenetic trees on personal computers.Bioinformatics, 1996, 12(4): 357–358DOI:10.1093/bioinformatics/12.4.357. |
| [26] | Librado P, Rozas J. DnaSP v5:a software for comprehensive analysis of DNA polymorphism data.Bioinformatics, 2009, 25(11): 1451–1452DOI:10.1093/bioinformatics/btp187. |
| [27] | Muhire BM, Varsani A, Martin DP. SDT:a virus classification tool based on pairwise sequence alignment and identity calculation.PLoS One, 2014, 9(9): e108277DOI:10.1371/journal.pone.0108277. |
| [28] | Pamilo P, Bianchi NO. Evolution of the Zfx and Zfy genes:rates and interdependence between the genes.Molecular Biology and Evolution, 1993, 10(2): 271–281. |
| [29] | Zhang CL, Gao R, Wang J, Zhang GM, Li XD, Liu HT. Molecular variability of Tobacco vein banding mosaic virus populations.Virus Research, 2011, 158(1/2): 188–198. |
| [30] | Nguyen HD, Tran HTN, Ohshima K. Genetic variation of the Turnip mosaic virus population of Vietnam:a case study of founder, regional and local influences.Virus Research, 2013, 171(1): 138–149DOI:10.1016/j.virusres.2012.11.008. |
| [31] | Wei TY, Yang JG, Liao FR, Liao FL, Gao FL, Lu LM, Zhang XT, Li F, Wu ZJ, Lin QY, Xie LH, Lin HX. Genetic diversity and population structure of rice stripe virus in China.Journal of General Virology, 2009, 90(4): 1025–1034DOI:10.1099/vir.0.006858-0. |
| [32] | Hudson RR. A new statistic for detecting genetic differentiation.Genetics, 2000, 155(4): 2011–2014. |
| [33] | Parameswari B, Bagyalakshmi K, Viswanathan R, Chinnaraja C. Molecular characterization of Indian Sugarcane streak mosaic virus isolate.Virus Genes, 2013, 46(1): 186–189DOI:10.1007/s11262-012-0827-5. |
| [34] | Gao FL, Shen JG, Shi FY, Fang ZG, Xie LH, Zhan JS. Detection and molecular variation of Potato virus Y CP Gene in China.Scientia Agricultura Sinica, 2013, 46(15): 3125–3133(in Chinese).高芳銮, 沈建国, 史凤阳, 方治国, 谢联辉, 詹家绥. 中国马铃薯Y病毒的检测鉴定及CP基因的分子变异.中国农业科学, 2013, 46(15): 3125–3133. |
